EGF triggers dephosphorylation of the mRNA-binding protein Grb7 in the nucleus, leading to its export, and it also stimulates FAK, which rephosphorylates Grb7 in the cytoplasm to release its mRNA cargo for translation.
Abstract
We report the first example of a coordinated dual action of epidermal growth factor (EGF) in stimulating the nuclear–cytoplasmic export and translation of a select messenger RNA (mRNA). The effect of EGF is mediated by the RNA-binding protein Grb7 (growth factor receptor–bound protein 7), which serves as an adaptor for a specific mRNA–protein export complex and a translational regulator. Using the κ–opioid receptor (OR [KOR]) as a model, we demonstrate that EGF activates nuclear SHP-2 (Src homology region 2–containing tyrosine phosphatase), which dephosphorylates Grb7 in the nucleus. Hypophosphorylated Grb7 binds to the KOR mRNA and recruits the Hu antigen R–exportin-1 (CRM1) complex to form a nuclear–cytoplasmic export complex that exports KOR mRNA. EGF also activates focal adhesion kinase in the cytoplasm to rephosphorylate Grb7, releasing KOR mRNA for active translation. In summary, this study uncovers a coordinated, dual activity of EGF in facilitating nuclear export of a specific mRNA–protein complex as well as translational activation of the exported mRNA.
Introduction
EGF is a mitogen that regulates multiple cellular processes, including proliferation, survival, and differentiation. EGF also exerts neurotrophic or neuromodulatory effects to regulate neuronal activity (Rosenberg and Noble, 1989; Goldshmit et al., 2004; Takebayashi et al., 2004; Kasai et al., 2005). EGF binds to the EGF receptor to initiate signal transduction through multiple downstream signal mediators. For neurons, it has become increasingly clear that regulation of protein expression frequently occurs at the level of posttranscription, such as RNA mobilization and local translation (Lin and Holt, 2007; Bassell and Warren, 2008; Costa-Mattioli et al., 2009). Given the documented action of EGF in neurons, it is unclear whether and how EGF may regulate gene and protein expression at the posttranscriptional level.
As demonstrated in studies of opioid receptor (OR) gene knockout models, activation of ORs could modulate pain sensation, cognition functions, and animal behavior (Gavériaux-Ruff and Kieffer, 2002; Kieffer and Gavériaux-Ruff, 2002). There are three ORs, which are designated μ, δ, and κ. All three ORs are detected in the developing central and peripheral nerve systems (Zhu et al., 1998; Land et al., 2008). κ-OR (KOR) signaling affects multiple physiological processes, including diuresis, dysphoria, stress response, and pain sensations (Kieffer and Gavériaux-Ruff, 2002; Ko et al., 2003; Ansonoff et al., 2006; Bruchas et al., 2007). Previously, we demonstrated that KOR translation could be enhanced by the axon guidance cue netrin-1 through FAK and Grb7 (growth factor receptor–bound protein 7) signaling (Tsai et al., 2006, 2007). In this study, we identify a new mechanism underlying the stimulating effect of EGF on KOR expression and demonstrate a unique signaling pathway of EGF in coordinately facilitating nuclear export and translation of a specific mRNA.
Results and discussion
EGF mediates Grb7-dependent activation of KOR levels and KOR mRNA nuclear export
We previously showed that axon guidance factor netrin-1 could regulate KOR mRNA translation through a FAK- and Grb7-dependent pathway (Tsai et al., 2006, 2007) and that KOR mRNA could be transported in neurons (Bi et al., 2006, 2007). The aim of this study was to identify regulatory signals for KOR posttranscriptional events, particularly in mobilizing mRNA, that could augment the functional KOR protein level in neurons. We found that EGF could play an important role. In a KOR ligand binding assay using 3H-U69,593 of rat dorsal root ganglion (DRG) neurons (Fig. 1 A), we found that EGF increased 3H-U69,593 binding to KOR, which was blocked by cold U69,593, dynorphin, or nor-binaltorphimine (nor-BNI). To determine the target of EGF action, we examined the level of EGF-stimulated KOR level in the presence of translational inhibitors (cycloheximide and puromycin) or a transcriptional inhibitor (actinomycin-D). Cycloheximide and puromycin reduced the amount of EGF-stimulated KOR protein, whereas actinomycin-D did not (Fig. 1 B). To determine the posttranscriptional target of EGF’s action in elevating the KOR level, we examined Grb7, a previously identified KOR mRNA–binding protein that recognizes its 5′ untranslated region (Tsai et al., 2007). First, we asked whether Grb7 was involved in the effect of EGF on KOR expression using siRNA against Grb7 (Fig. 1 C). The data showed that Grb7 knockdown reduced the effect of EGF on KOR expression, which was rescued by expressing siRNA-insensitive Flag-Grb7 (see Materials and methods).
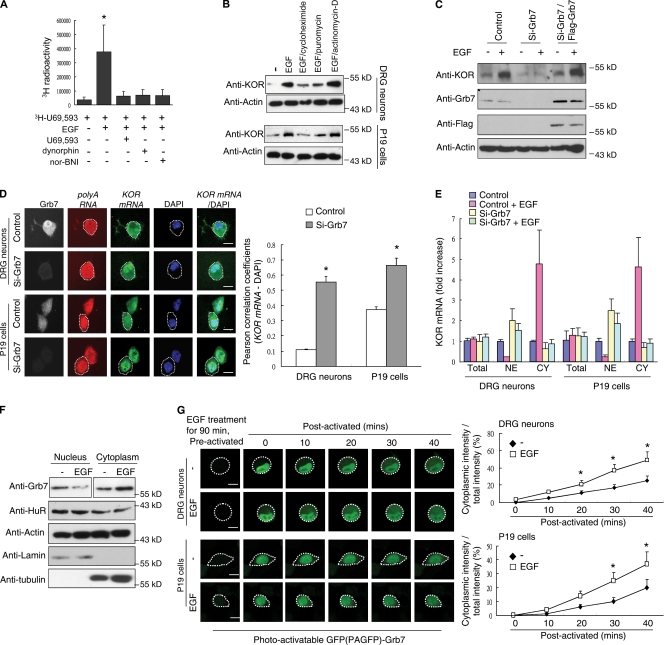
Figure 1.
Grb7 mediates EGF-induced KOR levels and KOR mRNA nuclear export. (A) Quantitative KOR ligand binding assay of primary rat DRG neurons with 3H-U69,593 in the presence or absence of EGF. Cold KOR agonists dynorphin and U69,593 and the antagonist nor-BNI were used for competition (*, P < 0.05). (B) Western blot analysis of primary rat DRG neurons (top) or P19 cells (bottom) in the presence or absence of EGF. Cycloheximide, puromycin, or actinomycin-D was applied along with EGF. (C) Western blot analysis of P19 cells transfected with control siRNA, Grb7 siRNA, or Grb7 siRNA plus an siRNA-insensitive Flag-Grb7 in the presence or absence of EGF. (D) FISH of DRG neurons (top) and P19 cells (bottom) with a fluorescein-12–labeled probe specific to KOR mRNA. Cells were pretransfected with control (first and third rows) or Grb7 siRNA (second and fourth rows). Oligo (dT) was used as a control to hybridize total poly(A) RNA. Silencing of Grb7 was monitored by immunohistochemistry using an anti-Grb7 antibody. Colocalization of KOR mRNA and DAPI was quantified using the Pearson correlation coefficients and is shown on the right (*, P < 0.05). (E) RT-qPCR of KOR mRNA from nuclear or cytoplasmic fractions of rat DRG neurons or P19 cells. Control siRNA– or Grb7 siRNA–transfected cells in the presence or absence of EGF treatment are shown. CY, cytoplasmic fraction; NE, nuclear fraction. (F) Western blot analysis of nuclear and cytoplasmic fractions from P19 cells in the presence or absence of EGF. α-Tubulin and lamin B served as the fractionation controls. (G) PAGFP-fused Grb7 was transfected into DRG neurons (top) and P19 cells (bottom). The images were obtained with a confocal microscope at the indicated time points. The quantified, relative cytoplasmic PAGFP-Grb7 signal from 50 cells is shown on the right (*, P < 0.05). (D and G) White dotted lines outline the cytoplasm of individual cells. (A, D, E, and G) Error bars represent SDs. Bars, 25 µm.
We then examined the possibility that Grb7 might regulate KOR mRNA distribution using FISH. As shown in Fig. 1 D, KOR mRNA was concentrated in the nuclei of Grb7-specific siRNA–transfected DRG neurons and P19 cells but not in control cells, suggesting a possible role of Grb7 in KOR mRNA export. The effect of EGF on nuclear and cytoplasmic distribution of endogenous KOR mRNA in DRG neurons and P19 cells was confirmed to be Grb7 dependent (Fig. 1 E and Fig. S1). Next, we determined whether the subcellular distribution of Grb7 was also affected by EGF and found a reduction in nuclear Grb7 and an elevation in cytoplasmic Grb7 upon EGF treatment (Fig. 1 F). We then determined whether EGF enhanced the nuclear export of Grb7 by using a photoactivatable GFP (PAGFP)–fused Grb7. As shown in Fig. 1 G, export of selectively activated nuclear PAGFP-Grb7 to the cytoplasm was significantly faster in EGF-treated cells than in control cells. Together, these data suggest that EGF triggers KOR mRNA nuclear export in a Grb7-dependent manner.
Hu antigen R (HuR) is required for Grb7 nuclear export
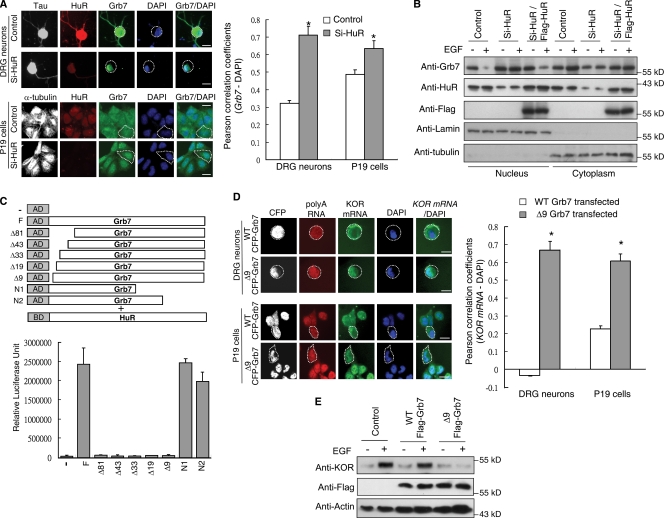
Grb7 does not mobilize RNA directly. However, Grb7 interacts with HuR (Tsai et al., 2008), a ubiquitously expressed RNA-binding protein which can be associated with export complexes (Brennan et al., 2000; Rebane et al., 2004) to regulate the nuclear–cytoplasmic shuttling of mRNA (Doller et al., 2008). We found that HuR-specific siRNA reduced Grb7 export (Fig. 2 A) and blocked EGF-stimulated cytoplasmic accumulation of Grb7 (Fig. 2 B), which was rescued by expressing siRNA off-target HuR (see Materials and methods). The HuR-interacting domain of Grb7 was previously mapped to the N-terminal segment (Tsai et al., 2008). Because the RNA-binding domain of Grb7 is also located in its N terminus (Tsai et al., 2007), a more detailed mapping was performed to determine the minimal HuR-interacting motif (Fig. 2 C). A dominant-negative mutant of Grb7 was made by deleting this HuR-interacting motif, called Δ9. The Δ9 mutant was detected in HuR interaction yet retained the KOR mRNA–binding ability (Fig. S2). The Δ9 and wild-type (WT) Grb7s were each fused to CFP and tested in FISH experiments. It appeared that WT CFP-Grb7 could be exported together with KOR mRNA, whereas Δ9 CFP-Grb7 remained trapped in the nucleus along with KOR mRNA in both DRG neurons and P19 cells (Fig. 2 D). The amount of KOR protein was not elevated by EGF treatment in P19 cells expressing Δ9 Flag-Grb7 but was effectively elevated by EGF in control cells and cells expressing WT Flag-Grb7 (Fig. 2 E). These results confirmed that the HuR–Grb7 complex is involved in exporting KOR mRNA to the cytoplasm.
Figure 2.
EGF induces HuR-dependent nuclear export of Grb7. (A) Immunohistochemistry of rat DRG neurons (top) and P19 cells (bottom) transfected with control or HuR siRNA. Colocalization of Grb7 and DAPI was quantified with the Pearson correlation coefficients and is shown on the right (*, P < 0.05). (B) Western blot analysis of nuclear and cytoplasmic fractions from P19 cells transfected as indicated in the presence or absence of EGF. (C) Mammalian two-hybrid assay in P19 cells transfected with Gal4-HuR and serial VP16-Grb7 deletion constructs. N-terminal deletions are named according to the number of deleted amino acid residues. (D) FISH detecting endogenous KOR mRNA in DRG neurons (top) and P19 cells (bottom) transfected with WT CFP-Grb7 or Δ9 CFP-Grb7. Colocalization of KOR mRNA and DAPI was quantified with the Pearson correlation coefficients and is shown on the right (*, P < 0.05). (A and D) White dotted lines outline the cytoplasm of the individual cells. (A, C, and D) Error bars represent SDs. (E) Western blots of P19 cells transfected as indicated, in the presence or absence of EGF. Bars, 25 µm.
Nuclear export of Grb7 is mediated by HuR and CRM1
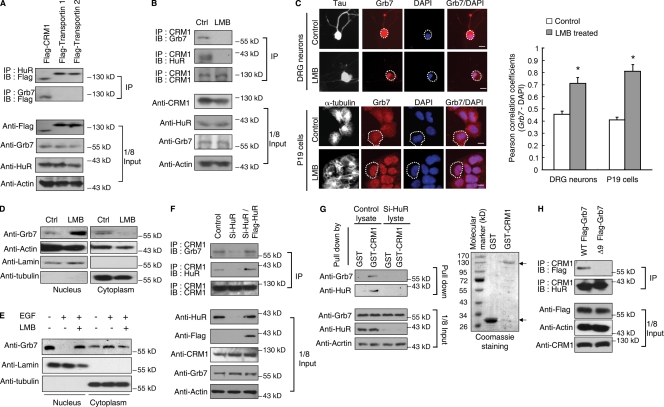
HuR utilizes three different complexes for nuclear export, including exportin-1 (also known as CRM1) and transportin 1 and 2 (Brennan et al., 2000; Rebane et al., 2004). Therefore, we wanted to determine the specific export complex associated with HuR–Grb7 using coimmunoprecipitation assays (Fig. 3 A). CRM1 was found in association with Grb7 and HuR, and the association was almost completely abolished upon treatment with the CRM1 inhibitor leptomycin B (LMB; Fig. 3 B), suggesting a specific interaction between CRM1 and HuR–Grb7 (Fig. 3 B). This finding was confirmed by immunohistochemical assays in the DRG neurons and P19 cells treated with LMB, in which there was a reduced cytoplasmic localization of Grb7 (Fig. 3 C). Fractionation experiments showed that LMB indeed reduced cytoplasmic accumulation of Grb7 in both control (Fig. 3 D) and EGF-treated (Fig. 3 E) P19 cells. The level of the Grb7–CRM1 immunocomplex was drastically decreased by HuR knockdown (Fig. 3 F, top), which is consistent with GST pull-down data (Fig. 3 G). Importantly, Δ9 Grb7 did not coimmunoprecipitate with CRM1 (Fig. 3 H), which confirmed the functional role of HuR in mediating the formation of a Grb7–CRM1 complex.
Figure 3.
Nuclear export of Grb7 is mediated by CRM1 and HuR. (A) Western blot analysis of coimmunoprecipitation of Flag-tagged CRM1–, transportin 1–, or transportin 2–transfected P19 cell lysates with anti-Grb7 or anti-HuR antibody. (B) Western blot analysis of coimmunoprecipitation of Grb7 and HuR from P19 cells with an anti-CRM1 antibody. (C) Immunohistochemistry of DRG neurons (top) and P19 cells (bottom) treated with or without LMB. White dotted lines outline the cytoplasm of individual cells. Colocalization of Grb7 and DAPI was quantified with Pearson correlation coefficients and is shown on the right (*, P < 0.05). Error bars represent SDs. (D) Western blot analysis of the nuclear and cytoplasmic fractions of P19 cells from cultures treated with or without LMB. (E) Western blot analysis of the nuclear and cytoplasmic fractions of P19 cells treated with EGF or EGF plus LMB. (D and E) α-Tubulin and lamin B served as the fractionation controls. (F) Western blot analysis of CRM1 immunoprecipitation from P19 cells transfected as indicated. (G) Western blot analysis of control or HuR-silenced P19 cell lysates pulled down by GST or GST-CRM1. The GST protein input is shown on the right. Predicted GST proteins are marked with arrows. (H) Western blot analysis of anti-CRM1 precipitated from P19 cells transfected as indicated. (A, B, and F–H) A one-eighth input control is shown on the bottom. Ctrl, control; IB, immunoblot; IP, immunoprecipitation. Bars, 25 µm.
Nuclear SHP-2 mediates the formation of an EGF-induced complex of Grb7, HuR, and CRM1
We have shown that the interaction between Grb7 and HuR is regulated by tyrosine phosphorylation of Grb7 (Tsai et al., 2008). Thus, we examined the tyrosine phosphorylation status of Grb7 in the nucleus and cytoplasm upon EGF treatment. LMB was used to block Grb7 export to obtain a quantitative measure of phosphorylated Grb7 in nucleus versus cytoplasm. Fig. 4 A showed reduced and increased tyrosine phosphorylation of nuclear and cytoplasmic Grb7, respectively, at 30 min and 90 min of EGF treatment. Without LMB, a significant redistribution of Grb7 (Fig. 4 A, bottom left) and KOR mRNA (Fig. 4 A, right) occurred after 90 and 120 min of EGF treatment, which is consistent with the time point at which nuclear Grb7 is dephosphorylated. Interestingly, the KOR protein level was readily elevated after only 30 min of EGF treatment, with or without LMB. These data suggest that the initial effect of EGF is to first enhance KOR translation, which is independent of the effect of EGF on KOR mRNA export that occurs later (Fig. 5, A and B).
Figure 4.
EGF activates nuclear SHP-2 to mediate complex formation of Grb7–HuR–CRM1. (A) Western blot analysis detecting nuclear or cytoplasmic Grb7 as well as tyrosine phosphorylated Grb7 in P19 cells treated with (top) or without (bottom) LMB and EGF as indicated. KOR was monitored, and α-tubulin and lamin B provided the fractionation controls. The RT-qPCR result of KOR mRNA from nuclear (top right) or cytoplasmic (bottom right) fractions of P19 cells treated with EGF is shown on the right (*, P < 0.05). (B) Phosphatase assay from immunoprecipitated nuclear and cytoplasmic SHP-2 from control or EGF-treated P19 cells (*, P < 0.05). The precipitation efficiency and input control are shown on the bottom. (C) Western blot analysis detecting phospho-tyrosine and Flag from the nuclear fractions of P19 cells transfected as indicated with or without EGF treatment. α-Tubulin and lamin B served as the fractionation controls. (D) Autoradiography of in vitro phosphatase assay on in vitro–prepared, anti-Flag antibody–precipitated WT or DM Flag-Grb7 in the presence or absence of SHP-2 or -1 and the SHP inhibitor NSC-87877. A similar amount of input protein was confirmed by Western blotting with an anti-Flag antibody and is shown on the bottom. The predicted signals of Flag-Grb7 are indicated by an arrow, and the nonspecific signal is marked by pound sign. (E) Western blot analysis of Grb7 or CRM1 immunoprecipitation (IP) from nuclear extracts of P19 cells transfected as indicated. A one-eighth input is shown on the bottom. (F) Western blots analysis of nuclear and cytoplasmic fractions (top) and RT-qPCR assay of KOR (bottom left) or actin (bottom right) mRNA from P19 cells transfected as indicated with or without EGF treatment. (A, B, and F) Error bars represent SDs. CY, cytoplasmic fraction; IB, immunoblot; NE, nuclear fraction.
Figure 5.
EGF activates cytoplasmic FAK to enhance KOR translation in the cytoplasm. (A) Western blot analysis detecting KOR, Grb7, and anti–phospho-tyrosine–immunoprecipitated Grb7 from the cytoplasmic (top) or nuclear (bottom) fractions of P19 cells. FAK, phospho-FAK (pFAK), Flag, Grb7, and actin serve as the controls for transfection and input. RT-qPCR assays of KOR and actin mRNA are shown on the right. Error bars represent SDs. CY, cytoplasmic fraction; NE, nuclear fraction. (B) Western blot analysis of P19 cells transfected as indicated and in the presence or absence of EGF. (C) Model of EGF’s dual action to elevate the level of KOR. Without EGF, nuclear SHP-2 and cytoplasmic FAK are inactive (i–SHP-2 and i-FAK). Upon EGF stimulation, unknown pathways activate nuclear SHP-2 (a–SHP-2), which dephosphorylates nuclear Grb7. Dephosphorylated nuclear Grb7 recruits KOR mRNA and HuR to form a nuclear export complex, thereby shuttling KOR mRNA to the cytoplasm. EGF also activates cytoplasmic FAK (a-FAK), which phosphorylates the preexisting cytoplasmic and nuclear-exported Grb7, releasing KOR mRNA for translation.
To identify the phosphatase responsible for EGF-induced dephosphorylation of Grb7 in the nucleus, we examined a direct interaction between the SHP-2 (Src homology region 2–containing phosphatase; Keegan and Cooper, 1996) and Grb7 (Fig. S3) and performed a tyrosine phosphatase assay using immunoprecipitated SHP-2 extracted from untreated or EGF-treated P19 cells. Fig. 4 B showed that SHP-2 activity was detected in both the nucleus and the cytoplasm, but only nuclear SHP-2 was activated upon EGF treatment. Because activation of SHP-2 would depend on its interacting proteins (Poole and Jones, 2005), selective nuclear activation of SHP-2 might result from the formation of a specific complex containing SHP-2 in the nucleus. This remains to be determined. EGF-stimulated dephosphorylation of nuclear Grb7 was found to occur at residues phosphorylated by FAK because WT Flag-Grb7 but not the dominant-negative phosphorylation mutant (DM Flag-Grb7; Tsai et al., 2007) was effectively dephosphorylated by EGF treatment (Fig. 4 C). The specificity of SHP-2 to Grb7 was further validated by an in vitro phosphatase assay (Fig. 4 D) and immunoprecipitation experiments (Fig. S3). SHP-2 but not SHP-1 lowered the level of tyrosine phosphorylation on Grb7 to 11%. Dephosphorylation of Grb7 was blocked by the SHP inhibitor NSC-87877 (Zhan et al., 2009), supporting the specificity of SHP-2 in dephosphorylating Grb7.
To validate SHP-2 as the activated phosphatase in response to EGF stimulation, the effect of SHP-2 on Grb7 phosphorylation and complex formation of Grb7, HuR, and CRM1 was examined by siRNA knockdown of SHP-2 in P19 cells. Fig. 4 E demonstrated that EGF stimulated tyrosine dephosphorylation of Grb7 in the nucleus and enhanced complex formation of Grb7, HuR, and CRM1. Silencing of SHP-2 blocked this complex formation (Fig. 4 E) and nuclear export of Grb7 and KOR mRNA (Fig. 4 F), which could be rescued by expressing siRNA off-target Flag–SHP-2 (see Materials and methods). The interaction between HuR–CRM1 was not affected, confirming the specificity of SHP-2 in dephosphorylating Grb7 and its subsequent complex formation with the HuR–CRM1 export complex.
EGF activates cytoplasmic FAK-dependent KOR translation
Hypophosphorylated Grb7 was shown to bind KOR mRNA and silence its translation. FAK could phosphorylate Grb7 and thereby release KOR mRNA for translation (Tsai et al., 2007). As shown in Fig. 5 A, EGF induced an increase in the phosphorylation of cytoplasmic Grb7, which was blocked by FAK inhibitor PF 573228 (Slack-Davis et al., 2007) and FAK inhibitor 14 (Golubovskaya et al., 2008) as well as by silencing FAK. The effect of silencing FAK was rescued by expressing siRNA-insensitive WT Flag-FAK (see Materials and methods) but not by the dominant-negative Flag-FAK-Y397F (Tsai et al., 2008). Importantly, blocking FAK function failed to inhibit EGF-stimulated Grb7 and KOR mRNA export (Fig. 5 A, right), suggesting the specificity of FAK in phosphorylating cytoplasmic Grb7. Finally, EGF enhanced the protein level of KOR in cells expressing WT Grb7 but not in cells expressing a phosphorylation mutant of Grb7 (DM Flag-Grb7; Fig. 5 B), concluding that EGF-stimulated KOR translation is mediated by FAK phosphorylation of cytoplasmic Grb7.
Collectively, this study establishes a dual action of EGF through a novel mechanism that couples stimulated nuclear–cytoplasmic export of a KOR mRNA–protein complex and enhanced KOR translation in the cytoplasm (Fig. 5 C). A genetic approach would be needed to evaluate this pathway in the context of whole animals in the future. Although a previous study showed posttranscriptional regulation in neurons by a cytoplasmic kinase (Hüttelmaier et al., 2005), this study provides the first example demonstrating mRNA mobilization and translation regulated by the dephosphorylation and phosphorylation of the same RNA-binding protein. Although HuR is known to use three different nuclear export machineries, Grb7 is only associated with CRM1 through interaction with HuR. These data suggest that other factors or molecules might be involved in either promoting KOR mRNA–Grb7–HuR interaction with CRM1 or prohibiting the interaction of Grb7 with the two transportin complexes. Because the nuclear and cytoplasmic distribution of SHP-2 was not affected by EGF treatment, the pathways transmitting the membrane EGF signal to the nucleus remain to be examined. Also, nuclear export of HuR is not affected by EGF, suggesting an interesting possibility that the nuclear export of a specific mRNA engages only certain specific RNA-associated proteins (in this case through Grb7). Upon stimulation, these specific RNA-associated protein complexes recruit the constitutively active housekeeping nuclear–cytoplasmic shuttling machinery. Presumably, it would be more economical for cells to dynamically modulate specific proteins (and mRNAs) that may be required to react to certain physiological needs instead of altering the global export system.
Materials and methods
Antibodies and reagents
The antibodies were purchased from Santa Cruz Biotechnology, Inc. (anti-Grb7, anti-HuR, anti–SHP-2, anti–lamin B, anti–α-tubulin, antiactin, anti-CRM1, anti-FAK, anti-EGF, anti–EGF receptor, and anti–phospho-FAK), Millipore (anti-GST and anti–phospho-tyrosine), Abcam (anti-KOR), and Sigma-Aldrich (anti-Tau and anti-Flag). EGF, rapamycin, LMB, cycloheximide, puromycin, actinomycin-D, DAPI, U69,593, nor-BNI, and dynorphin were purchased from Sigma-Aldrich. PF 573228 and FAK inhibitor 14 were obtained from Tocris Bioscience. NSC-87877 was obtained from EMD. 3H-U69,593 was obtained from PerkinElmer.
Plasmid constructs, siRNAs, restoration experiments, and transfection
The generation of GST-Grb7, GST-HuR, WT Flag-Grb7, DM Flag-Grb7, WT Flag-FAK, Flag-FAK-Y397F, and Flag-HuR was described previously (Tsai et al., 2007, 2008). Flag–SHP-2 was constructed by subcloning SHP-2 cDNA (provided by C.-K. Qu, Case Western Reserve University, Cleveland, OH) into Flag-tagged pCMX. Gal4-HuR was constructed by inserting full-length HuR cDNA into EcoRI–XbaI-digested pM (Takara Bio Inc.). Full-length and deleted Grb7 constructs were cloned into EcoRI–XbaI-digested pVP16 (Takara Bio Inc.). The 9-aa deleted (Δ9) Flag-Grb7 and GST-Grb7 were cloned into BamHI–NheI-digested pCMX and BamHI–NotI-digested pGEX-2T (GE Healthcare). CFP-tagged WT Grb7, Δ9 Grb7, and DM Grb7 were constructed by amplifying the Flag-tagged constructs and cloning them into BglII–BamHI-digested pECFP-C1 (Takara Bio Inc.). PAGFP-fused WT Grb7 was constructed by amplifying Flag-tagged constructs and cloning them into BglII–BamHI-digested pPAGFP-C1 (Addgene). Flag-tagged CRM1 and transportin 1 and 2 were cloned into BamHI–NheI-digested pCMX. GST-CRM1 was constructed by inserting full-length CRM1 cDNA into BamHI–HindIII-digested pGEX-2T (GE Healthcare). siRNAs were all purchased from QIAGEN. The targeting sequences are as follows: Grb7, 5′-AGGATTCGAAATCAAATCCAA-3′ and 5′-TAGCCGCTTCATCTTCCGTAA-3′; HuR, 5′-CAGAAACATTTGAGCATTGTA-3′ and 5′-AAAGTATATATTAAAGTGAAA-3′; SHP-2, 5′-CAGAGTATTAAAGAAATATAA-3′ and 5′-AAGAGTTACATTGCCACTCAA-3′; and SHP-1, 5′-CCCTTCTATTCTTGTAAATAA-3′ and 5′-TGGGAGGAGTTTGAGAGTCTA-3′ or were previously described (Tsai et al., 2007). The siRNA-insensitive Flag-Grb7 and Flag-FAK were made by mutating the cDNA sequences to reduce their complementarity to siRNAs without changing their protein sequences (Tsai et al., 2007). In the cases of siRNA off-target Flag-HuR and Flag–SHP-2, siRNAs were made to target the untranslated regions of the endogenous genes without affecting full-length Flag construct expression. Plasmid transfections were performed using Lipofectamine 2000 (Invitrogen), and siRNA transfections were performed by Hyperfect reagent (QIAGEN).
Cell culture and nuclear–cytoplasmic fractionation
P19 cells and DRG neurons were cultured as previously described (Tsai et al., 2006) with slight modification. In DRG culture, to exclude extrinsic effects on KOR expression, NGF and N2 supplements were excluded from the medium during DRG culture. All experiments were performed within 5 d after initial plating to avoid neuronal death because of the lack of NGF. On the second day after plating, EGF was added at a final concentration of 100 µg/ml. Nuclear–cytoplasmic fractionation was conducted using the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Immunoprecipitation, GST pull-down, Western blotting, and ligand binding assay
For immunoprecipitation, cells were sonicated and incubated with antibodies for 1 h at 4°C and then with protein G beads for another 1 h. After washing three times, the samples were subjected to SDS-PAGE analysis followed by Western blotting. In the assays, IgG was used as a control antibody. For GST pull-down, GST-Grb7 proteins were bound to glutathione beads and incubated with total protein extract of P19 for 1 h. After washing, the samples were subjected to SDS-PAGE and Western blotting analysis. For Western blotting after SDS-PAGE, the gel was transferred onto a polyvinylidene fluoride membrane. After blocking with a 5% milk solution, the membrane was incubated with the primary antibody for 2 h at room temperature, followed by a 30-min washing with PBS. The membrane was then incubated with an HRP-conjugated secondary antibody for 1 h, followed by another 30-min washing. The membrane was then developed with a SuperSignal West Pico kit (Thermo Fisher Scientific). For ligand binding, primary DRG neurons were first treated with or without EGF for 6 h, followed by the addition of 1 µM 3H-U69,593 along with or without nor-BNI, dynorphin, or unlabeled U69,593 for another 20 min. After washing, the cell pellet was collected, and 3H counts were determined by a liquid scintillation counter (LS 6500; Beckman Coulter).
FISH and in vitro transcription
RNA probes for FISH were verified and synthesized as previously described (Bi et al., 2006) with fluorescein-12–UTP (Roche). FISH in DRG neurons or P19 cells with a synthesized probe or oligo (dT) was described previously (Tsai et al., 2009).
Immunohistochemistry and image acquisition
DRG neurons or P19 cells were fixed with ice-cold buffer (3% paraformaldehyde and 5% sucrose in PBS) for 15 min followed by permeabilization with 0.5% Triton X-100 in PBS for 5 min. After blocking (1% BSA in PBS) for 30 min, primary antibodies were diluted 1:200 with blocking buffer and applied to the fixed cells overnight at 4°C. After washing three times with PBS, Cy3-, Cy5-, or FITC-conjugated secondary antibodies were diluted 1:500 and applied to the cells for another 2 h at room temperature in the dark. After additionally washing the cells three times and a 5-min DAPI stain, the cells were observed on an inverted fluorescence microscope (IX70; Olympus). Images were acquired by Image ProPlus software (Media Cybernetics) and merged by Photoshop (Adobe). To track PAGFP-fused Grb7, transfected DRG neurons or P19 cells were treated with EGF and supplied with a CO2-independent medium (Invitrogen) before acquiring the images. After EGF treatment for 90 min, the cells were placed on a heating plate set at 37°C. The nucleus of the selected cell (nucleus was determined under brightfields) was excited at 405 nm and observed with a confocal microscope (FluoView FV1000; Olympus) equipped with a 60× NA 1.42 Plan-Apochromat oil immersion lens at 488 nm.
Quantification of digital images
The GFP signal from PAGFP-Grb7–transfected cells was directly measured by FV1000 viewer software (Olympus) simultaneously as the images were being taken. Three areas from an individual cell, and a total 20 cells, were analyzed. To quantify colocalization of the overlaid images, three independent sets of experiments with a total of 50 ± 5 cells were analyzed. Z stacks (optical sections) of the images were collected by a FluoView FV1000 confocal microscope at an optical thickness of 0.2 µm. Colocalization of multiple sets of optical images was quantified by WCIF ImageJ software (National Institutes of Health), where Pearson’s r values (Pearson correlation coefficients) were obtained (Leil et al., 2004; Segret et al., 2007).
Tyrosine phosphatase assay and in vitro kinase/phosphatase assay
The phosphatase activity assay was performed according to the manufacture’s protocol (Tyrosine Phosphatase Assay System; Promega). In brief, immunoprecipitated SHP-2 from P19 cells was incubated in the supplied reaction buffer with 1 mM tyrosine phosphopeptides for 20 min. The reaction was stopped by adding a molybdate dye mixture, and the free phosphate was detected at 630 nm. The kinase reaction was performed by using immunoprecipitated FAK, and the complex was washed (50 mM Tris–HCl, pH 7.5, 3 mM MnCl2, 3 mM MgCl2, and 100 mM NaCl). Flag-Grb7 was incubated with precipitated FAK in the presence of γ-[32P]ATP for 30 min at 37°C and analyzed by SDS-PAGE, followed by PhosphoImager (Molecular Dynamics) detection. The phosphatase assays were performed by adding immunoprecipitated SHP-2 or -1 to the kinase reaction for another 30 min with or without the SHP inhibitor NSC-87877. The final reaction mixture was then subjected to SDS-PAGE and analyzed by PhosphorImager.
Mammalian two-hybrid assay and quantitative RT-PCR (RT-qPCR)
The mammalian two-hybrid assay was performed by transfecting cells with 0.1 µg pBD-Gal4-HuR together with a series of 0.1 µg of N terminus–deleted pAD-Gal4-Grb7 constructs. All cotransfections also included 0.25 µg of the Gal4-tk-luciferase reporter and 0.05 µg of a CMV-lacZ internal control. 30 h after transfection, cells were lysed, and the luciferase activity was determined. For RT-qPCR, P19 cells or DRG neurons were fractionated into cytoplasmic and nuclear fractions with the NE-PER Nuclear and Cytoplasmic Extraction Reagents kit. Total RNA was then extracted from the fractions with TRIZOL reagent (Invitrogen). The purified RNA was subjected to reverse transcription reactions with a cDNA synthesis kit (QIAGEN). The cDNA was used for real-time PCR using a SYBR green quantitative PCR kit (Agilent Technologies). The specific primer pairs for real-time PCR were as follows: KOR, 5′-CATCATCAGGAAACTGCA-3′ and 5′-TGGTCATGTTTGTCATC-3′; and α-actin, 5′-TGGCCTTAGGGTGCAGGG-3′ and 5′-GTGGGCCGCTCTAGGCACCA-3′.
Statistical analysis
The data in this study are presented as the mean ± SD and were analyzed with the Student’s t test, where P < 0.05 was considered significant.
Online supplemental material
Fig. S1 shows the corresponding control of actin mRNA and Grb7 silencing efficiency of Fig. 1 E. Fig. S2 reveals that Δ9 Grb7 lost its affinity for HuR but retained its RNA-binding ability. Fig. S3 confirms that Grb7 directly interacts with SHP-2 and that SHP-2 but not SHP-1 mediates nuclear dephosphorylation of Grb7 after EGF treatment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200910083/DC1.
Acknowledgments
We would like to thank Dr. Cheng-Kui Qu for providing the SHP-2 constructs.
This work is supported in part by National Institutes of Health grants DA11190, DA11806, DK54733, DK60521, and K02-DA13926.
Footnotes
Abbreviations used in this paper:
- DRG
- dorsal root ganglion
- HuR
- Hu antigen R
- KOR
- κ-OR
- LMB
- leptomycin B
- nor-BNI
- nor-binaltorphimine
- OR
- opioid receptor
- PAGFP
- photoactivatable GFP
- RT-qPCR
- quantitative RT-PCR
- WT
- wild type
References
- Ansonoff M.A., Zhang J., Czyzyk T., Rothman R.B., Stewart J., Xu H., Zjwiony J., Siebert D.J., Yang F., Roth B.L., Pintar J.E. 2006. Antinociceptive and hypothermic effects of Salvinorin A are abolished in a novel strain of kappa-opioid receptor-1 knockout mice. J. Pharmacol. Exp. Ther. 318:641–648 10.1124/jpet.106.101998 [DOI] [PubMed] [Google Scholar]
- Bassell G.J., Warren S.T. 2008. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 60:201–214 10.1016/j.neuron.2008.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Tsai N.P., Lin Y.P., Loh H.H., Wei L.N. 2006. Axonal mRNA transport and localized translational regulation of kappa-opioid receptor in primary neurons of dorsal root ganglia. Proc. Natl. Acad. Sci. USA. 103:19919–19924 10.1073/pnas.0607394104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Tsai N.P., Lu H.Y., Loh H.H., Wei L.N. 2007. Copb1-facilitated axonal transport and translation of kappa opioid-receptor mRNA. Proc. Natl. Acad. Sci. USA. 104:13810–13815 10.1073/pnas.0703805104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.M., Gallouzi I.E., Steitz J.A. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1–14 10.1083/jcb.151.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruchas M.R., Land B.B., Aita M., Xu M., Barot S.K., Li S., Chavkin C. 2007. Stress-induced p38 mitogen-activated protein kinase activation mediates kappa-opioid-dependent dysphoria. J. Neurosci. 27:11614–11623 10.1523/JNEUROSCI.3769-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M., Sossin W.S., Klann E., Sonenberg N. 2009. Translational control of long-lasting synaptic plasticity and memory. Neuron. 61:10–26 10.1016/j.neuron.2008.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doller A., Pfeilschifter J., Eberhardt W. 2008. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell. Signal. 20:2165–2173 10.1016/j.cellsig.2008.05.007 [DOI] [PubMed] [Google Scholar]
- Gavériaux-Ruff C., Kieffer B.L. 2002. Opioid receptor genes inactivated in mice: the highlights. Neuropeptides. 36:62–71 10.1054/npep.2002.0900 [DOI] [PubMed] [Google Scholar]
- Goldshmit Y., Greenhalgh C.J., Turnley A.M. 2004. Suppressor of cytokine signalling-2 and epidermal growth factor regulate neurite outgrowth of cortical neurons. Eur. J. Neurosci. 20:2260–2266 10.1111/j.1460-9568.2004.03698.x [DOI] [PubMed] [Google Scholar]
- Golubovskaya V.M., Nyberg C., Zheng M., Kweh F., Magis A., Ostrov D., Cance W.G. 2008. A small molecule inhibitor, 1,2,4,5-benzenetetraamine tetrahydrochloride, targeting the y397 site of focal adhesion kinase decreases tumor growth. J. Med. Chem. 51:7405–7416 10.1021/jm800483v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttelmaier S., Zenklusen D., Lederer M., Dictenberg J., Lorenz M., Meng X., Bassell G.J., Condeelis J., Singer R.H. 2005. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 438:512–515 10.1038/nature04115 [DOI] [PubMed] [Google Scholar]
- Kasai A., Shima T., Okada M. 2005. Role of Src family tyrosine kinases in the down-regulation of epidermal growth factor signaling in PC12 cells. Genes Cells. 10:1175–1187 10.1111/j.1365-2443.2005.00909.x [DOI] [PubMed] [Google Scholar]
- Keegan K., Cooper J.A. 1996. Use of the two hybrid system to detect the association of the protein-tyrosine-phosphatase, SHPTP2, with another SH2-containing protein, Grb7. Oncogene. 12:1537–1544 [PubMed] [Google Scholar]
- Kieffer B.L., Gavériaux-Ruff C. 2002. Exploring the opioid system by gene knockout. Prog. Neurobiol. 66:285–306 10.1016/S0301-0082(02)00008-4 [DOI] [PubMed] [Google Scholar]
- Ko M.C., Willmont K.J., Lee H., Flory G.S., Woods J.H. 2003. Ultra-long antagonism of kappa opioid agonist-induced diuresis by intracisternal nor-binaltorphimine in monkeys. Brain Res. 982:38–44 10.1016/S0006-8993(03)02938-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land B.B., Bruchas M.R., Lemos J.C., Xu M., Melief E.J., Chavkin C. 2008. The dysphoric component of stress is encoded by activation of the dynorphin kappa-opioid system. J. Neurosci. 28:407–414 10.1523/JNEUROSCI.4458-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leil T.A., Chen Z.W., Chang C.S., Olsen R.W. 2004. GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. J. Neurosci. 24:11429–11438 10.1523/JNEUROSCI.3355-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A.C., Holt C.E. 2007. Local translation and directional steering in axons. EMBO J. 26:3729–3736 10.1038/sj.emboj.7601808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole A.W., Jones M.L. 2005. A SHPing tale: perspectives on the regulation of SHP-1 and SHP-2 tyrosine phosphatases by the C-terminal tail. Cell. Signal. 17:1323–1332 10.1016/j.cellsig.2005.05.016 [DOI] [PubMed] [Google Scholar]
- Rebane A., Aab A., Steitz J.A. 2004. Transportins 1 and 2 are redundant nuclear import factors for hnRNP A1 and HuR. RNA. 10:590–599 10.1261/rna.5224304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg A., Noble E.P. 1989. EGF-induced neuritogenesis and correlated synthesis of plasma membrane gangliosides in cultured embryonic chick CNS neurons. J. Neurosci. Res. 24:531–536 10.1002/jnr.490240411 [DOI] [PubMed] [Google Scholar]
- Segret A., Rücker-Martin C., Pavoine C., Flavigny J., Deroubaix E., Châtel M.A., Lombet A., Renaud J.F. 2007. Structural localization and expression of CXCL12 and CXCR4 in rat heart and isolated cardiac myocytes. J. Histochem. Cytochem. 55:141–150 10.1369/jhc.6A7050.2006 [DOI] [PubMed] [Google Scholar]
- Slack-Davis J.K., Martin K.H., Tilghman R.W., Iwanicki M., Ung E.J., Autry C., Luzzio M.J., Cooper B., Kath J.C., Roberts W.G., Parsons J.T. 2007. Cellular characterization of a novel focal adhesion kinase inhibitor. J. Biol. Chem. 282:14845–14852 10.1074/jbc.M606695200 [DOI] [PubMed] [Google Scholar]
- Takebayashi M., Hayashi T., Su T.P. 2004. Sigma-1 receptors potentiate epidermal growth factor signaling towards neuritogenesis in PC12 cells: potential relation to lipid raft reconstitution. Synapse. 53:90–103 10.1002/syn.20041 [DOI] [PubMed] [Google Scholar]
- Tsai N.P., Bi J., Loh H.H., Wei L.N. 2006. Netrin-1 signaling regulates de novo protein synthesis of kappa opioid receptor by facilitating polysomal partition of its mRNA. J. Neurosci. 26:9743–9749 10.1523/JNEUROSCI.3014-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai N.P., Bi J., Wei L.N. 2007. The adaptor Grb7 links netrin-1 signaling to regulation of mRNA translation. EMBO J. 26:1522–1531 10.1038/sj.emboj.7601598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai N.P., Ho P.C., Wei L.N. 2008. Regulation of stress granule dynamics by Grb7 and FAK signalling pathway. EMBO J. 27:715–726 10.1038/emboj.2008.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai N.P., Tsui Y.C., Wei L.N. 2009. Dynein motor contributes to stress granule dynamics in primary neurons. Neuroscience. 159:647–656 10.1016/j.neuroscience.2008.12.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y., Counelis G.J., O’Rourke D.M. 2009. The protein tyrosine phosphatase SHP-2 is required for EGFRvIII oncogenic transformation in human glioblastoma cells. Exp. Cell Res. 315:2343–2357 10.1016/j.yexcr.2009.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Hsu M.S., Pintar J.E. 1998. Developmental expression of the mu, kappa, and delta opioid receptor mRNAs in mouse. J. Neurosci. 18:2538–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]