Abstract
Histopathological grading of astrocytic tumours based on current WHO criteria offers a valuable but simplified representation of oncological reality and is often insufficient to predict clinical outcome. In this study we report a new astrocytic tumour microarray gene expression dataset (n=65). We have used a simple Artificial Neural Network (ANN) algorithm to address grading of human astrocytic tumours, derive specific transcriptional signatures from histopathological subtypes of astrocytic tumours and asses whether these molecular signatures define survival prognostic subclasses. 59 classifier genes were identified and found to fall within three distinct functional classes namely angiogenesis, cell differentiation and lower grade astrocytic tumour discrimination. These gene classes were found to characterize three molecular tumour subtypes denoted ANGIO, INTER and LOWER. Grading of samples using these subtypes agreed with prior histopathological grading both for our dataset (96.15%) as well as an independent dataset. Six tumours were particularly challenging to diagnose histopathologically. We present an ANN grading for these samples, and offer an evidence-based interpretation of grading results using clinical metadata to substantiate findings. The prognostic value of the three identified tumour subtypes was found to outperform histopathological grading as well as tumour subtypes reported in other studies, indicating a high survival prognostic potential for the 59 gene classifiers. Finally, 11 gene classifiers that differentiate between primary and secondary glioblastomas were also identified.
Keywords: Astrocytic tumours, grading, classifier genes, PEA15, artificial neural networks
Introduction
Astrocytic tumours of malignancy grades II to IV are collectively termed diffusely infiltrating astrocytomas, and include diffuse astrocytoma (malignancy grade II, abbreviated ‘A’), anaplastic astrocytoma (malignancy grade III, abbreviated ‘AA’) and glioblastoma (malignancy grade IV, abbreviated ‘GB’). A total of four malignancy grades are recognised by the World Health Organization (WHO) system with grade I and IV tumours being the biologically least and most aggressive tumour grades respectively (1, 2). Glioblastoma commonly occur de novo (also called primary glioblastoma) but may also result from the progression of lower grade tumours to higher malignancy grades. Glioblastoma shows the greatest range of genetic abnormalities, with common changes in the de novo tumours including homozygous deletion of CDKN2A, CDKN2B, and p14ARF (9p21), loss of one allele and mutation of the retained allele of PTEN (10q23) and amplification of the EGFR gene (7p12) (2).
Use of expression microarray data in brain tumour classification/clustering (3) and survival prognosis (4-6) has received significant interest in the last few years. Approaches include statistical methods for gene set identification and tumour classification (7), principal component analysis and t-test for the selection of differentially expressed genes involved in astrocytoma progression (8), k-means along with multidimensional scaling for discriminating between glioblastomas, lower grade astrocytomas and other glioma types such as oligodendrogliomas (9), hierarchical clustering (3, 4, 9, 10), k-nearest-neighbour for classification of high-grade gliomas and outcome prognosis (5), gene voting for survival prediction of the diffusely infiltrating gliomas (4) and others. Expression profiling has identified molecular as well as genetic subtypes associated with tumour grade, progression, and patient survival (8, 10). While astrocytic tumours continue to be defined by histological criteria, reports that expression profiles predict survival better than histological grade (4, 5, 11) provide support for the hypothesis that tumours defined morphologically represent a mix of molecular genetic subtypes. Most of these studies, however, have compared diffusely infiltrating astrocytomas to tumours of mixed or non-astrocytic origin, have not included lower grade (II) tumours (4, 5, 7, 11) or have limited their efforts to a single tumour grade (3, 6). Several of these studies have also compared tumour tissue to normal brain, a task of arguable relevance when taking into account the vast differences in cellular composition between the two tissues. Moreover, studies have often focused on questions related more to the use of expression data towards general brain tumour classification rather than malignancy grading of diffusely infiltrating astrocytic tumours per se. Finally, discordances between histopathology and expression-based tumour classification for a given tumour set have seldom been interpreted or substantiated with thorough clinical and/or molecular evidence.
Using a new gene expression dataset originating from 65 highly annotated tumours and a simple artificial neural network (ANN) algorithm in the form of a single-layer perceptron, we address grading of human astrocytic tumours, derive specific transcriptional signatures from histopathological subtypes of astrocytic tumours and asses whether these molecular signatures define survival prognostic subclasses. We validate our approach with a number of independent datasets and offer valuable insight into the tumour biology and gene expression-based grading of astrocytomas.
Materials and Methods
Tumour samples, RNA isolation and Hybridization to Affymetrix U133A GeneChips
The tumour set consisted of 2 pilocytic astrocytomas (WHO grade I, ‘PA’) 5 diffuse astrocytomas (WHO grade II, ‘A’), 15 anaplastic astrocytomas (WHO grade III, ‘AA’) and 39 glioblastomas (WHO grade IV, ‘GB’). This sample distribution reflects tissue availability and relative frequency of diagnosis per tumour grade. 4 additional samples graded as AA that were exceptionally challenging to grade by histopathology were treated as separate “problem” cases. Histopathological diagnoses were made according to WHO criteria (1) by V.P.C. RNA from the 65 human astrocytic tumour samples was extracted using guanidine isothiocyanate ultracentrifugation as described previously (12). RNA quality was assessed using an Agilent Bioanalyzer 2100 (Agilent technologies). For each tumour sample, 7 μg of RNA were used to generate double stranded cDNA which was subsequently in vitro transcribed to produce biotin-labelled cRNA using the ENZO BioArray HighYield kit. cRNA (15 μg) was fragmented and hybridized to Affymetrix HG-U133A genechips (Affymetrix, Inc, Santa Clara, CA). GeneChips were washed, stained and scanned as described in the manufacturer’s manual. Quality of pre- and post-fragmentation cRNA was assessed using an Agilent Bioanalyzer 2100 (Agilent technologies).
Expression microarray data analysis
Raw data (CEL files) were imported into ‘R’, a freely available environment for statistical computing(13). Normalization and computation of expression measures was performed using the justRMA function within the Affy package of Bioconductor (14). All expression data has been submitted to GEO (15) in a MIAME-compliant fashion (accession number GSE1993). Annotation of probe set lists was performed using EASE (16).
Validation of results using Quantitative QPCR (QPCR)
QPCR was performed on a LightCycler machine (Roche) using DNA master SYBR Green I (Roche Molecular Biochemicals, or Sigma) according to the manufacturer’s protocol. Primers were ordered from MWG. Double stranded cDNA used as a template was the same as that used for cRNA target preparation. 1 μl of this cDNA was diluted 1:200 for generation of the final template used. Validation was performed on a subset of 23 tumours (15 GB, 8 AA) that were part of the original tumour dataset assessed. Assays were performed in duplicate. The raw data produced by QPCR referred to the number of cycles required for reactions to reach exponential phase as determined using the RelQuant software (Roche). Expression of MYO1C was used for normalisation of the QPCR data. Mean expression fold change differences between tumour groups were calculated using the 2−ΔΔCT method (17). Primer sequences: PEA15, 5′-GAGCAGCCAGCGTTAGATGC-3′, 3′-GGAGGTGTTCACAAGACCAGGG-5′; ADM,5′-GCAGAAGAATCCGAGTGTTTGC-3′, 3′-AATCAGTTTGTGGGCGAGCACG-5′.
Tissue array generation and Immunohistochemistry
Cores (n=2, for 57 tumours from our dataset) of 0.6 mm diameter were taken from paraffin embedded tumour tissue and arrayed into a fresh paraffin block using a manual tissue arrayer (Beecher Instruments, Silver Spring, MD, USA). Areas identified on hematoxylin and eosin-stained sections to have high tumour cell content were used. 10 non-neoplastic tissue cores with minimal or no tumour cell content were also included. Immunohistochemistry for ADM (1:50, Abcam, ab18092) and PEA15 (1:500) was performed as previously described (18, 19).
The ANN Model and statistical analysis
A single-layer perceptron was used for grading the tumour tissue samples. The number of inputs was equal to the number of classifier genes and the output layer consisted of a single neuron with a sigmoidal activation function. Initial weight values were chosen randomly and training was performed using a standard gradient descent learning rule (or Delta rule) with learning rate η = 0.05. Calibration was performed via leave-one-out cross-validation. The weight values were updated after every sample and the calibration was terminated after 100 passes (epochs) through the entire training set. The resulting parameters for a completed training define a “model” (also see online supplement). The source code for the ANN and visualization methods is available from: http://www.imbb.forth.gr/people/poirazi/software.html.
The Kaplan-Meier method was used to estimate the survival distributions(20). Log rank tests were used to test the difference between survival groups. For all of the analyses, a p < 5.0e−2 was accepted as significant. Statistical analyses were carried out with the freely available software package R.
Results
SELECTION OF CLASSIFIER GENES
Training the ANN to distinguish between different astrocytic tumour grades and concurrent selection of classifier genes
In order to train the neural network, tumour samples were randomly split into two sets in a way that approximately preserves the sample distribution across each tumour grade. The first 20 GB, 10 AA and 3 A were used as a training set and 19 GB, 5 AA and 2 A were used as a test set. A further test group of 6 astrocytic tumours comprised 4 AA that had proved difficult to grade histopathologically and two samples belonging to grade I, pilocytic astrocytomas (PA).
Training/calibration was performed in an all-pairs approach whereby the single problem of learning to differentiate between 3 grades (GB, AA, and A) was narrowed down to multiple 2-grade problems (Figure A online supplement). The 33 training samples were split into three sample groups each comprising 2 tumour grades, namely (a) GB-AA, (b) AA-A and (c) GB-A. Three different types of ANN models (A, B and C) were then trained, each corresponding to their respective sample groups. For each of these model types, genes that showed differential expression between the two grades in question were selected using the signal-to-noise method (21) on the entire U133A chip genome. Training performance and optimum number of genes required for grading were evaluated using leave-one-out cross-validation. For every leave-one-out run, genes were ranked according to signal-to-noise (taken over all but the left out sample) and then the grading success rate was determined using increasing numbers of these ranked genes. Leave-one-out cross-validation success rates optimized to 93.3%, 84.6% and 95.6% using a total of 44, 9 and 7 probe sets for the GB-AA, AA-A and GB-A grade comparisons respectively (see leave-one-out plots, Figure B online supplement).
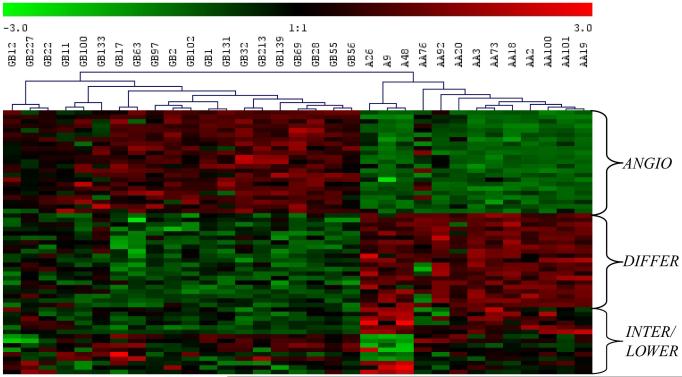
Pooling of all probe sets and elimination of redundancies resulted in a total of 59 unique probe sets. As anticipated, hierarchical clustering of all training samples using the above probe sets revealed clear distinctions between the GB, AA and A tumour grades and further defined 3 functional gene classes that delineate 3 molecular tumour subtypes. (Figure 1 - see next section for details). The trained/calibrated ANN models (see Materials and Methods) were subsequently used for grading of the test set.
Figure 1.
Hierarchical clustering of 33 training samples (20GB, 10 AA and 3 A) using 59 probe sets selected by S2N. MeV (33) was used to perform hierarchical clustering using Euclidean distance and complete linkage algorithm. Samples are labelled with their respective grades and genes are labelled according to the molecular tumour subtype(s) (ANGIO, DIFFER or INTER/LOWER) that they characterize. Gene expression values are standardized to a mean of 0 and standard deviation of 1. Red represents higher expression relative to green.
Expression profiles of gene classifiers selected during training define three molecular tumour subtypes
A thorough examination of the selected gene classifiers, most of which were also identified using empirical Bayesian analysis (Table 1 and online supplement), revealed two interesting features. Firstly, classifying genes were found to fall within three main functional classes and secondly, these functional classes could discriminate between three molecular tumour subtypes. The first subtype showed significant increased expression of genes involved in: i) wound-healing (ADM, PDGFa, EFEMP2), ii) extracellular matrix constituents and remodelling machinery (LGALS1 and 3, PLAT, TIMP1, COL5A2) and iii) cell adhesion (PARD3, DAG1, Kindlin1, ZYX and ALCAM). As all of these functions are necessary for the angiogenic properties of cells, this subtype was labelled ANGIO and was characteristic of the grade IV, GB samples. The next group was a mixture of histopathological and molecular subtypes and showed increased expression of genes involved in: i) cell-signaling and growth (BMP2, ABI1, REPS2, ADCY2, NET1), ii) protein biosynthesis (RPL22, ZMYND11) and the iii) cell cycle (PARD3, ZMYND11, CLASP2). This group, labelled DIFFER, characterizes the grade II and grade III, samples, which while active in growth and neuronal differentiation, have not yet acquired angiogenic properties. This group was further analyzed using a set of genes coding for ankyrin repeat proteins (ANK3, ANKS1B), solute carrier proteins (SLCO1A2, SLC34A1), a protein involved in apoptosis (DNAJA3) and PEA15, a cytostatic and anti-apoptotic phosphoprotein enriched in astrocytes (22). This analysis lead to the separation of the DIFFER group into the INTER (Intermediate) subtype, which was characteristic of the grade III samples and the LOWER subtype which was characteristic of grade II samples.
Table 1.
Three sets of selected genes each derived form one of the three pairwise tumour grade comparisons: a) GB-AA (ANGIO/DIFFER genes), b) GB-A (INTER/LOWER genes), c) AA-A (INTER/LOWER genes)
| a) | ||||
|---|---|---|---|---|
|
| ||||
| Gene Symbol | Gene Name |
Bayesian p value |
Mean expression fold change |
Gene class |
| ADM | Adrenomedullin | 2.62E-05 | 11.79 | ANGIO |
| TIMP1 | tissue inhibitor of metalloproteinase 1 | 1.51E-08 | 11.56 | ANGIO |
| FABP5 | fatty acid binding protein 5 | 1.85E-04 | 9.41 | ANGIO |
| EMP3 | epithelial membrane protein 3 | 5.27E-07 | 7.58 | ANGIO |
| PDPN | Podoplanin | 3.22E-05 | 6.00 | ANGIO |
| LGALS3 | lectin galactoside-binding soluble 3 (galectin 3) | 1.02E-05 | 5.86 | ANGIO |
| LGALS1 | lectin galactoside-binding soluble 1 (galectin 1) | 1.02E-05 | 4.41 | ANGIO |
| PDGFA | platelet-derived growth factor alpha polypeptide | 2.03E-05 | 4.09 | ANGIO |
| PLAT | plasminogen activator tissue | 6.60E-05 | 3.97 | ANGIO |
| EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 | 2.20E-06 | 3.92 | ANGIO |
| COL5A2 | collagen type V alpha 2 | 3.71E-05 | 3.72 | ANGIO |
| COL5A2 | collagen type V alpha 2 | 1.02E-05 | 3.60 | ANGIO |
| DDA3 | differential display and activated by p53 | 2.06E-05 | 3.53 | ANGIO |
| TAGLN2 | transgelin 2 | 3.15E-05 | 3.19 | ANGIO |
| DUSP6 | dual specificity phosphatase 6 | 5.49E-05 | 3.14 | ANGIO |
| LDHA | lactate dehydrogenase A | 7.84E-05 | 2.84 | ANGIO |
| PLP2 | proteolipid protein 2 | 6.34E-05 | 2.74 | ANGIO |
| EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 | 7.34E-05 | 2.43 | ANGIO |
| CENTD3 | centaurin delta 3 | 2.57E-04 | 2.42 | ANGIO |
| KIAA0495 | KIAA0495 | 1.69E-04 | 2.15 | ANGIO |
| DAG1 | dystroglycan 1 (dystrophin-associated glycoprotein 1) | 2.73E-05 | 1.80 | ANGIO |
| ZYX | Zyxin | 1.46E-04 | 1.78 | ANGIO |
| OSBPL10 | oxysterol binding protein-like 10 | 2.63E-04 | 1.75 | ANGIO |
| CUTC | cutC copper transporter homolog | 1.74E-05 | −1.59 | DIFFER |
| TNKS2 | TRF1-interacting ankyrin-related ADP-ribose polymerase 2 | 6.60E-05 | −1.67 | DIFFER |
| HSA9761 | dimethyladenosine transferase | 8.27E-05 | −1.71 | DIFFER |
| KIAA1279 | KIAA1279 | 2.03E-05 | −1.76 | DIFFER |
| RPL22 | ribosomal protein L22 | 2.83E-04 | −1.81 | DIFFER |
| ENAH | enabled homolog | 6.26E-05 | −1.82 | DIFFER |
| ZMYND11 | zinc finger MYND domain containing 11 | 4.53E-05 | −1.87 | DIFFER |
| HNRPH3 | heterogeneous nuclear ribonucleoprotein H3 | 3.65E-05 | −1.88 | DIFFER |
| RPL22 | ribosomal protein L22 | 2.62E-05 | −1.93 | DIFFER |
| CLASP2 | cytoplasmic linker associated protein 2 | 2.81E-04 | −2.05 | DIFFER |
| USH1C | Usher syndrome 1c (autosomal recessive severe) | 1.02E-05 | −2.10 | DIFFER |
| RAP2A | RAP2A | 7.90E-04 | −2.10 | DIFFER |
| ALCAM | activated leukocyte cell adhesion molecule | 4.72E-03 | −2.14 | DIFFER |
| ABI1 | abl-interactor 1 | 8.27E-05 | −2.16 | DIFFER |
| PARD3 | par-3 partitioning defective 3 homolog | 3.43E-06 | −2.30 | DIFFER |
| CRYAB | crystallin, alpha B | 5.80E-05 | −2.72 | DIFFER |
| NAP1L3 | nucleosome assembly protein 1-like 3 | 1.77E-04 | −2.88 | DIFFER |
| NET1 | neuroepithelial cell transforming gene 1 | 2.20E-06 | −2.93 | DIFFER |
| C20ORF42 | chromosome 20 open reading frame 42 | 2.87E-04 | −3.09 | DIFFER |
| BMP2 | bone morphogenetic protein 2 | 5.49E-05 | −3.27 | DIFFER |
| ADCY2 | adenylate cyclase 2 (brain) | 3.05E-05 | −3.79 | DIFFER |
| b) | ||||
|---|---|---|---|---|
|
| ||||
| Gene Symbol | Gene Name |
Bayesian p value |
Mean expression fold change |
Gene class |
| SLC34A1 | solute carrier family 34 (sodium phosphate) member 1 | 1.46E-02 | −1.35 | LOWER |
| RSNL2 | restin-like 2 | 1.88E-02 | −1.39 | LOWER |
| REPS2 | RALBP1 associated EPS domain containing 2 | 1.53E-03 | −1.94 | LOWER |
| SLCO1A2 | solute carrier organic anion transporter family member 1A2 | 4.46E-03 | −2.06 | LOWER |
| PEA15 | phosphoprotein enriched in astrocytes 15 | 1.93E-03 | −2.12 | LOWER |
| USH1C | Usher syndrome 1C | 1.53E-03 | −3.49 | LOWER |
| c) | ||||
|---|---|---|---|---|
|
| ||||
| Gene Symbol | Gene Name |
Bayesian p value |
Mean expression fold change |
Gene
class |
| B2M | beta-2-microglobulin | 3.69E-01 | 2.50 | INTER |
| SCP2 | sterol carrier protein 2 | 4.79E-01 | 1.81 | INTER |
| DDOST | dolichyl-diphosphooligosaccharide-protein glycosyltransferase |
5.68E-01 | 1.79 | INTER |
| NPTN | neuroplastin | 8.49E-01 | 1.39 | INTER |
| TAP2 | transporter 2 ATP-binding cassette sub-family B | 6.92E-01 | 1.39 | INTER |
| DNAJA3 | DnaJ (hsp40) homolog subfamily A, member 3 | 6.48E-01 | 1.25 | INTER |
| ANKS1b | ankyrin repeat and sterile alpha motif domain containing 1b | 5.17E-01 | −1.21 | LOWER |
| --- | DKFZp434M083 | 5.68E-01 | −1.22 | LOWER |
| ANK3 | ankyrin 3 node of Ranvier | 9.07E-02 | −1.93 | LOWER |
Gene classifiers of particular biological interest
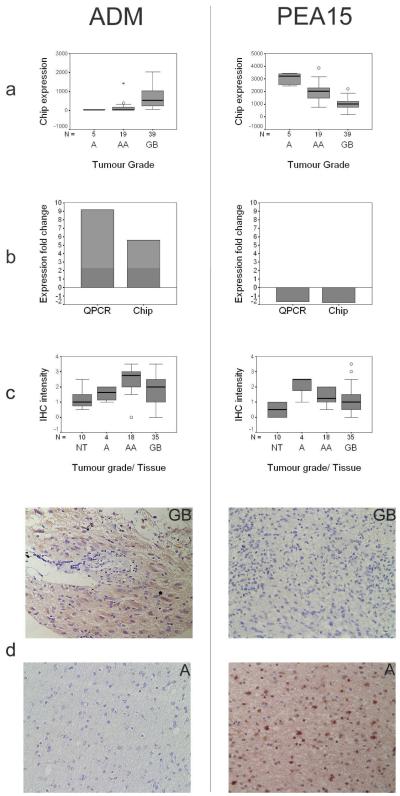
In addition to the identification of three molecular tumour subtypes, two classifiers, the phosphoprotein enriched in astrocytes 15 (PEA15, 1q21.1 - LOWER) and adrenomedullin (ADM, 11p15.4 - ANGIO) were of particular biological interest and/or novelty. These genes were also found to be differentially expressed between the GB-A and/or GB-AA tumour grades by empirical Bayesian analysis. Expression changes were validated by both QPCR and immunohistochemistry (IHC) (Figure 2). A further 23 differentially expressed genes identified by Bayesian analysis were successfully validated by QPCR. The correlation (R2) between Affymetrix and QPCR GB/AA expression fold changes for these genes was over 0.8 (data not shown).
Figure 2.
Expression of ADM and PEA15 changes with astrocytic tumour progression at both transcript and protein levels. (a) GeneChip (‘chip’) expression values for ADM and PEA15 across tumour grades. Samples PA68 and PA67 (grade I tumours – see text) have not been included in this analysis. Expression changes for both gene products are highly statistically significant (for the AA-GB and A-GB tumour grade comparisons respectively: ADM, p = 1.1e−6 and 1.6e−4; PEA15, p = 4.8e−5 and 8.3e−5). (b) Validation of expression changes using QPCR. Mean expression fold changes between GB and AA tumour grades (‘Expression fold change’) shown as assessed by both GeneChip (‘chip’) and QPCR expression technology. (c) Tissue array IHC immunoreactivity intensities for ADM and PEA15 across tumour grades and non-neoplastic brain (normal tissue, ‘NT’). 5-scale grading system used: ‘0’ no immunoreactivity, ‘4’ intense immunoreactivity. For each tumour grade and for the collection of normal tissues, an average immunoreactivity grade was obtained from replicate tissue cores available on the tissue array. Differences in IHC immunoreactivity (Mann-Whitney non-parametric test, p<5.0e−2) were significant for tumour group comparisons A-GB and A-AA (PEA15), and A-AA (ADM). (d) Representative IHC results for ADM and PEA15 on GB and A tumour sections. Immunoreactivity for both gene products was seen to be present in tumour cells only. ADM showed cytoplasmic staining and PEA15 showed both cytoplasmic and nuclear staining. The nuclear/cytoplasmic distribution of PEA15 immunoreactivity was not constant for all tumour cells in a given tumour sample.
GRADING USING TRAINED ANN MODELS
Grading of test samples using the trained ANN models into tumour subtypes agrees with prior histopathological grading
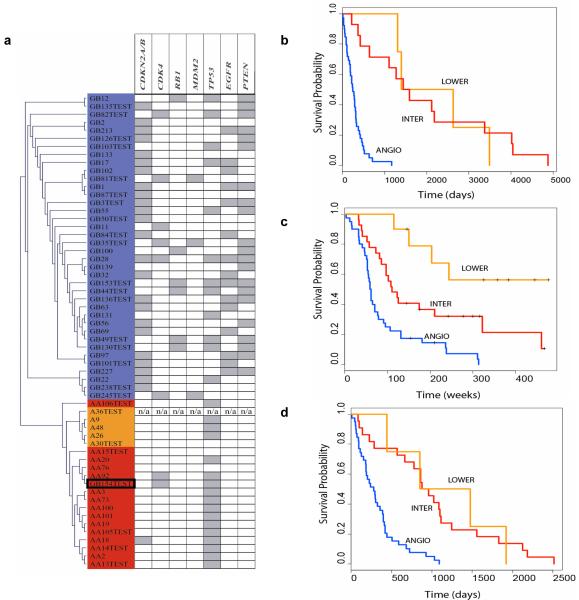
Grading of the test set (n=26) was performed by passing each test sample through all models saved during the training process (for details see online supplement). Through this way the 59 genes/probes selected during training can be put to the test of grading a ‘blind’ set of tumour samples. For each test sample, an initial voting was performed by the ANGIO/DIFFER trained models. The samples that were graded as DIFFER received a follow-up grading by the INTER/LOWER trained models to discriminate between INTER and LOWER subtypes (Table A - online supplement). Histopathological grading of the test samples was found to agree with the tumour subtypes observed during training. More specifically, all GB (except sample GB154) and all lower grade astrocytic tumours (A and AA) showed increased expression of ANGIO and DIFFER genes respectively. Furthermore, all A samples were distinguished from AA by the differential expression of INTER/LOWER genes. Overall our ANN-defined tumour subtypes were in agreement with prior histopathological grading, reporting 94.74%, 100% and 100% accuracies for ‘GB’, ‘AA’ and ‘A’ grading respectively Visualization of network outputs using available clustering algorithms (23, 24) for all train and test samples (including annotation with genomic metadata) is shown in Figure 3a (also see online supplement).
Figure 3.
(a)Visualization of network results for all 33 training and 26 test tumour samples (39GB, 15 AA and 5 A) using the 59 genes/probe sets selected during training. Hierarchical clustering of network outputs (Euclidean distance, single linkage algorithm). The visualization represents results from the propagation of all samples (training and test) through the trained models. Colour coding is according to the 3 molecular subtypes which best characterize the samples (blue: ANGIO, red: INTER, orange: LOWER). The only tumour where ANN subtyping does not agree with histopathology is GB154 (highlighted by a black box). AA106 appears to cluster separately from the rest of the AA but does not reside in the LOWER cluster. Tumours are annotated with genomic information for a total of 7 loci available from previously published work from our laboratory (34-38), known to be involved in astrocytic tumour genesis and/or progression. Grey boxes indicate homozygous deletion (CDKN2A/CDKN2B/p14ARF), amplification (CDK4, MDM2 and EGFR) or loss of one allele with mutation of the remaining allele (RB1, TP53 and PTEN). Test samples marked ‘TEST’. (b) Kaplan-Meier survival plot of our 59 astrocytic tumours as defined by our ANN grading results. ANGIO – blue line, INTER- red line, LOWER – orange line. (c) Kaplan-Meier survival plot of the 76 Phillips samples as defined by our ANN grading results using our 59 gene classifiers. (d) Kaplan-Meier survival plot of the 65 Freije samples. ANGIO subtype contained 38/50 GB, INTER was comprised of 6/15 AA and 12/50 GB, while the remaining 4/15 AA made up the LOWER subtype.
Grading of an independently published astrocytic tumour gene expression dataset using cross-chip gene classifiers
To further validate the grading capacity of our gene classifiers, we used an independent, astrocytic tumour gene expression dataset published by Shai et al in 2003 (9). Of the 59 probe sets selected during training from our HG-U133A genechips, 38 genes had >96% identity to probe sets on the U95Av2 genechip utilized by Shai et al, 2003 (9). Out of these, we selected 20 genes that appeared more than once in our leave-one-out cross-validation runs, thus ensuring that only the most significant probe sets were used in the cross-chip analysis. Of these, 17 genes were differentially expressed in the GB-AA comparison (comprised of ANGIO and DIFFER genes), 1 in the GB-A comparison (INTER/LOWER gene) and 2 in the AA-A comparison (INTER/LOWER genes). We re-trained the ANN models on our original training data utilizing these 20 probe sets (for gene names see online supplement). Due to the limited number of probe sets available for the GB-A assessment, we split the grading task into two pair-wise comparisons. ANN models of “Type 1” were trained to distinguish between grade IV and lower grade astrocytic tumours using the 17 ANGIO/DIFFER genes and models of “Type 2” were trained to distinguish between grade II and III tumours using the 3 INTER/LOWER genes. Only samples that were graded as lower grade DIFFER tumours by Type 1 models required follow-up grading by Type 2 models. The 23 (18 GB, 3 AA, 2 A) samples derived from the Shai et al, 2003 dataset were treated as a blind test set and were graded using our trained models. A remarkable consistency was observed between the two expression datasets using the 20 common probe sets, whereby histopathological and ANN-based subtyping resulted in an agreement accuracy of 100% (2/2), 100% (3/3) and 88.89% (16/18) for the A (graded as LOWER), AA (graded as INTER) and GB (graded as ANGIO) tumours of the Shai et al study respectively (Table C online supplement).
Grading of additional samples difficult to grade histopathologically and evaluation of ANN results using clinical, histopathological and genomic annotation
After verifying the grading power of our molecular signatures, we used them to identify the stage of certain samples that were particularly challenging to diagnose by histopathology. Histopathological identification of Pilocytic astrocytomas (grade I) and malignancy grading of astrocytic tumours that have been treated with irradiation and/or chemotherapy, can be extremely difficult. We therefore examined the expression data from 6 such problem cases using the trained ANN models.
The two pilocytic astrocytoma (PA) tumours (PA68 and PA67) were graded as ANGIO (GB-rich) and INTER (AA-rich) respectively by our trained ANN models (see Discussion). These tumours were histologically typical (25) and were derived from patients with excellent survival (alive at end of follow-up - see Table I online supplement). Samples AA49 and AA86, were difficult to grade as they had received irradiation and chemotherapy. Two other AA tumours, AA29 and AA93, were also difficult to grade histologically. Grading these samples using our trained ANN models did not concur with histopatholical grading and showed the grading of all 4 AA samples as ANGIO (GB-rich subtype) (Table B online supplement).
In order to investigate possible reasons for this discrepancy, we evaluated available annotation for all 4 ambiguous tumours in our dataset as well as for the miss-graded GB154 and the two grade I pilocytic astrocytomas. In addition to histopathological diagnosis, available annotation included i) clinical data (age at operation, gender, primary or secondary tumour, tumour location), ii) survival data and iii) previously published genomic information for a total of 9 genes (CDKN2A, CDKN2B, p14ARF, CDK4, RB1, MDM2, EGFR, PTEN and TP53) known to be affected in astrocytoma (see Table I online supplement).
The histology of tumours AA49 and AA86 was difficult to use for malignancy grading as previous treatment complicated the findings significantly. AA49 shows a clear GB genetic profile (homozygous deletion of CDKN2A, CDKN2B and p14ARF, EGFR amplification and no wild-type PTEN) while AA86 shows further genetic abnormalities commonly seen in glioblastoma: lack of wild-type CDKN2A, p14ARF, or TP53. In the case of tumour AA29, clinical, histopathological and genomic evidence indicated a significant resemblance to GB (suspicion of, but no frank necrosis found and no wild-type PTEN). Tumour AA93 had a histological and clinical appearance of an AA but shared the same classic GB-like genetic profile seen also for AA49. The only genetic difference between the two tumours related to the retention of one wild-type copy of PTEN.
The 4 ambiguous AA tumours classified by our ANN as ANGIO comprised 100% (2/2) of the EGFR amplifications, 100% (2/2) PTEN mutations and 66% (2/3) of the CDKNA/B nullizygosity found across all the AA samples assessed. With the exception of one INTER graded AA tumour with CDKN2A/B nullizygosity, lesions for the cyclin inhibitor locus were totally absent in all remaining AA of our dataset. All 3 of the AA cases where survival data was available, died within 2 years.
No apparent reasons for the disagreement between histopathology and ANN subtyping of the GB tumour (GB154) could be found. Although GB154 had some non-classic GB characteristics, the presence of amplification of CDK4, necrosis and microvascular proliferation, the latter being major histological criteria for glioblastoma, support the original histopathological diagnosis. Survival in this case was also under 2 years.
SURVIVAL ANALYSIS
Survival Analysis using the selected gene classifiers reveals a prognostic value for tumour subtypes
To investigate the survival prognostic capabilities of our gene classifiers we performed survival analysis on our 59 samples as graded by histopathology and then as defined by our trained ANN models into the three tumour subtypes. Although there was only a small difference between ANN- and histopathology-based grading efforts (difference of one sample - GB154), the survival analysis based on the ANN grading proved to be more significant (p = 8.76e−7) than that based on purely histopathological data (p = 2.088e−6), as defined by the log rank test (Figure 3b). Similar results were obtained from survival analysis of the Shai et al in 2003 (9) dataset. The prognostic value of our ANN defined subtypes was equally significant (p =6.0e−3) to that based on histopathology (p =6.0e−3).
Survival analysis substantiates grading of datasets where ANN defined subtypes do not concur with prior histopathological grading
To our surprise, for two other independently published datasets, the ANN failed to recapitulate histopathological grading. However, in both cases, survival analysis favoured the ANN-based grading.
The Phillips et al. 2006 (11) dataset comprised of 100 MDA samples (76 for which survival information was available). In that study the samples were divided into 3 “subclasses” representing the progression of astrocytic tumours. The subclasses were defined by the authors as Proneural (PN), proliferative (Prolif) and Mesenchymal (Mes), with increasing malignancy from PN to Mes. Since those samples consisted of grade III and IV tumours, we used the ANN models trained with ANGIO/DIFFER genes to classify the 100 MDA samples into the respective subtypes. The ANGIO subtype consisted of 50/76 GB and 4/24 AA while the DIFFER subtype was comprised of 22/76 GB and 12/24 AA. The ANGIO group consisted of 30/35 of the Phillips et al 2006 (11) Mes samples, in accordance to previously published results that show that Mes tumors display over-expression of angiogenic markers (11). The DIFFER group consisted of 33/37 of the PN samples, also in accordance to previous reports indicating that PN samples display over-expression of markers of neuronal differentiation and growth. Further analysis of the DIFFER survival samples using our INTER/LOWER genes partitioned them into the INTER subtype, which consisted of 18/76 GB and 4/76 AA, and the LOWER subtype, which consisted of 4/76 GB and 8/24 AA. This approach grouped the Phillips et al 2006 (11) samples into three very significant prognostic subclasses (Figure 3c, p = 1.922e−7), once again outperforming the previous subtyping defined in the Phillips et al 2006 (11) study (p = 1.0e−4). The Phillips et al 2006(11) Prolif samples, which according to their study represent the intermediate stage of the progression and are highly enriched for proliferative markers, were not so well defined by our tumour subtypes. However; 20/28 resided within the ANN-defined ANGIO subtype (which was rich in Mes samples) and 8/28 within our INTER subtype (rich in PN samples) (Table E - online supplement). This was in accordance with previous published results that show a very similar survival median for the Phillips et al 2006 (11) Mes and Prolif groups and a higher angiogenic index of the Prolif compared to the PN tumours (11). In addition, this concurs with the observation that the Prolif signature is less exclusive and the proportion of astocytic tumours with this signature varies across samples obtained from different institutions (11).
Finally, a probe comparison between the 59 gene classifiers utilized in this analysis and the final 35 probes identified in Phillips et al, 2006 showed that there were no common probes between the two gene/probe sets, once again highlighting the novelty of our gene classifiers.
Similar results were obtained for another independent dataset containing 65 astrocytic tumours (15 grade III and 50 grade IV) published by Frieje et al (4). More specifically, our subtyping significantly outperformed (Figure 3d, p = 8.13e−8) the final survival groups obtained by Freije et al (4) in the respective publication (p = 2.2e−4).
Genes predictive of survival versus genes predictive of histopathology
In order to investigate this unexpected performance on the Phillips et al. and Freije et al. datasets, whereby genes identified based on histopathology acted as prognostic signatures of survival, we decided to compare genes predictive of survival (survival-correlated genes) and genes predictive of histopathology (histopathology-based genes) within our own dataset as well as within the other two large datasets. We initially performed clustering using the top 80 positively- correlated and negatively-correlated genes to survival (Pearsons correlation of expression values versus survival times >0.55 or <−0.55), and observed three major clusters. We then recalibrated our ANN to optimize leave-one-out cross-validation runs for these clusters with survival correlated genes and resulted in an optimum set of 37 genes (see supplementary information Table G). We also performed the same histological based analysis on the two independent datasets as described earlier for our own dataset and selected the respective histopathology-based genes. Log rank tests using histopathology-based or survival-correlated genes are shown in table 2. Interestingly, we found that genes identified by signal-to-noise to be differentially expressed between histological grades were more successful than the respective survival-correlated genes in predicting survival in all datasets tested.
Table 2.
Comparison of histopathology genes and genes predicted by correlation to survival. p-values where predicted using the log-rank test after grouping the samples into survival groups
| Genes Used | ||||
|---|---|---|---|---|
| Our 59 histopathology genes | Phillips histopathology genesi | Freijer histopathology genesii | ||
| Dataset | Survival Our | p = 8.76e−7 | p = 4.258e−5 | p = 6.871e−6 |
| Survival Phillips | p = 1.922e−7 | p = 8.808e−8 | p = 1.133e−4 | |
| Survival Freijer | p = 8.13e−8 | p = 4.55e−8 | p = 2.318e−8 | |
| Genes Used | ||||
|---|---|---|---|---|
| Our 37 surv corr genes | Phillips 35 surv corr genesiii | Freijer 44 surv corr genesiv | ||
| Dataset | Survival Our | p = 4.377e−6 | p = 1.871e−4 | p = 4.431e−5 |
| Survival Phillips | p = 2.081e−5 | p = 1.0e−4 | p = 3.169e−5 | |
| Survival Freijer | p = 1.103e−5 | p = 1.191e−1 | p = 2.2e−4 | |
These p-values are the result of the grouping of the samples into 2 groups
These p-values are the result of the grouping of the samples into 2 groups
These p-values are the result of the grouping of the samples into 3 groups (23/35 genes were present on our gene chip as we only used the HGU133A gene chip)
These p-values are the result of the grouping of the samples into 3 groups (41/44 genes were present on our gene chip as we only used the HGU133A gene chip)
TP53 lesions further separate the grade IV GB into two survival groups
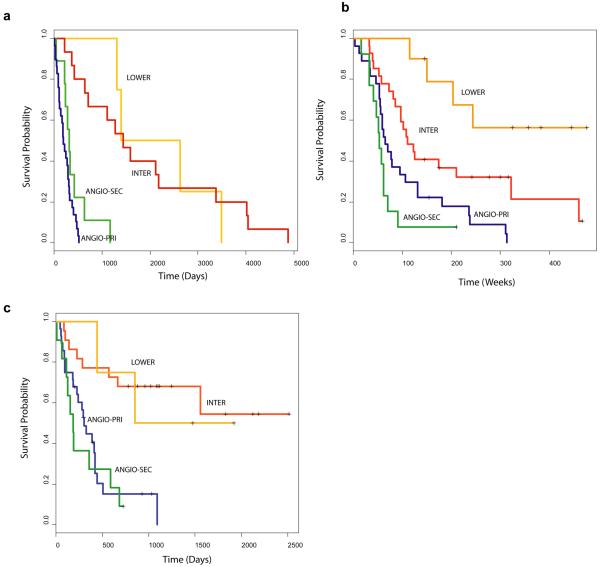
TP53 mutations are observed in over 65% of secondary GB and are considered a major hallmark that defines the separate molecular pathways, responsible for the development of the secondary GB and the primary (de novo) GB. In order to identify genes with distinct signatures for these two separate pathways, we performed a leave-one-out cross-validation using only the GB - separated into TP53 mutated and wild-type- and identified an optimum set of 11 probes (see supplementary information Table F). Using these genes, the ANN separated our ANGIO subtype into two groups denoted, ANGIO-PRI and ANGIO-SEC. This distinction was more significant for survival prediction (p = 3.325e−2), than the respective TP53 separation (p = 7.082e−1). In the Phillips dataset, we found that the ANGIO-SEC group consisted 16/28 Prolif samples and only 3/35 Mes samples while the ANGIO-PRI group consisted 12/28 Prolif and 32/35 Mes samples. This is in accordance to previous reports (26) showing that secondary GB undergo aggressive proliferation (as is the case with the Prolif samples) in contrast to primary GB, which show over-expression of angiogenic genes (as is the case with the Mes samples). Survival analysis using our 59 gene classifiers and the 11 gene signatures described here, for all three datasets, is shown in figure 4.
Figure 4.
Survival analysis of astrocytic tumours (including the 11 primary/secondary gene signature). (a) Kaplan-Meier survival plot of our 59 astrocytic tumours as defined by our ANN grading results. Primary-ANGIO – blue line, secondary-ANGIO – green INTER- red line, LOWER – orange line. (b) Kaplan-Meier survival plot of the 76 Phillips samples as defined by our ANN grading results using our 59 gene classifiers + 11 gene signature. (c) Kaplan-Meier survival plot of the 65 Freije samples using our 59 gene classifiers + 11 gene signature.
Discussion
In this study, we used a simple, ANN-based approach to derive specific transcriptional signatures from histopathological subtypes of astrocytic tumours and assesed whether these molecular signatures define survival prognostic subclasses. We found that the classifier genes selected fall into three distinct functional classes, which characterize three molecular tumour subtypes, denoted ANGIO, INTER and LOWER. ANN-based grading into the three tumour subtypes for our own as well as one independent dataset (9) was found to accurately match prior histopathogical grading. This was not the case for two other datasets (4, 11). In order to investigate this discrepancy we performed an extensive comparison between survival correlated genes and histopathology based genes. We showed that with respect to survival prediction (a) histopathology based genes outperform the respective survival-correlated genes in each dataset and (b) our histopathology based genes outperform survival-correlated genes, in all datasets tested. Finally, ANN analysis of TP53 mutated and wild-type samples identified a gene signature that appears to further separate the ANGIO subtype into two groups reflecting primary and secondary GB.
The prognostic nature of markers of angiogenesis and proliferation has previously been reported (27-30) with angiogenic markers (VEGF, flt1/VEGFR1, kdr/VEGFR2, PECAM1) and markers of proliferation (PCNA and TOP2A) commonly used by pathologists for astrocytic tumour grading. Here, we provide a novel set of genes that characterize the ANGIO subtype and appear to control angiogenesis. The general trend for grade IV, GB to reside within the ANGIO subtype is in accordance with these reports. The presence of most of the Phillips et al 2006 (11) defined Mes samples within the ANGIO subtype further substantiates findings as these samples have been reported to over-express angiogenic markers such as VEGF. The differentiating and developing nature of the lower grade AA and A, is consistent with the observation that these tumours reside within the DIFFER group. The general trend for the Phillips et al 2006 (11) PN samples to resemble the DIFFER samples is in accordance to reports that show that PN samples over-express markers of neurogenesis and neuronal differentiation (11). The Phillips et al 2006 (11) Prolif subtype is not defined on the basis of our tumour subtyping, but was partially defined by the 11 genes used to differentiate between the primary and secondary GB. The characteristic of the Phillips Prolif samples to be less clearly defined, confirms previous observations which report a less specific phenotype for these samples as well as a greater variability across samples obtained from different institutions(4, 11). Furthermore, we identified an interesting set of genes (including PEA), that appear to separate the DIFFER group (lower grade II, A and grade III, AA) into the INTER (grade III, AA) and the LOWER (grade II, A) subtypes and further define a prognostic class with the highest survival probability (LOWER).
Survival analysis suggests that histopathological grading, although categorical and oversimplified, provides a general trend by which genes predictive of survival can be identified, with prognostic value greater than histopathological grading per se. Survival prognosis can be achieved either independently or, as in the case of our dataset, in conjunction with histopathology prediction. A comparison of survival-correlated and histopathology-based genes showed that the latter were more efficient in survival prognosis. This was observed for our dataset as well as two other independent datasets tested. A possible explanation for this unintuitive finding relates to the methodology used to obtain survival prognostic groups. This involves the prediction of survival correlated genes and the concurrent clustering of the tumour samples using these genes. The clusters defined are considered as prognostic groups and a unique gene signature for each cluster is obtained. This methodology is highly dependent on clustering techniques and may be less accurate than using histopathological groups to define gene expression signatures. Other reasons include the numerous external factors that influence survival probability and do not directly relate to cancer, like the patient’s age, physical and neurologic performance, etc. Genes encoding such factors will appear highly-correlated with survival in small sample groups frequently used in microarray studies, despite having no association with cancer per se. However, such genes may have limited predictive capacity when applied to other datasets. Expression profiles of histopathology-associated genes on the other hand are directly linked to cancer and are expected to be more consistent among different patients, thus having a better predictive capacity. Although there is significant variability between different studies in specimen processing, analysis and tissue heterogeneity which is likely to affect the identification of classifier genes, our findings show that it is possible to use expression data to identify genes with predictive capacity that extents across multiple datasets.
Two genes of special interest have been selected for further analysis in this study, namely PEA15 and ADM. Tumour-suppressing functions for PEA15 have been suggested (31). PEA15 suppresses DISC-mediated caspase 8 activation, limits entry to the cell cycle and has not been previously associated with astrocytic tumour progression. Physiological levels of PEA15 expressed in cultured astrocytes are capable of restricting ERK to the cytosol, blocking ERK-dependent c-Fos transciption and cell proliferation (22). Candidate tumour suppressor genes, such as PEA, may act as major stalling points for tumour progression and perhaps the diminished expression of such genes may directly contribute to a cascade of events that lead to the progression of early grade tumours to later more malignant phenotypes. PEA15 was selected for further analysis in order to investigate its subcellular localization but also in a preliminary attempt to elucidate possible correlations between PEA15 expression and astrocytic tumour cell programmed cell death. ADM is a 52-aminoacid peptide suggested to be capable of affecting tumour growth by both direct tumour cell-related mitogenic effects and indirect vasculature-related angiogenic mechanisms(32). ADM expression in astrocytic tumours has been previously shown while its increased expression with tumour grade progression was recently suggested by Tso et al 2006 (26). ADM was selected in order to validate previous suggestions relating the peptide to regulation of angiogenesis and because very few publications commented on its exact subcellular or tissue localization.
This work presents a large, new expression profiling dataset of astrocytic tumours and employs a novel ANN-based grading of these tumours into molecular subtypes. We show that it is possible to derive transcriptome signatures from the tripartite histolopathological grading used to train the ANN-model. Moreover these signatures attain a more significant survival prognosis when compared to histopathological grading as well as tumour subtyping reports from other studies. We hope that the identification of the novel set of genes underlying this subtyping will enable tumour diagnosis to progress towards a more quantitative realm, where tumours are viewed within a malignancy spectrum that includes samples from all stages of tumour progression. We also believe that the interpretation of grading and classification efforts based on gene expression data must be performed using thorough tumour annotation on as many levels as possible. It is the integration of such work with clinical, genotypic and histopathological annotation that can maximize the value of gene expression data, increase our understanding of tumour pathology and further develop current diagnostic and therapeutic approaches.
Supplementary Material
Acknowledgements
The authors would like to thank François Renault-Mihara, INSERM, Chaire de Neuropharmacologie, Paris, France, for his most generous PEA15 antibody gift. This work was support by grants from Cancer Research UK, UK Medical Research Council, The Jacqueline Seroussi Memorial Foundation for Cancer Research, Samantha Dickson Research Trust, the Ludwig Institute for Cancer Research, the General Secretariat for Research and Technology, Hellas (project PENED 03ED842) and the EMBO Young Investigator program.
References
- 1.Kleihues P, Cavenee WK. Pathology and genetics of tumours of the nervous system. IARCPress; Lyon: 2000. [Google Scholar]
- 2.Ichimura K, Ohgaki H, Kleihues P, Collins VP. Molecular pathogenesis of astrocytic tumours. Journal of Neuro-Oncology. 2004;70:137–160. doi: 10.1007/s11060-004-2747-2. [DOI] [PubMed] [Google Scholar]
- 3.Mischel P, Cloughesy T, Nelson S. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat Rev Neurosci. 2004;10:782–92. doi: 10.1038/nrn1518. [DOI] [PubMed] [Google Scholar]
- 4.Freije WA, Castro-Vargas FE, Fang Z, et al. Gene Expression Profiling of Gliomas Strongly Predicts Survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 5.Nutt CL, Mani DR, Betensky RA, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63:1602–1607. [PubMed] [Google Scholar]
- 6.Liang Y, Diehn M, Watson N, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim S, Dougherty ER, Shmulevich I, et al. Identification of Combination Gene Sets for Glioma Classification. Mol Cancer Ther. 2002;1:1229–1236. [PubMed] [Google Scholar]
- 8.van den Boom J, Wolter M, Kuick R, et al. Characterization of Gene Expression Profiles Associated with Glioma Progression Using Oligonucleotide-Based Microarray Analysis and Real-Time Reverse Transcription-Polymerase Chain Reaction. Am J Pathol. 2003;163:1033–1043. doi: 10.1016/S0002-9440(10)63463-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shai R, Shi T, Kremen TJ, et al. Gene expression profiling identifies molecular subtypes of gliomas. Oncogene. 2003;22:4918–4923. doi: 10.1038/sj.onc.1206753. [DOI] [PubMed] [Google Scholar]
- 10.Rickman DS, Bobek MP, Misek DE, et al. Distinctive molecular profiles of high-grade and low-grade gliomas based on oligonucleotide microarray analysis. Cancer Res. 2001;61:6885–6891. [PubMed] [Google Scholar]
- 11.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Ekstrand AJ, James CD, Cavenee WK, Seliger B, Pettersson RF, Collins VP. Genes for epidermal growth factor receptor, transforming growth factor alpha, and epidermal growth factor and their expression in human gliomas in vivo. Cancer Res. 1991;51:2164–2172. [PubMed] [Google Scholar]
- 13.Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. Journal of Computational and Graphical Statistics. 1996;5:299–314. [Google Scholar]
- 14.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett T, Suzek TO, Troup DB, et al. NCBI GEO: mining millions of expression profiles--database and tools. Nucleic Acids Res. 2005;33:D562–566. doi: 10.1093/nar/gki022. Database Issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hosack D, Dennis G, Sherman B, Lane H, Lempicki R. Identifying biological themes within lists of genes with EASE. Genome Biology. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Oehler MK, Fischer DC, Orlowska-Volk M, et al. Tissue and plasma expression of the angiogenic peptide adrenomedullin in breast cancer. Br J Cancer. 2003;89:1927–1933. doi: 10.1038/sj.bjc.6601397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharif A, Renault F, Beuvon F, et al. The expression of PEA-15 (phosphoprotein enriched in astrocytes of 15 kDa) defines subpopulations of astrocytes and neurons throughout the adult mouse brain. Neuroscience. 2004;126:263–275. doi: 10.1016/j.neuroscience.2004.02.039. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan EMP. Nonparametric estimation from incomplete observations. J AmStat Assoc. 1958:457–481. [Google Scholar]
- 21.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 22.Renault F, Formstecher E, Callebaut I, Junier M-P, Chneiweiss H. The multifunctional protein PEA-15 is involved in the control of apoptosis and cell cycle in astrocytes. Biochemical Pharmacology. 2003;66:1581–1588. doi: 10.1016/s0006-2952(03)00514-8. [DOI] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burger PC, Scheithauer BW. Atlas of Tumor Pathology. Armed Forces Institute of Pathology; Washington, D.C.: 1994. [Google Scholar]
- 26.Tso CL, Freije WA, Day A, et al. Distinct transcription profiles of primary and secondary glioblastoma subgroups. Cancer Res. 2006;66:159–167. doi: 10.1158/0008-5472.CAN-05-0077. [DOI] [PubMed] [Google Scholar]
- 27.Ho DM, Hsu CY, Ting LT, Chiang H. MIB-1 and DNA topoisomerase IIa could be helpful for predicting long-term survival of patients with glioblastoma. Am J Clin Pathol. 2003;119:715–722. doi: 10.1309/UN4W-V65U-H94J-EWUV. [DOI] [PubMed] [Google Scholar]
- 28.Hsu SC, Volpert OV, Steck PA, et al. Inhibition of angiogenesis in human glioblastomas by chromosome 10 induction of thrombospondin-1. Cancer Res. 1996;56:5684–5691. [PubMed] [Google Scholar]
- 29.Osada H, Tokunaga T, Nishi M, et al. Overexpression of the neuropilin 1 (NRP1) gene correlated with poor prognosis in human glioma. Anticancer Res. 2004;24:547–552. [PubMed] [Google Scholar]
- 30.Godard S, Getz G, Delorenzi M, et al. Classification of human astrocytic gliomas on the basis of gene expression: a correlated group of genes with angiogenic activity emerges as a strong predictor of subtypes. Cancer Res. 2003;63:6613–6625. [PubMed] [Google Scholar]
- 31.Gaumont-Leclerc MF, Mukhopadhyay UK, Goumard S, Ferbeyre G. PEA-15 is inhibited by adenovirus E1A and plays a role in ERK nuclear export and Ras-induced senescence. J Biol Chem. 2004;279:46802–46809. doi: 10.1074/jbc.M403893200. [DOI] [PubMed] [Google Scholar]
- 32.Benes L, Kappus C, McGregor GP, Bertalanffy H, Mennel HD, Hagner S. The immunohistochemical expression of calcitonin receptor-like receptor (CRLR) in human gliomas. J Clin Pathol. 2004;57:172–176. doi: 10.1136/jcp.2003.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saeed A, Sharov V, White J, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 34.Ichimura K, Bolin MB, Goike HM, Schmidt EE, Moshref A, Collins VP. Deregulation of the p14ARF/MDM2/p53 pathway is a prerequisite for human astrocytic gliomas with G1-S transition control gene abnormalities. Cancer Res. 2000;60:417–424. [PubMed] [Google Scholar]
- 35.Reifenberger G, Reifenberger J, Ichimura K, Meltzer PS, Collins VP. Amplification of multiple genes from chromosomal region 12q13-14 in human malignant gliomas: preliminary mapping of the amplicons shows preferential involvement of CDK4, SAS, and MDM2. Cancer Res. 1994;54:4299–4303. [PubMed] [Google Scholar]
- 36.Ichimura K, Schmidt EE, Goike HM, Collins VP. Human glioblastomas with no alterations of the CDKN2A (p16INK4A, MTS1) and CDK4 genes have frequent mutations of the retinoblastoma gene. Oncogene. 1996;13:1065–1072. [PubMed] [Google Scholar]
- 37.Liu L, Ichimura K, Pettersson EH, Goike HM, Collins VP. The complexity of the 7p12 amplicon in human astrocytic gliomas: detailed mapping of 246 tumors. J Neuropathol Exp Neurol. 2000;59:1087–1093. doi: 10.1093/jnen/59.12.1087. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt EE, Ichimura K, Goike HM, Moshref A, Liu L, Collins VP. Mutational profile of the PTEN gene in primary human astrocytic tumors and cultivated xenografts. J Neuropathol Exp Neurol. 1999;58:1170–1183. doi: 10.1097/00005072-199911000-00007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.