After chromosome missegregation, the growth of nondiploid cells is inhibited thanks to a p53-dependent mechanism.
Abstract
Most solid tumors are aneuploid, and it has been proposed that aneuploidy is the consequence of an elevated rate of chromosome missegregation in a process called chromosomal instability (CIN). However, the relationship of aneuploidy and CIN is unclear because the proliferation of cultured diploid cells is compromised by chromosome missegregation. The mechanism for this intolerance of nondiploid genomes is unknown. In this study, we show that in otherwise diploid human cells, chromosome missegregation causes a cell cycle delay with nuclear accumulation of the tumor suppressor p53 and the cyclin kinase inhibitor p21. Deletion of the p53 gene permits the accumulation of nondiploid cells such that CIN generates cells with aneuploid genomes that resemble many human tumors. Thus, the p53 pathway plays an important role in limiting the propagation of aneuploid human cells in culture to preserve the diploid karyotype of the population. These data fit with the concordance of aneuploidy and disruption of the p53 pathway in many tumors, but the presence of aneuploid cells in some normal human and mouse tissues indicates that there are known exceptions to the involvement of p53 in aneuploid cells and that tissue context may be important in how cells respond to aneuploidy.
Introduction
Aneuploidy is defined as an abnormal number of chromosomes and is one of the most prominent features in solid tumors. Karyotypes of tumors frequently range from 40 to 60 chromosomes (http://cgap.nci.nih.gov/Chromosomes/Mitelman), and it has been proposed that this level of aneuploidy is generated through an elevated rate of single chromosome missegregation as seen in chromosomal instability (CIN; Lengauer et al., 1998). However, the relationship between CIN and aneuploidy is unclear as we recently showed that aneuploid human cells do not propagate efficiently when chromosome missegregation rates are elevated to levels that are equivalent to tumor cells with CIN (Thompson and Compton, 2008). Moreover, mice heterozygous for mutations in genes encoding the checkpoint proteins Mad1 and -2 and CENP-E have elevated chromosome missegregation rates but only form tumors late in life in selective tissues (Michel et al., 2001; Iwanaga et al., 2007; Weaver et al., 2007). Some groups argue that this delay in tumor formation is caused by a requirement for other cooperating mutations (Holland and Cleveland, 2009). Another possibility is that only specific chromosome combinations promote tumorigenesis. Not all chromosomes may have the same impact on cell viability, but data from others showing reduced growth rates in aneuploid strains of budding yeast (Torres et al., 2007) and mouse cell lines (Williams et al., 2008) led those investigators to suggest that growth retardation in response to aneuploidy is a general phenomenon (Torres et al., 2008). However, aneuploid cell lines isolated from tumors grow efficiently in culture, indicating that they have overcome this limitation. From these data, we hypothesized that aneuploid tumor cells only arise from otherwise diploid somatic cells when chromosome missegregation is coupled to the acquisition of tolerance for a nondiploid genome (Thompson and Compton, 2008). However, the mechanism underlying the intolerance for a nondiploid genome is unknown.
Results
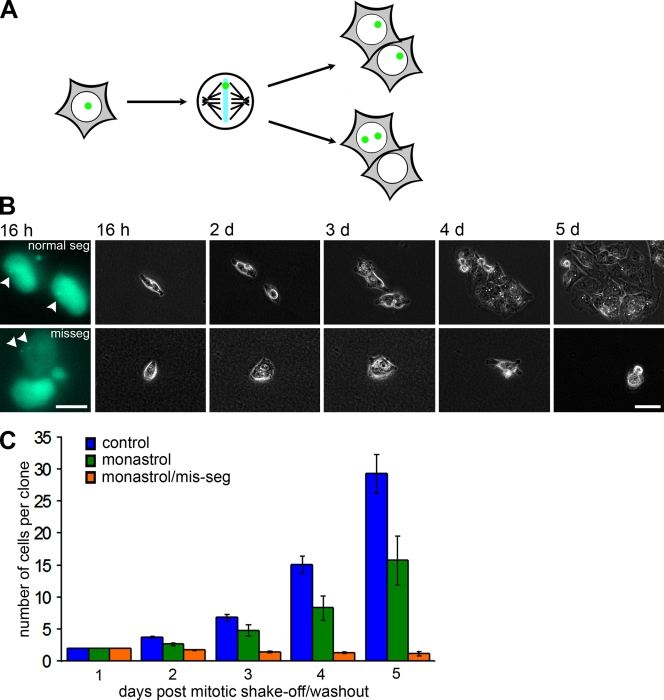
To follow the fate of live cells that missegregate chromosomes, we generated HCT116 cells that expressed LacIGFP and had multiple copies of lacO integrated into a single chromosomal locus (Robinett et al., 1996; Straight et al., 1996). We chose to use HCT116 cells for this purpose because it is an established near-diploid colon cancer cell line that faithfully segregates chromosomes to maintain a stable karyotype (Lengauer et al., 1997). Cells with integrated lacO and expressing LacIGFP remained near diploid. The expressed LacIGFP bound to the chromosomal lacO site, producing a single, bright fluorescent mark in each interphase nucleus and unique marks on sister chromatids of mitotic chromosomes (Fig. 1). Accurate segregation of this chromosome during mitosis yielded daughter cells with single marks in each nucleus, whereas missegregation yielded one daughter cell with no marks and the other with two (Fig. 2 A).
Figure 1.
HCT116 cells with a single marked chromosome. Interphase (top left) and anaphase (bottom left) cells and a chromosome spread (right) with LacIGFP (green) bound to multiple copies of lacO integrated at a single chromosomal locus. DNA is stained with DAPI (blue). Bar, 10 µm.
Figure 2.
Growth arrest after chromosome missegregation. (A) Schematic for proper segregation and missegregation of a single GFP-marked chromosome. (B) GFP (left) and phase-contrast images of daughter cells that either segregated the marked chromosome normally (top) or abnormally (bottom) at the indicated times after drug washout. Arrowheads point to the LacIGFP/lacO chromosome mark. (C) Number of cells per clone after no treatment, monastrol washout, and monastrol washout with missegregation (mis-seg) of the marked chromosome. Bars represent mean ± SEM (independent clones counted for control, n = 49; monastrol, n = 15; and monastrol/mis-seg, n = 22). Bars: (B, left) 10 µm; (B, right) 50 µm.
Using these chromosomally marked cells, we induced chromosome missegregation through a monastrol washout strategy (Cimini et al., 1999; Knowlton et al., 2006; Thompson and Compton, 2008) and followed the fate of individual cell clones. We previously showed that this washout strategy elevates chromosome missegregation to levels where ∼33% of cells missegregate at least one chromosome (Thompson and Compton, 2008). Mitotic cells were collected and plated at low density, and subsequent daughter cell pairs were identified using live cell fluorescent microscopy in which the marked chromosome either missegregated or segregated normally (Fig. 2 B). These cells were readily identifiable despite the low signal to noise ratio for the chromosomal spot when viewed under live imaging conditions (Fig. 2 B, arrowheads). As expected, untreated control cells that properly segregated the marked chromosome grew into large colonies over a 5-d period (Fig. 2, B and C). In contrast, cells that missegregated the marked chromosome failed to divide over the 5-d period (Fig. 2, B and C). In aggregate, colonies generated from cells that had undergone the drug washout strategy yet segregated the marked chromosome correctly showed an intermediate number of cells compared with colonies derived from untreated control cells and cells that missegregated the marked chromosome (Fig. 2 C) because they fall into two subsets. 5 out of 15 cell colonies formed one subset that contained clones with three or fewer cells at the end of the 5 d of imaging. Colonies in the remaining subset had cell numbers similar to the untreated controls, indicating that the initial drug washout step does not impair subsequent colony growth. Previously, we showed that HCT116 cells missegregate chromosomes at a rate of approximately one chromosome in every three cell divisions using this monastrol washout strategy (Thompson and Compton, 2008). Because one third of the colonies formed after monastrol washout did not grow beyond two to three cells, it stands to reason that their growth is delayed in response to the missegregation of a chromosome different from the one carrying the fluorescent mark.
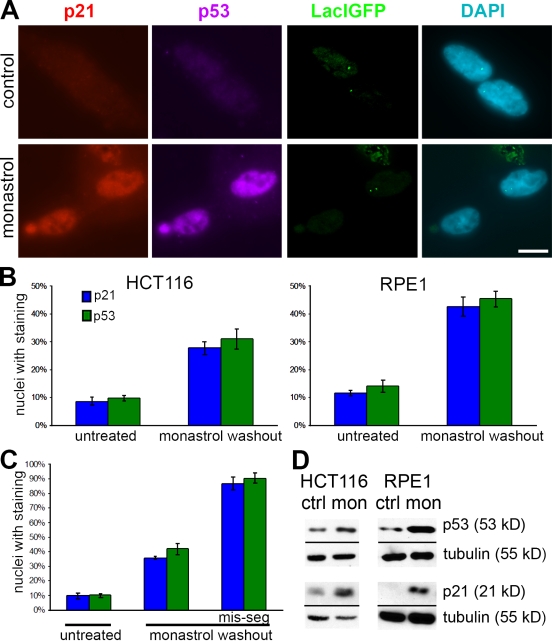
Cell cycle delay or arrest under various conditions is often associated with activation of the tumor suppressor protein p53 (Horn and Vousden, 2007). Using our chromosomally marked HCT116 cells, we induced chromosome missegregation with the monastrol washout strategy and then stained for p53 and one of its downstream targets, the cyclin-dependent kinase inhibitor p21 (Fig. 3). Staining intensities for both p53 and p21 were low in HCT116 cells that segregated the marked chromosome normally but were elevated in cells that missegregated the marked chromosome. Of note, p53 and p21 staining intensities were elevated in both daughter cells: one that gained the chromosome and its sibling that lost the chromosome (Fig. 3 A). In cell populations, 24 h after monastrol washout, elevated p53 and p21 staining was observed in ∼30% of HCT116 cells, which is consistent with the chromosome missegregation rate induced by this treatment (Fig. 3 B). Intensities for p53 and p21 were also elevated in 40% of immortalized RPE1 cells after monastrol washout, showing that similar effects occur in nontransformed cells (Fig. 3 B). When daughter cell pairs were directly examined, the specific subset of HCT116 cells that missegregated the marked chromosome showed that 87% of nuclei stained positive for p21 and >90% stained positive for p53, whereas only 40% of the cells that segregated the chromosome properly were positive (Fig. 3 C). The percentage of p53- and p21-positive cells in general populations (30%) was skewed downward from the isolated pairs of daughter cells (40%) because of ongoing cell division of some cells (presumably those with low levels of p21 and p53) during the 24 h period after monastrol washout in the population experiment. Similar results were observed using three different HCT116 clones, each derived from independent lacO integrations with a different chromosome labeled in each clone. This indicates that activation of the growth arrest/delay pathway involving the elevation of p21 and p53 levels is a general response to chromosome missegregation and is not confined to the missegregation of a specific chromosome. The increase in p21 and p53 after drug washout could also be detected by immunoblotting in both HCT116 and RPE1 cells (Fig. 3 D).
Figure 3.
p21 and p53 expression after chromosome missegregation. (A) HCT116 cells that have properly segregated (top) or missegregated (bottom) the marked chromosome 24 h after no treatment (top) or monastrol washout (bottom) were stained for p21, p53, and DAPI and with LacIGFP as indicated. (B) Percentage of nuclei staining positive for p21 and p53 24 h after control and monastrol washout in populations of HCT116 (left) and RPE1 (right) cells. Bars represent mean ± SEM of at least three experiments (>300 nuclei counted per condition per experiment). (C) Percentage of nuclei staining positive for p21 and p53 in chromosomally marked HCT116 cell line after control or monastrol washout in pairs of daughter cells that have either properly segregated or missegregated (mis-seg) the marked chromosome. Bars represent mean ± SEM of >180 cells in at least three experiments. (D) Immunoblots of untreated (ctrl) cells or cells after monastrol washout (mon) probed for p21, p53, and tubulin (loading control) in HCT116 and RPE1 cells. Bar, 10 µm.
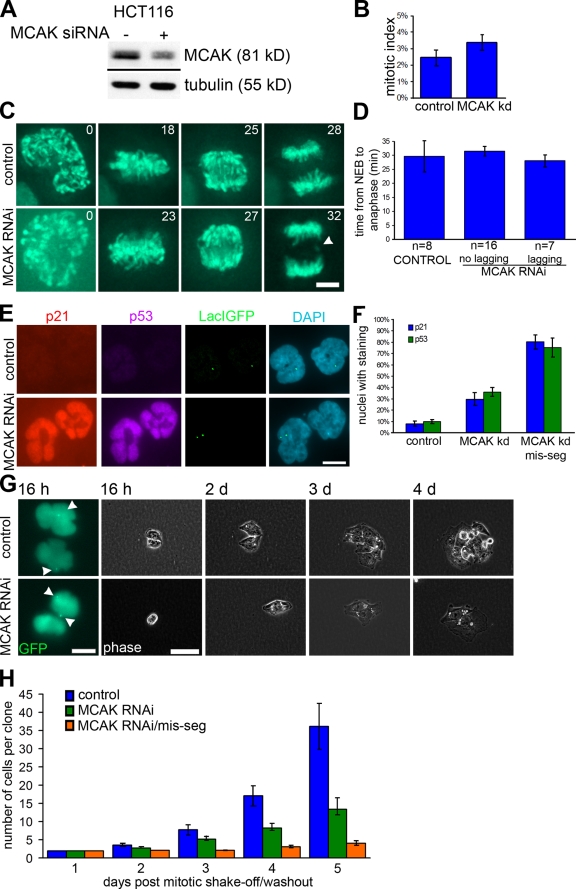
Because the drug washout strategy involves an extended mitotic arrest, we also induced chromosome missegregation using an independent strategy whereby we transfected cells with siRNA specific to the centromere-associated kinesin-13 protein MCAK (mitotic centromere–associated kinesin; Wordeman and Mitchison, 1995). Depletion of MCAK increases levels of merotely (attachment of single kinetochores to microtubules oriented toward both spindle poles) such that 30–40% of anaphase cells have lagging chromosomes, which promotes chromosome missegregation (Maney et al., 1998; Kline-Smith et al., 2004). 72 h after transfection of HCT116 cells with MCAK-specific siRNA, MCAK protein levels were reduced to ∼35% of control levels (Fig. 4 A), and ∼33% of anaphase cells had lagging chromosomes, compared with ∼10% in control untreated populations. MCAK depletion did not significantly increase the mitotic index of the cell population (P = 0.7376, χ2 test; Fig. 4 B). Moreover, live cell imaging showed that MCAK depletion did not significantly prolong the duration of mitosis (time from nuclear envelope breakdown [NEB] to anaphase onset) regardless of whether there were lagging chromosomes or not (Fig. 4, C and D). Staining intensities for both p53 and p21 were elevated in MCAK-deficient HCT116 cells that missegregated the marked chromosome (Fig. 4 E). Again, this was observed in both daughter cells. In the population of MCAK-deficient HCT116 cells that correctly segregated the marked chromosome, p21 and p53 staining intensity was elevated in ∼30% and ∼36% of nuclei, respectively, likely caused by missegregation of another chromosome (Fig. 4 F). In the subset of cells that missegregated the marked chromosome, 80% of nuclei had elevated p21 staining intensity, and 76% had elevated p53 (Fig. 4 F). As expected, most cells that properly segregated the marked chromosome after transfection with MCAK-specific siRNA grew into large colonies over several days (Fig. 4, G and H). In contrast, cells that missegregated the marked chromosome failed to divide over the same period (Fig. 4, G and H). As with the monastrol washout experiment, some cells that segregated the marked chromosome properly displayed only three or fewer cells after 5 d (4 out of 16 colonies). This most likely reflects a subset of cells that underwent growth arrest induced by missegregation of an unmarked chromosome (Fig. 4 H). Many of the remaining colonies that were transfected with MCAK-specific siRNA grew into colonies with cell numbers comparable with control cells. However, a subset of these colonies (58%) contained only 5–10 cells after 5 d of growth. Growth of cells in this subset of colonies may have been limited by missegregation of a chromosome after the first division because of the lingering deficiency of MCAK induced by ongoing RNA interference (Fig. 4 H). These data show that chromosome missegregation in mitosis stabilizes p53 and the cyclin kinase inhibitor p21 in the subsequent G1 phase of the cell cycle and delays cell cycle progression. This is consistent with the observation that rat embryonic fibroblasts undergo a p53-dependent arrest in G1 after the formation of micronuclei because micronuclei are often the consequence of chromosome missegregation caused by persistent merotelic attachment of microtubules to kinetochores (Sablina et al., 1998).
Figure 4.
p21 and p53 expression and growth arrest after chromosome missegregation induced by MCAK depletion. (A) Western blot of HCT116 cells 72 h after transfection with MCAK siRNA (+) or no siRNA (−) probing for MCAK and tubulin (loading control). (B) Mitotic indexes of HCT116 cells 72 h after MCAK or mock knockdown (kd). Bars represent mean ± SEM (three experiments; >500 cells in each experiment). (C) Still frames of control (top) or MCAK knockdown (bottom) in HCT116 cells expressing GFP-H2B, with time given in minutes after NEB. A lagging chromosome in MCAK knockdown anaphase is labeled with an arrowhead. (D) Timing (in minutes) from NEB to anaphase onset in HCT116 cells after control or MCAK knockdown. Bars represent mean ± SEM. (E) HCT116 cells that have properly segregated (top) or missegregated (bottom) the marked chromosome in control (top) or MCAK-deficient cells (bottom) stained for p21, p53, and DAPI and with LacIGFP as indicated. (F) Percentage of nuclei positive for p21 and p53 after normal or missegregation (mis-seg) of the marked chromosome in control or MCAK-depleted HCT116 cells. Bars represent mean ± SEM of >150 cells in at least three experiments. (G) GFP (left) and phase-contrast images of daughter cells that segregated the marked chromosome normally (top) or abnormally in MCAK-deficient cells (bottom) at the indicated times after mitotic shake-off. Arrowheads point to the LacIGFP/lacO chromosome mark. (H) Number of cells in each clone after control or MCAK knockdown or MCAK knockdown cells that have missegregated the marked chromosome. Bars represent mean ± SEM (independent clones counted for control, n = 7; MCAK, n = 16; and MCAK/mis-seg, n = 14). Bars: (C) 5 µm; (E and G [left]) 10 µm; (G, right) 60 µm.
To confirm that proliferation defects after chromosome missegregation are not specific to transformed HCT116 cells, we induced chromosome missegregation in primary fetal lung fibroblast IMR-90 cells by depleting MCAK. 72 h after transfection with MCAK-specific siRNA, IMR-90 cells displayed a sixfold increase in anaphases with lagging chromosomes, which is indicative of elevated rates of chromosome missegregation. Because IMR-90 cells are highly motile, we generated cell clones after transfection with MCAK-specific siRNA by limiting dilution into 96-well plates. We then counted the numbers of cells in each colony 5 d after plating. Under these conditions, the number of MCAK-deficient cells that failed to progress into colonies larger than one cell was significantly higher than that of control cells (Fig. S1). Although it is impossible to confirm chromosome missegregation in these primary fibroblasts, these results are consistent with the growth arrest we observed in HCT116 cells after chromosome missegregation.
DNA damage is a common mechanism to stabilize p53 (Kastan et al., 1991). Under our experimental conditions, DNA damage may be induced at centromeric DNA in metaphase or early anaphase by spindle forces pulling merotelic kinetochores toward opposite poles. To test for DNA damage, we examined the levels of phosphorylated histone γ-H2AX, a modified histone which rapidly appears at sites of double-strand breaks (Rogakou et al., 1998). Doxorubicin treatment induced DNA double-strand breaks and generated numerous γ-H2AX foci on mitotic chromosomes in anaphase cells (Fig. S2 A). However, γ-H2AX foci were not detected on lagging chromosomes that were induced using the monastrol washout strategy. We also examined interphase cells after doxorubicin treatment or 24 h after the monastrol washout (Fig. S2, B and C). As expected, nuclei in nearly all cells treated with doxorubicin stained positive for γ-H2AX. In contrast, nuclei from only a small fraction of cells treated with monastrol washout stained positive for γ-H2AX, and this fraction is much smaller than the fraction of cells that would have missegregated a chromosome using this strategy (Thompson and Compton, 2008) and is not different from control cells. Thus, p53 stabilization in cells that missegregate chromosomes is not the result of DNA damage caused by either the treatments used to induce missegregation or mechanical forces involved in missegregation.
If the stabilization of p53 contributes to the prolonged cell cycle delay after chromosome missegregation, loss of p53 would permit the growth of aneuploid cells. To test this idea, we induced chromosome missegregation using a single monastrol washout in HCT116 cells lacking both alleles of the p53 gene (Bunz et al., 2002). We then determined the chromosome content of these cells as they propagated without further treatment using FISH with chromosome-specific probes (Fig. 5). The percentage of untreated p53-null cells that were aneuploid in the population was low and equivalent to wild-type HCT116 cells. Immediately after washout of monastrol, there was an equivalent increase in the percentage of aneuploid cells in both populations. The percentage of aneuploid cells in HCT116 wild-type populations returned to basal levels by 6 d after treatment (Fig. 5), as previously shown (Thompson and Compton, 2008), indicating a failure of aneuploid cells to propagate efficiently. In contrast, the percentage of aneuploid cells in the population of HCT116 cells lacking p53 remained high 6 d after treatment, indicating that aneuploid cells in this population propagate and compete efficiently with diploid cells. This demonstrates that p53 is a key component of a pathway that provides intolerance to aneuploidy in somatic cultured human cells.
Figure 5.
p53-null HCT116 cells continue to proliferate after missegregation of chromosomes. p53 wild-type (left) and null (right) cells were treated with or without (control) monastrol, and mitotic cells were collected by shake-off, plated, and allowed to proliferate with no further treatment. At 4 h, 2 d, and 6 d, cells were harvested, and FISH was performed using probes specific for chromosomes 3, 7, and 15. For each time point/condition, 600 nuclei were counted per chromosome. Bars represent the mean percent deviation from the modal chromosome number, and error bars show SEM. *, P < 0.05; χ2 test.
It has recently been shown that cell cycle arrest is mediated by the p38 stress kinase in response to disruption of the cytoskeleton (Mikule et al., 2007; Uetake et al., 2007), so we tested whether the p38-dependent stress pathway participated in cell cycle delay in cells after chromosome missegregation. We induced chromosome missegregation in HCT116 cells as well as in immortalized but nontransformed RPE1 cells by using the monastrol washout strategy. Using FISH with chromosome-specific probes, we then determined the chromosome content of the populations as they propagated in the presence of either the p38 inhibitor SB203580 or the MAPK inhibitor PD98059 (Fig. 6). As expected, the percentage of aneuploid cells in populations of both HCT116 and RPE1 cells increased immediately after monastrol washout. In the presence of the MAPK inhibitor, the percentage of aneuploid cells in both populations declined to background levels over time (Fig. 6 B). In contrast, the percentage of aneuploid cells in both populations remained elevated in the presence of the p38 inhibitor (Fig. 6 A). As p38 is known to directly phosphorylate p53 in response to cellular stress (Bulavin et al., 1999), these data suggest that a stress response can induce a p53-dependent pathway to pause the cell cycle in response to chromosome missegregation.
Figure 6.
Inhibition of p38 stress kinase but not MAPK allows proliferation of aneuploid cells. (A and B) HCT116 cells with wild-type p53 and RPE1 cells were treated as described in Fig. 5. After shake-off, cells were grown in the presence of p38 inhibitor (SB203580; A) or MAPK inhibitor (PD98059; B) for the remainder of the experiment. For each time point/condition, 600 nuclei were counted per chromosome. Bars represent the mean percent deviation from the modal chromosome number, and error bars show SEM. *, P < 0.05; χ2 test.
Deficiency of p53 alone is not sufficient to generate aneuploidy or CIN in HCT116 cells (Bunz et al., 2002). To test whether combining p53 deficiency with elevated chromosome missegregation rates generates viable aneuploid and CIN cells, we induced chromosome missegregation for several generations in p53-deficient HCT116 cells. We quantified modal chromosome number and the percentage of cells deviating from that mode in clonal populations of p53 wild-type and null cells after 27 consecutive generations. Both p53 wild-type and null HCT116 cells maintained a disomic modal copy number with very little deviation from that mode for each chromosome analyzed in this clonal analysis (Table I). This confirms that these cells are near diploid and chromosomally stable and that p53 deficiency alone is insufficient to generate aneuploidy or CIN. Next, we used the monastrol washout strategy to increase chromosome missegregation rates at each generation during the growth of colonies of both cell lines. Viable colonies of p53 wild-type cells failed to grow under these conditions, which is consistent with our previous findings showing that elevating chromosome missegregation rates alone is insufficient to generate viable aneuploid or CIN cells (Thompson and Compton, 2008). In contrast, elevating chromosome missegregation rates did not inhibit colony growth from p53-null HCT116 cells. All of these colonies displayed deviation from the modal chromosome number that was significantly elevated for each chromosome analyzed in each individual colony (Table I). Because these percentages represent aneusomy for specific chromosomes, it is likely that the overall level of aneuploidy in these colonies was much higher. These data show that combining elevated chromosome missegregation rates with increased tolerance for a nondiploid genome through p53 deficiency permits the conversion of otherwise stable diploid cells into cells with the hallmarks of CIN.
Table I.
Modal chromosome number and percentage of cells that deviate from modal chromosome number in p53 wild-type and null clones
| Cell line | Mode (percent deviation) | |||
| Chr 2 | Chr 4 | Chr 7 | Chr 15 | |
| p53+/+ control (clone A) | 2 (3.0)a | 2 (2.7)a | 2 (2.7) | 2 (3.7)a |
| p53+/+ control (clone B) | 2 (2.7) | 2 (2.7) | 2 (4.0)a | 2 (3.7) |
| p53+/+ control (clone C) | 2 (3.0) | 2 (2.0) | 2 (2.7) | 2 (3.7) |
| p53−/− control (clone D) | 2 (3.3) | 2 (3.0) | 2 (2.3) | 2 (3.7) |
| p53−/− control (clone E) | 2 (3.3) | 2 (3.3) | 2 (2.7) | 2 (4.3) |
| p53−/− control (clone F) | 2 (2.0) | 2 (2.7) | 2 (2.0) | 2 (3.7) |
| p53−/− monastrol 27 d (clone G) | 2 (18.0)b | 2 (26.0)b | 2 (10.0)b | 2 (21.0)b |
| p53−/− monastrol 27 d (clone H) | 2 (8.0)b | 2 (18.7)b | 2 (18.7)b | 2 (23.0)b |
| p53−/− monastrol 27 d (clone I) | 2 (17.3)b | 2 (24.3)b | 2 (20.0)b | 2 (29.7)b |
| p53−/− monastrol 27 d (clone J) | 2 (13.3)b | 2 (33.7)b | 2 (20.3)b | 2 (23.7)b |
Chr, chromosome. 300 nuclei were counted per chromosome per clone.
Clone used for statistical analysis.
Deviation from mode is statistically significant (P < 0.05, χ2 test).
We next tested the long-term genomic stability of cells derived from these p53-deficient colonies with CIN. We derived subclones from each of the p53-deficient cell colonies shown in Table I by plating cells at low density. Subclones were grown for ∼27 more generations with no additional monastrol washout, and we used FISH to determine the percentage of cells in each subclone that displayed chromosome numbers that deviated from the chromosomal mode (Table II). With one exception (p53−/− H subclone 4), all of the subclones displayed significant deviation from the modal chromosome number for at least two of the chromosomes analyzed. One subclone (p53−/− J subclone 2) showed exceptionally high deviations from the mode for all four chromosomes analyzed (Table II) and had a highly variable karyotype (Fig. S3). Karyotypes of these subclones often deviated from the normal modal chromosome number of 45, indicating that they were aneuploid (HCT116 cells are disomic for all autosomes and have lost the Y chromosome; Fig. S3). Thus, these subclones are aneuploid in addition to being chromosomally unstable. It is not surprising that most subclones had karyotypes with modes near 45 despite being CIN because there is no selective pressure for specific chromosome combinations under in vitro culture conditions, as would be expected for a tumor that is growing in situ. In the absence of selective pressure, CIN induces significant deviation from modal chromosome numbers and broadens the distribution of karyotypes in the population (Fig. S3) without shifting the modal chromosome number of the population far from 45. Moreover, the inherent chromosome missegregation rate in these subclones is unlikely to reach the extremely high missegregation rate induced by monastrol washout (Thompson and Compton, 2008), explaining why these subclones have lower percentages of cells with chromosome numbers that deviate from the mode compared with the initial clones despite similar generation times. These data show that the acquisition of tolerance for aneuploidy through p53 deficiency coupled with initial chromosome missegregation renders cells in these subclones genetically unstable and intrinsically CIN.
Table II.
Modal chromosome number and percentage of nonmodal cells in subclones generated from p53-null clones induced to persistently missegregate chromosomes for 27 generations
| Cell line | Mode (percent deviation) | |||
| Chr 2 | Chr 4 | Chr 7 | Chr 15 | |
| p53−/− G subclone 1 | 2 (3.3)a | 2 (5.3) | 2 (8.3)a | 2 (13.3)a |
| p53−/− H subclone 1 | 2 (3.3) | 2 (11.3)a | 2 (9.0)a | 2 (9.0)a |
| p53−/− H subclone 2 | 2 (14.3)a | 2 (16.7)a | 2 (18.0)a | 2 (14.7)a |
| p53−/− H subclone 3 | 2 (10.3)a | 2 (11.0)a | 2 (14.0)a | 2 (13.0)a |
| p53−/− H subclone 4 | 2 (5.0) | 2 (4.7) | 2 (4.7) | 2 (6.3) |
| p53−/− I subclone 1 | 2 (8.7)a | 2 (16.7)a | 2 (11.7)a | 2 (12.3)a |
| p53−/− J subclone 1 | 2 (4.7) | 2 (7.0) | 2 (9.7)a | 2 (8.3)a |
| p53−/− J subclone 2 | 4 (55.3)a | 2 (67.3)a | 4 (62.0)a | 2 (51.0)a |
Chr, chromosome. 300 nuclei were counted per chromosome per subclone.
Deviation from mode is statistically significant compared with p53+/+ controls in Table I (P < 0.05, χ2 test).
To discount the possibility that the p53−/− HCT116 cells have acquired an adaptation that permits their tolerance for nondiploid genomes, we transfected our chromosomally marked HCT116 (p53+/+) cells with p53-specific siRNA (Fig. S4). We induced chromosome missegregation by simultaneously transfecting cells with MCAK-specific siRNA. Immunoblotting confirmed deficiency of p53 after siRNA transfection (Fig. S4 A), and an increase in lagging chromosomes at anaphase confirmed depletion of MCAK under these conditions (Fig. S4 B). As expected, cells that were deficient in p53 alone and that properly segregated the marked chromosome grew into large colonies over a 5-d period (Fig. S4 C). p53-deficient cells that missegregated the marked chromosome because of MCAK deficiency also grew into large colonies over a 5-d period (Fig. S4 C). Thus, acute loss of p53 function from otherwise wild-type cells generates tolerance for the propagation of aneuploid cells and confirms our data generated using a cell line that has been engineered to be deficient in p53.
Discussion
In summary, we show that the p53-dependent pathway is a mechanism to limit the growth of diploid human cells in culture after chromosome missegregation. Our data fit a pathway whereby chromosome missegregation generates a p38 kinase–dependent stress response that activates p53 to induce the cyclin kinase inhibitor p21, which delays cell cycle progression. The cellular stress that activates this pathway is most likely derived from the imbalance of products encoded by genes on the aneusomic chromosome. Trisomy has been shown to create gene dosage imbalances in both budding yeast and mammals (Torres et al., 2007; Williams et al., 2008), and, by logical extension, monosomy would also generate gene dosage imbalances. This provides a likely explanation for why our data show activation of the p53-dependent pathway in both the trisomic and the monosomic daughter cells after chromosome missegregation. The p53 pathway has been shown to promote CIN when associated with a compromised spindle assembly checkpoint (Burds et al., 2005). Our data show that the p53 pathway potently inhibits cell growth after missegregation of single chromosomes. The importance for this pathway to respond to imbalances in single chromosomes to prevent accumulation of aneuploid cells is readily apparent given that the human body contains millions of dividing cells at any given time (Alberts et al., 2002) and that a chromosome will spontaneously missegregate about once every 100 cell divisions (Cimini et al., 1999; Thompson and Compton, 2008).
The prolonged cell cycle delay that is imparted by the p53 pathway in response to chromosome missegregation represents one of several different cellular responses to aneuploidy. For example, in some contexts, trisomy merely slows cellular growth rates (Williams et al., 2008), and budding yeast lack a p53 gene, so the responses reported in aneuploid yeast (Torres et al., 2007) cannot involve a p53-dependent pathway. In addition, there is a growing body of literature showing that aneuploidy is common in human and mouse neural progenitor cells and neurons, and these probably retain a functional p53 pathway (Rehen et al., 2001, 2005; Kaushal et al., 2003; Yang et al., 2003; Kingsbury et al., 2005). For example, 33% of mouse neuroblasts are aneuploid compared with only 3.4% of adult lymphocytes (Rehen et al., 2001), and high levels of aneuploidy have also been seen in human brain cells (Rehen et al., 2005), indicating that those cell types tolerate an aneuploid state. Moreover, it has been reported that deletion of p53 in embryonic neural progenitor cells actually decreases the percentage of aneuploid cells, suggesting that p53-mediated arrest in response to aneuploidy is not the sole mechanism to inhibit aneuploid proliferation (McConnell et al., 2004). Also, levels of aneuploidy (often of sex chromosomes) in leukocytes increase as people age (Jacobs, et al., 1963), and there is no reported link to loss of p53 in these cases. Finally, aneuploidy increases in many tissues of mice that are heterozygous for mutations in genes encoding checkpoint proteins, and it is unlikely that these have all lost p53 function (Michel et al., 2001; Babu et al., 2003; Baker et al., 2004; Iwanaga et al., 2007; Jeganathan et al., 2007; Weaver et al., 2007). Interestingly, Vanneste et al. (2009) recently showed that human cleavage-stage embryos show a high level of aneuploidy, with 19 out of 23 embryos containing blastomeres with whole chromosome losses or gains. Embryonic stem cells have reduced levels of some checkpoint proteins and a shortened G1 phase, suggesting that the developing embryo may relax checkpoint control and sacrifice chromosome segregation accuracy to expedite proliferation (Egozi et al., 2007; Ambartsumyan and Clark, 2008). These embryologic adaptations to aneuploidy may underlie the viability of trisomic mouse cells (Williams et al., 2008) and humans (i.e., Down’s syndrome). In addition, mosaic embryos containing both aneuploid and diploid cells can result in chromosomally normal fetuses (Vanneste et al., 2009). This selective retention of diploid cells may explain why mice heterozygous for mutations in mitotic checkpoint genes are viable and have no obvious developmental defects (for review see Foijer et al., 2008). Collectively, these findings indicate that the cellular response to aneuploidy varies according to the specific cell context (i.e., tissue type, embryonic vs. somatic cell, etc.), and responses may vary depending on which chromosome has missegregated. Activation of the p53 pathway observed in this study appears to be critical to maintain the diploid karyotype of the population of cells grown under culture conditions. This may be the preferred response of somatic cells to prevent aneuploidy from contributing to the initiation or promotion of cancer that frequently arises in epithelial cell types. However, cells in other tissue contexts in humans and mice do not appear to respond to aneuploidy by activation of the p53 pathway.
p53 is one of the most highly mutated genes in cancer, and there is a strong correlation between aneuploid tumors and mutation of p53 (Campomenosi et al., 1998), supporting the idea that loss of p53 permits aneuploid cell propagation. For example, in some tumors and precancerous conditions including Barrett’s esophagus, p53 mutations arise in diploid cells before aneuploidy (Blount et al., 1994). Furthermore, diploid cells in culture often inactivate p53 and become aneuploid at the same time during the immortalization process (Harvey et al., 1993), and cells from nonmalignant tissues in p53-null mice are often aneuploid (Fukasawa et al., 1997). However, it remains controversial whether mutation of p53 is obligatory for aneuploid cell propagation. For example, aneuploid embryonic cells divide for multiple generations before embryonic lethality (Dobles et al., 2000; Kalitsis et al., 2000; Williams et al., 2008) in contrast to the growth inhibition we observe in cultured somatic cells. Moreover, p53 is not lost or mutated in many aneuploid tumor cell lines (www.sanger.ac.uk/perl/genetics/CGP/cosmic), and in some cancers, aneuploidy appears during tumorigenesis before p53 mutation (Baker et al., 1990). These tumors may harbor mutations in genes participating in other pathways that limit aneuploid cell growth or they may have impaired signaling upstream or downstream of p53. For example, the stress kinase pathway is inactivated in some tumors (Greenman et al., 2007; Ventura et al., 2007) and attenuated in transformed cells (Mikhailov et al., 2007), which would limit the capacity of aneuploidy-induced gene dosage imbalances to activate the p53 pathway. Alternatively, some tumor cells are wild type for p53 but have mutations in the downstream cyclin kinase inhibitor genes, limiting the functionality of the p53 pathway (e.g., MCF7 breast carcinoma cells; Berns et al., 1995), or have inactivated regulators upstream of p53 (e.g., p14ARF expression is often lost by deletion or promoter hypermethylation in many tumors, and this loss occurs at early stages of tumorigenesis in some tumors; Esteller et al., 2000; Vousden and Lu, 2002) or overexpress p53 inhibitors such as MDM2 or -4 (Toledo and Wahl, 2006).
The chromosome missegregation rate in human cancer cells with CIN is alarmingly high. CIN cells missegregate a chromosome every one to five divisions depending on the cell line (Thompson and Compton, 2008). These high missegregation rates are matched by treatments that elevate merotely (Thompson and Compton, 2008) and are likely to be matched by securin depletion (Pfleghaar et al., 2005). The cell cycle delay imposed by repeated activation of the p53 pathway in response to such frequent chromosome missegregation explains why otherwise diploid cells fail to survive when missegregation rates are elevated to levels equivalent to those of CIN cancer cells (Pfleghaar et al., 2005; Thompson and Compton, 2008). This view may explain the reported increases in the percentage of aneuploid cells in populations of HCT116 cells that are heterozygous for mutations in the Mad2 gene (Michel et al., 2001). Chromosome missegregation rates were not directly measured in Mad2+/− cells and may be lower than those of cells without securin or with elevated merotely because the percentage of aneuploid cells in populations of Mad2+/− cells after 25 generations is similar to that observed in cells without securin or with elevated merotely after just 2 to 5 generations (Pfleghaar et al., 2005; Thompson and Compton, 2008). Thus, loss of p53 function can provide tolerance necessary for cellular survival of otherwise diploid cells when chromosome missegregation rates reach those observed in human cancer cells with CIN.
Finally, our data fit a model whereby abnormal numbers of chromosomes disrupt the sensitive balance of mitotic proteins, which in turn undermines faithful chromosome segregation to produce further aneuploidy (Duesberg et al., 1998, 1999). If the dosage of any one of the many proteins involved in ensuring chromosome segregation fidelity is disrupted by the missegregation of the chromosome carrying that gene, the resulting imbalance could further compromise chromosome segregation accuracy. In this context, we demonstrate that by combining persistent chromosome missegregation with loss of the p53 pathway, we can convert stable diploid cells into aneuploid cells that have karyotypes reminiscent of many human tumors. Moreover, we show that otherwise diploid cells become intrinsically CIN once aneuploidy is induced in a genetic background that tolerates chromosome changes. Although these results fit some aspects of the model proposed by Duesberg et al. (1998, 1999), there is divergence in other aspects because we show that aneuploidy and CIN only become self-propagating in a genetic background in which the p53 pathway has been inactivated. Thus, conversion of diploid progenitor cells into cells that are aneuploid and CIN takes two steps; reduction in chromosome segregation fidelity and acquisition of tolerance for a nondiploid genome that can arise through inactivation of the p53 pathway.
Materials and methods
Cell culture
Cells were maintained at 37°C with 5% CO2 in Dulbecco’s modified medium (RPE1 and IMR-90) or McCoy’s 5a (HCT116) supplemented with 10% FBS, 50 IU/ml penicillin, and 50 µg/ml streptomycin. Paired isogenic HCT116 cell lines with wild-type p53 or null p53 were a gift from B. Vogelstein (Johns Hopkins Oncology Center, Baltimore, MD). IMR-90 cells were purchased from the American Type Culture Collection and used between passage 3 and 7 (after we obtained them) for all experiments.
Plasmid transfections
HCT116 cells were transfected with p3′-SS-GFP-lacI-NLS (LacIGFP plasmid; provided by W. Bickmore, Western General Hospital, Edinburgh, Scotland, UK; Chubb et al., 2002) using Lipofectamine 2000 (Invitrogen) and placed under hygromycin selection at 300 µg/ml for ∼20 d. Clones expressing LacIGFP were then transfected simultaneously with pJRC49 (plasmid with multiple lacO sites) and JCR41 (blasticidin resistance cassette; plasmids provided by W. Bickmore; Chubb et al., 2002) and placed under blasticidin selection at 2 µg/ml. Clones were isolated after ∼15–20 d and screened for LacIGFP expression and integration of lacO in a single locus, and selection was maintained by alternating blasticidin and hygromycin selection every 5–7 d. Three independent clones with the brightest chromosomal marks were examined for p21 and p53 expression in cells that missegregated the labeled chromosome after monastrol washouts and MCAK knockdown (see next section). Chromosome spreads were performed to verify that the labeled chromosome differed in each of the clones. One clone was chosen for live cell imaging (Fig. 2) because it had the highest signal to noise ratio.
RNA interference
MCAK was depleted using a published sequence (Cassimeris and Morabito, 2004; Applied Biosystems). p53 was depleted using the predesigned sequence 5′-GGGAGUUGUCAAGUCUUGC-3′ from Applied Biosystems. 200 nM double-stranded RNA was transfected into cells using Oligofectamine as previously described (Ganem and Compton, 2004). Cells were analyzed or used for subsequent experiments 60–72 h later.
GFP and phase-contrast live cell imaging over extended time period
For monastrol washouts, cells were treated with 100 µM monastrol or remained untreated (control) for 8 h. Mitotic cells were isolated by shake-off, washed with PBS, and plated at low density in 6-well plates. For MCAK knockdowns, cells were harvested 72 h after transfection as described in the previous section and plated at low density in 6-well plates. For both monastrol and MCAK experiments, 16 h after plating, pairs of daughter cells were screened for segregation of the marked chromosome by fluorescence microscopy, and a single focal plane GFP and phase image were acquired using a Plan Fluor 40× NA 0.6 ELW air objective on a microscope (TE 2000-E; Nikon) at 37°C equipped with a cooled charge-coupled device camera (Orca ER; Hamamatsu Photonics). The spatial position of each pair of daughter cells was recorded using Phylum Live software (PerkinElmer), and phase-contrast images were acquired for each position at 24-h intervals for a total of 5 d. Cell clones were included in histograms only if they could be distinguished as two separate daughter cells to exclude cells that may have failed cytokinesis. Autocontrast was applied to images using Photoshop CS2 (Adobe).
IMR-90 cells were transfected with MCAK siRNA as described in the previous section. 72 h after transfection, cells were split into 96-well plates at a concentration of one cell per every two wells. Cells were incubated for an additional 5 d, and the number of cells per well was counted.
Fixed cell imaging
Cells were fixed in 3.5% PFA and stained, and images were acquired as previously described (Thompson and Compton, 2008). For monastrol washouts, cells were grown in flasks, treated with 100 µM monastrol for 8 h, isolated by mitotic shake-off, washed with PBS, plated on coverslips, and incubated for 24 h. For MCAK knockdowns, cells were grown and transfected on coverslips and fixed at 72 h after siRNA transfection. 1 µM doxorubicin was added to cells 4 h before fixation (interphase) or at 0.5 µM 50 min before fixation for mitotic cells. Antibodies were specific for p21 (1:500; Santa Cruz Biotechnology, Inc.), p53 (1:1,000; Santa Cruz Biotechnology, Inc.), anticentromere antibody (1:1,000; provided by K. Sullivan, National University of Ireland, Galway, Ireland), and/or γ-H2AX (1:1,000; Novus Biologicals) and were detected with FITC, Texas red, and/or Cy5. Images were acquired on the aforementioned microscope with a Plan-Apochromat 60× NA 1.4 oil immersion objective. 0.25-µm optical sections in the z axis were collected. Iterative restoration was performed using Phylum Live software. Autocontrast using Photoshop CS2 was applied to images. Nuclei were scored positive for p21 and p53 accumulation if pixel intensities inside the nucleus were at least 1.5 times and 2.0 times, respectively, higher than pixel intensities in untreated cells. Cells were scored positive for γ-H2AX foci when pixel intensities of foci matched or were brighter than single foci in doxorubicin-treated cells.
Live cell imaging of HCT116 GFP-H2B
Live imaging of HCT116 cells expressing GFP-H2B was performed as previously described (Thompson and Compton, 2008). HCT116 cells expressing H2BGFP growing on coverslips were transfected with MCAK siRNA as described in RNA interference. At 48–72 h, coverslips were mounted on modified rose chambers, and mitotic cells were imaged at 37°C with the 60× objective on the aforementioned microscope by acquiring 27 0.5-µm optical sections in the z axis in the GFP channel at 1-min intervals. Images were deconvolved using Phylum Live, autocontrast was applied to images using Photoshop CS2, and still frames represent full volume projections. Time from NEB to anaphase was calculated by averaging movies from multiple MCAK knockdowns.
Immunoblotting
Total cell protein was separated by SDS-PAGE and transferred to a polyvinylidene fluoride membrane. Membranes were incubated with antibodies for p53 (1:2,500), p21 (1:500), MCAK (1:1,000), or tubulin (1:5,000; DM1-α; Sigma-Aldrich) in 1% milk in Tris-buffered saline and detected with HRP-conjugated secondary antibodies followed by chemiluminescence. Band intensities were quantified using ImageJ software (National Institutes of Health).
Monastrol washout time courses and CIN assay
Colony growth and FISH analyses were performed as previously described (Thompson and Compton, 2008). For clonal analyses, HCT116 p53+/+ and p53−/− cells were grown in flasks. Once cells reached ∼70–80% confluency, they were treated with 100 µM monastrol or untreated for 8 h. Mitotics were isolated by shake-off, washed with PBS, and plated at very low density in 100-mm plates in fresh medium with no drug. Cells were allowed to recover and finish dividing for 16 h and then treated with either monastrol or fresh media (control) for 8 h followed by two washes with PBS and once again incubated in fresh medium with no drug for 16 h. p53+/+ and p53−/− control cells and p53−/− monastrol washout cells were treated in this manner for a total of 27 generations. At ∼15–20 generations, clones were harvested by trypsinization of colonies using cloning rings. At 27 generations, cells were harvested for FISH. For subclones derived from p53−/− monastrol 27 d clones, at generation 27, a small percentage of the cells were plated at very low density, and subclones were grown for an additional 27 generations. Chromosome spreads were prepared from mitotic cells from populations of untreated p53+/+ and p53−/− HCT116 cells and p53−/− subclones grown for >50 generations treated with 150 ng/ml nocodazole for 16 h. Cells were collected by trypsinization, briefly resuspended in 75 mM potassium chloride, fixed, washed twice in 3:1 methanol/acetic acid mix, dropped onto wet slides, air dried, and stained with DAPI.
For analysis of aneuploidy in the populations, HCT116 and RPE1 cells were grown in flasks and treated with monastrol or fresh medium as described in the previous paragraph. Mitotic cells were isolated by shake-off, washed with PBS, and replated in 100-mm dishes. Cells were grown in fresh medium with no further treatment, and at 4 h, 2 d, and 6 d (HCT116) or 8 d (RPE1), cells were harvested for FISH analysis. For kinase inhibitor experiments, medium containing 10 µM SB203580 or 50 µM PD98059 was added to the cells immediately after washout of monastrol, and cells were maintained in the presence of the drug for the remainder of the experiment.
Online supplemental material
Fig. S1 shows the percentage of IMR-90 cells that did not form a colony in MCAK knockdown and control cells. Fig. S2 displays γ-H2AX staining in RPE1 cells after monastrol washout or treatment with doxorubicin. Fig. S3 displays histograms of karyotypes from untreated p53+/+ and p53−/− cells and p53−/− subclones derived from clones treated for 27 d with monastrol washout and representative images of karyotypes. Fig. S4 shows the fate of cell clones that have missegregated chromosomes in cells depleted of both MCAK and p53. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905057/DC1.
Acknowledgments
We thank Drs. W. Bickmore, K. Sullivan, and B. Vogelstein for reagents. We thank Emily Hood and Samuel Bakhoum for comments on the manuscript.
This work was supported by National Institutes of Health grants GM51542 (to D.A. Compton) and GM008704 (to S.L. Thompson).
Footnotes
Abbreviations used in this paper:
- CIN
- chromosomal instability
- MCAK
- mitotic centromere–associated kinesin
- NEB
- nuclear envelope breakdown
References
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. 2002. Molecular Biology of the Cell. Fourth edition Garland Science, New York: 1548 pp [Google Scholar]
- Ambartsumyan G., Clark A.T. 2008. Aneuploidy and early human embryo development. Hum. Mol. Genet. 17:R10–R15 10.1093/hmg/ddn170 [DOI] [PubMed] [Google Scholar]
- Babu J.R., Jeganathan K.B., Baker D.J., Wu X., Kang-Decker N., van Deursen J.M. 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J. Cell Biol. 160:341–353 10.1083/jcb.200211048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S.J., Preisinger A.C., Jessup J.M., Paraskeva C., Markowitz S., Willson J.K., Hamilton S., Vogelstein B. 1990. p53 gene mutations occur in combination with 17p allelic deletions as late events in colorectal tumorigenesis. Cancer Res. 50:7717–7722 [PubMed] [Google Scholar]
- Baker D.J., Jeganathan K.B., Cameron J.D., Thompson M., Juneja S., Kopecka A., Kumar R., Jenkins R.B., de Groen P.C., Roche P., van Deursen J.M. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat. Genet. 36:744–749 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- Berns E.M., Klijn J.G., Smid M., van Staveren I.L., Gruis N.A., Foekens J.A. 1995. Infrequent CDKN2 (MTS1/p16) gene alterations in human primary breast cancer. Br. J. Cancer. 72:964–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount P.L., Galipeau P.C., Sanchez C.A., Neshat K., Levine D.S., Yin J., Suzuki H., Abraham J.M., Meltzer S.J., Reid B.J. 1994. 17p allelic losses in diploid cells of patients with Barrett’s esophagus who develop aneuploidy. Cancer Res. 54:2292–2295 [PubMed] [Google Scholar]
- Bulavin D.V., Saito S., Hollander M.C., Sakaguchi K., Anderson C.W., Appella E., Fornace A.J., Jr 1999. Phosphorylation of human p53 by p38 kinase coordinates N-terminal phosphorylation and apoptosis in response to UV radiation. EMBO J. 18:6845–6854 10.1093/emboj/18.23.6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F., Fauth C., Speicher M.R., Dutriaux A., Sedivy J.M., Kinzler K.W., Vogelstein B., Lengauer C. 2002. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 62:1129–1133 [PubMed] [Google Scholar]
- Burds A.A., Lutum A.S., Sorger P.K. 2005. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc. Natl. Acad. Sci. USA. 102:11296–11301 10.1073/pnas.0505053102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campomenosi P., Assereto P., Bogliolo M., Fronza G., Abbondandolo A., Capasso A., Bellomo P.F., Monaco R., Rapallo A., Sciutto A., et al. 1998. p53 mutations and DNA ploidy in colorectal adenocarcinomas. Anal. Cell. Pathol. 17:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L., Morabito J. 2004. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol. Biol. Cell. 15:1580–1590 10.1091/mbc.E03-07-0544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chubb J.R., Boyle S., Perry P., Bickmore W.A. 2002. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12:439–445 10.1016/S0960-9822(02)00695-4 [DOI] [PubMed] [Google Scholar]
- Cimini D., Tanzarella C., Degrassi F. 1999. Differences in malsegregation rates obtained by scoring ana-telophases or binucleate cells. Mutagenesis. 14:563–568 10.1093/mutage/14.6.563 [DOI] [PubMed] [Google Scholar]
- Dobles M., Liberal V., Scott M.L., Benezra R., Sorger P.K. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 101:635–645 10.1016/S0092-8674(00)80875-2 [DOI] [PubMed] [Google Scholar]
- Duesberg P., Rausch C., Rasnick D., Hehlmann R. 1998. Genetic instability of cancer cells is proportional to their degree of aneuploidy. Proc. Natl. Acad. Sci. USA. 95:13692–13697 10.1073/pnas.95.23.13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P., Rasnick D., Li R., Winters L., Rausch C., Hehlmann R. 1999. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 19:4887–4906 [PubMed] [Google Scholar]
- Egozi D., Shapira M., Paor G., Ben-Izhak O., Skorecki K., Hershko D.D. 2007. Regulation of the cell cycle inhibitor p27 and its ubiquitin ligase Skp2 in differentiation of human embryonic stem cells. FASEB J. 21:2807–2817 10.1096/fj.06-7758com [DOI] [PubMed] [Google Scholar]
- Esteller M., Tortola S., Toyota M., Capella G., Peinado M.A., Baylin S.B., Herman J.G. 2000. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res. 60:129–133 [PubMed] [Google Scholar]
- Foijer F., Draviam V.M., Sorger P.K. 2008. Studying chromosome instability in the mouse. Biochim. Biophys. Acta. 1786:73–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukasawa K., Wiener F., Vande Woude G.F., Mai S. 1997. Genomic instability and apoptosis are frequent in p53 deficient young mice. Oncogene. 15:1295–1302 10.1038/sj.onc.1201482 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Compton D.A. 2004. The KinI kinesin Kif2a is required for bipolar spindle assembly through a functional relationship with MCAK. J. Cell Biol. 166:473–478 10.1083/jcb.200404012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature. 446:153–158 10.1038/nature05610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey M., Sands A.T., Weiss R.S., Hegi M.E., Wiseman R.W., Pantazis P., Giovanella B.C., Tainsky M.A., Bradley A., Donehower L.A. 1993. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 8:2457–2467 [PubMed] [Google Scholar]
- Holland A.J., Cleveland D.W. 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10:478–487 10.1038/nrm2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H.F., Vousden K.H. 2007. Coping with stress: multiple ways to activate p53. Oncogene. 26:1306–1316 10.1038/sj.onc.1210263 [DOI] [PubMed] [Google Scholar]
- Iwanaga Y., Chi Y.H., Miyazato A., Sheleg S., Haller K., Peloponese J.M., Jr., Li Y., Ward J.M., Benezra R., Jeang K.T. 2007. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 67:160–166 10.1158/0008-5472.CAN-06-3326 [DOI] [PubMed] [Google Scholar]
- Jacobs P.A., Brunton M., Court Brown W.M., Doll R., Goldstein H. 1963. Change of human chromosome count distribution with age: evidence for a sex differences. Nature. 197:1080–1081 10.1038/1971080a0 [DOI] [PubMed] [Google Scholar]
- Jeganathan K., Malureanu L., Baker D.J., Abraham S.C., van Deursen J.M. 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J. Cell Biol. 179:255–267 10.1083/jcb.200706015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P., Earle E., Fowler K.J., Choo K.H. 2000. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 14:2277–2282 10.1101/gad.827500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B., Onyekwere O., Sidransky D., Vogelstein B., Craig R.W. 1991. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51:6304–6311 [PubMed] [Google Scholar]
- Kaushal D., Contos J.J.A., Treuner K., Yang A.H., Kingsbury M.A., Rehen S.K., McConnell M.J., Okabe M., Barlow C., Chun J. 2003. Alteration of gene expression by chromosome loss in the postnatal mouse brain. J. Neurosci. 23:5599–5606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury M.A., Friedman B., McConnell M.J., Rehen S.K., Yang A.H., Kaushal D., Chun J. 2005. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc. Natl. Acad. Sci. USA. 102:6143–6147 10.1073/pnas.0408171102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith S.L., Khodjakov A., Hergert P., Walczak C.E. 2004. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol. Biol. Cell. 15:1146–1159 10.1091/mbc.E03-08-0581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowlton A.L., Lan W., Stukenberg P.T. 2006. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr. Biol. 16:1705–1710 10.1016/j.cub.2006.07.057 [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K.W., Vogelstein B. 1997. Genetic instability in colorectal cancers. Nature. 386:623–627 10.1038/386623a0 [DOI] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K.W., Vogelstein B. 1998. Genetic instabilities in human cancers. Nature. 396:643–649 10.1038/25292 [DOI] [PubMed] [Google Scholar]
- Maney T., Hunter A.W., Wagenbach M., Wordeman L. 1998. Mitotic centromere–associated kinesin is important for anaphase chromosome segregation. J. Cell Biol. 142:787–801 10.1083/jcb.142.3.787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell M.J., Kaushal D., Yang A.H., Kingsbury M.A., Rehen S.K., Treuner K., Helton R., Annas E.G., Chun J., Barlow C. 2004. Failed clearance of aneuploid embryonic neural progenitor cells leads to excess aneuploidy in the Atm-deficient but not the Trp53-deficient adult cerebral cortex. J. Neurosci. 24:8090–8096 10.1523/JNEUROSCI.2263-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V.V.S., Benezra R. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 409:355–359 10.1038/35053094 [DOI] [PubMed] [Google Scholar]
- Mikhailov A., Patel D., McCance D.J., Rieder C.L. 2007. The G2 p38-mediated stress-activated checkpoint pathway becomes attenuated in transformed cells. Curr. Biol. 17:2162–2168 10.1016/j.cub.2007.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikule K., Delaval B., Kaldis P., Jurcyzk A., Hergert P., Doxsey S. 2007. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat. Cell Biol. 9:160–170 10.1038/ncb1529 [DOI] [PubMed] [Google Scholar]
- Pfleghaar K., Heubes S., Cox J., Stemmann O., Speicher M.R. 2005. Securin is not required for chromosomal stability in human cells. PLoS Biol. 3:e416 10.1371/journal.pbio.0030416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen S.K., McConnell M.J., Kaushal D., Kingsbury M.A., Yang A.H., Chun J. 2001. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc. Natl. Acad. Sci. USA. 98:13361–13366 10.1073/pnas.231487398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen S.K., Yung Y.C., McCreight M.P., Kaushal D., Yang A.H., Almeida B.S.V., Kingsbury M.A., Cabral K.M.S., McConnell M.J., Anliker B., et al. 2005. Constitutional aneuploidy in the normal human brain. J. Neurosci. 25:2176–2180 10.1523/JNEUROSCI.4560-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., Belmont A.S. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Sablina A.A., Ilyinskaya G.V., Rubtsova S.N., Agapova L.S., Chumakov P.M., Kopnin B.P. 1998. Activation of p53-mediated cell cycle checkpoint in response to micronuclei formation. J. Cell Sci. 111:977–984 [DOI] [PubMed] [Google Scholar]
- Straight A.F., Belmont A.S., Robinett C.C., Murray A.W. 1996. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr. Biol. 6:1599–1608 10.1016/S0960-9822(02)70783-5 [DOI] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180:665–672 10.1083/jcb.200712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo F., Wahl G.M. 2006. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 6:909–923 10.1038/nrc2012 [DOI] [PubMed] [Google Scholar]
- Torres E.M., Sokolsky T., Tucker C.M., Chan L.Y., Boselli M., Dunham M.J., Amon A. 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 317:916–924 10.1126/science.1142210 [DOI] [PubMed] [Google Scholar]
- Torres E.M., Williams B.R., Amon A. 2008. Aneuploidy: cells losing their balance. Genetics. 179:737–746 10.1534/genetics.108.090878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Loncarek J., Nordberg J.J., English C.N., La Terra S., Khodjakov A., Sluder G. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176:173–182 10.1083/jcb.200607073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste E., Voet T., Le Caignec C., Ampe M., Konings P., Melotte C., Debrock S., Amyere M., Vikkula M., Schuit F., et al. 2009. Chromosome instability is common in human cleavage-stage embryos. Nat. Med. 15:577–583 10.1038/nm.1924 [DOI] [PubMed] [Google Scholar]
- Ventura J.J., Tenbaum S., Perdiguero E., Huth M., Guerra C., Barbacid M., Pasparakis M., Nebreda A.R. 2007. p38α MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat. Genet. 39:750–758 10.1038/ng2037 [DOI] [PubMed] [Google Scholar]
- Vousden K.H., Lu X. 2002. Live or let die: the cell’s response to p53. Nat. Rev. Cancer. 2:594–604 10.1038/nrc864 [DOI] [PubMed] [Google Scholar]
- Weaver B.A.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Williams B.R., Prabhu V.R., Hunter K.E., Glazier C.M., Whittaker C.A., Housman D.E., Amon A. 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 322:703–709 10.1126/science.1160058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L., Mitchison T.J. 1995. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J. Cell Biol. 128:95–104 10.1083/jcb.128.1.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang A.H., Kaushal D., Rehen S.K., Kriedt K., Kingsbury M.A., McConnell M.J., Chun J. 2003. Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J. Neurosci. 23:10454–10462 [DOI] [PMC free article] [PubMed] [Google Scholar]