Abstract
Caspases are a conserved family of cell death proteases that cleave intracellular substrates at Asp residues to modify their function and promote apoptosis. In this report we identify the integrin β4 subunit as a novel caspase substrate using an expression cloning strategy. Together with its α6 partner, α6β4 integrin anchors epithelial cells to the basement membrane at specialized adhesive structures known as hemidesmosomes and plays a critical role in diverse epithelial cell functions including cell survival and migration. We show that integrin β4 is cleaved by caspase-3 and -7 at a conserved Asp residue (Asp1109) in vitro and in epithelial cells undergoing apoptosis, resulting in the removal of most of its cytoplasmic tail. Caspase cleavage of integrin β4 produces two products, 1) a carboxyl-terminal product that is unstable and rapidly degraded by the proteasome and 2) an amino-terminal cleavage product (amino acids 1–1109) that is unable to assemble into mature hemidesmosomes. We also demonstrate that caspase cleavage of integrin β4 sensitizes epithelial cells to apoptosis and inhibits cell migration. Taken together, we have identified a previously unrecognized proteolytic truncation of integrin β4 generated by caspases that disrupts key structural and functional properties of epithelial cells and promotes apoptosis.
Apoptosis is a genetically regulated cellular suicide program activated by diverse signals that results in a series of stereotypical events culminating in cell death. These events include membrane blebbing, nuclear and DNA fragmentation, loss of cell adhesion, dismantling of the cytoskeleton, and packaging the nuclear and cytoplasmic remnants into apoptotic bodies which are engulfed and degraded by adjacent cells. Caspases are highly conserved cell death proteases that execute many of these stereotypical events by cleaving target proteins at aspartic acid residues (1). For example, caspases trigger the internucleosomal fragmentation of DNA by cleaving ICAD, an inhibitor of the caspase-activated DNase (CAD), thereby releasing ICAD from the inactive ICAD·CAD complex and activating CAD (2, 3). Caspases dismantle the nuclear envelope and intermediate filament cytoskeleton by specifically proteolyzing the nuclear lamins, cytokeratins 14, 18, and 19 in epithelial cells, vimentin in mesenchymal cells, and desmin in muscle cells (4–10). Caspases also disrupt the actin microfilament network by proteolyzing and activating the actin-severing protein gelsolin and by cleaving actin directly (11, 12). In addition, caspases proteolyze several components of adherens junctions and desmosomes, intercellular junctions attached to the actin microfilament or cytokeratin intermediate filament networks, respectively, which play a critical role in cell-cell adhesion in epithelial tissues. Indeed, classical (E-cadherin) and desmosomal cadherins (desmoglein-1 and -3 and desmocollin-3), β-catenin, plakoglobin, and desmosomal plaque proteins (plakophilin-1, desmoplakin-1 and -2) are all caspase substrates (13–16). Clearly, adherens junctions and desmosomes and their associated cytoskeletal networks have been extensively targeted for degradation by caspases. The proteolytic disassembly of these structures by caspases likely contributes to the disruption of cell-cell adhesion and the dramatic cytoskeletal reorganization that typifies apoptosis.
In an effort to systematically decipher the molecular mechanisms by which caspases induce apoptosis, we have used a small pool expression cloning approach to isolate caspase substrates (9, 17–20). Here, we report the identification of the integrin β4 subunit as a novel caspase substrate. Integrins are a family of heterodimeric cell surface receptors composed of an α and β subunit that adhere cells to the extracellular matrix (ECM) and transmit signals from the ECM that regulate cell survival, proliferation, differentiation, and migration (21). The integrin β4 subunit is distinguished from other integrin subunits by virtue of several unique characteristics. In contrast to the short intracellular domains of other integrins, integrin β4 has a large cytoplasmic tail spanning ~1000 amino acids that consists of two pairs of fibronectin type III (FNIII)2 repeats separated by a connecting sequence (22, 23). Moreover, the integrin β4 subunit and its α6 partner (α6β4 integrin) are receptors for the extracellular matrix protein laminin-5, recently designated laminin-332 by a laminin nomenclature committee (24). α6β4 integrin is organized into multiprotein cytoplasmic plaques called hemidesmosomes that anchor cells in the basal layer of epithelial tissues, including skin and the mammary gland, to the basement membrane. Hemidesmosomes are attached to the cytokeratin network, whereas other integrins are attached to actin microfilaments (22, 23). Additional protein components of hemidesmosomes are BP180 (BPAG2), a type II transmembrane protein, BP230 (BPAG1), and the linker protein plectin. Importantly, the cytoplasmic tail of integrin β4 plays a key role in the localization of α6β4 integrin to hemidesmosomal structures and in the recruitment of other hemidesmosomal proteins to these structures. Specifically, the first and/or second FNIII repeat and a portion of the connecting sequence are required for assembly of α6β4 integrin into hemidesmosomes and plectin binding, whereas the third FNIII repeat and part of the connecting sequence mediate BP180 binding (25–27). BP230 interacts with the third and fourth FNIII repeats and the carboxyl terminus of integrin β4 (28). The paramount importance of hemidesmosomes in maintaining the structural integrity of certain epithelial tissues is evident from patients with junctional epidermolysis bullosa (JEB) with congenital pyloric atresia, a fatal skin blistering disease characterized by abnormal hemidesmosomes, detachment of epithelial cells from the basement membrane, and mutations in the β4 or α6 subunit genes (29, 30). Indeed, mice with targeted deletion of the entire integrin β4 gene or the cytoplasmic domain lack hemidesmosomes and develop a JEB-like syndrome notable for increased detachment and death of epithelial cells (31–33). Moreover, deletion of integrin β4 or its carboxyl-terminal tail in keratinocytes results in profound abnormalities in cell migration and cell survival that likely impair wound healing (34, 35). Taken together, these findings point to a critical role for the cytoplasmic tail of integrin β4 in the structural integrity, function, and survival of certain epithelial tissues.
In this report we demonstrate for the first time that the integrin β4 subunit is cleaved by caspase-3 and -7 at Asp1109 in vitro and in cells undergoing apoptosis, thereby removing much of its cytoplasmic tail, including all four FNIII repeats. Caspase proteolysis of integrin β4 generates a carboxyl-terminal product that is unstable and rapidly degraded by the proteasome and an amino-terminal cleavage product that is unable to assemble into mature hemidesmosomes. In addition, caspase cleavage of integrin β4 sensitizes epithelial cells to apoptosis and inhibits cell migration. Collectively, our results point to a novel caspase-mediated truncation of integrin β4 that disrupts key structural and functional properties of epithelial cells and promotes apoptosis.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
cDNAs encoding wild-type (WT) full-length integrin β4 or its cytoplasmic tail (amino acids 734–1752) were constructed by PCR amplifying the human integrin β4 cDNA using the primer pairs 5′-ccgggaattcATGGCAGGGCCACGCCCCAGCCCA-3′ and 5′-ccggctcgagTCAAGTTTGGAAGAACTGTTGGTCC-3′ (WT) or 5′-ccgggaattcATGGGGAAGTACTGTGCCTGCTGC-3′ and 5′-ccggctcgagTCAAGTTTGGAAGAACTGTTGGTCC-3′ (tail). The PCR products were then digested with EcoRI and XhoI and subcloned into pcDNA3 (Invitrogen). A D1109E mutant full-length and tail β4 construct in which the putative caspase cleavage site at Asp1109 was replaced with a Glu residue were generated using the QuikChange site-directed mutagenesis kit (Stratagene) using the primers 5′-GACCCAGATGAACTGGAGCGGAGCTTCACGAGTCA-3′ and 5′-TGACTCGTGAAGCTCCGCTCCAGTTCATCTGGGTC-3′. cDNAs encoding carboxyl-terminal GFP-tagged, full-length integrin β4 (WT-β4) or the amino-terminal caspase cleavage fragment composed of amino acids 1–1109 (caspase-truncated β4, designated Tr-β4) were generated by PCR amplification of the wild-type human integrin β4 cDNA using the primers 5′-ggccctcgaggccaccATGGCAGGGCCACGCCCC-3′ and 5′-ggggctcgagAGTTTGGAAGAACTGTTGGTCC-3′ (WT-β4) or 5′-ggccctcgaggccaccATGGCAGGGCCACGCCCC-3′ and 5′-ggggctcgagGTCCAGTTCATCTGGGTCCC-3′ (Tr-β4). The PCR products were digested with XhoI and subcloned into pLEGFP-N1 (BD Biosciences). Each cDNA was verified by DNA sequence analysis.
Small Pool Expression Cloning
cDNAs encoding putative caspase substrates were isolated from a human prostate adenocarcinoma cDNA library (Invitrogen) by small pool expression cloning as described previously (9, 17–20).
Caspase Cleavage of Integrin β4 in Vitro
Full-length integrin β4, the WT integrin β4 cytoplasmic tail, or mutant D1109E integrin β4 tail were 35S-labeled with [35S]methionine using the TNT T7 Quick Coupled Transcription/Translation system (Promega). 35S-Labeled full-length integrin β4 was incubated with buffer or 2.5 ng of caspase-3 for 1 h at 37 °C, whereas the 35S-labeled tail proteins were incubated with buffer or 2.5 or 25 ng of caspase-1, -2, -3, -5, -6, -7, -8, or -9 for 1 h at 37 °C; cleavage reactions were analyzed as described previously (19, 36).
Cell Culture and Apoptosis Experiments
Immortalized, non-transformed human MCF-10A mammary epithelial cells (37) and human MDA-MB-435 breast cancer cells were purchased from the ATCC. MCF-10A cells were cultured in Dulbecco’s modified Eagle’s medium/F-12 medium (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/ml epidermal growth factor (Sigma), 0.5 mg/ml hydrocortisone (Sigma), 100 ng/ml cholera toxin (Sigma), 10 μg/ml insulin (Sigma), and penicillin/streptomycin (Invitrogen) as described (38). MDA-MB-435 cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum. Immortalized integrin β4-deficient keratinocytes derived from a patient with JEB with pyloric atresia (34) were kindly provided by M. Peter Marinkovich, Stanford University School of Medicine. JEB keratinocytes were cultured in defined keratinocyte-serum-free medium (Invitrogen) and penicillin/streptomycin (Invitrogen). For MCF-10A cells, apoptosis was induced by adding 1 μg/ml recombinant tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), which was produced as described previously (39, 40), or 1 μM staurosporine (Sigma). To verify that the apoptotic cleavage of integrin β4 was mediated by caspases, cells were pretreated for 1 h with vehicle or 50 μM zVAD-fmk, a broad spectrum caspase inhibitor, and then treated with TRAIL or staurosporine. In some experiments cells were pretreated for 1 h with vehicle or 100 nM epoxomicin (Calbiochem), a proteasome inhibitor, and then treated with TRAIL to determine whether the apoptotic cleavage product of integrin β4 was degraded by the proteasome. Apoptosis was induced in MDA-MB-435 breast cancer cells by adding 1 μg/ml TRAIL in combination with 1 μg/ml cycloheximide (Sigma); the latter agent sensitizes these cells to TRAIL-induced apoptosis. Apoptotic nuclei (fragmented or condensed) were scored by staining with 10 μg/ml Hoescht 33258 (Sigma) as described previously (41). Two hundred cells per treatment condition were scored.
Immunoblotting
Whole cell lysates were prepared from both floating and adherent cells using SDS sample buffer consisting of 8 M urea, 1% SDS in 10 mM Tris-HCl, pH 6.8, and 15% β-mercaptoethanol. Protein concentration was measured using the NI protein assay (Genotech). Samples were separated by SDS-PAGE, transferred to nitrocellulose and immunoblotted as described previously (36) using antibodies to integrin β4 subunit (BD Biosciences), α-tubulin, α-actin (Sigma), GFP (Roche Applied Science), or protein kinase Cδ (Santa Cruz Biotechnology).
Transfection and Retroviral Transduction
MDA-MB-435 carcinoma cells were transiently transfected with 1 μg of pcDNA3 plasmids containing full-length WT or mutant D1109E integrin β4 using Lipofectamine PLUS reagent (Invitrogen). For retroviral transduction of JEB keratinocytes, 10 μg of pLEGFP-N1 vector (BD Biosciences) or pLEGFP-N1 plasmid containing WT integrin β4 (WT-β4) or caspase-truncated integrin β4 (Tr-β4) were transfected into the Phoenix amphotrophic retrovirus packaging cell line (ATCC), and the retroviral supernatant was prepared as described previously (42). JEB cells were incubated with retrovirus for 48 h, and pools were then generated by selection in 125 μg/ml G418 (Invitrogen) for 10 days. The expression of proteins in these pools was confirmed by immunoblotting with a GFP antibody. Apoptosis was induced in JEB pools by treatment with 5 μg/ml TRAIL, and apoptosis was scored by nuclear morphology as described under “Cell Culture and Apoptosis Experiments” in this section.
Cell Surface Expression of Integrin β4
JEB keratinocytes stably expressing GFP vector or GFP-tagged WT-β4 or Tr-β4 were collected, washed with PBS, and incubated with an integrin β4 extracellular domain antibody (3E1, Chemicon) for 45 min at RT. Cells were then washed, incubated with goat anti-mouse Cy5 secondary antibody for 30 min at RT, washed, and fixed in 0.5% paraformaldehyde for 5 min at RT. Cell surface expression of integrin β4 was determined by fluorescence-activated cell sorting using a DakoCytomation CyAn Flow Cytometer.
Microscopy
JEB cells stably expressing integrin β4 constructs were cultured on glass coverslips, washed with PBS, fixed with 3.7% paraformaldehyde for 20 min at RT, and permeabilized with prechilled 0.5% Triton X-100 for 15 min. Slides were incubated with primary antibodies against plectin (43) (1:50 dilution) or BP180 (44) (1:50 dilution) for 1 h at 37 °C. After washing, slides were incubated with secondary antibodies, Alexa Fluor 594 F(ab′)2 fragments of either goat anti-mouse or anti-rabbit IgGs (Molecular Probes) (1:200 dilution) for 1 h at 37 °C. Slides were incubated with 0.5 ng/ml 4′,6-diamidino-2-phenylindole (Sigma) for 15 min at RT and then washed with PBS. Coverslips were mounted with ProLong Gold antifade reagent (Invitrogen). Confocal images of planes near the basal cell-substrate surface were obtained with a Zeiss LSM510 UV META confocal microscope. Images were analyzed with Zeiss LSM5 software. For localization and colocalization studies, 100 cells in each of two experiments were scored.
Wound Closure Assay
JEB cells stably expressing GFP constructs were sorted by flow cytometry to ensure GFP expression, plated in 6-well plates, and grown overnight. The following day cells were washed with PBS, scraped with a pipette tip, washed again with PBS, and grown in fresh medium (medium was replenished every 24 h). Forty-eight hours later wounds were photographed using a Nikon ECLIPSE TE 2000-U microscope, and wound closure was measured with MetaMorph software (Version 6.1r6, Universal Imaging). For immunoblotting, confluent JEB cells stably expressing GFP-tagged WT-β4 were scraped with a pipette tip in a grid-like pattern (multi-scratch wound assay) to increase the percentage of migrating cells for analysis.
RESULTS
Identification of the Integrin β4 Subunit as a Caspase-3 Substrate in Vitro by Small Pool Expression Cloning
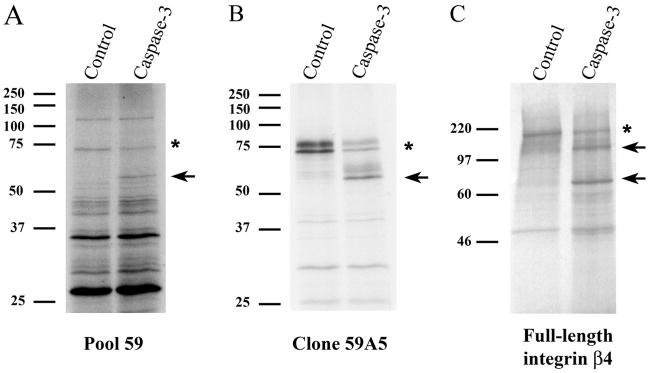
We have previously described an expression cloning method to systematically identify cDNAs encoding putative caspase substrates from small pools of a cDNA library (9, 17–20). In these experiments caspase-3 was added to 35S-labeled protein pools derived from small cDNA pools (48 cDNAs in each pool) of a human prostate adenocarcinoma cDNA library (Invitrogen). Pools with protein bands that were cleaved by caspase-3 were identified. An ~75-kDa protein (Fig. 1A, indicated by an asterisk) in 35S-labeled pool 59 was proteolyzed by caspase-3 into a fragment of ~58 kDa (Fig. 1A, indicated by the arrow) in vitro. To identify the caspase-3 substrate corresponding to the ~75-kDa band in protein pool 59, we divided cDNA pool 59 into progressively smaller pools, 35S-labeled these pools, and incubated them with caspase-3. In this way, a single cDNA (59A5) encoding an ~75-kDa protein (Fig. 1B, doublet, indicated by an asterisk) was cleaved by caspase-3 into a ~58-kDa product (Fig. 1B, doublet, indicated by an arrow) in vitro. Sequence analysis of cDNA 59A5 revealed that it encoded amino acids 593–1266 of the integrin β4 subunit (45). The cDNA contains two ATG codons near the 5′-end, which likely results in the observed doublet of the 35S-labeled protein (Fig. 1B). Importantly, 35S-labeled full-length integrin β4 (Fig. 1C, indicated by the asterisk) was proteolyzed by caspase-3 into products of ~130 and ~79 kDa (Fig. 1C, indicated by arrows). Taken together, these results indicate that the integrin β4 subunit is proteolyzed by caspase-3 at a single site in vitro.
FIGURE 1. Identification of the integrin β4 subunit as a novel caspase-3 substrate in vitro by small pool expression cloning.
A, a protein of ~75 kDa (denoted by an asterisk) in 35S-labeled protein pool 59 is cleaved by caspase-3 into a ~58-kDa fragment (indicated by an arrow); this fragment was not observed when protein pool 59 was incubated with buffer control. 35S-Labeled protein pools were generated from small pools of a human prostate adenocarcinoma cDNA library and incubated with 2.5 ng of recombinant caspase-3 as described (9, 17–20). B, the cDNA encoding the ~75-kDa protein that is proteolyzed by caspase-3 into a ~58-kDa fragment was isolated from cDNA pool 59 by subdividing the pool and retesting smaller pools by the same approach. Clone 59A5 was sequenced and identified as a partial integrin β4 subunit cDNA. C, the full-length integrin β4 subunit is cleaved by caspase-3 in vitro. Full-length 35S-labeled integrin β4 (denoted by an asterisk) is cleaved by 2.5 ng of caspase-3 into two products of ~130 and ~79 kDa (indicated by arrows).
The Cytoplasmic Tail of the Integrin β4 Subunit Is Cleaved at Asp1109 by Caspases-3 and -7 in Vitro
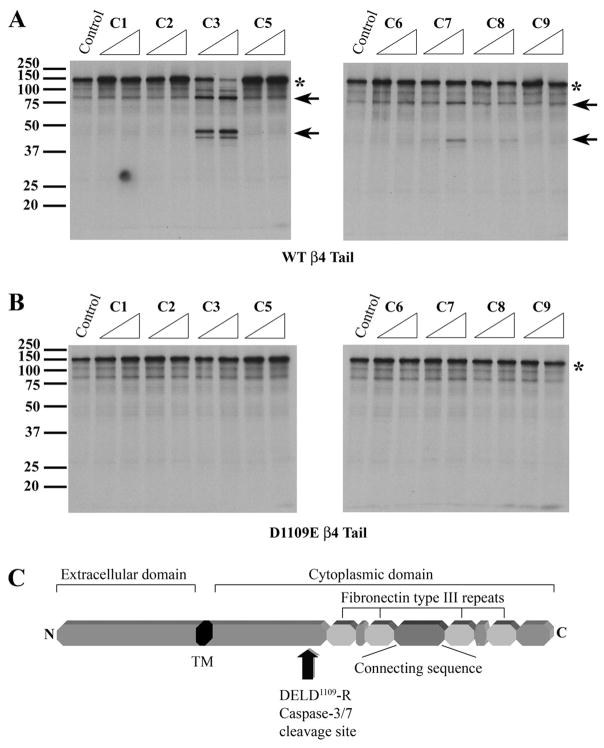
Because caspases are intracellular proteases that do not have access to the extracellular and transmembrane domains of integrin β4, we examined the sensitivity of the 35S-labeled cytoplasmic tail of integrin β4 (amino acids 734–1752) to a panel of recombinant caspases. Incubation of the 35S-labeled β4 cytoplasmic tail with caspase-3 generated prominent fragments of ~79 and ~43 kDa (Fig. 2A, indicated by arrows) and reduced the amount of the intact cytoplasmic tail (Fig. 2A, denoted by an asterisk). Caspase-7, which cleaves substrates at a similar DEXD motif as caspase-3 (46), produced the same sized fragments but to a lesser extent. A faint band corresponding to the ~43-kDa product was also observed when the 35S-labeled β4 cytoplasmic tail was incubated with caspase-8. However, caspases-1, -2, -5, -6, and -9 did not proteolyze the β4 cytoplasmic tail. Of note, the observed ~79-kDa product was similar in size to the fragment produced by caspase-3 cleavage of full-length 35S-labeled β4 (Fig. 1C). These results indicate that the cytoplasmic tail of integrin β4 is preferentially proteolyzed by caspase-3 and -7 in vitro.
FIGURE 2. The cytoplasmic tail of the integrin β4 subunit is cleaved at Asp1109 by caspases-3 and -7 in vitro.
A, the 35S-labeled cytoplasmic tail of integrin β4 (denoted by an asterisk) is preferentially cleaved by caspases-3 and -7 into a ~79- and a ~43-kDa product (indicated by arrows) in vitro. The 35S-labeled integrin β4 cytoplasmic tail was incubated with buffer control or 2.5 or 25 ng of caspase-1, -2, -3, -5, -6, -7, -8, or -9 (C1-C9) for 1 h at 37 °C. The reaction products were separated by SDS-PAGE and detected by autoradiography. B, a mutant integrin β4 cytoplasmic tail in which Asp1109 is replaced with a Glu residue (D1109E) is resistant to caspase-3 proteolysis in vitro. 35S-Labeled mutant D1109E integrin β4 cytoplasmic tail was incubated with buffer control or 2.5 or 25 ng of caspase-1, -2, -3, -5, -6, -7, -8, or -9 (C1-C9) for 1 h at 37 °C. C, a protein domain map of the integrin β4 subunit showing the caspase cleavage site DELD1109↓R in the cytoplasmic domain.
A potential caspase-3 and -7 cleavage motif (DEXD) (46) was identified in the β4 cytoplasmic tail (DELD1109↓R) that would be expected to yield a ~79-kDa carboxyl-terminal cleavage product. To determine whether integrin β4 was indeed cleaved by caspase-3 and -7 at this site in vitro, we replaced Asp1109 with a Glu residue and tested the sensitivity of the mutant D1109E β4 cytoplasmic tail to caspase proteolysis. Unlike the WT β4 cytoplasmic tail (Fig. 2A), the 35S-labeled D1109E β4 tail was not cleaved by caspase-3 or -7 (Fig. 2B), indicating that Asp1109 is the bona fide caspase-3 and -7 cleavage site in vitro. A domain map of the integrin β4 protein reveals that caspase proteolysis at Asp1109 removes much of the cytoplasmic tail, including all four fibronectin type III repeats and the connecting sequence (Fig. 2C). Taken together, these findings indicate that β4 inte-grin is cleaved by caspases-3 and -7 at Asp1109 in its cytoplasmic tail, thereby removing several domains involved in hemidesmosome assembly and/or signaling.
Integrin β4 Is Proteolyzed at Asp1109 by Caspases in Cells Undergoing Apoptosis and Produces an Unstable Cleavage Product That Is Degraded by the Proteasome
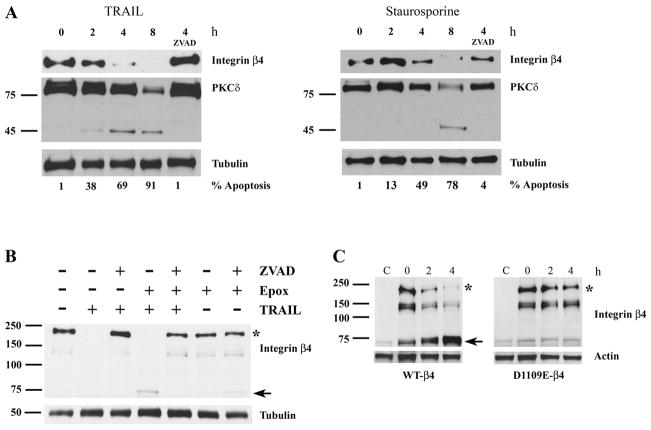
We next examined whether the integrin β4 subunit is cleaved in epithelial cells undergoing apoptosis. For these experiments we treated MCF-10A mammary epithelial cells, which express integrin β4 and assemble mature hemidesmosomes (47, 48), with 1 μg/ml TRAIL for 0–8 h. The amount of full-length integrin β4 detected with an antibody directed against its carboxyl terminus decreased progressively over time, and there was no detectable full-length β4 8 h after TRAIL treatment (Fig. 3A, left panel). Although no β4 cleavage product was observed (data not shown), the reduction in the amount of full-length β4 in response to TRAIL was completely suppressed by the broad spectrum caspase inhibitor zVAD-fmk, suggesting that caspases were responsible for the observed reduction in full-length β4. The caspase substrate protein kinase Cδ was used as a positive control; it was cleaved into its expected size apoptotic fragment (49) within 2 h of TRAIL treatment, and its apoptotic proteolysis was blocked by zVAD-fmk. A reduction in full-length β4 was also observed in MCF-10A cells treated with staurosporine, and this reduction was suppressed by zVAD-fmk (Fig. 3A, right panel). These data suggest that caspases cleave the integrin β4 subunit during apoptosis in vivo and produce an unstable proteolytic product. Indeed, caspase proteolysis of several proteins has been demonstrated to yield unstable cleavage products that are rapidly degraded by the proteasome (50–53). Hence, we postulated that the integrin β4 carboxyl-terminal cleavage product was similarly degraded. Consistent with this hypothesis, a ~72-kDa β4 cleavage product was observed in TRAIL-treated MCF-10A cells preincubated for 1 h with 100 nM epoxomicin, a proteasome inhibitor (Fig. 3B). This proteolytic product is similar in size to the ~79-kDa 35S-labeleded β4 cleavage product generated by caspase cleavage in vitro (Fig. 2A). Importantly, both the disappearance of full-length β4 and the appearance of the ~72-kDa cleavage product in MCF-10A cells treated with epoxomicin and TRAIL were inhibited by zVAD-fmk. These findings provide unequivocal evidence that caspases proteolyze the integrin β4 subunit into an unstable carboxyl-terminal cleavage product that is subsequently degraded by the proteasome in epithelial cells undergoing apoptosis.
FIGURE 3. Integrin β4 is proteolyzed at Asp1109 by caspases in cells undergoing apoptosis and produces an unstable cleavage product which is degraded by the ubiquitin-proteasome system.
A, full-length integrin β4 is rapidly degraded in human MCF-10A mammary epithelial cells treated with TRAIL (top panel) by a caspase-dependent mechanism. MCF-10A cells were treated with 1 μg/ml TRAIL or 1 μM staurosporine for 0 – 8 h. For the caspase inhibitor experiments, MCF-10A cells were preincubated for 1 h with 50 μM zVAD-fmk, a pan-caspase inhibitor, and then treated with 1 μg/ml TRAIL or 1 μM staurosporine for 4 h. Immunoblotting was performed with antibodies recognizing the cytoplasmic tail of integrin β4, protein kinase Cδ (PKCδ) or tubulin. The percentage of apoptotic nuclei was determined in parallel experiments. B, the apoptotic integrin β4 caspase cleavage product is degraded by the proteasome. MCF-10A cells were preincubated with 20 μM ZVAD-fmk, 100 nM epoxomicin (Epox), a proteasome inhibitor, or both for 1 h and then treated with 1 μg/ml TRAIL (or untreated) for 6 h. C, mutant D1109E integrin β4 is not cleaved during apoptosis. MDA-MB-435 breast cancer cells (which lack integrin β4) were transiently transfected with either WT or mutant D1109E integrin β4 cDNA. Twenty-four hours after transfection cells were treated with 0.5 μg/ml TRAIL and 1 μg/ml cycloheximide. In B and C, full-length integrin β4 (asterisks) and the ~72-kDa cleavage product (arrow) are indicated.
To verify that integrin β4 is cleaved by caspases at Asp1109 in apoptotic cells, we transiently transfected human MDA-MB-435 breast carcinoma cells, which lack endogenous integrin β4 subunit, with a full-length WT-β4 or D1109E-β4 cDNA (Fig. 3C). Twenty-four hours later cells were treated with 1 μg/ml TRAIL and 1 μg/ml cycloheximide to induce apoptosis (the latter drug sensitizes carcinoma cells to TRAIL). Full-length WT-β4 (denoted by the asterisk) was rapidly cleaved into the ~72-kDa product; some cleavage was also evident at t = 0 due to the toxicity of the transfection reagents (a prominent more rapidly migrating band that was also detected by the carboxyl-terminal integrin β4 Ab was also cleaved). Of note, in these transiently transfected cells, the ~72-kDa caspase cleavage product was detectable in the absence of proteasome inhibition, perhaps due to the high levels of the ectopically expressed β4. Conversely, D1109E-β4 was resistant to proteolysis with little production of the ~72-kDa fragment in TRAIL-treated cells. These results indicate that integrin β4 is cleaved by caspases at Asp1109 in cells undergoing apoptosis.
Caspase Cleavage of Integrin β4 Disrupts Hemidesmosome Assembly
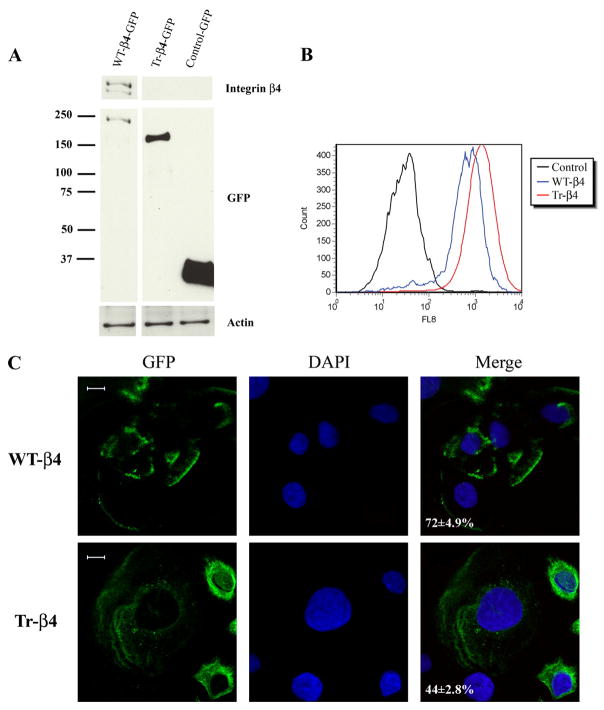
Because caspase cleavage of β4 integrin removes cytoplasmic domains required for its localization to hemidesmosomes and its binding to other hemidesmosomal proteins (25–27), we examined the ability of GFP-tagged full-length WT-β4 and caspase-truncated β4 (amino acids 1–1109, Tr-β4) to assemble into hemidesmosomes in integrin β4-deficient keratinocytes. These keratinocytes were derived from a patient with JEB and contain all of the protein components of hemidesmosomes except the integrin β4 subunit (34). Importantly, the addition of GFP to the carboxyl terminus of WT integrin β4 does not alter its ligand binding, localization to hemidesmosomes, or signaling (54–56). JEB keratinocytes stably expressing GFP WT-β4, GFP Tr-β4, or control-GFP vector were generated by retroviral infection and selection in G418. Stable expression of each construct was confirmed by immunoblotting, as was the absence of full-length β4 in JEB cells stably expressing Tr-β4 or GFP vector (Fig. 4A). We next performed fluorescence-activated cell sorting analysis using an antibody that recognizes the extracellular domain of integrin β4. Both WT-β4 and Tr-β4 were expressed on the surface of retrovirally transduced JEB cells (Fig. 4B). Confocal microscopy images of planes near the cell-substrate surface revealed WT-β4 in polarized basal structures characteristic of hemidesmosomal proteins in 72 ± 4.9% of cells (Fig. 4C). In contrast, Tr-β4 was often more diffusely distributed and was present in polarized basal structures in only 44 ± 2.8% of cells. These results indicate that although Tr-β4 is expressed on the cell surface, it often fails to incorporate into hemidesmosome-like structures.
FIGURE 4. Caspase-truncated integrin β4 is impaired in its localization to polarized basal structures characteristic of hemidesmosomes.
A, stable expression of GFP-tagged WT integrin β4 (WT-β4) or caspase-truncated integrin β4 (Tr-β4) in integrin β4-deficient JEB keratinocytes. JEB cells stably expressing GFP WT-β4, GFP Tr-β4 (amino acids 1–1109, mimicking caspase cleavage), or control-GFP vector were generated by retroviral infection and selection in G418. Lysates were analyzed by immunoblotting with antibodies recognizing the carboxyl terminus of integrin β4, GFP, or actin. B, stably expressed WT and caspase-truncated integrin β4 (Tr-β4) are present on the cell surface. JEB cells stably expressing GFP vector, GFP WT-β4, or GFP Tr-β4 were immunostained with an antibody that recognizes the extracellular domain of integrin β4 and analyzed by fluorescence-activated cell sorting. C, confocal microscopy images of planes near the cell-substrate surface reveal WT-β4 in polarized basal structures characteristic of hemidesmosomal proteins, whereas Tr-β4 is often diffusely distributed. The percentage of cells with WT-β4 or Tr-β4 present in polarized basal structures is indicated (100 cells scored in each of two experiments). DAPI, 4′,6-diamidino-2-phenylindole. Bar, 10 μm.
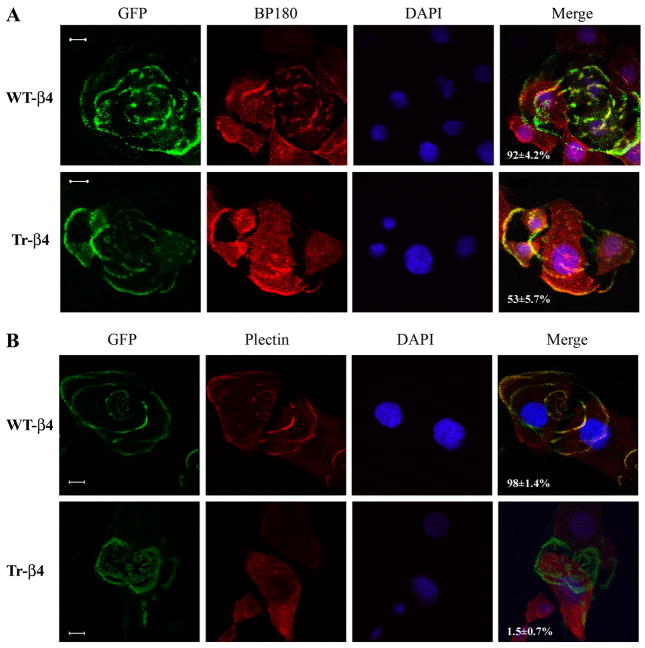
We next examined whether WT-β4 or Tr-β4, when localized in polarized basal structures, supported the assembly of mature hemidesmosomes by recruiting BP180 and plectin to these hemidesmosome-like structures. In JEB keratinocytes stably expressing WT-β4, polarized basal WT-β4 colocalized with BP180 in 92 ± 4.2% of cells (Fig. 5A) and plectin in 98 ± 1.4% of cells (Fig. 5B). In contrast, polarized basal Tr-β4 colocalized with BP180 in only 53 ± 5.7% of cells (Fig. 5A) and rarely colocalized with plectin (only 1.5 ± 0.7% of cells, Fig. 5B). Collectively, these data indicate that caspase cleavage of integrin β4 disrupts the assembly of mature hemidesmosomes.
FIGURE 5. Caspase-truncated integrin β4 is unable to form mature hemidesmosomes.
A, WT-β4 colocalizes with the hemidesmosomal protein BP180, whereas Tr-β4 only partly colocalizes with BP180. JEB keratinocytes stably expressing GFP WT-β4 or GFP Tr-β4 were examined by confocal microscopy in focal planes close to the cell-substrate surface. BP180 (red) was detected by indirect immunofluorescence as detailed under “Experimental Procedures.” The percentage of cells in which polarized basal WT-β4 or Tr-β4 colocalized with BP180 is indicated (100 cells scored in each of two experiments). Bar, 10 μm. B, WT-β4 colocalizes with the hemidesmosomal protein plectin, whereas Tr-β4 does not colocalize with plectin. Confocal microscopy was performed as in A, and plectin (red) was detected by indirect immunofluorescence as described under “Experimental Procedures.” The percentage of cells in which polarized WT-β4 or Tr-β4 colocalized with plectin is indicated (100 cells scored in each of two experiments). DAPI, 4′,6-diamidino-2-phenylindole. Bar, 10 μm.
Caspase Proteolysis of Integrin β4 Promotes Apoptosis
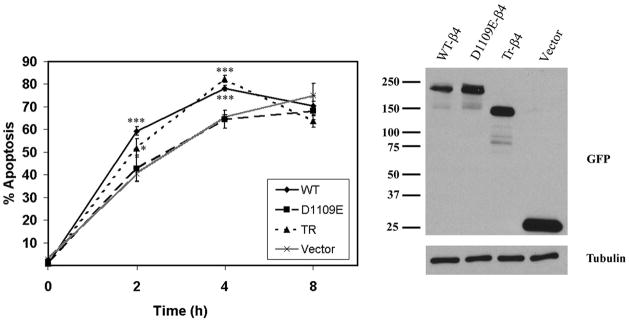
To determine whether caspase cleavage of integrin β4 alters the sensitivity of epithelial cells to apoptosis induction, we treated JEB pools stably expressing GFP vector or GFP-tagged WT-β4, D1109E-β4, or Tr-β4 with 5 μg/ml TRAIL for 0–8 h. Both Tr-β4 and WT-β4, but not cleavage-resistant D1109E-β4, sensitized JEB keratinocytes to TRAIL-induced apoptosis 2 and 4 h after treatment (Fig. 6). However, by 8 h the induction of apoptosis was similar in each of the JEB pools. WT-β4 was cleaved by caspases as early as 2 h after TRAIL treatment (data not shown). These results indicate that caspase cleavage of integrin β4 sensitizes keratinocytes to TRAIL-induced apoptosis.
FIGURE 6. Caspase proteolysis of integrin β4 promotes apoptosis.
JEB pools stably expressing GFP vector or GFP-tagged WT-β4, D1109E-β4, or Tr-β4 were treated with 5 μg/ml TRAIL for the indicated times, and apoptotic nuclei were scored as described under “Experimental Procedures.” Data are presented as the mean ± S.E. of three experiments (left panel). *, p < 0.05 versus D1109E; ***, p < 0.001 versus D1109E. The expression of each construct in JEB pools before TRAIL treatment was determined by immunoblotting using a GFP antibody (right panel).
Caspase Cleavage of Integrin β4 Inhibits Keratinocyte Migration
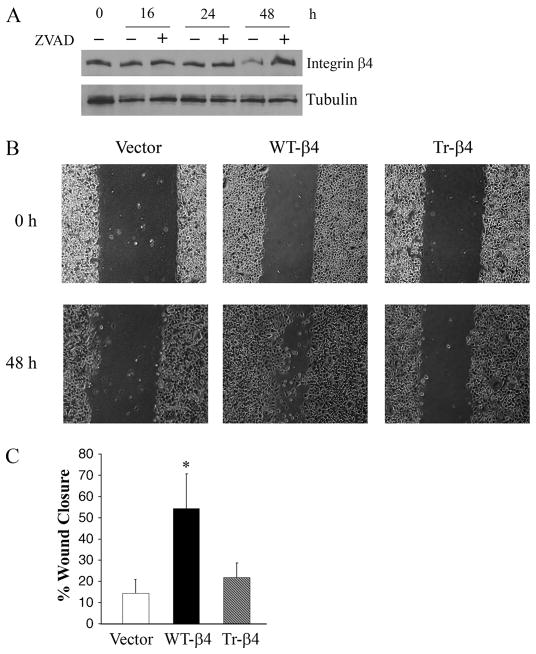
Because caspase-3 has been implicated in cell migration in non-apoptotic epithelial cells (57), we examined whether integrin β4 was cleaved in migrating keratinocytes. To this end, confluent JEB cells stably expressing GFP-tagged WT-β4 were scraped with a pipette tip in a grid-like pattern (multi-scratch wound assay). A reduction in the amount of full-length WT-β4 was observed at 48 h in this assay, and this reduction was inhibited by zVAD-fmk (Fig. 7A). These results indicate that integrin β4 is cleaved by caspases in migrating keratinocytes. To determine the effects of integrin β4 cleavage on cell migration, confluent JEB cells stably expressing GFP vector or GFP-tagged WT-β4 or Tr-β4 were scraped with a pipette tip, and wound closure was measured 48 h later. Although WT-β4 cells were highly motile in this assay, Tr-β4 cells exhibited little if any migration and were not significantly different from β4-deficient vector-transduced JEB cells (Fig. 7, B and C). Importantly, these migration differences were not due to differences in cell number (data not shown). These results suggest that caspase cleavage of integrin β4 inhibits keratinocyte migration.
FIGURE 7. Caspase cleavage of integrin β4 inhibits keratinocyte migration.
A, immunoblot analysis of JEB cells stably expressing GFP-tagged WT-β4 that were subjected to a multi-scratch wound assay as described under “Experimental Procedures” in the absence or presence of 50 μM zVAD-fmk. Lysates were prepared 0 – 48 h after scratching. B, photomicrograph of a wound closure experiment. Confluent JEB cells stably expressing GFP vector or GFP-tagged WT-β4 or Tr-β4 were scraped with a pipette tip, and wound closure was assessed at 48 h as described under “Experimental Procedures.” C, data are presented as the mean ± S.E. of three experiments. *, p < 0.05 versus vector control.
DISCUSSION
We have demonstrated that the integrin β4 subunit is a novel caspase substrate that is cleaved by caspases-3 and -7 in vitro and in apoptotic epithelial cells. Caspases-3 and -7 proteolyze integrin β4 at a DEXD consensus caspase-3 and -7 cleavage motif (46), DELD1109↓R, in its cytoplasmic tail. Interestingly, this aspartic acid residue and the entire cleavage motif in integrin β4 has been highly conserved during evolution (DELD-R in dogs and cattle and DETD-R in mice and rats), suggesting that caspase proteolysis of β4 may occur at this same site during apoptosis in other species as well. As illustrated in Fig. 2C, caspase cleavage of β4 removes much of its cytoplasmic tail, including all four of the FNIII repeats and the connecting sequence. The resulting carboxyl-terminal caspase cleavage product is unstable and is rapidly degraded by the proteasome. Indeed, the amino-terminal amino acid of this cleavage fragment (Arg1110) is a destabilizing residue that likely targets this cleavage product for degradation by the ubiquitin-dependent N-end rule pathway (58). Caspase proteolysis of NF-κB and the Drosophila inhibitor of apoptosis protein DIAP1 also generate unstable cleavage products that are degraded by the N-end rule pathway (51, 52), suggesting that caspases and the ubiquitin-proteasome pathway may collaborate to efficiently degrade certain substrates during the execution of apoptosis.
We have also demonstrated that removal of the cytoplasmic tail of integrin β4 by caspases, resulting in a truncated β4 integrin (amino acids 1–1109), has important structural and functional consequences. Although arbitrary deletional analyses of the cytoplasmic tail of β4 have demonstrated its importance in hemidesmosome assembly and diverse functions (25–27, 33, 35), our results provide the first evidence that β4 indeed undergoes proteolytic removal of its cytoplasmic tail in vivo during a normal cellular process, namely, apoptosis. Regarding the structural consequences, caspase-truncated β4, unlike full-length β4, is unable to assemble mature hemidesmosomal structures, which play a critical role in adhering epithelial cells to the basement membrane. Specifically, we have demonstrated that although Tr-β4 is present on the cell surface, it is often diffusely distributed rather than localized in polarized basal structures characteristic of hemidesmosomes. We also observed that Tr-β4 was completely impaired in its ability to recruit the hemidesmosomal protein plectin to hemidesmosome-like structures, whereas BP180 recruitment was partly impaired. One potential explanation for the more severe defect in plectin recruitment to hemidesmosomal structures by Tr-β4 compared with BP180 is that BP180 is a membrane spanning protein that also interacts with α6 integrin and laminin-5 (59–61). Hence, α6 integrin and laminin-5 may contribute to BP180 recruitment to hemidesmosomes even in the absence of the cytoplasmic tail of β4. In contrast, plectin is dependent on the first and/or second FNIII repeat and a portion of the connecting sequence of β4 (26, 27), domains that are absent from Tr-β4, for incorporation into hemidesmosomes. Collectively, our results suggest that caspase cleavage of integrin β4 disrupts mature hemidesmosomes, an event that may promote the detachment of epithelial cells from the basement membrane and subsequent extracellular matrix detachment-induced apoptosis or anoikis.
Consistent with this idea, we have shown that caspase cleavage of integrin β4 sensitizes keratinocytes to apoptosis. Specifically, we demonstrated that JEB cells stably expressing Tr-β4 or WT-β4 (which was cleaved in response to TRAIL treatment), but not caspase cleavage-resistant D1109E-β4, were more sensitive to apoptosis at early time points after treatment with TRAIL. These findings are concordant with reports that deletion of various domains in the cytoplasmic tail of integrin β4 result in enhanced apoptosis in keratinocytes and mammary epithelial cells (35, 62, 63). Because the cytoplasmic tail of integrin β4 is required for laminin-5-induced activation of cell survival pathways such as NF-κB and phosphatidylinositol 3-kinase (35, 62, 63), proteolytic removal of this domain by caspases might promote apoptosis by abrogating these survival signals.
In addition, we observed that integrin β4 is cleaved by caspases in migrating keratinocytes during wound healing, a finding that is consistent with a recent report implicating caspase-3 in cell migration of non-apoptotic epithelial cells (57). We have also demonstrated that proteolysis of integrin β4 has important functional consequences for cell migration. Although caspase-truncated β4 was unable to promote keratinocyte migration in a wound closure assay, full-length β4 robustly enhanced cell motility. These results indicate that caspase cleavage of integrin β4 inhibits cell migration. Our results are in agreement with previous reports demonstrating that integrin β4-deficient JEB keratinocytes identical to those used in these experiments or keratinocytes expressing a less extensively truncated β4 (amino acids 1–1355) are impaired in cell migration, an important component of wound healing (34, 35). These defects in cell migration have been attributed to impaired Rac1, NF-κB, and/or c-Jun NH2-terminal kinase activation in β4-deficient keratinocytes or keratinocytes expressing truncated β4. Indeed, it seems likely that caspase cleavage of integrin β4 alters its downstream signaling to these and other pathways by removing key signaling residues/domains, an hypothesis that will be systematically explored in future studies. Collectively, the findings presented here demonstrate a novel proteolytic mechanism that profoundly alters integrin β4 localization and function. Together with the recent demonstration that the intracellular domains of other receptors such as epidermal growth factor receptor and HER-2/ErbB2 are cleaved by caspases (64 – 66), these findings suggest that caspases may participate more broadly in regulating receptor-mediated signaling.
Acknowledgments
We are indebted to Dr. M. Peter Marinkovich for providing JEB keratinocytes.
Footnotes
The abbreviations used are: FNIII, fibronectin type III; JEB, junctional epidermolysis bullosa; WT, wild type; TRAIL, tumor necrosis factor-related apoptosis-inducing ligand; GFP, green fluorescent protein; PBS, phosphate-buffered saline; RT, room temperature; zVAD-fmk, benzyloxycarbonyl-VAD-fluoromethyl ketone.
This work was supported in part by National Institutes of Health Grants R01CA097198 (to V. L. C.), P50CA89018 (Specialized Programs of Research Excellence (SPORE) in Breast Cancer (to V. L. C.)), R01AR054184 (to J. C. R. J.), and T32DK07169 (to M. E. W.), by Dept. of Defense Breast Cancer Research Program DAMD17-03-1-0426 (to V. L. C.), and by the Avon Foundation Breast Cancer Research and Care Program (to V. L. C.).
References
- 1.Cryns V, Yuan J. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 2.Sakahira H, Enari M, Nagata S. Nature. 1998;391:96–99. doi: 10.1038/34214. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, Zou H, Slaughter C, Wang X. Cell. 1997;89:175–184. doi: 10.1016/s0092-8674(00)80197-x. [DOI] [PubMed] [Google Scholar]
- 4.Lazebnik YA, Takahashi A, Moir RD, Goldman RD, Poirier GG, Kaufmann SH, Earnshaw WC. Proc Natl Acad Sci U S A. 1995;92:9042–9046. doi: 10.1073/pnas.92.20.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao L, Perez D, White E. J Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caulín C, Salvesen GS, Oshima RG. J Cell Biol. 1997;138:1379–1394. doi: 10.1083/jcb.138.6.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ku NO, Liao J, Omary MB. J Biol Chem. 1997;272:33197–33203. doi: 10.1074/jbc.272.52.33197. [DOI] [PubMed] [Google Scholar]
- 8.Morishima N. Genes Cells. 1999;4:401–414. doi: 10.1046/j.1365-2443.1999.00270.x. [DOI] [PubMed] [Google Scholar]
- 9.Byun Y, Chen F, Chang R, Trivedi M, Green KJ, Cryns VL. Cell Death Differ. 2001;8:443–450. doi: 10.1038/sj.cdd.4400840. [DOI] [PubMed] [Google Scholar]
- 10.Chen F, Chang R, Trivedi M, Capetanaki Y, Cryns VL. J Biol Chem. 2003;278:6848–6853. doi: 10.1074/jbc.M212021200. [DOI] [PubMed] [Google Scholar]
- 11.Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, Chu K, McGarry TJ, Kirschner MW, Koths K, Kwiatkowski DJ, Williams LT. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 12.Kayalar C, Örd T, Testa MP, Zhong LT, Bredesen DE. Proc Natl Acad Sci U S A. 1996;93:2234–2238. doi: 10.1073/pnas.93.5.2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinhusen U, Weiske J, Badock V, Tauber R, Bommert K, Huber O. J Biol Chem. 2001;276:4972–4980. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- 14.Weiske J, Schoneberg T, Schroder W, Hatzfeld M, Tauber R, Huber O. J Biol Chem. 2001;276:41175–41181. doi: 10.1074/jbc.M105769200. [DOI] [PubMed] [Google Scholar]
- 15.Herren B, Levkau B, Raines EW, Ross R. Mol Biol Cell. 1998;9:1589–1601. doi: 10.1091/mbc.9.6.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dusek RL, Getsios S, Chen F, Park JK, Amargo EV, Cryns VL, Green KJ. J Biol Chem. 2006;281:3614–3624. doi: 10.1074/jbc.M508258200. [DOI] [PubMed] [Google Scholar]
- 17.Chen F, Arseven OK, Cryns VL. J Biol Chem. 2004;279:27542–27548. doi: 10.1074/jbc.M400971200. [DOI] [PubMed] [Google Scholar]
- 18.Chen F, Kamradt M, Mulcahy M, Byun Y, Xu H, McKay MJ, Cryns VL. J Biol Chem. 2002;277:16775–16781. doi: 10.1074/jbc.M201322200. [DOI] [PubMed] [Google Scholar]
- 19.Cryns VL, Byun Y, Rana A, Mellor H, Lustig KD, Ghanem L, Parker PJ, Kirschner MW, Yuan J. J Biol Chem. 1997;272:29449–29453. doi: 10.1074/jbc.272.47.29449. [DOI] [PubMed] [Google Scholar]
- 20.Lustig KD, Stukenberg PT, McGarry TJ, King RW, Cryns VL, Mead PE, Zon LI, Yuan J, Kirschner MW. Methods Enzymol. 1997;283:83–99. doi: 10.1016/s0076-6879(97)83009-1. [DOI] [PubMed] [Google Scholar]
- 21.Giancotti FG, Ruoslahti E. Science. 1999;285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 22.Nievers MG, Schaapveld RQ, Sonnenberg A. Matrix Biol. 1999;18:5–17. doi: 10.1016/s0945-053x(98)00003-1. [DOI] [PubMed] [Google Scholar]
- 23.Jones JC, Hopkinson SB, Goldfinger LE. BioEssays. 1998;20:488–494. doi: 10.1002/(SICI)1521-1878(199806)20:6<488::AID-BIES7>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 24.Aumailley M, Bruckner-Tuderman L, Carter WG, Deutzmann R, Edgar D, Ekblom P, Engel J, Engvall E, Hohenester E, Jones JC, Kleinman HK, Marinkovich MP, Martin GR, Mayer U, Meneguzzi G, Miner JH, Miyazaki K, Patarroyo M, Paulsson M, Quaranta V, Sanes JR, Sasaki T, Sekiguchi K, Sorokin LM, Talts JF, Tryggvason K, Uitto J, Virtanen I, von der Mark K, Wewer UM, Yamada Y, Yurchenco PD. Matrix Biol. 2005;24:326–332. doi: 10.1016/j.matbio.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 25.Spinardi L, Ren YL, Sanders R, Giancotti FG. Mol Biol Cell. 1993;4:871–884. doi: 10.1091/mbc.4.9.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niessen CM, Hulsman EH, Oomen LC, Kuikman I, Sonnenberg A. J Cell Sci. 1997;110:1705–1716. doi: 10.1242/jcs.110.15.1705. [DOI] [PubMed] [Google Scholar]
- 27.Schaapveld RQ, Borradori L, Geerts D, van Leusden MR, Kuikman I, Nievers MG, Niessen CM, Steenbergen RD, Snijders PJ, Sonnenberg A. J Cell Biol. 1998;142:271–284. doi: 10.1083/jcb.142.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopkinson SB, Jones JC. Mol Biol Cell. 2000;11:277–286. doi: 10.1091/mbc.11.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vidal F, Aberdam D, Miquel C, Christiano AM, Pulkkinen L, Uitto J, Ortonne JP, Meneguzzi G. Nat Genet. 1995;10:229–234. doi: 10.1038/ng0695-229. [DOI] [PubMed] [Google Scholar]
- 30.Ruzzi L, Gagnoux-Palacios L, Pinola M, Belli S, Meneguzzi G, D’Alessio M, Zambruno G. J Clin Investig. 1997;99:2826–2831. doi: 10.1172/JCI119474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dowling J, Yu QC, Fuchs E. J Cell Biol. 1996;134:559–572. doi: 10.1083/jcb.134.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Neut R, Krimpenfort P, Calafat J, Niessen CM, Sonnenberg A. Nat Genet. 1996;13:366–369. doi: 10.1038/ng0796-366. [DOI] [PubMed] [Google Scholar]
- 33.Murgia C, Blaikie P, Kim N, Dans M, Petrie HT, Giancotti FG. EMBO J. 1998;17:3940–3951. doi: 10.1093/emboj/17.14.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russell AJ, Fincher EF, Millman L, Smith R, Vela V, Waterman EA, Dey CN, Guide S, Weaver VM, Marinkovich MP. J Cell Sci. 2003;116:3543–3556. doi: 10.1242/jcs.00663. [DOI] [PubMed] [Google Scholar]
- 35.Nikolopoulos SN, Blaikie P, Yoshioka T, Guo W, Puri C, Tacchetti C, Giancotti FG. Mol Cell Biol. 2005;25:6090–6102. doi: 10.1128/MCB.25.14.6090-6102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cryns VL, Bergeron L, Zhu H, Li H, Yuan J. J Biol Chem. 1996;271:31277–31282. doi: 10.1074/jbc.271.49.31277. [DOI] [PubMed] [Google Scholar]
- 37.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 38.Debnath J, Muthuswamy SK, Brugge JS. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 39.Kamradt MC, Lu M, Werner ME, Kwan T, Chen F, Strohecker A, Oshita S, Wilkinson JC, Yu C, Oliver PG, Duckett CS, Buchsbaum DJ, LoBuglio AF, Jordan VC, Cryns VL. J Biol Chem. 2005;280:11059–11066. doi: 10.1074/jbc.M413382200. [DOI] [PubMed] [Google Scholar]
- 40.Lu M, Kwan T, Yu C, Chen F, Freedman B, Schafer JM, Lee EJ, Jameson JL, Jordan VC, Cryns VL. J Biol Chem. 2005;280:6742–6751. doi: 10.1074/jbc.M411519200. [DOI] [PubMed] [Google Scholar]
- 41.Kamradt MC, Chen F, Cryns VL. J Biol Chem. 2001;276:16059–16063. doi: 10.1074/jbc.C100107200. [DOI] [PubMed] [Google Scholar]
- 42.Moyano JV, Evans JR, Chen F, Lu M, Werner ME, Yehiely F, Diaz LK, Turbin D, Karaca G, Wiley E, Nielsen TO, Perou CM, Cryns VL. J Clin Investig. 2006;116:261–270. doi: 10.1172/JCI25888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skalli O, Jones JC, Gagescu R, Goldman RD. J Cell Biol. 1994;125:159–170. doi: 10.1083/jcb.125.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hopkinson SB, Riddelle KS, Jones JC. J Investig Dermatol. 1992;99:264–270. doi: 10.1111/1523-1747.ep12616615. [DOI] [PubMed] [Google Scholar]
- 45.Hogervorst F, Kuikman I, von dem Borne AE, Sonnenberg A. EMBO J. 1990;9:765–770. doi: 10.1002/j.1460-2075.1990.tb08171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, Houtzager VM, Nordstrom PA, Roy S, Vaillancourt JP, Chapman KT, Nicholson DW. J Biol Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 47.Stahl S, Weitzman S, Jones JC. J Cell Sci. 1997;110:55–63. doi: 10.1242/jcs.110.1.55. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez AM, Otey C, Edlund M, Jones JC. J Cell Sci. 2001;114:4197–4206. doi: 10.1242/jcs.114.23.4197. [DOI] [PubMed] [Google Scholar]
- 49.Emoto Y, Manome Y, Meinhardt G, Kisaki H, Kharbanda S, Robertson M, Ghayur T, Wong WW, Kamen R, Weichselbaum R, Kufe D. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Breitschopf K, Haendeler J, Malchow P, Zeiher AM, Dimmeler S. Mol Cell Biol. 2000;20:1886–1896. doi: 10.1128/mcb.20.5.1886-1896.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, Meier P. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 52.Rathore N, Matta H, Chaudhary PM. J Biol Chem. 2004;279:39358–39365. doi: 10.1074/jbc.M406712200. [DOI] [PubMed] [Google Scholar]
- 53.Demontis S, Rigo C, Piccinin S, Mizzau M, Sonego M, Fabris M, Brancolini C, Maestro R. Cell Death Differ. 2006;13:335–345. doi: 10.1038/sj.cdd.4401744. [DOI] [PubMed] [Google Scholar]
- 54.Geuijen CA, Sonnenberg A. Mol Biol Cell. 2002;13:3845–3858. doi: 10.1091/mbc.02-01-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsuruta D, Hopkinson SB, Jones JC. Cell Motil Cytoskeleton. 2003;54:122–134. doi: 10.1002/cm.10089. [DOI] [PubMed] [Google Scholar]
- 56.Tsuruta D, Hopkinson SB, Lane KD, Werner ME, Cryns VL, Jones JC. J Biol Chem. 2003;278:38707–38714. doi: 10.1074/jbc.M301637200. [DOI] [PubMed] [Google Scholar]
- 57.Zhao X, Wang D, Zhao Z, Xiao Y, Sengupta S, Zhang R, Lauber K, Wesselborg S, Feng L, Rose TM, Shen Y, Zhang J, Prestwich G, Xu Y. J Biol Chem. 2006;281:29357–29368. doi: 10.1074/jbc.M513105200. [DOI] [PubMed] [Google Scholar]
- 58.Varshavsky A. Nat Cell Biol. 2003;5:373–376. doi: 10.1038/ncb0503-373. [DOI] [PubMed] [Google Scholar]
- 59.Hopkinson SB, Baker SE, Jones JC. J Cell Biol. 1995;130:117–125. doi: 10.1083/jcb.130.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hopkinson SB, Findlay K, deHart GW, Jones JC. J Investig Dermatol. 1998;111:1015–1022. doi: 10.1046/j.1523-1747.1998.00452.x. [DOI] [PubMed] [Google Scholar]
- 61.Tasanen K, Tunggal L, Chometon G, Bruckner-Tuderman L, Aumailley M. Am J Pathol. 2004;164:2027–2038. doi: 10.1016/S0002-9440(10)63762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weaver VM, Lelievre S, Lakins JN, Chrenek MA, Jones JC, Giancotti F, Werb Z, Bissell MJ. Cancer Cell. 2002;2:205–216. doi: 10.1016/s1535-6108(02)00125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zahir N, Lakins JN, Russell A, Ming W, Chatterjee C, Rozenberg GI, Marinkovich MP, Weaver VM. J Cell Biol. 2003;163:1397–1407. doi: 10.1083/jcb.200302023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tikhomirov O, Carpenter G. J Biol Chem. 2001;276:33675–33680. doi: 10.1074/jbc.M101394200. [DOI] [PubMed] [Google Scholar]
- 65.Benoit V, Chariot A, Delacroix L, Deregowski V, Jacobs N, Merville MP, Bours V. Cancer Res. 2004;64:2684–2691. doi: 10.1158/0008-5472.can-03-2914. [DOI] [PubMed] [Google Scholar]
- 66.He YY, Huang JL, Chignell CF. Oncogene. 2006;25:1521–1531. doi: 10.1038/sj.onc.1209184. [DOI] [PubMed] [Google Scholar]