Sisters on the loose (SOLO) protein functions as a meiotic cohesin component critical for correct chromosome segregation in Drosophila.
Abstract
Sister chromatid cohesion is essential to maintain stable connections between homologues and sister chromatids during meiosis and to establish correct centromere orientation patterns on the meiosis I and II spindles. However, the meiotic cohesion apparatus in Drosophila melanogaster remains largely uncharacterized. We describe a novel protein, sisters on the loose (SOLO), which is essential for meiotic cohesion in Drosophila. In solo mutants, sister centromeres separate before prometaphase I, disrupting meiosis I centromere orientation and causing nondisjunction of both homologous and sister chromatids. Centromeric foci of the cohesin protein SMC1 are absent in solo mutants at all meiotic stages. SOLO and SMC1 colocalize to meiotic centromeres from early prophase I until anaphase II in wild-type males, but both proteins disappear prematurely at anaphase I in mutants for mei-S332, which encodes the Drosophila homologue of the cohesin protector protein shugoshin. The solo mutant phenotypes and the localization patterns of SOLO and SMC1 indicate that they function together to maintain sister chromatid cohesion in Drosophila meiosis.
Introduction
Meiosis is a specialized cell division that functions in sexual reproduction to generate haploid gametes from diploid precursor cells. It consists of two divisions preceded by a single round of DNA replication. Meiosis I is a reductional division in which homologous chromosomes (homologues) segregate to opposite spindle poles. Meiosis II is an equational division in which sister chromatids separate (Page and Hawley, 2003).
Two key differences in chromosome behavior underlie the different segregation patterns in meiosis I and II. One is the manner in which segregating chromosomes are connected. Stable connections between segregating chromosomes are essential to prevent them from separating prematurely and to provide the tension required to enable the chromosomes to achieve bipolar alignment on the spindle. In meiosis II, as in mitosis, the critical connections are cohesion between sister centromeres. Cohesion is established during replication and preserved throughout the cell cycle until its removal at the onset of anaphase (anaphase II of meiosis). In meiosis I, stable connections between homologues must be established. In most organisms, these connections take the form of chiasmata, which derive from crossovers between homologous chromatids and which are stabilized by cohesion between sister chromatid arms distal to the crossover sites. Thus, sister chromatid cohesion underlies the connections between segregating chromosomes in both meiotic divisions (Petronczki et al., 2003). However, in some eukaryotes, such as Drosophila melanogaster males, homologue exchange and chiasmata are absent (Wolf, 1994). In Drosophila, homologue connections are provided by the male meiosis-specific chromosomal proteins stromalin in meiosis (SNM) and mod(mdg4) in meiosis (MNM; Thomas et al., 2005).
Cohesion is mediated by a conserved cohesin complex consisting of one member each of the SMC1, SMC3, SCC1/RAD21/REC8, and SCC3/SA families. Proteolytic cleavage of the SCC1 subunit (or its meiotic paralogue REC8) of cohesin at anaphase by separase triggers chromosome segregation during mitosis and at both meiotic divisions. In meiosis, this requires two separate rounds of separase activation: one round at anaphase I to cleave arm cohesins, release chiasmata, and allow homologues to segregate, and a second round at anaphase II to cleave centromere cohesin and allow sister chromatids to segregate (Petronczki et al., 2003). Conserved centromeric proteins called shugoshins function to protect centromeric cohesins from premature cleavage by separase during anaphase I (Watanabe, 2005).
A second critical difference between meiosis I and II is the orientation adopted by sister centromeres. In meiosis II, as in mitosis, sister centromeres orient back to back and establish separate kinetochores that make independent attachments to spindle poles. In meiosis I, sister centromeres adopt a side by side orientation and collaborate in forming a single functional kinetochore, ensuring that only two functional kinetochores are present per bivalent despite the presence of four chromatids. This enables sister centromeres to coorient (become attached to spindle fibers emanating from the same pole), which in turn enables homologous centromeres to biorient (Hauf and Watanabe, 2004). The mechanism of sister centromere coorientation is not well understood. In Saccharomyces cerevisiae, it depends on a centromeric meiosis I–specific complex called monopolin. The role of cohesin in centromere orientation is unclear (Tóth et al., 2000; Monje-Casas et al., 2007). In Schizosaccharomyces pombe, coorientation requires both meiosis-specific Rec8 cohesin and MoaI, a specialized centromere protein that appears to function primarily by stabilizing occupancy of centromere core sequences by Rec8 cohesin (Watanabe and Nurse, 1999; Watanabe et al., 2001; Yokobayashi and Watanabe, 2005). Recently, it has been shown that provision of an artificial tether between sister centromere core sequences suffices for preferential sister centromere coorientation in the absence of Rec8 or MoaI (Sakuno et al., 2009). The mechanism of coorientation in higher eukaryotes is not known in any detail, but the fact that rec8 mutations disrupt centromere orientation in several model eukaryotes suggests a role for cohesin (Yu and Dawe, 2000; Cai et al., 2003; Chelysheva et al., 2005; Xu et al., 2005).
Although there is considerable evidence that the aforementioned two-stage, cohesin-based meiotic segregation mechanism is widely conserved (Pasierbek et al., 2001; Cai et al., 2003; Petronczki et al., 2003; Chelysheva et al., 2005; Xu et al., 2005), the role of cohesin in meiotic cohesion in Drosophila remains unclear. This is due in large part to the absence of a functional rec8 orthologue and of meiosis-specific cohesin mutations. In addition to the four mitotic cohesins, the fly genome encodes two meiosis-specific cohesin paralogues: C(2)M, an SCC1/RAD21 paralogue required for homologue synapsis and recombination in female meiosis, and SNM, an SCC3/SA paralogue required for stable homologue pairing in male meiosis (Manheim and McKim, 2003; Heidmann et al., 2004; Thomas et al., 2005). However, despite their homology to cohesin proteins, both C(2)M and SNM are dispensable for sister chromatid cohesion in meiosis. Although the orientation disruptor (ord) gene is required for meiotic sister chromatid cohesion, ORD has no homology to cohesins or to any other known proteins, and its subcellular localization pattern differs from that of cohesin. Thus, the relationship between ORD and cohesin and the precise role of ORD in cohesion are unclear (Miyazaki and Orr-Weaver, 1992; Bickel et al., 1996, 1997; Balicky et al., 2002).
Two lines of evidence support a role for cohesin in Drosophila meiosis. First, immunocytological studies have localized SMC1 to centromeres in both male and female meiosis I and to synaptonemal complexes in female meiosis (Thomas et al., 2005; Khetani and Bickel, 2007). Second, mutations in the Drosophila shugoshin homologue mei-S332 cause precocious sister chromatid separation (PSCS) and high frequencies of meiosis II nondisjunction (NDJ; Davis, 1971; Kerrebrock et al., 1992), which is consistent with a possible role of MEI-S332 in protection of centromeric cohesin at anaphase I. However, the molecular function of mei-S332 has not been established, and the inviability of cohesin component mutants has thus far prevented their meiotic roles from being characterized. Thus, the molecular basis for meiotic cohesion in Drosophila remains poorly defined.
In this study, we describe a novel Drosophila protein, sisters on the loose (SOLO), which is required for sister centromere cohesion and SMC1 localization to centromeres throughout meiosis and colocalizes with SMC1 on centromeres from the onset of meiosis until both proteins disappear at anaphase II. In addition to randomizing chromatid segregation in meiosis II, solo mutations result in a unique “random 2::2” segregation pattern at meiosis I that reflects complete loss of sister centromere coorientation but partial maintenance of bivalent structure and function. Our data indicate that SOLO plays a direct role in sister chromatid cohesion during Drosophila meiosis and suggest that it does so in close association with cohesin.
Results
solo mutations cause NDJ of homologous and sister chromatids
Three alleles of solo were identified among a group of EMS-induced mutations that interfere with paternal transmission of the fourth chromosome (Koundakjian et al., 2004; Wakimoto et al., 2004). All three mutations were found to cause high frequencies of sex chromosome NDJ in male meiosis, averaging 55.8% (Table I). NDJ frequencies were similar in homozygous (unpublished data) and hemizygous males, indicating that all three alleles are genetic nulls. The NDJ products included XX sperm, XY sperm, and sperm with no sex chromosomes (nullo-XY sperm) at frequencies of 4.4%, 10.7%, and 36%, respectively. This pattern is consistent with the premature loss of sister chromatid cohesion before anaphase I. A similar pattern has been previously reported for mutations in ord (Miyazaki and Orr-Weaver, 1992; Bickel et al., 1997). solo males also exhibited high sister chromatid and homologue NDJ frequencies for the autosomal second and fourth chromosomes (Table S1 and unpublished data). High frequencies of meiotic chromatid NDJ are also seen in solo females (unpublished data).
Table I.
Sex chromosome NDJ in solo males
| Male genotype | X | Y + YY | XY | XX | O | n | %NDJ | %sis |
| Z2-0198/Df | 438 | 505 | 172 | 104 | 658 | 1,877 | 56.4 | ND |
| Z2-3534/Df | 357 | 309 | 181 | 53 | 551 | 1,443 | 58.0 | ND |
| Z2-0338/Df | 472 | 478 | 199 | 76 | 720 | 1,945 | 55.1 | ND |
| Z2-0198/Z2-3534 | 94 | 105 | 58 | 14 | 132 | 403 | 54.1 | ND |
| Total solo | 1,361 | 1,397 | 610 | 247 | 2,061 | 5,676 | 55.8 | 44.7 |
| Gamete frequency (%) | 24.0 | 24.6 | 10.7 | 4.4 | 36.3 | 100 | ND | ND |
+/BsYy+ males with the indicated second chromosome genotypes were crossed singly to 2-3 C(1)RM/0, y2 su(wa) wa females. Df = Df (2L)A267 (35B1; 35C1), in which the solo locus is completely deleted. n, total number of progeny scored; %sis, (percent sister chromatid NDJ/total NDJ) = 100 x (2 x XX)/((2 x XX) + XY).
To gain insight into the mechanism of NDJ, we first compared spermatocytes from solo and wild-type (WT) males that had been stained with DAPI to visualize DNA and with an antitubulin antibody to visualize spindles. Although no major abnormalities in meiosis I chromosome morphology or segregation were observed (Fig. S1, A and B), meiosis II proved to be visibly abnormal. More than 90% of solo spermatocytes in metaphase II exhibited DAPI-stained (DNA) masses that were both smaller and more numerous than in WT males, which is indicative of PSCS (Fig. S1 C). Chromosome segregation at anaphase II appeared disorganized. Laggards were observed in ∼38% of anaphase II nuclei, and meiosis II poles exhibited clearly unequal amounts of DNA in 88% of nuclei, indicating high rates of meiosis II NDJ (Fig. S1 D).
Sister centromere coorientation is disrupted by solo mutations
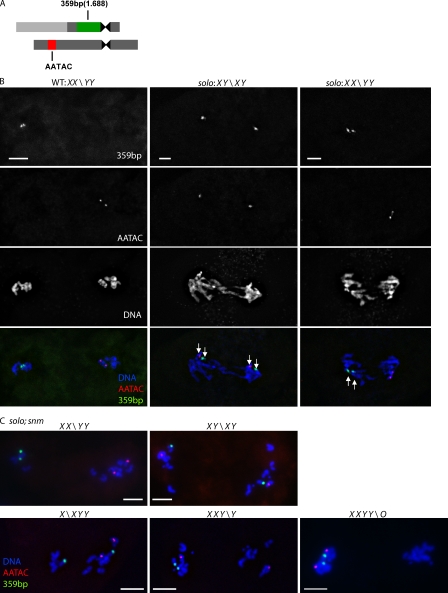
To examine the effects of solo mutations on meiosis I in more depth, we performed dual FISH using two probes that recognize X chromosome–specific (359 bp) and Y chromosome–specific (AATAC) repeat arrays (Fig. 1 A). This allowed us to track the segregation patterns of the X and Y chromosomes. As expected, in WT meiosis I, the X and Y chromosomes segregated reductionally, i.e., to opposite poles. The 359 bp and AATAC signals were at opposite poles in all 46 anaphase I spermatocytes (Fig. 1 B and Table II), and 128 metaphase II spermatocytes showed either 359 bp or AATAC signals but never both (Fig. 2 A and Table II). The 359 bp and AATAC loci behaved differently with respect to sister chromatid cohesion. All metaphase II nuclei that stained positively for AATAC exhibited two clearly separated AATAC spots, indicating absence of cohesion of the Y chromosome long arm. However, 66/67 metaphase II spermatocytes exhibited either a single 359 bp spot or two 359 bp spots in very close proximity, suggesting a much higher degree of cohesion. This difference might reflect the much greater proximity of the 359 bp repeats than the AATAC repeats to their respective centromeres.
Figure 1.
X-Y segregation patterns at anaphase I in solo and solo; snm males. X and Y chromatids were recognized by probes against 359 bp (green) and AATAC (red) satellite DNA repeats, respectively. DNA was stained with DAPI. Sum projections of 3D-deconvolved z series stacks were performed. (A) Schematic representation of probes on sex chromosomes used in FISH analysis. The 359 bp repeat probe detects a large heterochromatic region proximal to the X centromere; the AATAC repeat probe hybridizes to the middle of the long arm of the Y chromosome. (B) Representative segregation patterns in WT and soloZ2-0198/Df(2L)A267 spermatocytes. (left) Reductional segregation of X and Y chromatids in WT spermatocytes as indicated by 359 bp and AATAC probe signals at opposite poles. (middle and right) Equational and reductional (respectively) segregation of X and Y chromatids in solo spermatocytes. White arrows indicate fully separate sister chromatid probe signals. (C) Representative segregation patterns in solo; snm (soloZ2-0198/soloZ2-3534; snmZ3-2138/snmZ3-0317) spermatocytes. Merged images only are shown. Bars, 5 µm.
Table II.
Quantification of chromatid segregation patterns in solo, snm, solo; snm, and controls
| Chromatids | WT | solo | snm | solo; snm |
| Anaphase I | ||||
| XX\YY | 46 (100%) | 23 (31%) | 41 (58%) | 13 (14%) |
| XY\XY | 0 | 48 (65%) | 0 | 41 (43%) |
| XXY\Y | 0 | 2 (2.7%) | 4 (5.6%) | 19 (20%) |
| XYY\X | 0 | 1 (1.3%) | 4 (5.6%) | 16 (17%) |
| XXYY\O | 0 | 0 | 22 (31%) | 6 (6%) |
| Total | 46 | 74 | 71 | 95 |
| Metaphase II | ||||
| XX | 67 (52%) | 15 (20%) | ND | ND |
| YY | 61 (48%) | 14 (18%) | ND | ND |
| XY | 0 | 47 (62%) | ND | ND |
| X or XYY | 0 | 0 | ND | ND |
| Y or XXY | 0 | 0 | ND | ND |
| O or XXYY | 0 | 0 | ND | ND |
| Total | 128 | 76 | ND | ND |
Genotypes: WT, yw; solo, soloZ2-0198/Df(2L)A267; snm, snmZ3-2138/snmZ3-0317; and solo; snm, soloZ2-0198/soloZ2-3534; snmZ3-2138/snmZ3-0317. Chromatid constitution was determined primarily by numbers of 359 bp and AATAC repeat spots per pole or nucleus. For 359 bp repeat spots in WT and snm, some poles or nuclei exhibited only one 359 bp spot as a result of cohesion. In these cases, the karyotype determination was based on the number of X chromatids per chromosome revealed by DAPI staining. Percentages of each karyotype within each genotype are shown in parentheses.
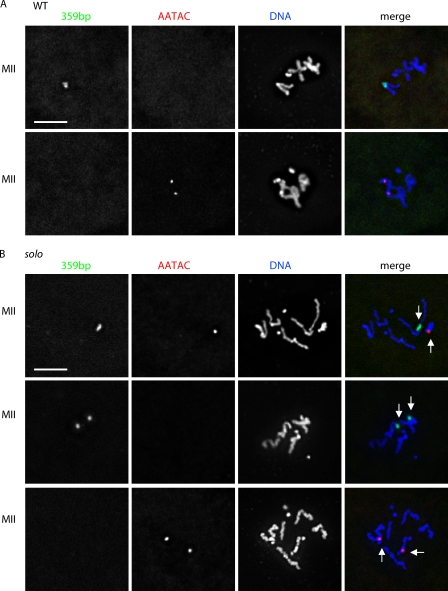
Figure 2.
PSCS in solo metaphase II spermatocytes. X and Y chromosomes were recognized by probes against 359 bp (green) and AATAC (red) satellite DNA repeats, respectively. DNA was stained with DAPI. Sum projections of 3D-deconvolved z series stacks were performed. (A) WT metaphase II (MII) nuclei all result from reductional meiosis I segregation and exhibit intact sister centromere cohesion. 128 spermatocytes were observed: 66 with either one bright 359 bp spot or two closely adjacent 359 bp spots and no AATAC spots (top), 61 with two AATAC spots and no 359 spots (bottom), and one with two well-separated 359 spots and no AATAC spots. (B) solo metaphase II nuclei exhibit PSCS and result from a mix of equational and reductional meiosis I segregation. Spermatocytes from adult soloZ2-0198/Df(2L)A267 were examined. 359 bp and AATAC signals are indicated by white arrows. (top) Nucleus with one 359 bp spot and one AATAC spot reflecting equational meiosis I segregation. (middle and bottom) Nuclei with two spots of 359 bp and no AATAC repeats or two spots of AATAC repeats and no 359 bp repeats, respectively, reflecting reductional meiosis I segregation. Bars, 5 µm.
solo spermatocytes differed from WT spermatocytes in two respects. First, cohesion was absent in all metaphase II solo spermatocytes (Fig. 2). This is evident both from the full separation of chromatids revealed by DAPI staining and from the presence of two separated FISH spots in all metaphase II spermatocytes, even those with only 359 bp signals (unpublished data). Thus, as suggested by the cross data (Table I) and the DAPI images in Fig. S1, solo causes extensive PSCS before or during meiosis II.
Second, solo mutants exhibited a more complex meiosis I segregation pattern than in WT (Fig. 1 and Table II). Reductional (normal) sex chromosome segregation was observed in only about one third of anaphase I spermatocytes (23/74). The most common meiosis I segregation pattern was equational segregation of both chromosomes, i.e., segregation of one X and one Y chromatid to each pole. 359 bp and AATAC signals (single spots of each) were present at both poles in 48/74 anaphase I solo spermatocytes. In contrast, spermatocytes resulting from 3::1 segregations of the X and Y chromatids were rare (only 3/74 anaphase I spermatocytes), and 4::0 segregations were completely absent. Thus, nearly all of the spermatocytes exhibited two chromatids at each pole: either two X chromatids at one pole and two Y chromatids at the other pole (reductional) or an X and a Y chromatid at each pole (equational). A similar pattern was observed in metaphase II spermatocytes (Fig. 2 and Table II).
The observed ratio of reductional to equational segregations at anaphase I (23:48) conformed to the 1:2 ratio that would be expected if the four chromatids of the X-Y bivalent segregate randomly at meiosis I subject to a constraint that two chromatids nearly always orient to each pole (2::2). This would yield a 1:2 ratio of reductional to equational segregations because there are two distinct 2::2 equational orientations (X1Y1::X2Y2 and X1Y2::X2Y1) but only one reductional orientation (X1X2::Y1Y2).
These results indicate that in the great majority of meiosis I spermatocytes, both members of the X-Y pair segregate in the same manner, i.e., either both reductionally or both equationally, although the reductional/equational choice appears to be random. This strongly suggests that centromere orientation patterns are coordinated in some manner between the two chromosomes. How might such coordination be achieved? Two possible contributing factors could be cohesion between centromeres and/or pairing between homologues.
Sister centromere cohesion is lost before prometaphase I in solo spermatocytes
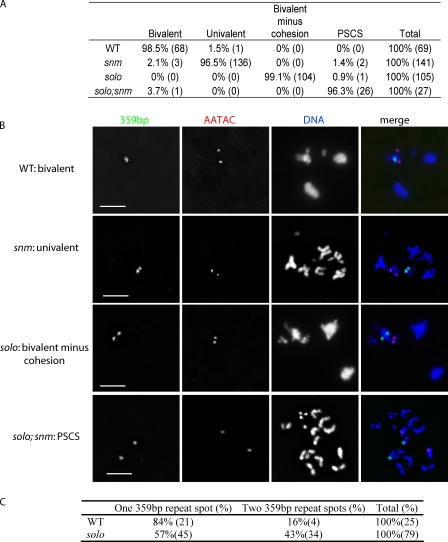
To examine centromere cohesion when orientation patterns are established, we extended the FISH analysis to earlier stages of meiosis I. 43% of solo prometaphase I spermatocytes showed two separate 359 bp signals compared with only 16% of WT spermatocytes (Fig. 3). The observation of a low level of separation of sister 359 bp loci in WT is consistent with previous results (Balicky et al., 2002). It could be due either to the FISH squash procedure or the fact that the 359 bp repeats are outside, albeit adjacent to, the centromere proper. Nevertheless, our results indicate that cohesion proximal to the X centromere is reduced in solo mutants by 27% more than in WT spermatocytes. Similar observations were obtained using a probe against the dodeca satellite locus, a repetitive locus near the centromere of chromosome 3 (Fig. S2). At prometaphase I, WT third chromosome bivalents typically exhibited only two dodeca signals, whereas four signals were typically present in solo bivalents at the same stage. At anaphase I, one dodeca signal was present at each pole in WT, but two signals were usually seen at each pole in solo mutants.
Figure 3.
Sister chromatid cohesion and homologue pairing at prometaphase I in solo; +, +; snm, and solo; snm mutants. X and Y chromosomes were recognized by probes against the 359 bp (green) and AATAC (red) satellite DNA repeats, respectively. DNA was stained with DAPI. Sum projections of 3D-deconvolved z series stacks were performed. (A) Quantification of cohesion and pairing patterns. Table shows the percent and number of nuclei (in parentheses) exhibiting indicated pairing and cohesion patterns. (B) Representative images. Bivalents versus univalents were judged from DAPI panels. Presence or absence of cohesion was based on separation between sister chromatids as judged from DAPI images and on separation between sister FISH signals. Bars, 5 µm. (C) Quantification of 359 bp signal separation patterns. See Table II for genotypes.
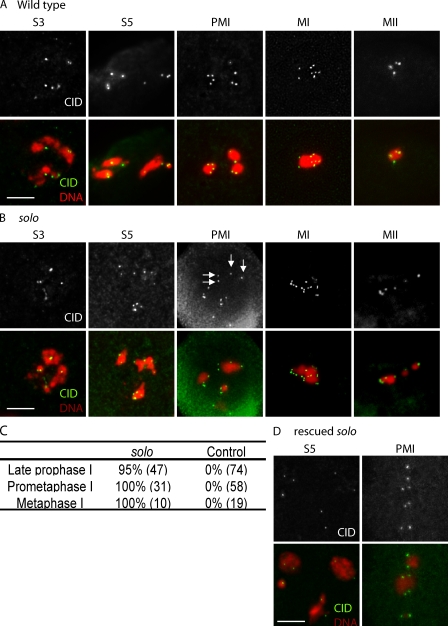
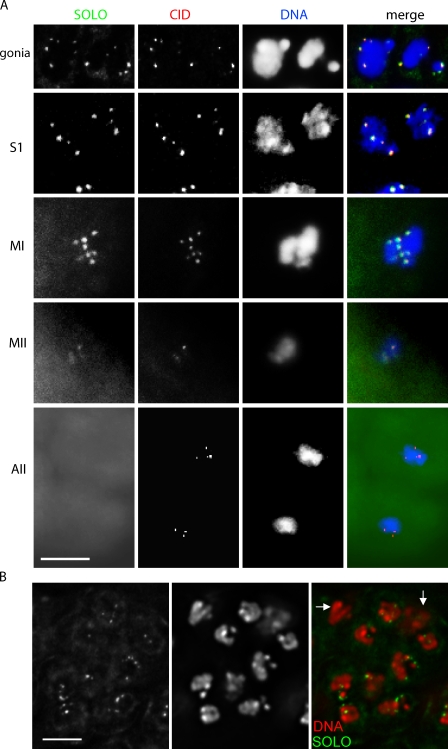
To examine the cohesive status of sister centromeres during meiosis I more globally, we made use of an antibody against centromere identifier (CID), a centromere-specific histone H3–like protein (Ahmad and Henikoff, 2001; Blower and Karpen, 2001) that enables visualization of all centromeres simultaneously (Fig. 4). In WT spermatocytes, the number of anti-CID foci per nucleus never exceeded the number of homologous chromosomes (eight in meiosis I and four in meiosis II). During late prophase I when the four bivalents occupy well-separated territories, two CID spots could often be seen in each chromosome territory.
Figure 4.
Cohesion of sister centromeres is lost by late prophase I in solo mutants. (A, B, and D) Testes from WT (A), solo/Df(2L)A267 (B), and rescued solo (solo/Df(2L)A267; {UASp-Venus::SOLO}/{nos-GAL4::VP16}) (D) males stained with anti-CID antibody to identify centromeres and with DAPI to visualize DNA. Sum or maximum projections of 3D-deconvolved z series stacks were performed to obtain CID signals. No more than eight CID spots are present in WT meiosis I at any stage, whereas solo spermatocytes show more than eight CID spots at late prophase I, prometaphase I, and metaphase I (11, 13, and 15 spots, respectively, in the nuclei shown). Arrows indicate a bivalent with four fully separated sister centromeres. S3, mid–prophase I; S5, late prophase I; PMI, prometaphase I; MI, metaphase I; MII, metaphase II. Bars, 5 µm. (C) Quantification of CID spots in (soloZ2-0198/Df(2L)A267) at different stages. The percent of spermatocytes with more than eight spots is shown. The number of nuclei scored is in parentheses.
The numbers of anti-CID foci in solo mutants were similar to WT throughout early and mid–prophase I (stages S1–S4; Cenci et al., 1994). However, from late prophase I (stages S5 and S6) through metaphase I, many bivalents in solo spermatocytes exhibited three or four spots instead of the normal two. Virtually all nuclei exhibited more than eight spots, with the number ranging up to 16, which is the number of sister centromeres in a diploid nucleus (Fig. 4 C). solo mutants also exhibited too many CID spots during meiosis II, up to eight instead of four. These observations indicate that sister centromere cohesion is lost well before prometaphase I in solo mutants. In many nuclei in late prophase I and prometaphase I, the distance between CID spots that appeared to represent sister centromeres (Fig. 4 B, white arrows) considerably exceeded the diameter of CID spots, suggesting that solo mutations may lead to loss of cohesion of heterochromatic domains that flank the centromeres as well as of the centromeres themselves.
Bivalent structure coordinates centromere orientation patterns
Inspection of DAPI-stained late prophase I and prometaphase I spermatocytes revealed that bivalents are almost always intact in solo mutants despite the absence of centromere cohesion (Fig. 3). Metaphase I configurations also appear normal and anaphase I segregation regular in most cells (Fig. S1 A). These observations suggest that the proteins responsible for maintaining pairing of homologues during meiosis I, SNM, and MNM likely carry out their functions and maintain bivalent stability in the absence of SOLO and centromere cohesion. Indeed, spermatocytes from solo males stained with anti-SNM antibody exhibited chromosomal SNM foci of normal intensity and distribution, including the prominent X-Y focus at the rDNA-pairing region (Thomas et al., 2005; and unpublished data). As a further test of this, we compared chromosome morphology in solo; snm double mutants with those in solo and snm single mutants. As is evident from Fig. 3 B, loss of SNM in a solo mutant background abrogates all remaining connections among chromatids, leading to the presence of up to 16 fully separated single chromatids by prometaphase I. In contrast, loss of SNM in a WT background disrupts bivalents but leaves cohesion intact, resulting in the presence of up to eight univalents, each with two chromatids. Thus, the residual connections among chromatids responsible for intact bivalents in solo mutants are dependent on SNM.
We next asked whether SNM plays a role in coordinating chromatid segregation in the absence of SOLO by carrying out FISH analysis of X-Y segregation using the 359 bp and AATAC probes described previously. More than 43% of anaphase I spermatocytes from solo; snm males segregated either 3::1 (X::XYY or Y::YXX) or 4::0 compared with <4% of such segregations in sibling solo single mutants (Fig. 1 C and Table II). We conclude that preferential 2::2 segregation in solo males is largely a consequence of intact bivalent structure maintained by the SNM–MNM homologue conjunction complex.
SOLO is not required for arm cohesion or for mitotic chromatid segregation
In WT males, cohesion between sister chromatid arms is maintained throughout the early stages of prophase I (S1 and S2), as shown by fusion of sister GFP-LacI foci bound to chromosomally inserted lacO arrays (Vazquez et al., 2002). Using this assay, solo mutants exhibited normal frequencies of arm cohesion during early prophase I (Fig. S3). These experiments also provided evidence that solo is dispensable for cohesion and sister chromatid segregation in premeiotic gonia. Mutations that cause mitotic NDJ in gonial nuclei frequently result in the presence of four LacI-GFP spots during late prophase I in lacO hemizygotes instead of the normal two, reflecting trisomy for chromosome 2. No spermatogonia or spermatocytes with more than two GFP-LacI spots were observed in solo males hemizygous for the lacO array (unpublished data).
SOLO is a novel protein encoded by an alternative splice product of vasa
solo was mapped by deficiency complementation to the vasa (vas) locus at 35B on chromosome arm 2L (Fig. 5 A). vas encodes a conserved DEAD box RNA helicase involved in germline establishment and axis specification in oocytes and early embryos (Styhler et al., 1998; Tinker et al., 1998). Cohesion defects have not been previously described in vas mutants. Consistent with this, our DNA sequence analysis revealed no mutations in the vas coding sequences in any of the solo alleles. However, sequence alterations were found within the third intron of vas, which contains two large exons. Each of the three solo alleles exhibited a single-base substitution that creates a premature stop codon in one of those exons.
Figure 5.
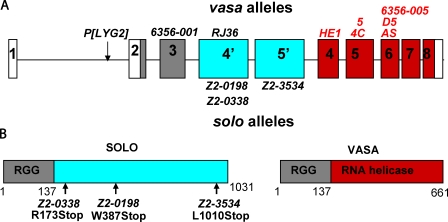
Molecular characterization of solo. (A) The genomic structure of solo and vas. The solo and vas transcription units share exons 1–3. Gray shading represents shared translated sequences, and white represents the 5′ and 3′ untranslated region. Exons 4’ and 5′ (blue) are unique to solo, and exons 4–8 (red) are unique to vas. Mutations above the locus are vas alleles, those in red fully complement solo, and those in black fail to complement solo. solo mutations are shown below the locus. (B) Predicted structures of SOLO and VASA proteins and mutation sites of solo alleles.
To characterize the solo transcription unit, we sequenced a nearly full-length cDNA as well as several RT-PCR and 5′ and 3′ rapid amplification of cDNA ends (RACE) fragments that include part or all of the intronic exons. Those analyses revealed that in addition to the two intronic exons, solo transcripts also include the three upstream vas exons, which encode several RGG repeats found in RNA-binding proteins (Alex and Lee, 2005) but lack the five downstream vas exons, which encode the RNA helicase domain. The three upstream vas exons and the two intronic exons are spliced together to create a continuous ORF that extends from the translation start site of vas in exon 2 to a stop codon in the downstream intronic exon and that could encode a protein 1,031 amino acids in length (Fig. 5 B).
Complementation analysis between solo and vas mutations confirmed our proposed exon structure of solo (Fig. 5 A). solo alleles complemented all vas alleles containing mutations in any of the five C-terminal exons (Liang et al., 1994), which encode the VASA helicase domain, indicating that the C terminus of VASA is not shared by SOLO. However, vas mutations that map upstream of the SOLO-specific exons, including one nonsense mutation in exon 3, vas6356-001 (Tinker et al., 1998), failed to complement the solo alleles, indicating that the 137 amino acids encoded by the upstream exons are present in both proteins. It is unlikely that the SOLO-specific exons are expressed independently of vas in addition to being expressed as a fusion product with the N terminus of VASA, as vas6356-001 behaves as a null allele of solo, giving X-Y NDJ frequencies of 41–44% in trans with solo alleles. We conclude that solo encodes a protein that includes the N-terminal 137 amino acids of VASA fused to 894 amino acids encoded within the third intron of vas.
Single homologues of SOLO were identified by BLAST analysis in all 12 sequenced Drosophila genomes (http://flybase.org/blast/). Overall conservation is fairly low; Drosophila SOLO exhibits only ∼30% amino acid identity with its homologues in Drosophila virilis and Drosophila pseudoobscura. However, in all of the Drosophila genomes, the solo sequences are nested within a large intron upstream of the exons that encode the helicase domain of VASA, and SOLO appears capable of being expressed by the same alternative splice mechanism used in Drosophila.
No homologues of SOLO were identified outside of the genus Drosophila, not even in the genome of the mosquito Anopheles gambiae. Although it is possible that solo exists in A. gambiae but is unrecognizable because of divergence, it would have to be located elsewhere in the genome, as there are no large exons nested within introns of the A. gambiae vas gene. Other than the RGG motifs in the common N terminus, SOLO exhibits no significant homologies with other proteins in the sequence database.
SOLO colocalizes with CID and SMC1 from early prophase I until anaphase II
To study the intracellular localization pattern of SOLO, transgenic insertions of a solo cDNA tagged at its N or C terminus with Venus (an enhanced yellow fluorescent protein) were generated. Expression of the fusion proteins was induced by the GAL4::VP16 transcription activator under control of the Drosophila nanos (nos) promoter, which is active in most male germ cells. Two third chromosome insertions of Venus::SOLO and one third chromosome insertion of SOLO::Venus were tested for ability to complement the meiotic phenotypes of solo mutants. One copy of each SOLO transgene sufficed for virtually complete rescue of solo meiotic phenotypes. Sex chromosome NDJ was reduced to background levels, and cytological analysis indicated that meiosis II segregation is regular (unpublished data). Venus::SOLO also suppressed the centromere cohesion defect of solo mutants (Fig. 4 D). These data indicate that the tagged SOLO proteins function similarly to endogenous SOLO in male meiosis.
Using nos-GAL4::VP16 to induce Venus::SOLO expression, bright Venus foci were seen in nuclei of spermatocytes of all stages up to and including metaphase II but were absent at anaphase II and subsequent stages (Fig. 6 A). Both the numbers of Venus::SOLO foci per nucleus (up to eight in primary spermatocytes and four in secondary spermatocytes) and the fact that they overlapped CID foci at all stages indicate that Venus::SOLO localizes to centromeres. In late prophase I nuclei, the centromeric Venus::SOLO foci were sometimes larger and more diffuse (Fig. S4 A, stage S6) than CID foci at similar stages (Figs. 4 and 6), suggesting that SOLO localizes to centric heterochromatic regions that are broader than the centromeres proper. A similar localization pattern was observed using a transgene containing a 2.7-kb fragment from the region immediately upstream of exon 1 of the vas/solo locus that has previously been shown to contain the vas regulatory elements (Sano et al., 2002) fused to SOLO::Venus (unpublished data). Curiously, in these males, SOLO::Venus foci are also present in nuclei of premeiotic spermatogonial cells despite the aforementioned evidence that solo is dispensable for spermatogonial chromatid segregation. However, no expression of UPS-SOLO::Venus was detected in the somatic cyst cells of testes (Fig. 6 B). Centromeric SOLO::Venus foci were also present in meiotic cells of UPS-SOLO::Venus females, which is consistent with a requirement for solo in female meiotic chromosome segregation, but were absent in somatic follicle cells (Fig. S5). Thus, SOLO appears to be germline specific but not meiosis specific in its expression pattern, although only meiotic phenotypes have been detected in solo mutants.
Figure 6.
Localization pattern of SOLO. (A) Colocalization of Venus::SOLO and CID on meiotic centromeres in WT spermatocytes. {UASp-Venus::SOLO}/{nos-GAL4::VP16}) WT males were stained with anti-CID and DAPI. (B) SOLO::Venus localizes to spermatogonia (gonia) but not somatic cyst cells in an eight-cell cyst from UPS-SOLO:Venus males. Arrows indicate cyst nuclei. Venus::SOLO and SOLO::Venus were detected by native fluorescence. S1, early prophase I; MI, metaphase I; MII, metaphase II; AII, anaphase II. Bars, 5 µm.
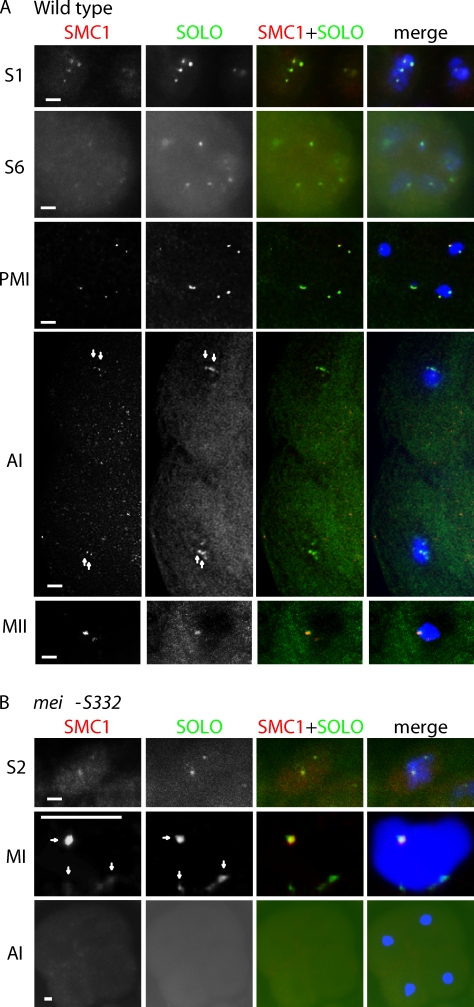
To determine whether SOLO functions together with a member of the cohesin complex, we compared the localization patterns of SOLO and the cohesin protein SMC1 in spermatocytes from males expressing Venus::SOLO and stained with anti-SMC1 antibody (Fig. 7 A). Anti-SMC1 foci were present at all stages of meiosis I and at metaphase II. In all spermatocytes we examined, anti-SMC1 and Venus::SOLO foci colocalized throughout meiosis until anaphase II, when both proteins became undetectable. These data strongly suggest that SOLO and SMC1 function together to maintain cohesion between sister centromeres in male meiosis. The absence of noncentromeric SMC1 foci during middle and late prophase I was expected from prior observations (Vazquez et al. 2002) that arm cohesion is absent during those stages.
Figure 7.
Colocalization of Venus::SOLO and SMC1 foci on centromeres in WT and mei-S332 spermatocytes. Venus::SOLO and SMC1 foci were detected by anti-GFP and anti-SMC1, respectively, and DNA was stained with DAPI. (A and B) Venus::SOLO and SMC1 foci colocalize until anaphase II in WT (A) but are lost by anaphase I in mei-S332 (B). Arrows point to colocalizing foci. Mutant spermatocytes are from mei-S3324/mei-S3328; {UASp-Venus::SOLO}/{nos-GAL4::VP16} males. All images are sum projections of 3D-deconvolved z series planes. S1 and S2, early prophase I; S6, late prophase I; PMI, prometaphase I; AI, anaphase I; MI, metaphase I; MII, metaphase II. Bars, 1 µm.
Premature loss of SOLO and SMC1 foci in mei-S332 mutants
In mei-S332 mutants, centromere cohesion is lost prematurely at anaphase I (Kerrebrock et al., 1992). MEI-S332 is a homologue of yeast shugoshin proteins, mutations that cause similar phenotypes because of premature removal of centromeric cohesin at anaphase I (Watanabe, 2005). To test whether mei-S332 mutations cause premature loss of SMC1 and/or SOLO, we compared the localization patterns of Venus::SOLO and SMC1 in mei-S332 mutant spermatocytes with those in WT spermatocytes (Fig. 7 B and Fig. S4). Venus::SOLO and SMC1 foci were present through metaphase I in mei-S332 spermatocytes and were morphologically similar to those in WT spermatocytes. However, unlike WT spermatocytes in which Venus::SOLO and SMC1 foci were present at anaphase I (13/13) and metaphase II, no foci of either protein were detected at anaphase I (24/24) or later stages of meiosis in mei-S332 spermatocytes. Therefore, we conclude that persistence of SOLO and SMC1 on meiotic centromeres after metaphase I is dependent on the shugoshin protein MEI-S332. This result provides further evidence that SMC1 and SOLO collaborate in maintaining centromeric cohesion in meiosis. It also provides the first direct evidence that MEI-S332 functions to stabilize a cohesin protein on meiotic centromeres between anaphase I and II like yeast shugoshins.
Interactions among solo, ord, and SMC1
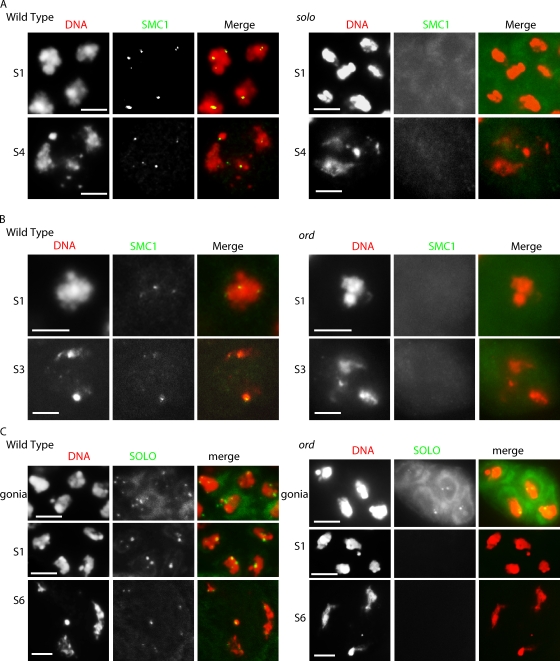
In light of the strong colocalization of SOLO and SMC1, we used an anti-SMC1 antibody to test for effects of solo mutations on SMC1 localization (Fig. 8 A). Unlike WT spermatocytes, which exhibited SMC1 foci throughout meiosis, no distinct anti-SMC1 foci were detected at any stage of meiosis in solo spermatocytes. This absence of centromere staining is not caused by a failure of solo spermatocytes to produce SMC1 protein, as a Western blot analysis revealed only a slight (∼10%) reduction in SMC1 amount in extracts from solo testes compared with WT controls (unpublished data). We conclude that SOLO is required for localization of SMC1 to centromeres from the beginning of prophase I in male meiosis and suggest that SOLO may be required not only for maintenance of cohesion at centromeres but also for its establishment.
Figure 8.
Interactions among SOLO, ORD, and SMC1. (A and B) Absence of SMC1 foci in solo (A) and ord (B) mutants. SMC1 foci were detected by anti-SMC1, and DNA was stained with DAPI. Mutant spermatocytes are from soloZ2-0198/soloZ2-0198 (A) and ord5/Df(2R)WI370 (B) adult males. Centromeric SMC1 foci are visible throughout prophase I in WT but are completely absent in solo and ord spermatocytes. (C) Effect of ord hemizygosity on Venus::SOLO localization. Venus::SOLO was detected by native fluorescence (FITC channel), and DNA was stained with DAPI using spermatocytes from ord5/Df(2R)WI370; {UASp-Venus::SOLO}/{nos-GAL4::VP16} adult males. Venus::SOLO foci are robust in spermatogonia (gonia) but absent in early and late prophase I meiotic stages in ord mutants. All images are sum projections of 3D-deconvolved z series planes. S1, early prophase I; S3 and S4, mid–prophase I; S6, late prophase I. Bars, 5 µm.
The strong similarity in effects of solo and ord mutations on chromosome behavior prompted us to investigate the effect of ord mutations on localization of SMC1 using our anti-SMC1 antibody. The results indicated that ord is also required for centromere localization of SMC1. As in solo mutants, no discrete SMC1 signals were detected at any stage of meiosis in an ord-null genotype (Fig. 8 B).
We then examined the effect of loss of ord function on SOLO localization (Fig. 8 C). Except for very weak Venus::SOLO signals in a minority of S1 (very early prophase I) spermatocytes, no Venus signals were seen at any stage of meiosis in ord males, although strong centromere signals were present in a normal pattern in heterozygous sibling controls. Thus, ord is required for stable centromere localization of both SMC1 and SOLO.
Discussion
Sister centromere cohesion and coorientation in solo mutants
solo mutants exhibit premature loss of centromere cohesion and high NDJ at both meiosis I and II. Centromere cohesion is strongly impaired by stage S5 of prophase I long before centromere orientation patterns are established at prometaphase I. Although the premature loss of centromere cohesion is likely the underlying cause of NDJ at both divisions, the mechanisms of meiosis I and meiosis II NDJ nevertheless differ in important ways. During meiosis II, sister chromatids are fully separated at metaphase II, and anaphase II segregation appears to involve random assortment of fully independent chromatids to the two poles. However, during meiosis I, fully separated sister chromatids are rarely observed, and bivalents containing the four chromatids of a homologous pair remain intact throughout the division. Moreover, at least for the X-Y pair, chromatid segregation is not fully random. Although random assortment would lead to numerically unequal segregation (3:1 or 4:0) in 62.5% of meiosis I divisions, in solo males, >95% of anaphase I cells exhibit two chromatids at each pole. This restriction probably applies to autosomes as well because in DAPI-stained preparations, >90% of anaphase I spermatocytes exhibit poles with roughly equal DNA content (Fig. S1, A and B). Nevertheless, segregation is very abnormal, indeed random in a more limited sense. Unlike WT spermatocytes in which sister chromatids always cosegregate at meiosis I, in solo spermatocytes X and Y chromatids exhibit no preference for or against their sister as a segregation partner. The result is a 2:1 ratio of equational (XY::XY) to reductional (XX::YY) segregations. Thus, bivalents in solo males retain their gross structure and the ability to segregate in an orderly fashion but lose sister-specific connections and with them the ability to distinguish sister from homologous chromatids. The resulting bivalents have four functional kinetochores instead of the normal two, and these orient independently of each other yet are somehow constrained to orient two to each pole.
How might SOLO perform its role in sister centromere orientation? One possibility is a role similar to the monopolin complex in S. cerevisiae or MoaI in S. pombe, proteins that function specifically in coorientation (Tóth et al., 2000; Shonn et al., 2002; Yokobayashi and Watanabe, 2005). However, mutations in these proteins do not disrupt sister centromere cohesion, whereas solo mutations disrupt both cohesion and coorientation. Therefore, a more parsimonious idea is that the primary role of SOLO is in centromere cohesion and that cohesion is required for coorientation. SOLO would thus be more similar to REC8, a meiotic cohesin component that is also required for both cohesion and coorientation in S. pombe (Watanabe and Nurse, 1999; Watanabe et al., 2001; Yokobayashi et al., 2003; Sakuno et al., 2009). It remains to be determined whether other proteins analogous to monopolin or MoaI are also required for centromere coorientation in Drosophila.
Homologue pairing and centromere orientation
Homologue connections, in the form of recombination-generated chiasmata, have been shown in both S. cerevisiae and S. pombe to promote fidelity of sister centromere coorientation to varying degrees both in WT cells and in cells deficient for other centromere orientation factors (Shonn et al., 2002; Yamamoto and Hiraoka, 2003; Yokobayashi and Watanabe, 2005). Drosophila males lack chiasmata but use the SNM–MNM complex to maintain homologue pairing until anaphase I. Our data indicate that SNM (likely in complex with MNM) serves to coordinate chromatid segregation patterns in the absence of centromere cohesion but has only a minimal effect on sister centromere orientation by itself. The fact that the reductional/equational segregation ratio in solo mutants almost exactly matches the random expectation makes it unlikely that SNM does anything to actively promote reductional segregations. The main effect of the loss of SNM in a SOLO-deficient background is abrogation of the restriction against unequal segregations. More than 40% of anaphase I cells in solo; snm males exhibit numerically unequal segregations compared with <5% in solo males. Although the ratio of equational to reductional 2::2 segregations increases somewhat in solo; snm mutants relative to solo mutants (Table II), for reasons that are not clear, reductional segregations are nevertheless preserved and indeed occur at approximately the expected random frequency (12.5%). This stands in sharp contrast to spo13 or Moa1 mutants (in S. cerevisiae and S. pombe, respectively), which exhibit mixed reductional/equational meiosis I segregation patterns similar to solo but which revert to 100% equational segregation when homologue connections are removed by spo11 mutations (Shonn et al., 2002; Yokobayashi and Watanabe, 2005). The basis for this difference is that spo13 and Moa1 interfere with sister centromere orientation without disrupting cohesion before anaphase I so that loss of homologue connections leaves most chromosomes still connected at sister centromeres. However, solo mutations ablate sister chromatid cohesion so leave no basis for regular equational segregation.
How does SNM–MNM promote regular chromatid segregation? A plausible scenario is that SNM–MNM provides nonspecific connections among all four chromatids at homologue-pairing sites such as the rDNA locus of the X-Y pair. Although inadequate to direct centromere orientation, such connections would preserve bivalent stability and could provide the resistance necessary for generation of tension on the meiosis I spindle. The 2::2 segregation bias could reflect a checkpoint mechanism that serves to monitor and balance such tension. Alternatively, it could reflect a rigidity of bivalent structure that tends to discourage unbalanced orientations. Further research will be required to understand the basis for the unique meiosis I segregation pattern in solo.
Meiotic cohesion complexes in Drosophila
In S. cerevisiae and S. pombe, multiple meiotic cohesion functions are performed by cohesin complexes that include meiosis-specific subunits such as REC8, which replaces the mitotic kleisin subunit RAD21. REC8 is widely conserved among eukaryotes and has been shown in several model plants and animals to be critical for many of the same meiotic functions identified in yeast (Pasierbek et al., 2001; Cai et al., 2003; Petronczki et al., 2003; Chelysheva et al., 2005; Xu et al., 2005). However, in Drosophila, no true REC8 homologue has been identified, and the role of cohesin in meiotic cohesion has been unclear.
Our data strongly suggest that SOLO and SMC1 function as partners in mediating centromere cohesion in Drosophila meiosis. First, anti-SMC1 and Venus::SOLO foci overlap extensively on centromeres throughout meiosis until anaphase II when both proteins disappear. Second, both Venus::SOLO and anti-SMC1 foci disappear prematurely at anaphase I in mei-S332 mutants, which is consistent with a role of MEI-S332 to protect meiotic cohesin from proteolytic cleavage by separase. Third, centromere localization of SMC1 is abolished at all meiotic stages in solo spermatocytes. Finally, we have recently obtained evidence for a physical interaction between SMC1 and SOLO in ovaries (unpublished data).
Another protein with an essential role in Drosophila meiotic cohesion is ORD (Miyazaki and Orr-Weaver, 1992; Bickel et al., 1996). The phenotypes of solo and ord mutations are very similar, including missegregation of both homologous and sister chromatids and ablation of centromeric SMC1 foci. Like SOLO, ORD is a centromere protein, but there are significant differences in the localization patterns of the two proteins. SOLO localizes to centromeres from the earliest stages of prophase I and remains on the centromeres until anaphase II. ORD has been reported to localize predominantly to interchromosomal spaces in early prophase I nuclei in male meiosis, then to the chromosome arms in late prophase I, finally accumulating on centromeres at prometaphase I where it remains until anaphase II (Balicky et al., 2002). Nevertheless, the striking phenotypic similarity of solo and ord mutants strongly suggests that both ORD and SOLO are intimately involved in establishing and maintaining cohesion in Drosophila meiosis.
The exact role of SOLO (and ORD) in meiotic cohesion remains to be determined. One possibility is that SOLO is a regulatory protein required for stable localization of cohesin to centromeres. Several known cohesin cofactors are required for specific aspects of cohesin function, such as chromosomal loading, establishment of cohesion, removal of cohesin during prophase, protection of centromeric cohesin, etc. (Petronczki et al., 2003; Uhlmann, 2004). SOLO appears to play a more general role than most of these cofactors: it is involved both in stable chromosome association of cohesin and in the establishment and maintenance of cohesion throughout meiosis. Moreover, unlike the known cofactors that associate with cohesin during certain stages of the cell cycle, SOLO colocalizes with SMC1 throughout meiosis. Thus, except for the lack of homology to any of the four families of cohesin proteins, our data are consistent with the possibility that SOLO is a novel component of a meiosis-specific cohesin complex. It will be of considerable interest to determine the composition of the meiotic cohesin complexes in Drosophila.
Materials and methods
Fly stocks, special chromosomes, and Drosophila culture methods
The solo mutations were obtained from the Zuker-2 (Z2) collection of EMS-mutagenized second chromosomes (Koundakjian et al., 2004). The lines used in this study were identified in a screen for paternal fourth chromosome loss and were provided by B. Wakimoto (University of Washington, Seattle, WA; Wakimoto et al., 2004). vas alleles were obtained from M. Ashburner (University of Cambridge, Cambridge, England, UK), P. Lasko (McGill University, Montreal, Quebec, Canada), D. Montell (Johns Hopkins University, Baltimore, MD), and the Bloomington Drosophila Stock Center at Indiana University. mei-S332 alleles were donated by T. Orr-Weaver (Whitehead Institute, Massachusetts Institute of Technology, Cambridge, MA). ord5 and the deficiency Df(2R)WI370 were donated by S. Bickel (Dartmouth College, Hanover, NH). All flies were maintained at 23°C. Compound chromosomes and markers are described in Flybase. Unless otherwise specified, tested males were crossed singly to two or three females in shell vials. Crosses were incubated at 23°C on cornmeal/molasses/yeast/agar medium. Parents were removed from the vial on day 10, and progeny were counted between days 13 and 22.
Sex chromosome NDJ assays
To measure X-Y NDJ and discriminate between NDJ of homologues and sister chromatids, +/BsYy+ males were crossed singly to 2–3 C(1)RM, y2 wa su(wa)/0 females in which both X chromosomes are attached to a single centromere. These females produce eggs that are diplo-X and nullo-X at approximately equal frequencies. Fertilization of nullo-X eggs yields progeny derived from both XX and XY sperm (+ females and BS males, respectively), which are diagnostic of sister chromatid and homologue NDJ, respectively. Fertilization of diplo-X eggs by nullo-XY (O) sperm, which can result from either sister chromatid or homologue NDJ, yields y2 wa su(wa) females. The cross can also yield progeny from XXY, XYY, and XXYY sperm, but such progeny were recovered only very rarely and were neglected in the analysis.
Immunostaining
α-Tubulin/DAPI staining of testes was performed as described previously (Thomas et al., 2005). Immunostaining was performed with modification according to Bonaccorsi et al. (2000). The following primary antibodies were used: 1:500 anti-CID (chicken; provided by G. Karpen), 1:1,000 anti-CID (rabbit; Abcam), 1:250 anti-SNM C terminal (rabbit; Thomas et al., 2005), 1:250 anti-GFP (rabbit; Invitrogen), and 1:250 anti-SMC1 (rabbit; Thomas et al., 2005). The following secondary antibodies were used: Alexa Fluor 555 goat anti–chicken IgG (H+L; Invitrogen), Alexa Fluor 546 goat anti–rabbit IgG (H+L; Invitrogen), and Alexa Fluor 647 goat anti–rabbit IgG (H+L; Invitrogen). Except where specified, Venus::SOLO expression was induced by nos-GAL4::VP16 (Van Doren et al., 1998), and fluorescent signals were detected in the FITC channel.
FISH analysis and probe preparation
FISH experiments were performed according to Balicky et al. (2002) with modification (Thomas et al., 2005). The 359 bp repeat probe was amplified by PCR according to Dernburg (2000) and labeled using the Fluorescein-High Prime kit (Roche). The AATAC and dodeca repeat probes were synthesized as a single-stranded oligonucleotide (IDT Biophysics) and labeled with Alexa Fluor 546 (Invitrogen) using terminal deoxynucleotidyl transferase (Promega).
Staging of meiotic cells
Meiosis I and II spermatocytes were distinguished by several criteria that are independent of chromosome number, DNA content, and cohesion status, which are factors affected by solo and other mutants used in this study. For FISH analyses, the most useful criteria were cell size and cell number per cyst (16 or 32 for meiosis I or meiosis II cysts, respectively). In experiments in which spindles were stained with anti–α-tubulin, spindle size and the presence or absence of duplicated but undivided spindle pole bodies (diagnostic of meiosis I spindles) were also used. Criteria for distinguishing meiotic substages were described previously (Cenci et al., 1994).
Microscopy and image processing
All images were collected using a microscope (Axioplan; Carl Zeiss, Inc.) equipped with a 100-W mercury lamp (HBO; Carl Zeiss, Inc.), Plan Neofluar 100×/1.30 NA oil immersion lenses (Carl Zeiss, Inc.), and a high resolution charge-coupled device camera (Roper Industries) at room temperature. Image data were collected and merged using MetaMorph software (MDS Analytical Technologies). For some images, maximum or sum projections of deconvolved z series were obtained using MetaMorph. Images were processed with Photoshop (CS2; Adobe).
Mapping and identification of solo mutations
solo alleles were mapped by deficiency complementation against the chromosome 2 deficiency kit obtained from the Bloomington Drosophila Stock Center using the X-Y NDJ phenotype. solo was mapped to the vas locus in the 35B region of chromosome arm 2L using deficiencies supplied by M. Ashburner (Ashburner et al., 1990).
All exons and the third intron of vas were amplified from genomic DNA of flies homozygous for each of the three solo mutations and sequenced using a cycle sequencing kit (ABI BigDye Terminator version 3.1; Applied Biosystems). No mutations were detected in vas exons, but the two large exons in the third intron of vas contained single mutations in each of the three solo alleles, each of which result in a nonsense mutation (Fig. 5). Sequencing also showed that the preexisting vasRJ36 allele (Tinker et al., 1998) has an 8-bp insertion in the first intronic exon, resulting in a frame shift mutation. The sequence of the SOLO cDNA reported in this paper has been deposited in GenBank as accession no. DQ851162.
Characterization of solo transcripts
To characterize the solo transcription unit, total RNA was prepared from WT (strain Zuker-2, cn bw) adults using TRI reagent (Sigma-Aldrich). After DNase treatment, the total RNA was used for cDNA synthesis using Superscript First-Strand Synthesis system (Invitrogen). solo cDNAs were amplified by PCR using the following primers: 5′-GTGAGAACTTTGTCACTCGG-3′ and 5′-TTTATGGGAGGCAGTAAGGC-3′. A nested PCR reaction was performed using the following primers: 5′-CAATTCGAGTAGTGGTCAGC-3′ and 5′-GAATCCGAATACCCTGTTGC-3′. This procedure yielded a specific amplification product of 972 bp that contains parts of the two large ORFs from intron 3 of vas spliced together to generate a continuous reading frame. To identify the 5′ and 3′ ends of the solo transcript, 5′ and 3′ RACE reactions were performed (SMART RACE cDNA Amplification kit; BD), and a cDNA (EST clone AT08465) obtained from Berkeley Drosophila Genome Project was sequenced. These experiments revealed that the second intronic ORF terminates at a stop codon located 92 bases upstream of a poly A site and 294 bases upstream of the fourth exon of vas. At the 5′ end, AT08645 includes all sequences in the first three exons of vas except for the first 22 bp of exon 1. It is not clear whether this difference reflects different transcription start sites for the two genes or whether AT08645 is incomplete at the 5′ end. The primer sequences used in the RACE experiments are available upon request.
Construction of SOLO fusion clones and generation of transgenic flies
Two SOLO fusion constructs, UASp-Venus::SOLO and UASp-SOLO::Venus, were made. For Venus::SOLO, the solo coding sequence and 3′ untranslated region were amplified from the EST clone AT08465 using Pfx polymerase (Invitrogen) and primers 5′-CACCATGTCTGACGACTGGGATG-3′ and 5′-CACCCGACATAGATGCCTCG-3′. For SOLO::Venus, the following primers were used: 5′-CACCATGTCTGACGACTGGGATG-3′ and 5′-GAGCAGCCCGAAAAATCTACC-3′. The PCR products were cloned into the pENTR/D-TOPO entry vector (Invitrogen), and the resulting products were sequenced.
Both entry constructs were recombined into Gateway P-element vectors pPVW and pPWV (obtained from the Drosophila Genomics Resource Center), generating the germline transformation vectors P{w+mC UASp-Venus::SOLO and P{w+mC UASp-SOLO::Venus. Both vectors include Venus, upstream activation sequences for transcriptional activation by GAL4, and mini-white to detect germline transformants. Both constructs were transformed into w1118 flies (BestGene Inc.). Transformants were mapped by standard procedures.
For the upstream activation sequence UPS-SOLO::Venus construct, a 2.7-kb fragment of vas located immediately upstream of exon 1, which contains complete vas regulatory elements (Sano et al. 2002), was cloned upstream of SOLO cDNA into the pENTR/D-TOPO entry vector, recombined into pPWV, and thereafter transformed into flies by the aforementioned methods.
Online supplemental material
Fig. S1 shows cytological analysis of meiosis in WT and solo spermatocytes. Fig. S2 shows premature loss of centromeric cohesion of chromosome 3 in solo spermatocytes. Fig. S3 shows that SOLO is not required for arm cohesion or mitotic chromatid segregation. Fig. S4 shows diffuse SOLO staining during late prophase I and that SOLO is not protected in mei-S332 mutants. Fig. S5 shows that SOLO overlaps with CID in female germline cells. Table S1 shows homologous and sister chromatid NDJ of chromosome 2 in solo males. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200904040/DC1.
Acknowledgments
We thank B. Wakimoto, P. Lasko, M. Ashburner, D. Montell, T. Orr-Weaver, and S. Bickel for providing stocks of mutant Drosophila and G. Karpen for providing the anti-CID antiserum.
This work was supported by National Institutes of Health (grant R01 GM40489) to B.D. McKee.
Footnotes
Abbreviations used in this paper:
- CID
- centromere identifier
- MNM
- mod(mdg4) in meiosis
- NDJ
- nondisjunction
- ORD
- orientation disruptor
- PSCS
- precocious sister chromatid separation
- RACE
- rapid amplification of cDNA ends
- SNM
- stromalin in meiosis
- SOLO
- sisters on the loose
- WT
- wild type
References
- Ahmad K., Henikoff S. 2001. Centromeres are specialized replication domains in heterochromatin. J. Cell Biol. 153:101–110 10.1083/jcb.153.1.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alex D., Lee K.A. 2005. RGG-boxes of the EWS oncoprotein repress a range of transcriptional activation domains. Nucleic Acids Res. 33:1323–1331 10.1093/nar/gki270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Thompson P., Roote J., Lasko P.F., Grau Y., el Messal M., Roth S., Simpson P. 1990. The genetics of a small autosomal region of Drosophila melanogaster containing the structural gene for alcohol dehydrogenase. VII. Characterization of the region around the snail and cactus loci. Genetics. 126:679–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balicky E.M., Endres M.W., Lai C., Bickel S.E. 2002. Meiotic cohesion requires accumulation of ORD on chromosomes before condensation. Mol. Biol. Cell. 13:3890–3900 10.1091/mbc.E02-06-0332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel S.E., Wyman D.W., Miyazaki W.Y., Moore D.P., Orr-Weaver T.L. 1996. Identification of ORD, a Drosophila protein essential for sister chromatid cohesion. EMBO J. 15:1451–1459 [PMC free article] [PubMed] [Google Scholar]
- Bickel S.E., Wyman D.W., Orr-Weaver T.L. 1997. Mutational analysis of the Drosophila sister-chromatid cohesion protein ORD and its role in the maintenance of centromeric cohesion. Genetics. 146:1319–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower M.D., Karpen G.H. 2001. The role of Drosophila CID in kinetochore formation, cell-cycle progression and heterochromatin interactions. Nat. Cell Biol. 3:730–739 10.1038/35087045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaccorsi S., Giansanti M.G., Cenci G., Gatti M. 2000. Cytological analysis of spermatocyte growth and male meiosis in Drosophila melanogaster. Drosophila Protocols. Sullivan W., Ashburner M., Hawley R. Scott, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 87–109 [Google Scholar]
- Cai X., Dong F., Edelmann R.E., Makaroff C.A. 2003. The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. J. Cell Sci. 116:2999–3007 10.1242/jcs.00601 [DOI] [PubMed] [Google Scholar]
- Cenci G., Bonaccorsi S., Pisano C., Verni F., Gatti M. 1994. Chromatin and microtubule organization during premeiotic, meiotic and early postmeiotic stages of Drosophila melanogaster spermatogenesis. J. Cell Sci. 107:3521–3534 [DOI] [PubMed] [Google Scholar]
- Chelysheva L., Diallo S., Vezon D., Gendrot G., Vrielynck N., Belcram K., Rocques N., Márquez-Lema A., Bhatt A.M., Horlow C., et al. 2005. AtREC8 and AtSCC3 are essential to the monopolar orientation of the kinetochores during meiosis. J. Cell Sci. 118:4621–4632 10.1242/jcs.02583 [DOI] [PubMed] [Google Scholar]
- Davis B.K. 1971. Genetic analysis of a meiotic mutant resulting in precocious sister-centromere separation in Drosophila melanogaster. Mol. Gen. Genet. 113:251–272 10.1007/BF00339546 [DOI] [PubMed] [Google Scholar]
- Dernburg A.F. 2000. In situ hybridization to somatic chromosomes. Drosophila Protocols. Sullivan W., Ashburner M., Hawley R. Scott, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: 25–55 [DOI] [PubMed] [Google Scholar]
- Hauf S., Watanabe Y. 2004. Kinetochore orientation in mitosis and meiosis. Cell. 119:317–327 10.1016/j.cell.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Heidmann D., Horn S., Heidmann S., Schleiffer A., Nasmyth K., Lehner C.F. 2004. The Drosophila meiotic kleisin C(2)M functions before the meiotic divisions. Chromosoma. 113:177–187 10.1007/s00412-004-0305-5 [DOI] [PubMed] [Google Scholar]
- Kerrebrock A.W., Miyazaki W.Y., Birnby D., Orr-Weaver T.L. 1992. The Drosophila mei-S332 gene promotes sister-chromatid cohesion in meiosis following kinetochore differentiation. Genetics. 130:827–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khetani R.S., Bickel S.E. 2007. Regulation of meiotic cohesion and chromosome core morphogenesis during pachytene in Drosophila oocytes. J. Cell Sci. 120:3123–3137 10.1242/jcs.009977 [DOI] [PubMed] [Google Scholar]
- Koundakjian E.J., Cowan D.M., Hardy R.W., Becker A.H. 2004. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 167:203–206 10.1534/genetics.167.1.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L., Diehl-Jones W., Lasko P. 1994. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development. 120:1201–1211 [DOI] [PubMed] [Google Scholar]
- Manheim E.A., McKim K.S. 2003. The Synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr. Biol. 13:276–285 10.1016/S0960-9822(03)00050-2 [DOI] [PubMed] [Google Scholar]
- Miyazaki W.Y., Orr-Weaver T.L. 1992. Sister-chromatid misbehavior in Drosophila ord mutants. Genetics. 132:1047–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje-Casas F., Prabhu V.R., Lee B.H., Boselli M., Amon A. 2007. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 128:477–490 10.1016/j.cell.2006.12.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. 2003. Chromosome choreography: the meiotic ballet. Science. 301:785–789 10.1126/science.1086605 [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., Loidl J. 2001. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 15:1349–1360 10.1101/gad.192701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petronczki M., Siomos M.F., Nasmyth K. 2003. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 112:423–440 10.1016/S0092-8674(03)00083-7 [DOI] [PubMed] [Google Scholar]
- Sakuno T., Tada K., Watanabe Y. 2009. Kinetochore geometry defined by cohesion within the centromere. Nature. 458:852–858 10.1038/nature07876 [DOI] [PubMed] [Google Scholar]
- Sano H., Nakamura A., Kobayashi S. 2002. Identification of a transcriptional regulatory region for germline-specific expression of vasa gene in Drosophila melanogaster. Mech. Dev. 112:129–139 10.1016/S0925-4773(01)00654-2 [DOI] [PubMed] [Google Scholar]
- Shonn M.A., McCarroll R., Murray A.W. 2002. Spo13 protects meiotic cohesin at centromeres in meiosis I. Genes Dev. 16:1659–1671 10.1101/gad.975802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styhler S., Nakamura A., Swan A., Suter B., Lasko P. 1998. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 125:1569–1578 [DOI] [PubMed] [Google Scholar]
- Thomas S.E., Soltani-Bejnood M., Roth P., Dorn R., Logsdon J.M., Jr., McKee B.D. 2005. Identification of two proteins required for conjunction and regular segregation of achiasmate homologs in Drosophila male meiosis. Cell. 123:555–568 10.1016/j.cell.2005.08.043 [DOI] [PubMed] [Google Scholar]
- Tinker R., Silver D., Montell D.J. 1998. Requirement for the vasa RNA helicase in gurken mRNA localization. Dev. Biol. 199:1–10 10.1006/dbio.1998.8941 [DOI] [PubMed] [Google Scholar]
- Tóth A., Rabitsch K.P., Gálová M., Schleiffer A., Buonomo S.B., Nasmyth K. 2000. Functional genomics identifies monopolin: a kinetochore protein required for segregation of homologs during meiosis i. Cell. 103:1155–1168 10.1016/S0092-8674(00)00217-8 [DOI] [PubMed] [Google Scholar]
- Uhlmann F. 2004. The mechanism of sister chromatid cohesion. Exp. Cell Res. 296:80–85 10.1016/j.yexcr.2004.03.005 [DOI] [PubMed] [Google Scholar]
- Van Doren M., Williamson A.L., Lehmann R. 1998. Regulation of zygotic gene expression in Drosophila primordial germ cells. Curr. Biol. 8:243–246 10.1016/S0960-9822(98)70091-0 [DOI] [PubMed] [Google Scholar]
- Vazquez J., Belmont A.S., Sedat J.W. 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12:1473–1483 10.1016/S0960-9822(02)01090-4 [DOI] [PubMed] [Google Scholar]
- Wakimoto B.T., Lindsley D.L., Herrera C. 2004. Toward a comprehensive genetic analysis of male fertility in Drosophila melanogaster. Genetics. 167:207–216 10.1534/genetics.167.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. 2005. Shugoshin: guardian spirit at the centromere. Curr. Opin. Cell Biol. 17:590–595 10.1016/j.ceb.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Nurse P. 1999. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 400:461–464 10.1038/22774 [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Yokobayashi S., Yamamoto M., Nurse P. 2001. Pre-meiotic S phase is linked to reductional chromosome segregation and recombination. Nature. 409:359–363 10.1038/35053103 [DOI] [PubMed] [Google Scholar]
- Wolf K.W. 1994. How meiotic cells deal with non-exchange chromosomes. Bioessays. 16:107–114 [DOI] [PubMed] [Google Scholar]
- Xu H., Beasley M.D., Warren W.D., van der Horst G.T., McKay M.J. 2005. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell. 8:949–961 10.1016/j.devcel.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Yamamoto A., Hiraoka Y. 2003. Monopolar spindle attachment of sister chromatids is ensured by two distinct mechanisms at the first meiotic division in fission yeast. EMBO J. 22:2284–2296 10.1093/emboj/cdg222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S., Watanabe Y. 2005. The kinetochore protein Moa1 enables cohesion-mediated monopolar attachment at meiosis I. Cell. 123:803–817 10.1016/j.cell.2005.09.013 [DOI] [PubMed] [Google Scholar]
- Yokobayashi S., Yamamoto M., Watanabe Y. 2003. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol. Cell. Biol. 23:3965–3973 10.1128/MCB.23.11.3965-3973.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.G., Dawe R.K. 2000. Functional redundancy in the maize meiotic kinetochore. J. Cell Biol. 151:131–142 10.1083/jcb.151.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]