The clockwise turning of neurites is caused by the rotations of filopodia as they extend and sweep across the substratum.
Abstract
The direction of neurite elongation is controlled by various environmental cues. However, it has been reported that even in the absence of any extrinsic directional signals, neurites turn clockwise on two-dimensional substrates. In this study, we have discovered autonomous rotational motility of the growth cone, which provides a cellular basis for inherent neurite turning. We have developed a technique for monitoring three-dimensional motility of growth cone filopodia and demonstrate that an individual filopodium rotates on its own longitudinal axis in the right-screw direction from the viewpoint of the growth cone body. We also show that the filopodial rotation involves myosins Va and Vb and may be driven by their spiral interactions with filamentous actin. Furthermore, we provide evidence that the unidirectional rotation of filopodia causes deflected neurite elongation, most likely via asymmetric positioning of the filopodia onto the substrate. Although the growth cone itself has been regarded as functionally symmetric, our study reveals the asymmetric nature of growth cone motility.
Introduction
In the developing nervous system, neurons can extend long processes toward their targets. Elongating neurites, tipped by growth cones, explore guidance cues in their environment for appropriate paths (Tessier-Lavigne and Goodman, 1996). It has been assumed that the growth cone becomes functionally and morphologically asymmetric upon encountering a graded or spatially defined expression of guidance cues, thereby turning toward a new direction (Dent and Gertler, 2003; Mortimer et al., 2008). However, even in the absence of such extrinsic directional signals, neurites turn rightward or clockwise on 2D substrates of poly-lysine (Heacock and Agranoff, 1977). Cellular and molecular mechanisms underlying this inherent turning behavior of neurites have remained unknown. In this study, we demonstrate a unidirectional rotation of growth cone filopodia that renders the growth cone inherently asymmetric and causes the deflected elongation of neurites.
Results
Growth cone filopodia rotate in the right-screw direction
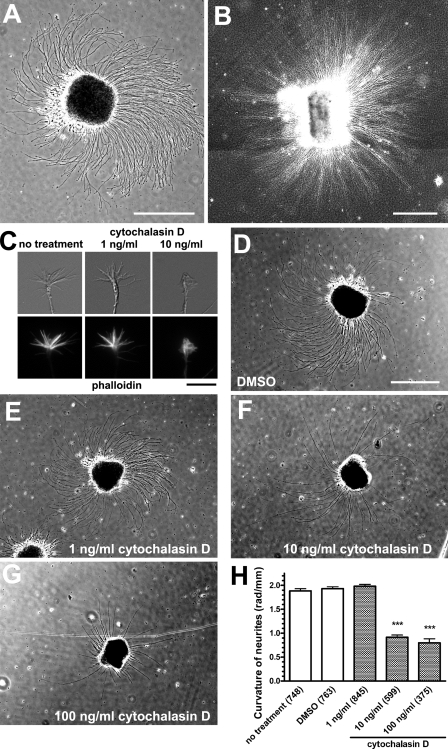
Because adhesive interactions of growth cones with their environment play a crucial role in neurite elongation, we first examined the neurite growth linearity on different 2D substrates. When hippocampal explants from rat embryos were cultured on coverslips coated with poly-d-lysine (PDL) and laminin, the explants extended neurites turning rightward or clockwise from the top view (Fig. 1 A), which is in accordance with a previous study using retinal explants (Heacock and Agranoff, 1977). We also confirmed that neurites of cerebellar granule cells showed rightward turning on PDL/laminin substrates (unpublished data). Such inherent turning behavior of neurites was also observed in dissociated cultures of neocortical neurons (Romijn et al., 1980), retinal ganglion cells (Schwartz and Agranoff, 1981), and hippocampal neurons (unpublished data). Neurites from hippocampal explants turned rightward on other 2D substrates (coverslips coated with N-cadherin or the cell adhesion molecule L1; unpublished data). In contrast, neurites grew practically straight when explants were embedded in 3D substrates composed of a collagen gel (Fig. 1 B), suggesting that the dimension of culture substrates influences the neurite linearity. We next tested for the involvement of the cytoskeleton, actin filaments and microtubules, in neurite turning. Treatment with 10 ng/ml cytochalasin D, an inhibitor of actin polymerization, reduced the number of filopodia (Fig. 1 C) and inhibited the rightward turning of neurites on PDL/laminin substrates (Fig. 1, D–H). It was reported that, when treated with higher concentrations of cytochalasin B (5–10 µg/ml), growth cones lose both filopodia and lamellipodia and wander around, often forming neurite loops (Marsh and Letourneau, 1984). However, treatment with paclitaxel, a microtubule-stabilizing drug, had no detectable effect on rightward neurite turning (unpublished data). These results suggest that the inherent turning behavior of neurites depends on actin filaments but not on microtubules.
Figure 1.
The linearity of neurite elongation depends on the dimension of culture substrates and actin filaments. (A and B) Phase-contrast images of hippocampal explants. Neurites from an explant turned rightward on a 2D substrate of PDL/laminin (A) but grew practically straight in a 3D substrate of collagen gels (B). B is a composite of four photomicrographs. (C) DIC and phalloidin fluorescent images of growth cones in the absence (no treatment) or presence of cytochalasin D. Treatment with 10 ng/ml cytochalasin D inhibited the formation of filopodia. (D–G) Hippocampal explants on PDL/laminin substrates in the absence (D) or presence of cytochalasin D at concentrations of 1 ng/ml (E), 10 ng/ml (F), or 100 ng/ml (G). (H) The y axis represents the curvature of neurites on PDL/laminin substrates, with positive values indicating rightward turning. Numbers in parentheses indicate the total number of neurites examined. Data represent mean ± SEM. ***, P < 0.001 versus no treatment (Bonferroni’s multiple comparison test). Bars: (A, B, and D–G) 500 µm; (C) 10 µm.
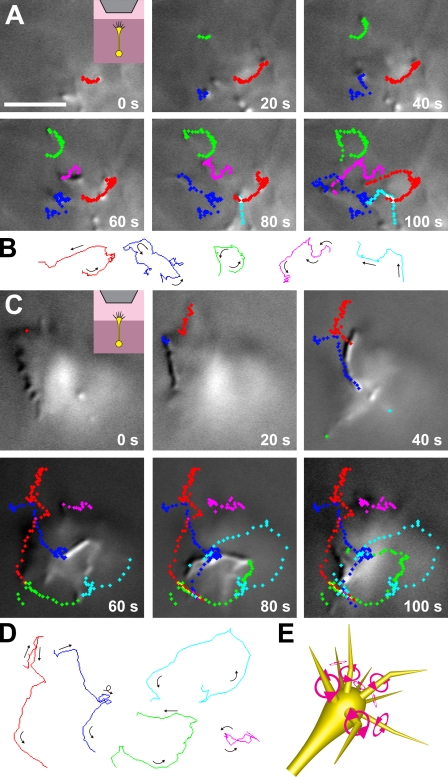
To explain the dependence of autonomous neurite turning on the dimension of culture substrates, we hypothesized that the growth cone filopodia rotate in the direction of a right-handed screw from the viewpoint of the neurite shaft. This form of filopodial movement could generate a leftward force onto the 2D substrate, e.g., via preferential filopodia–substrate attachments on the right of the growth cone followed by filopodial contraction along their own longitudinal axes. As a result, the growth cone would receive the rightward reaction force that deflects the neurite from its straight course. In contrast, a growth cone in a 3D substrate would not receive net biased force, resulting in straight elongation of the neurite. We examined this hypothesis by direct observation of filopodial movement. Hippocampal neurons were embedded in a collagen gel, and a single neurite elongating vertically toward the objective lens was observed under an upright microscope. A growth cone in the 3D substrate exhibited a conical shape with filopodia extended forward. When we observed a growth cone migrating toward the objective lens in a collagen gel, the filopodia tended to rotate counterclockwise from the observer’s point of view (Fig. 2, A and B; and Video 1). The rotation of individual filopodia was eccentric rather than concentric. We quantified the rotational motility of all of the filopodial tips (n = 29) of this growth cone that were present in the focal plane of differential interference contrast (DIC) imaging (see Materials and methods for calculations): the mean angular velocity (ω) = 0.065 rad/s (counterclockwise direction) and the mean velocity (v) = 0.135 µm/s. We also observed a growth cone that had protruded from a collagen gel into a liquid medium (Fig. 2, C and D; and Video 2) and quantified the rotational motility of all of its filopodial tips (n = 56) in the focal plane: ω = 0.053 rad/s and v = 0.279 µm/s. Filopodia in liquid media tended to move faster than those in collagen gels, perhaps because the filopodial motility was not hindered by the meshwork of collagen fibers. These results indicate that an individual filopodium autonomously rotates in the right-screw direction on its own longitudinal axis (Fig. 2 E). We have also observed this right-screw rotation of filopodia in neocortical neurons (Video 3), thalamic neurons, and cerebellar granule cells (not depicted), suggesting that this form of filopodial motility is a general phenomenon in the brain. We also found that in hippocampal neurons on 2D PDL/laminin substrates, the filopodia rotate in the right-screw direction when detached from the substrate (Video 4).
Figure 2.
Individual filopodia rotate in the direction of a right-handed screw. (A) Time-lapse DIC images of filopodia of a single growth cone in a 3D collagen gel. (inset) Cartoon showing the experimental setup of a growth cone migrating in the gel (dark pink) toward the objective lens (gray). The focal plane was positioned at the level of the filopodial tips. Representative trajectories of the filopodial tips are indicated by series of color-coded diamonds. Each color corresponds to the single filopodium tracked for a maximum of 100 s. Each diamond represents the position of the tip at 1-s intervals. (B) The line traces indicate the paths of the single filopodial tips shown in A. Arrows indicate the direction of movement, showing that the filopodia display a general tendency to rotate counterclockwise from the observer’s perspective. (C) Time-lapse DIC images and representative trajectories of filopodia of a growth cone that has protruded from a collagen gel (dark pink) into a liquid medium (light pink). Each color corresponds to the single filopodium tracked for a maximum of 100 s. Each diamond represents the position of the tip at 1-s intervals. (D) The line traces indicate the paths of the single filopodial tips shown in C. Arrows indicate the direction of movement. (E) Schematic diagram of a growth cone showing individual filopodia rotating in the right-screw direction on their longitudinal axis. Original image data of A and C can be found in DataViewer. Bar, 5 µm.
The filopodial rotation involves myosin V
Next we investigated molecular mechanisms underlying the filopodial rotation. Because filopodia are composed predominantly of actin filaments, the rotation might be powered by actomyosin interactions. Actin filaments are aligned unidirectionally within a filopodium, with their plus end pointing toward the filopodial tip (Lewis and Bridgman, 1992). It has been shown that several classes of dimeric myosins move spirally along an actin filament. Myosin V (Cheney et al., 1993) is a left-handed spiral motor toward the plus end of actin filaments (Ali et al., 2002). When the movement of myosin V is hindered, for example, via interactions with static cytosolic molecules or organelles, the immobilized myosin V would move actin filaments toward their minus end as well as rotate them in the right-screw direction from the viewpoint of their minus end. In contrast, myosin II is a plus end-directed, right-handed spiral motor (Nishizaka et al., 1993) and would rotate actin filaments in the opposite direction. Both myosin V and myosin II are expressed in growth cones (Miller et al., 1992; Wang et al., 1996; Evans et al., 1997).
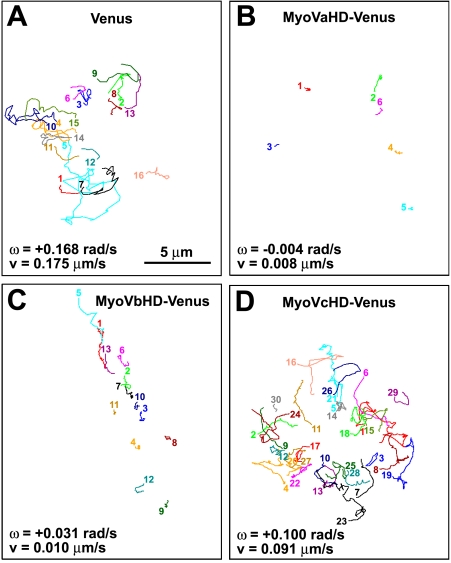
To test our hypothesis that the right-screw rotation of filopodia involves myosin V, we monitored the movement of the filopodial tips of hippocampal neurons that overexpressed a truncated mutant of myosin V consisting of its head domain only. The mutant should interfere with the binding between actin filaments and endogenous myosin V in a dominant-negative manner. To visualize transfected cells, the YFP Venus was fused to the C terminus of the head domain of the three members in the class V myosins, myosin Va, Vb, and Vc. These three constructs are designated in this paper as MyoVaHD-, MyoVbHD-, and MyoVcHD-Venus, respectively. Growth cone filopodia of Venus-transfected neurons (Fig. 3 A and Video 5) exhibited the right-screw rotation. In contrast, the filopodial rotation was blocked in neurons transfected with either MyoVaHD-Venus (Fig. 3 B and Video 6) or MyoVbHD-Venus (Fig. 3 C). However, MyoVcHD-Venus had no substantial effect on the filopodial rotation (Fig. 3 D). Given the structural similarity of the head domain among different forms of myosin, the observed inhibition of filopodial rotation by MyoVaHD- and MyoVbHD-Venus may have been mediated by dominant-negative effects on other myosin motors. Therefore, we tested whether overexpression of full-length myosin V rescues the filopodial rotation in neurons transfected with MyoVaHD-Venus. As expected, the filopodial rotation was rescued partially by cotransfection with either myosin Va (Fig. 4 A and Video 7) or myosin Vb (Fig. 4 B) using a vector for bicistronic expression of full-length myosin V and monomeric RFP (mRFP) designated as MyoVa/internal ribosomal entry site (IRES)/mRFP or MyoVb/IRES/mRFP, respectively. In contrast, full-length myosin Vc plus mRFP (MyoVc/IRES/mRFP) failed to rescue the filopodial rotation in neurons transfected with MyoVaHD-Venus (Fig. 4 C). As a control, cotransfection with mRFP alone had no detectable effect (Fig. 4 D and Video 8). Furthermore, the filopodial rotation was analyzed in neurons expressing the Venus-fused head domain of myosin IIa, IIb, or IIc. None of these three proteins showed a detectable effect on filopodial rotation (Figs. S1–S3).
Figure 3.
The head domain of myosin Va or Vb inhibits the filopodial rotation. (A–D) The line sketches in each panel show trajectories of filopodial tips of a single growth cone that expresses Venus (A), MyoVaHD-Venus (B), MyoVbHD-Venus (C), or MyoVcHD-Venus (D). Each color corresponds to a single filopodium. All of the filopodial tips that appeared in the focal plane for a period of 5-min imaging were included in this study. The numbered end of each line represents the point where a filopodial tip first appeared in the focal plane, and the other end of the line is the point at which it moved out of the focal plane. The mean angular velocity (ω) and the mean velocity (v) of filopodial tips for each growth cone are shown. Positive and negative values of the angular velocity indicate right- and left-screw rotation, respectively.
Figure 4.
Full-length myosin Va or Vb rescues the filopodial rotation in neurons expressing the myosin Va head domain. (A–D) The line sketches in each panel show trajectories of filopodial tips of a single growth cone that expresses MyoVaHD-Venus plus MyoVa/IRES/mRFP (A), MyoVaHD-Venus plus MyoVb/IRES/mRFP (B), MyoVaHD-Venus plus MyoVc/IRES/mRFP (C), or MyoVaHD-Venus plus mRFP (D). Each color corresponds to a single filopodium. All of the filopodial tips that appeared in the focal plane for a period of 5-min imaging were included in this study. The numbered end of each line represents the point where a filopodial tip first appeared in the focal plane, and the other end of the line is the point at which it moved out of the focal plane. The mean angular velocity (ω) and the mean velocity (v) of filopodial tips for each growth cone are shown. Positive values of the angular velocity indicate right-screw rotation.
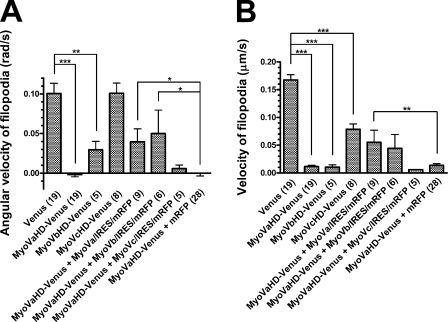
We analyzed the trajectories of individual filopodial tips and calculated their mean angular velocity (radians/second, with positive values indicating right-screw rotation; Fig. 5 A) and their mean velocity (micrometers/second; Fig. 5 B) as measures of rotation and overall motility within the x–y plane, respectively. In comparison with Venus-transfected neurons, both parameters were drastically reduced in neurons transfected with either MyoVaHD- or MyoVbHD-Venus, indicating that the head domain of myosins Va and Vb inhibited the rotation and overall motility of filopodia. In contrast, MyoVcHD-Venus had no detectable effect on the angular velocity (Fig. 5 A) but caused a partial reduction in the velocity (Fig. 5 B). More importantly, the inhibitory effect of MyoVaHD-Venus was canceled partially by cotransfecting neurons with MyoVa/IRES/mRFP or MyoVb/IRES/mRFP but not with MyoVc/IRES/mRFP. Therefore, we concluded that the right-screw rotation of filopodia is driven by myosins Va and Vb.
Figure 5.
The involvement of myosin V in filopodial rotation. (A and B) The y axis represents the filopodial angular velocity (A) or the filopodial velocity (B) in growth cones that express the indicated transgene products. Positive and negative values of the angular velocity indicate right- and left-screw rotation, respectively. Numbers in parentheses indicate the total number of growth cones examined. Data represent mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Bonferroni’s multiple comparison test).
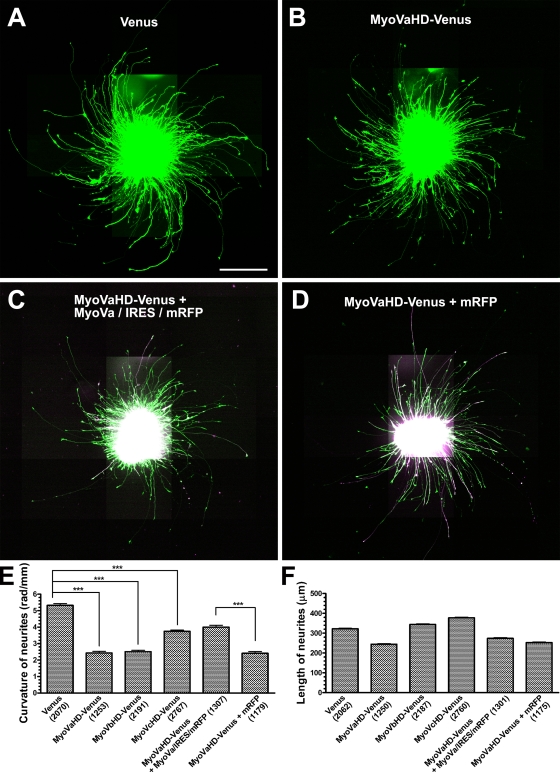
Next we examined the effect of the head domain of myosin V on the growth linearity of neurites on 2D substrates. Dissociated hippocampal neurons were transfected, reaggregated, and incubated for 3 d until the expression level of transgenes became sufficiently high. Then, the reaggregates were replated on 2D PDL/laminin substrates and cultured for an additional 1 d. Examples of a reaggregate transfected with Venus (Fig. 6 A), MyoVaHD-Venus (Fig. 6 B), MyoVaHD-Venus plus MyoVa/IRES/mRFP (Fig. 6 C), or MyoVaHD-Venus plus mRFP (Fig. 6 D) are shown. Turning was evaluated in all of the neurites that had Venus fluorescence or, in case of double transfection, both Venus and mRFP fluorescence. As a measure of neurite turning, we calculated the curvature (radians/millimeter) averaged within the distal 100-µm arc of each neurite (Fig. 6 E) because the proximal neurites were often detached from the substrate and fasciculated. Neurites from Venus-transfected reaggregates turned rightward. In comparison with this control, neurites from reaggregates transfected with either MyoVaHD- or MyoVbHD-Venus grew straighter, indicating that the head domain of myosins Va and Vb inhibited the rightward turning of neurites. MyoVcHD-Venus inhibited the rightward turning of neurites to a lesser degree. Cotransfection with full-length myosin Va (MyoVa/IRES/mRFP) but not with mRFP alone partially rescued the rightward turning of neurites that expressed MyoVaHD-Venus. We also measured the length of neurites that had Venus fluorescence or, in case of double transfection, both Venus and mRFP fluorescence (Fig. 6 F). A correlation between the length of neurites and their turning activity will be evaluated in the following experiment.
Figure 6.
The involvement of myosin V in inherent neurite turning. (A–D) Hippocampal neurons, which had been transfected with cDNAs for Venus (A), MyoVaHD-Venus (B), MyoVaHD-Venus plus MyoVa/IRES/mRFP (C), or MyoVaHD-Venus plus mRFP (D), were reaggregated and plated on 2D PDL/laminin substrates. Each panel is a composite of 30 fluorescent images acquired under the same optical conditions, which causes both saturation and stray light artifacts in the central images where the neuronal cell bodies are concentrated. In the cases of double transfection (C and D), Venus fluorescence (green) and mRFP fluorescence (magenta) have been superimposed. (E) The y axis represents the curvature of the distal 100 µm of neurites that express the indicated transgene products. Positive values represent rightward turning. ***, P < 0.001 (Bonferroni’s multiple comparison test). (F) The y axis represents the estimated length of neurites that express the indicated transgene products. (E and F) Numbers in parentheses indicate the total number of neurites examined. Data represent mean ± SEM. Bar, 200 µm.
Causal relationship between filopodial rotation and neurite turning
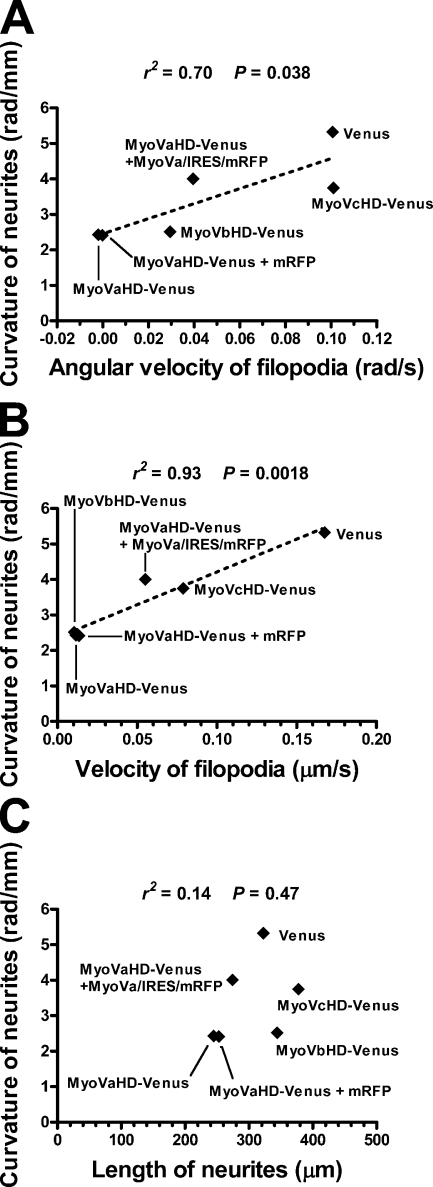
To test whether right-screw filopodial rotation is associated with rightward neurite turning on 2D substrates, we analyzed the relationship between these two forms of motile behavior. We compared the neurite curvature (Fig. 6 E) with either the filopodial angular velocity (Fig. 5 A) or the filopodial velocity (Fig. 5 B). There was a linear correlation between the filopodial angular velocity and the neurite curvature (Fig. 7 A), indicating that a neurite turns more sharply when filopodia rotate more quickly. We also found a linear correlation between the filopodial velocity and the neurite curvature (Fig. 7 B), with the velocity values reflecting not only the rotation but also the random movement of filopodia. These results are consistent with our hypothesis that the right-screw rotation of filopodia drives the rightward turning of neurites on 2D substrates.
Figure 7.
Correlation between filopodial dynamics and neurite turning. (A) A scatter plot of the curvature of neurites (data in Fig. 6 E) versus the angular velocity of filopodia (data in Fig. 5 A) expressing the indicated transgene products. The correlation was statistically significant (P < 0.05). (B) A scatter plot of the curvature of neurites (data in Fig. 6 E) versus the velocity of filopodia (data in Fig. 5 B). The correlation was statistically significant (P < 0.01). (A and B) The broken lines show the linear regression fit of the data. (C) A scatter plot of the curvature of neurites (data in Fig. 6 E) versus the length of neurites (data in Fig. 6 F). The correlation was not statistically significant (P = 0.47).
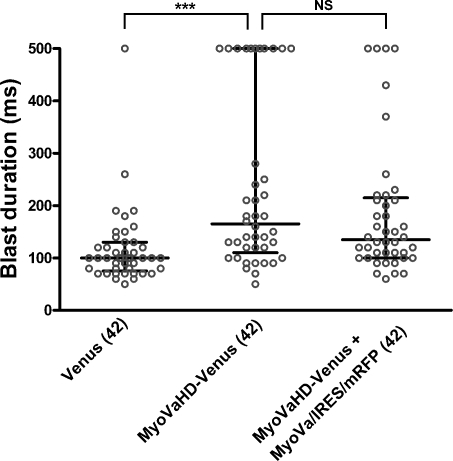
We still observed rightward neurite turning on 2D substrates even under the conditions in which filopodial rotation in 3D substrates was almost completely blocked (Fig. 7). It remains unclear whether this discrepancy was caused by different experimental conditions between 2D and 3D cultures or by the presence of an additional mechanism that affects the linearity of neurite elongation. For example, some growth-promoting machinery of neurites may have another type of inherent asymmetry that causes rightward turning. Therefore, one possible interpretation of our data is that inhibition of neurite elongation by cytochalasin D or myosin V mutants is associated with the reduced activity of such machinery responsible for neurite turning. To examine this possibility, we analyzed the relationship between the neurite curvature (Fig. 6 E) and neurite length (Fig. 6 F). As shown in Fig. 7 C, the two parameters exhibited no significant correlation, suggesting that the inherent turning of neurites is independent of their growth activity. Another possibility is that transfection of myosin V mutants reduces growth cone–substrate adhesion, thereby preventing the growth cone from receiving biased force for the rightward deflection. Therefore, we quantified growth cone–substrate adhesiveness by blasting assays described previously (Gundersen and Barrett, 1980; Lemmon et al., 1992). The blast duration required to dislodge a growth cone was used as a measure of adhesiveness. The adhesiveness of growth cones expressing MyoVaHD-Venus was not weaker than that of growth cones expressing Venus alone or MyoVaHD-Venus plus MyoVa/IRES/mRFP (Fig. 8), excluding the possibility that the inhibition of myosin Va causes straighter neurite elongation via reduced growth cone–substrate adhesion.
Figure 8.
Quantitative analysis of growth cone–substrate adhesiveness. The adhesiveness of growth cones expressing the indicated transgene products was measured by blasting assays. Blast durations required to dislodge growth cones are shown as dot blots. The duration value was taken as 500 ms when a growth cone had not been dislodged by a 500-ms blast. Each circle corresponds to a single growth cone. Horizontal bars indicate the median and the interquartile range. Numbers in parentheses indicate the total number of growth cones examined. ***, P < 0.001 (Dunn’s multiple comparison test).
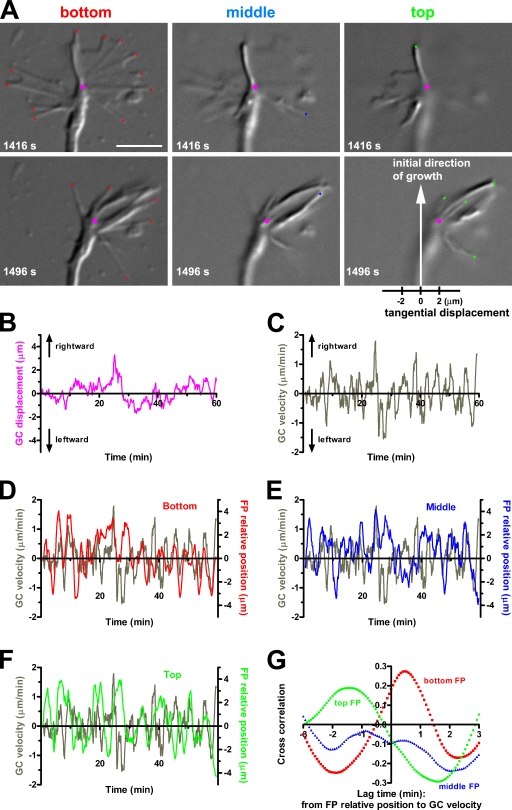
The causal relationship between filopodial dynamics and growth cone deflection was assessed by long-term time-lapse imaging of a single growth cone on a 2D substrate. In our simplified model, a rotating filopodium moves rightward when unattached to the substrate. Then, the filopodium adheres to the substrate and pulls the growth cone rightward by means of filopodial contraction along its own longitudinal axis. If this model is the case, the growth cone deflection should be correlated with asymmetric positioning of filopodia onto the substrate. This hypothesis was tested by quantitative analysis of a growth cone showing multiple rightward and leftward deflections rather than that showing continued rightward turning. DIC images were acquired at three different focal planes: bottom, middle, and top (Fig. 9 A and Video 9). The bottom plane was kept focused on the top surface of the coverglass. The filopodial tips in each of the focal planes are marked with color-coded diamonds (Fig. 9 A). The magenta circle indicates the growth cone center that was defined as the middle point of the distal edge of the central domain (Fig. 9 A; Ooashi et al., 2005). During 1 h time-lapse imaging, this growth cone exhibited multiple deflections, as indicated by displacement (Fig. 9 B) and velocity (Fig. 9 C) of the magenta circle along the horizontal axis shown in Fig. 9 A. We also monitored the centroid of all of the filopodial tips in each of the three focal planes: bottom (Fig. 9 D), middle (Fig. 9 E), and top (Fig. 9 F). The colored lines in these graphs represent relative filopodial position, which was defined as a distance along the horizontal axis between the centroid of filopodial tips and the growth cone center at any given time. It appeared that the pattern of fluctuation in the growth cone tangential velocity (Fig. 9, D–F, gray lines) resembled that in the relative filopodial position in the bottom plane (Fig. 9 D) but not in the middle (Fig. 9 E) and top (Fig. 9 F) planes. Using cross-correlation analysis, we estimated the degree to which the relative filopodial position and the growth cone tangential velocity were correlated (Fig. 9 G). In the bottom focal plane, the two parameters showed a single peak of correlation at a lag time of 28 s. The correlation at this lag time was statistically significant at the P < 0.0001 level, indicating that the centroid deviation of the bottom filopodial tips is followed by the lateral movement of the growth cone. This result supports the idea that deviated filopodia–substrate adhesion to the right of the growth cone causes rightward neurite turning.
Figure 9.
Temporal correlation between filopodial deviation and growth cone deflection. (A) DIC images of a single growth cone at two time points at three focal planes, each separated by 1 µm in the z direction. All of the filopodial tips in the bottom plane, focused at the level of the coverglass, are marked with red diamonds (left), those in the middle plane with blue diamonds (middle), and those in the top plane with green diamonds (right). The growth cone center is marked with a magenta circle. The horizontal axis shown below the lower right panel was used to determine the tangential components of displacement/velocity of the growth cone center and relative position of the filopodial tips. (B) Tangential displacement of the growth cone (GC) shown in A, with positive and negative values indicating rightward and leftward displacement, respectively. (C) Tangential velocity of the same growth cone. Central difference quotients of the tangential displacement with 80-s intervals are shown. (D–F) Superimposition of the growth cone tangential velocity and the relative position of filopodia (FP) in the indicated focal planes. The filopodia relative position was defined as the tangential component of the distance between the growth cone center and the centroid of filopodia tips. The colored lines represent 80-s central moving averages of the filopodia relative position. (G) The cross-correlation function between the filopodia relative position and the growth cone velocity. Bar, 5 µm.
We propose that filopodial rotation drives inherent neurite turning independently of extrinsic guidance signals. To examine this hypothesis, we quantified the speed of filopodial rotation in the absence or presence of semaphorin 3F (Sema3F), which chemorepels hippocampal axons (Chédotal et al., 1998). Consistent with this previous study, long neurites of hippocampal neurons elongated away from reaggregates of Sema3F-secreting COS-7 cells (Fig. S4 A), which was in strong contrast to those co-cultured with reaggregates of control COS-7 cells (Fig. S4 B). There was no statistically significant difference in the angular velocity and the velocity of filopodial tips between the two culture conditions (Fig. S4, C and D), indicating that Sema3F does not affect filopodial rotation and motility. These results are consistent with our idea that filopodial rotation is not controlled by extrinsic guidance signals but depends primarily on the inherent polarization/structure of filamentous actin and myosin V causing their spiral interactions.
Discussion
In this study, we have discovered a novel form of growth cone motility: the right-screw rotation of filopodia. We further revealed that this motility depends on myosins Va and Vb but not on myosin Vc. Myosins Va and Vb show processive interactions with actin filaments, allowing its two heads to make multiple steps toward the plus end of actin filaments without release from them (Mehta et al., 1999; Watanabe et al., 2006). In contrast, myosin Vc is a nonprocessive motor protein (Takagi et al., 2008). Although it is widely accepted that myosin V is involved in organelle transport along actin filaments (Bridgman, 2004), no experimental evidence has been provided that myosin V pulls and translocates actin filaments instead of traveling on the filaments. However, the movement of myosin V can be hindered via several mechanisms, e.g., binding of myosin Va to neurofilaments (Rao et al., 2002) or an interaction of myosin Va–containing organelles with microtubules via kinesin (Bridgman, 1999; Huang et al., 1999). Because myosin V is a plus end–directed, left-handed spiral motor (Ali et al., 2002), the immobilized myosin V would pull actin filaments toward their minus end as well as rotate them in the right-screw direction from the viewpoint of their minus end. If this physical interaction occurs at the base of filopodia where the expression of myosin V is abundant (Evans et al., 1997), actin filaments in the filopodia flow toward the growth cone central domain (Forscher and Smith, 1988) and rotate in the right-screw direction. As we have observed, filopodia are not absolutely straight but often curved; therefore, the axial rotation of actin filaments results in a swirling of their distal portion. We propose that, through spiral interactions with actin filaments, myosins Va and Vb drive the filopodial rotation that causes the deflected elongation of neurites.
A technique for reversing the direction of filopodial rotation should be useful to explore its functional significance in vivo. It was reported that treating hippocampal neurons with concanavalin A causes leftward or counterclockwise turning of neurites on poly-l-lysine substrates (Farmer et al., 1992). However, we have not been able to show that concanavalin A reverses the direction of either filopodial rotation in 3D substrates or neurite turning on 2D substrates (unpublished data). We also tested whether the direction of filopodial rotation can be reversed by overexpression of full-length myosin II, a right-handed spiral motor (Nishizaka et al., 1993) which would rotate actin filaments in the opposite direction. However, myosin II overexpression failed to reverse the direction of filopodial rotation (unpublished data). It has been proposed that the property of myosin V as a spiral motor depends on its step size (Ali et al., 2002), raising the possibility that the deflection angle and even the direction of spiral movement of myosin V can be manipulated. The generation of myosin V mutants that have altered spiral interactions with actin filaments, especially those that rotate in the opposite direction, will be an important approach for studying the role of filopodial rotation in the nervous system.
There are at least two possibilities of in vivo significance of the filopodial rotation. Considering that the filopodia are antenna-like protrusions that sample the external environment for guidance cues (Bentley and Toroian-Raymond, 1986), the rotatory motion would increase the volume of extracellular matrix that the growth cone is able to probe with each filopodium. In this way, filopodial rotation might be involved in the precise perception of environmental cues by the growth cone. Another possibility is that filopodial rotation influences the direction of neurite elongation in a similar way as neurite turning on 2D substrates in vitro. When a growth cone is migrating along an interface between adhesive and nonadhesive environments, the filopodial rotation would result in the deflected neurite elongation. For example, an elongating neurite could twine around a preexisting neuronal process in a right-handed spiral configuration, forming a tight fascicle. Such unidirectionally deflected elongation of neurites in both sides of the nervous system would break the left–right symmetry. In summary, the present study reveals for the first time the asymmetric nature of growth cone motility that is driven by spiral interactions of myosins Va and Vb with actin filaments.
Materials and methods
Expression vectors
cDNAs encoding for the class V myosins were provided by the Kazusa DNA Research Institute, FANTOM (Functional Annotation of the Mouse) in RIKEN Genomic Science Center, or the IMAGE consortium. Full-length cDNA for myosin Va (GenBank/EMBL/DDBJ accession no. X57377) was obtained by connecting the FANTOM clone 0610009A05 with an RT-PCR fragment amplified using the following primer pair: 5′-GTCGACGCCACCATGGCCGCGTCC-3′ and 5′-CAGATTGACAGCCACACCAC-3′. Full-length cDNA for myosin Vb (GenBank/EMBL/DDBJ accession no. AB032945) was provided by the Kazusa DNA Research Institute (clone ORK01152). Full-length cDNA for myosin Vc (MGI accession no. 2442485) was obtained by connecting the FANTOM clone C330029I24 with the IMAGE clone 5251351. These cDNAs were subcloned into pCAGGS, a mammalian expression vector under the control of the CAG promoter (provided by J. Miyazaki, Osaka University, Suita, Osaka, Japan; Niwa et al., 1991), followed by sequences of IRES and mRFP (provided by R.Y. Tsien, University of California, San Diego, La Jolla, CA; Campbell et al., 2002). The resulting constructs enabled bicistronic expression of full-length myosin V and mRFP. A cDNA fragment corresponding to the head domain of myosin Va (aa 1–763), Vb (1–761), or Vc (1–753) was subcloned into the pCAGGS vector. Venus cDNA (provided by A. Miyawaki, RIKEN Brain Science Institute, Wako, Saitama, Japan; Nagai et al., 2002) was also subcloned into the vector adjacent to the 3′ end of the head domain insert such that the vector drove the expression of a fusion protein consisting of the myosin V head domain and Venus.
Animals
Timed-pregnant Wistar (Nihon-SLC) rats were used for this study. The day on which the plug was detected was designated as embryonic day (E) 0. Embryos were removed from pregnant rats that had been deeply anesthetized with 50 mg/kg body weight of sodium pentobarbitone (Nembutal; Abbott). All experiments were conducted in compliance with the Guidelines for Use of Laboratory Animals of RIKEN.
Explant cultures
Hippocampuses were dissected from E18 rat embryos and sliced into small explants (<500 µm in diameter) with fine scissors in cold Hanks’ BSS (−) (Invitrogen). As a substrate for 2D cultures, coverslips that had been cleaned with nitric acid were coated sequentially with 0.5 mg/ml PDL (Sigma-Aldrich) and 50 µg/ml laminin (Invitrogen) and rinsed with L15 (Invitrogen) supplemented with penicillin and streptomycin (L15/P/S) immediately before use. Explants were plated on the coverslips in a small volume of L15/P/S, incubated for ∼30 min until they attached to the coverslip, and cultured in an additional volume of L15/P/S for 2 d at 37°C in air. Cytochalasin D (Sigma-Aldrich) or paclitaxel (EMD) was applied to some cultures.
For 3D cultures, collagen gels were prepared as described previously (Shirasaki et al., 1995). In brief, 160 µl of collagen solution extracted from rat tail tendons was mixed with 20 µl of 10× DME/F12 medium (Sigma-Aldrich) and 20 µl of alkaline solution (0.14 M NaHCO3 and 0.23 N NaOH). Hippocampal explants were placed on coverslips, embedded in the mixed collagen solution, incubated in a CO2 incubator until the gel had set, and cultured in L15/P/S supplemented with 10% FBS (L15/P/S/FBS) for 2 d at 37°C in air.
Dissociated cell cultures
Hippocampuses dissected from E18 rat embryos were digested with 0.1% trypsin/1 mM EDTA/Hanks’ BSS (−) for 10–20 min at 37°C and suspended in DME/F12 medium supplemented with 3.85 mg/ml glucose and 10% FBS (DME/F12/P/S/FBS). The dissociated cells were centrifuged, resuspended in DME/F12/P/S/FBS, filtrated with a cell strainer (40-µm mesh), and diluted to 1–4 × 107 cells/ml. For cDNA transfection, the dissociated cells were nucleofected with 10 µg of expression vectors using Nucleofector (Lonza). The cells were suspended in the mixed collagen gel solution at the density of 1–4 × 106 cells/ml and poured on glass-based dishes. After gelation, the embedded cells were cultured in L15/P/S/FBS for 2–3 d at 37°C in air.
Reaggregated cell cultures
Dissociated hippocampal cells were transfected with expression vectors as described in the previous section. Then, 3 × 103 cells were incubated in each well of 96-well plates (Celltight Spheroid; SUMILON) in 200 µl L15/P/S/FBS supplemented with B27 (Invitrogen) for reaggregate formation. After 3-d incubation, the reaggregates were harvested, rinsed twice with L15/P/S, plated on PDL/laminin-coated coverslips, and cultured in L15/P/S supplemented with B27 for 24 h at 37°C in air.
Phalloidin labeling
Dissociated hippocampal cells cultured on PDL/laminin substrates were fixed with 4% paraformaldehyde for 30 min, permeabilized and blocked with 0.1% Triton X-100 and 10% horse serum for 60 min, and incubated with 5 U/ml Alexa Fluor 594–conjugated phalloidin (Invitrogen) for 20 min.
Imaging and quantitative analysis of 3D cultures
A glass-based dish containing hippocampal neurons embedded in a collagen gel was placed in a heat chamber (Tokai Hit) on the stage of an upright microscope (BX61WI; Olympus). Neurons were observed with a 100× NA 1.0 water-dipping objective lens, and a growth cone migrating toward the objective lens was chosen for time-lapse imaging. When observing cells transfected with myosin cDNAs, expression of the transgenes was confirmed by fluorescence of Venus and/or mRFP. Time-lapse DIC images of growth cones were acquired every second for a total of 5 min with a cooled charge-coupled device (CCD) camera (ORCA-ER; Hamamatsu Photonics; CCD binning set at 1 × 1) under the control of MetaMorph software (Universal Imaging). On average, the tip of a single filopodium remained in the focal plane for 157 s after its initial appearance.
For quantification of filopodial motility, all of the filopodial tips present in the focal plane of DIC imaging were traced for the entire duration of the time lapse. Then, coordinates of their positions were extracted using MetaMorph software. Local velocity and local angular velocity were calculated from these coordinates using a custom program written with C++ (Visual C++; Microsoft). The definition of local values was as follows: let P(t) denote a position at time point t. Vectors and were defined as and where Δt is time interval. Local velocity at time point t, v(t), is given as Local angular velocity at time point t, ω(t), is given as
If a filopodial tip deflects counterclockwise, ω(t) has a positive value. To calculate the velocity and angular velocity of an individual filopodial tip, the local values were averaged during the whole period of its appearance in time-lapse imaging. The mean filopodial velocity for an individual growth cone was defined as a time-weighted average of the velocity of all of the observed filopodia of that growth cone. Similarly, the mean filopodial angular velocity for an individual growth cone was defined as a time-weighted average of the angular velocity of all of the observed filopodia of that growth cone.
Image analysis of 2D cultures
After fixation with 4% paraformaldehyde, phase-contrast images of explant cultures were acquired with a 4× NA 0.13 objective lens and a CCD camera (CoolSNAP-HQ; Roper Industries; CCD binning set at 1 × 1) on an inverted microscope (TE300; Nikon) under the control of MetaMorph software. Neurite turning (Fig. 1 H) was quantified by calculating the curvature of all of the individual neurites. The curvature of a neurite was given as an inverse of the radius (R) of the circle that passes through the center of the explant (G) and the distal tip of the neurite (P), with its tangential line matching the vector (V) representing the direction of neurite elongation at the distal tip (Fig. S5 A). The value was calculated as 2sinθ/D, where θ is an angle between line GP and vector V, and D is a distance between point G and point P.
After fixation with 4% paraformaldehyde, DIC and fluorescent images of reaggregate cultures of transfected cells were acquired with a 20× NA 0.5 water-dipping objective lens and an ORCA-ER CCD camera (CCD binning set at 1 × 1) on a BX61WI upright microscope under the control of MetaMorph software. Neurites expressing fluorescent proteins were included for the measurements of curvature (Fig. 6 E) and length (Fig. 6 F). Because the proximal neurites near the reaggregate were often detached from the substrate and fasciculated with neighboring neurites, the curvature and length were determined from the distal 100-µm segment of each neurite (Fig. S5 B). A neurite was traced backward from its tip (P) to the point 100 µm along its own length (B). The curvature was given as an angular difference (Δθ) between the tangential lines at point P and point B divided by the arc length (ΔS = 100 µm). With the assumption that a neurite trajectory was an arc of the circle that passes through points G, B, and P, the neurite length was given as the length of arc GP subtracted by the approximated radius of reaggregate:
where θ is an angle between line GB and line BP, D is a distance between point G and point P, and Aagr is an area of the reaggregate.
Quantitative 3D imaging of growth cone motility on 2D substrate
Time-lapse DIC images of a growth cone in an explant culture (Fig. 9) were acquired with a 100× NA 1.0 water-dipping objective lens and an ORCA-ER CCD camera (CCD binning set at 1 × 1) on a BX61WI upright microscope under the control of MetaMorph software. Each set of three images at different focal planes (bottom, middle, and top; each separated by 1 µm) was obtained every 4 s for a total of 60 min. The bottom plane was kept focused on the glass surface. All of the filopodial tips in each focal plane and the growth cone center (the midpoint of the distal edge of the central domain) were traced for the entire time lapse. Using the first frame, we determined the initial position of the growth cone center and the horizontal axis that was perpendicular to the initial direction of neurite growth. Tangential displacement of the growth cone center was defined as a distance between its initial position and its position at any given time along the horizontal axis. Tangential velocity of the growth cone center was defined as central difference quotients of the tangential displacement with 80-s intervals. Relative position of filopodia at any given time was defined as an 80-s central moving average of the tangential component of the vector from the position of the growth cone center to the centroid of all of the filopodial tips in each focal plane. Using R software (http://www.r-project.org), cross-correlation function was calculated between the relative position of filopodia and the tangential velocity of the growth cone center.
Assay of growth cone adhesiveness
The adhesiveness of growth cones on PDL/laminin-coated coverslips was measured using a modification of growth cone blasting assays described previously (Gundersen and Barrett, 1980; Lemmon et al., 1992). Transfected growth cones in reaggregate cultures were selected with a 20× NA 0.45 objective lens on a TE300 inverted florescence microscope and used to correspond to the neurite curvature data (Fig. 6). The blasting apparatus consisted of a pneumatic picopump (PV820; World Precision Instruments; 10-psi driving pressure) and a borosilicate glass pipette (Sutter Instrument Co.; 1.0 mm in outer diameter and 0.78 mm in inner diameter) that had been pulled to a 3-µm opening and back-filled with culture medium. The pipette was angled at 40° to the coverslip, and its tip was positioned 20 µm above the glass surface and 20 µm directly in front of a single growth cone. The medium was pressure ejected from the pipette toward the growth cone for a duration of 10 ms. At 2-s intervals, the duration was incrementally increased by 10 ms with each ejection. This process was continued until the growth cone was completely dislodged from its original position or until a blast duration of 500 ms was reached. The duration of the final blast was recorded as a measure of growth cone–substrate adhesiveness. A single pipette was used repeatedly to blast the same number of growth cones in each of all of the experimental groups. Subsequently, we confirmed that the opening of that pipette remained unchanged by light microscopy.
Co-cultures with Sema3F-secreting COS-7 cells
As previously reported (Yamauchi et al., 2009), a plasmid containing Sema3F cDNA under the control of the CAG promoter and the same plasmid without an insert (control) were used to transfect COS-7 cells. Then, 2 × 104 cells were incubated overnight in each well of 96-well plates for reaggregate formation. The reaggregate was harvested and co-cultured in a collagen gel with dissociated hippocampal cells. Filopodial dynamics were examined in growth cones that were migrating toward the objective lens within a horizontal distance of 300 µm from the edge of COS-7 cell reaggregates. Time-lapse images of the growth cones were acquired every second for a total of 100 s. The velocity and angular velocity of filopodial tips were determined as described in the previous section.
Statistics
Statistical analyses were performed using Prism version 4.0 software (GraphPad Software, Inc.). P-values <0.05 were judged statistically significant.
Online supplemental material
Figs. S1–S3 show trajectories of filopodial tips of hippocampal neuronal growth cones that express MyoIIaHD-, MyoIIbHD-, and MyoIIcHD-Venus, respectively. Fig. S4 shows analysis of filopodial rotation in Sema3F-guided growth cones. Fig. S5 illustrates methods for measuring neurite curvature and length in explant/reaggregate cultures. Videos 1 and 2 show filopodial dynamics in a hippocampal growth cone that is migrating in a gel toward the observer and that has protruded into a liquid medium, respectively. Video 3 shows filopodial dynamics of neocortical neurons in a gel. Video 4 shows filopodial dynamics in a hippocampal growth cone on a 2D substrate. Videos 5–8 show filopodial dynamics in a hippocampal growth cone that has been transfected with Venus cDNA, MyoVaHD-Venus, MyoVaHD-Venus plus MyoVa/IRES/mRFP, and MyoVaHD-Venus plus mRFP, respectively. Video 9 shows motility of a hippocampal growth cone and its filopodial tips on a 2D substrate. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200906043/DC1.
Acknowledgments
We are grateful to J. Miyazaki, R.Y. Tsien, and A. Miyawaki for providing DNA constructs. We also thank A.T. Guy for valuable comments on this manuscript.
This work was partially supported by Grants-in-Aid for Scientific Research (C) (15650060 and 18500255 to A. Tamada and 19500335 to H. Kamiguchi).
Footnotes
Abbreviations used in this paper:
- CCD
- charge-coupled device
- DIC
- differential interference contrast
- IRES
- internal ribosomal entry site
- mRFP
- monomeric RFP
- PDL
- poly-d-lysine
- Sema3F
- semaphorin 3F
References
- Ali M.Y., Uemura S., Adachi K., Itoh H., Kinosita K., Jr., Ishiwata S. 2002. Myosin V is a left-handed spiral motor on the right-handed actin helix. Nat. Struct. Biol. 9:464–467 10.1038/nsb803 [DOI] [PubMed] [Google Scholar]
- Bentley D., Toroian-Raymond A. 1986. Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia by cytochalasin treatment. Nature. 323:712–715 10.1038/323712a0 [DOI] [PubMed] [Google Scholar]
- Bridgman P.C. 1999. Myosin Va movements in normal and dilute-lethal axons provide support for a dual filament motor complex. J. Cell Biol. 146:1045–1060 10.1083/jcb.146.5.1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgman P.C. 2004. Myosin-dependent transport in neurons. J. Neurobiol. 58:164–174 10.1002/neu.10320 [DOI] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882 10.1073/pnas.082243699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chédotal A., Del Rio J.A., Ruiz M., He Z., Borrell V., de Castro F., Ezan F., Goodman C.S., Tessier-Lavigne M., Sotelo C., Soriano E. 1998. Semaphorins III and IV repel hippocampal axons via two distinct receptors. Development. 125:4313–4323 [DOI] [PubMed] [Google Scholar]
- Cheney R.E., O’Shea M.K., Heuser J.E., Coelho M.V., Wolenski J.S., Espreafico E.M., Forscher P., Larson R.E., Mooseker M.S. 1993. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 75:13–23 [DOI] [PubMed] [Google Scholar]
- Dent E.W., Gertler F.B. 2003. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 40:209–227 10.1016/S0896-6273(03)00633-0 [DOI] [PubMed] [Google Scholar]
- Evans L.L., Hammer J., Bridgman P.C. 1997. Subcellular localization of myosin V in nerve growth cones and outgrowth from dilute-lethal neurons. J. Cell Sci. 110:439–449 [DOI] [PubMed] [Google Scholar]
- Farmer L.M., Hagmann J., Dagan D., Matus A.I., Levitan I.B. 1992. Directional control of neurite outgrowth from cultured hippocampal neurons is modulated by the lectin concanavalin A. J. Neurobiol. 23:354–363 10.1002/neu.480230403 [DOI] [PubMed] [Google Scholar]
- Forscher P., Smith S.J. 1988. Actions of cytochalasins on the organization of actin filaments and microtubules in a neuronal growth cone. J. Cell Biol. 107:1505–1516 10.1083/jcb.107.4.1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen R.W., Barrett J.N. 1980. Characterization of the turning response of dorsal root neurites toward nerve growth factor. J. Cell Biol. 87:546–554 10.1083/jcb.87.3.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock A.M., Agranoff B.W. 1977. Clockwise growth of neurites from retinal explants. Science. 198:64–66 10.1126/science.897684 [DOI] [PubMed] [Google Scholar]
- Huang J.D., Brady S.T., Richards B.W., Stenolen D., Resau J.H., Copeland N.G., Jenkins N.A. 1999. Direct interaction of microtubule- and actin-based transport motors. Nature. 397:267–270 10.1038/16722 [DOI] [PubMed] [Google Scholar]
- Lemmon V., Burden S.M., Payne H.R., Elmslie G.J., Hlavin M.L. 1992. Neurite growth on different substrates: permissive versus instructive influences and the role of adhesive strength. J. Neurosci. 12:818–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A.K., Bridgman P.C. 1992. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 119:1219–1243 10.1083/jcb.119.5.1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L., Letourneau P.C. 1984. Growth of neurites without filopodial or lamellipodial activity in the presence of cytochalasin B. J. Cell Biol. 99:2041–2047 10.1083/jcb.99.6.2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A.D., Rock R.S., Rief M., Spudich J.A., Mooseker M.S., Cheney R.E. 1999. Myosin-V is a processive actin-based motor. Nature. 400:590–593 10.1038/23072 [DOI] [PubMed] [Google Scholar]
- Miller M., Bower E., Levitt P., Li D., Chantler P.D. 1992. Myosin II distribution in neurons is consistent with a role in growth cone motility but not synaptic vesicle mobilization. Neuron. 8:25–44 10.1016/0896-6273(92)90106-N [DOI] [PubMed] [Google Scholar]
- Mortimer D., Fothergill T., Pujic Z., Richards L.J., Goodhill G.J. 2008. Growth cone chemotaxis. Trends Neurosci. 31:90–98 10.1016/j.tins.2007.11.008 [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. 2002. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20:87–90 10.1038/nbt0102-87 [DOI] [PubMed] [Google Scholar]
- Nishizaka T., Yagi T., Tanaka Y., Ishiwata S. 1993. Right-handed rotation of an actin filament in an in vitro motile system. Nature. 361:269–271 10.1038/361269a0 [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 108:193–199 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- Ooashi N., Futatsugi A., Yoshihara F., Mikoshiba K., Kamiguchi H. 2005. Cell adhesion molecules regulate Ca2+-mediated steering of growth cones via cyclic AMP and ryanodine receptor type 3. J. Cell Biol. 170:1159–1167 10.1083/jcb.200503157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao M.V., Engle L.J., Mohan P.S., Yuan A., Qiu D., Cataldo A., Hassinger L., Jacobsen S., Lee V.M., Andreadis A., et al. 2002. Myosin Va binding to neurofilaments is essential for correct myosin Va distribution and transport and neurofilament density. J. Cell Biol. 159:279–290 10.1083/jcb.200205062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romijn H.J., Mud M.T., Wolters P.S., Corner M.A. 1980. Neurite formation in dissociated cerebral cortex in vitro: evidence for clockwise outgrowth and autotopic contacts. Brain Res. 192:575–580 10.1016/0006-8993(80)90910-5 [DOI] [PubMed] [Google Scholar]
- Schwartz M., Agranoff B.W. 1981. Outgrowth and maintenance of neurites from cultured goldfish retinal ganglion cells. Brain Res. 206:331–343 10.1016/0006-8993(81)90535-7 [DOI] [PubMed] [Google Scholar]
- Shirasaki R., Tamada A., Katsumata R., Murakami F. 1995. Guidance of cerebellofugal axons in the rat embryo: directed growth toward the floor plate and subsequent elongation along the longitudinal axis. Neuron. 14:961–972 10.1016/0896-6273(95)90334-8 [DOI] [PubMed] [Google Scholar]
- Takagi Y., Yang Y., Fujiwara I., Jacobs D., Cheney R.E., Sellers J.R., Kovács M. 2008. Human myosin Vc is a low duty ratio, nonprocessive molecular motor. J. Biol. Chem. 283:8527–8537 10.1074/jbc.M709150200 [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne M., Goodman C.S. 1996. The molecular biology of axon guidance. Science. 274:1123–1133 10.1126/science.274.5290.1123 [DOI] [PubMed] [Google Scholar]
- Wang F.S., Wolenski J.S., Cheney R.E., Mooseker M.S., Jay D.G. 1996. Function of myosin-V in filopodial extension of neuronal growth cones. Science. 273:660–663 10.1126/science.273.5275.660 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Mabuchi K., Ikebe R., Ikebe M. 2006. Mechanoenzymatic characterization of human myosin Vb. Biochemistry. 45:2729–2738 10.1021/bi051682b [DOI] [PubMed] [Google Scholar]
- Yamauchi K., Mizushima S., Tamada A., Yamamoto N., Takashima S., Murakami F. 2009. FGF8 signaling regulates growth of midbrain dopaminergic axons by inducing semaphorin 3F. J. Neurosci. 29:4044–4055 10.1523/JNEUROSCI.4794-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]