Abstract
Bladder cancer is one of the most prevalent cancers worldwide. Furthermore, nonmuscle invasive bladder cancer has a 70% rate of recurrence, making it a considerable strain to the healthcare system. Patients with bladder cancer require repeat cystoscopic examinations of the bladder to monitor for tumor recurrence. The reason these patients have to undergo these costly, painful, invasive procedures is owing to the absence of accurate urine-based assays to detect the presence of bladder cancer noninvasively. Consequently, the development of a urine-based test to detect bladder cancer would be of tremendous benefit to both patients and healthcare systems. This article reports some of the more prominent urine markers in use today. In addition, the article will highlight some new technologies that are used to investigate novel urinary markers.
Keywords: bladder cancer, genomics, marker, NMP 22, proteomics, urinary cytology, urine
In 2008, a total of 68,810 Americans were diagnosed with bladder cancer and 14,100 succumbed to bladder cancer, ranking bladder cancer among the top ten causes of cancer-related deaths in both men and women in the USA [1]. Urothelial tumors can be classified into two groups based on histopathology. At presentation, more than 80% of bladder tumors are nonmuscle invasive papillary tumors (Ta or T1). The remaining 20% of tumors are muscle invasive at the time of diagnosis, which portends a much less favorable prognosis. Radical surgery is required for these invasive bladder tumors; however, the noninvasive bladder tumors can be treated more conservatively by transurethral resection (with or without adjuvant intravesical therapy). Despite therapy, more than 70% of patients with nonmuscle invasive papillary tumors (Ta or T1) confined to the mucosa will recur during the first 2 years [2]. Taking into consideration the high incidence of bladder cancer and the high recurrence rate of bladder cancer yields a prevalence of approximately 500,000 cases of bladder cancer in the USA [101]. Extensive and long-term surveillance is required to prevent progression to the invasive, lethal cancer.
The gold standard for initial clinical diagnosis and surveillance of bladder cancer involves upper tract images (e.g., CT scan of the abdomen and pelvis with contrast or intravenous pyelogram) and cystoscopic examination of the bladder together with cytologic examination for malignant cells in the urine [3,4]. Cystoscopy is an uncomfortable, invasive and costly procedure [5]. Visualized tumors will be resected and subjected to histopathological evaluation. Cystoscopy is performed every 3 months for 2 years, then every 6 months for 2 years and then every year thereafter [3]. Owing to the rigorous surveillance schema, high prevalence of bladder cancer in addition to other factors related to invasive bladder cancer, it is estimated that the total Medicare payments per patient is highest for bladder cancer compared with other cancers [6].
Although cystoscopy is the gold standard for the detection of bladder cancer, the false-negative results associated with cystoscopy can range from 10 to 40% [7]. With respect to carcinoma in situ (CIS), cystoscopy may miss up to 20% of the lesions because of its flat nature or resemblance to erythema from benign urologic conditions [8]. Furthermore, in a study assessing follow-up compliance of patients with bladder cancer, only 40% of patients adhered to all the tests recommended by current guidelines [9]. Consequently, the development of noninvasive urine-based assays using reliable diagnostic markers would be of tremendous benefit to both patients and healthcare systems. Herein, we will discuss some of the more prominent urine markers in use today, as well as highlighting some technologies used to develop and investigate new urine markers.
Urinary cytology
Urinary cytology relies on the presence of shed cancer cells in voided urine. Normal bladders and bladders with small tumors or low-grade tumors are less likely to shed cells spontaneously into the urine because of the strong intercellular attachments. As a result of the inability of low-grade tumors to shed cancer cells into the urine, sensitivity for detecting low-grade tumors range from 20 to 60%, whereas sensitivities associated with detecting high-grade tumors are reported to be over 80% [10–12]. Overall sensitivities reported for urinary cytology are in the order of 25–65% [10,12,13]. Multiple studies have demonstrated that urine cytology has a high specificity with a low false-positive interpretation [13–15]. Since not every voided urine specimen contains cancer cells, some have advocated collecting and analyzing three serial first-morning specimens for urinary cytology to improve detection rates. Thus, the serial examination in this case may reduce the sampling error associated with voided urinary cytology [16].
In accordance with accepted nomenclature, final cytologic testing results are classified by the cytopathologists into one of four categories: normal, atypical/indeterminate, suspicious or malignant [17]. Unfortunately, variability in the interpretation of cytology can lead to needless invasive testing [18]. For example, urinary tract infections or other inflammatory conditions of the bladder can also produce an incorrect result [19]. Owing to the high specificity, inexpensive equipment necessary for interpretation of urinary cytology, no patient preparation and very little time for interpretation, urinary cytology has been the cornerstone of urine-based bladder cancer assays for the past 50 years. Since urine cytology is not a laboratory test, it relies on the interpretation by a trained pathologist, specifically, cytopathologist who will assess the morphological changes within intact bladder cells. It is imperative that the sample is properly collected and immediately fixed for optimal results.
NMP 22
NMP 22 is a nuclear mitotic apparatus protein, which is present in all cells and is responsible for the distribution of chromatin to daughter cells during mitosis [20]. NMP 22 is routinely elevated up to 25-times the normal in the urine of bladder cancer patients. Furthermore, other nuclear matrix proteins have been reported to be elevated in a host of other cancers (e.g., breast and colon cancer). NMP 22 can be elevated in pyuria, urolithiasis, in the presence of foreign bodies, hematuria or cystitis [21].
Initially designed as a quantitative assay, NMP 22 has undergone a metamorphosis into an immunochromographic, qualitative point-of-care assay that requires only four drops of voided urine. After 20–50 min, positive or negative results are available. The qualitative assay, which is US FDA approved, makes it useful in the clinical setting, as its interpretation does not rely on a trained pathologist. Other advantages for NMP 22 include inexpensive assay, no patient preparation, is not dependent on intact cells and requires very little time to for interpretation [21,101].
Grossman and others investigated whether NMP 22 could enhance the detection of bladder cancer in voided urine specimens in 1331 subjects. The diagnosis of bladder cancer was based on cytoscopy with bladder biopsy. Urinary cytologies were obtained for comparison [22]. The extremely low sensitivity may be due to the lack of standardization of cytologic assessment and, thus, must be critically analyzed in other studies. NMP 22 had a sensitivity of 56% (95% CI: 4–67) whereas cytology had a sensitivity of 16% (95% CI: 7–24). Specificity of NMP 22 in this study was 86% (95% CI: 84–88) compared with 99% (95% CI: 98–100) for urinary cytology [22]. When NMP 22 was used for surveillance, NMP 22 had a sensitivity of 50% (95% CI: 40–60). Specificity of NMP 22 in this study was 87% (95% CI: 84–90) compared with 94% (95% CI: 88–98) for urinary cytology [23].
A clinical dilemma that is not unique to NMP 22 is a positive assay and negative urinary cytology, cystoscopy and imaging. These patients tend to be at high risk of developing clinical overt cancer and must be monitored closely. Although sensitivity is still not ideal and specificity is lower than urinary cytology, the NMP 22 assay is a major advancement in the identification and adoption of a urine-based tumor marker into today’s clinical practice [13,15].
BTAstat™ & BTA-TRAK™
The original bladder tumor antigen (BTA) test is a latex agglutination test that detects basement membrane degradation complexes. Two new versions of this assay, BTAstat™ and BTATRAK ™ are designed to detect complement factor H-related protein in voided urine. The complement factor H-related protein, which is produced by bladder cancer cells, has an almost identical sequence and function to human complement factor H-related protein, which is responsible for the protection of cells from complement activation [24,25]. BTAstat is FDA approved for surveillance (monitoring) of bladder cancer in conjunction with cystoscopy, but not for screening/diagnosis. BTAstat is an immunochromographic, qualitative point-of-care assay similar to NMP 22 that utilizes only five drops of urine. Anytime from 5–30 min later, positive or negative results are available. Similarly, the BTA-TRAK test is a standard ELISA test that measures human complement factor H-related protein in a quantitative fashion. This assay requires urine stabilization. The acquisition of rapid results makes BTAstat more useful in the clinical setting. In addition, the interpretation of both BTAstat and BTA-TRAK do not rely on trained personnel. Lastly, both assays are not expensive, do not require intact cells and do not require patient preparation.
BTAstat has a median sensitivity of 58% (range: 29–74%) with improved sensitivity in high-grade tumors. Median specificity is 73% (range: 56–86%), significantly lower than urological cytology. As for BTA-TRAK, median sensitivity is 71% (range: 60–83%) with improved sensitivity in high-grade tumors similar to BTAstat. Median specificity is 66% (range: 60–79%), which is similar to BTAstat, but lower than urinary cytology. In healthy individuals, specificities of BTAstat and BTA-TRAK are extremely high, however, in individuals with benign urologic conditions, false-positive results occur in over 25% of the cases, tempering the acceptance of these assays as screening tools for bladder cancer [26–31].
UroVysion™
UroVysion™ is FDA approved as a urine marker for the diagnosis of bladder cancer as well as surveillance of bladder cancer. Previously, cytogenetic studies have demonstrated aneuploidy of chromosomes 3, 7 and 17, and loss of 9p21 locus in bladder cancers [32]. A multicolored, multiprobe FISH assay has been developed by Abbott Laboratories (Abbott Park, IL, USA) that stains exfoliated cells in urine specimens for these chromosomal alterations. The criteria for detecting bladder cancer include finding more than five urinary cells with gains of more than two chromosomes, or more than ten cells with a gain of a single chromosome. However, different institutions could use different cutoff criteria to consider a sample as positive.
As an initial modality with cytoscopy to diagnose bladder cancer, UroVysion offers some compelling data. In CIS of the bladder, UroVysion sensitivity is approximately 100% and specificity ranges from 89 to 96% [33–34]. For low-grade and low-stage bladder cancer, reported sensitivity for UroVysion ranged from 36 to 65%. By contrast, the sensitivity of UroVysion in the detection of high-grade and high-stage bladder cancers ranges from 83 to 97%. Overall, the specificity of UroVysion is reported to be 70–78% [13,35]. Unfortunately, the lower specificity observed with UroVysion compared with urinary cytology has the potential to subject patients to further invasive testing and added anxiety.
The capability of UroVysion to detect recurrent disease despite negative cystoscopic finding has been well documented by multiple studies. A positive UroVysion test in the setting of a normal cystoscopic exam can predict disease recurrence in 35–63% of patients. Furthermore, if a positive UroVysion was reported prior to resection of the bladder tumor and UroVysion remains positive after complete resection, there is a 2.5–4.5 increased likelihood of imminent disease recurrence [36]. After previous bacillus Calmette–Guerin (BCG) therapy, urinary cytology may give inconclusive results, whereas UroVysion has the potential to distinguish changes associated with BCG therapy from disease recurrence s[36,37]. At this time, FISH has not been able to conclusively stratify between high versus low risk of recurrence based on the chromosomal abnormality. Limitations of wide-spread adoption of UroVysion includes increase cost compared with urinary cytology, need for a trained cytopathologist to detect the levels of polysomy of the chromosomes, expensive equipment required for the visualization of fluorescent probes and it is not a point-of-care assay.
Hyaluronic acid & hyaluronidase
Hyaluronic acid and hyaluronidase have been developed in tandem as urinary markers for the detection of bladder cancer. Hyaluronic acid is a tissue matrix component that regulates cell adhesion and proliferation. It was found to be elevated in colon, lung and breast cancers [38]. Bladder cancer cells cocultured with fibroblasts have been shown to induce production of hyaluronic acid [39]. Hyaluronic acid is an immunoassay that needs a specialized laboratory for interpretation. Preliminary results are encouraging with sensitivity and specificity of 92 and 93%, respectively. Furthermore, unlike urinary cytology, sensitivity associated with the detection of bladder cancer with hyaluronic acid is reported to be over 75% [39]. Hyaluronidase is produced by the liver and cleaves hyaluronic acid [40]. It has poor sensitivity for low-grade bladder tumors, but is more sensitive than hyaluronic acid for detecting high-grade bladder tumors [41]. Therefore, when used in combination, the two markers have sensitivity and specificity of 91 and 84%, respectively [42].
Other tumor marker assays (ImmunoCyt®, survivin, sFas, UBC, BLCA-1/4, FGFR3 mutations and methylation markers) are currently in the early phase of development.
Future advances
Recently, Lokeshwar and colleagues convened a panel to critically analyze current bladder tumor markers (Table 1) and elaborated on how to critically evaluate new biomarkers. The ideal biomarker would be a point-of-care assay that is technically simple with a low variability and a high accuracy rate. Furthermore, biomarker development must be standardized into the following four phases. Phase 1 revolves around assay development and evaluation of clinical prevalence (e.g., feasibility study). It is imperative that the prevalence and expression of the markers are examined in a target population. Phase 2 evaluates the biomarker for its clinical utility in a larger cohort and further optimizes the biomarker. The crucial objectives of this phase include refining hypotheses and defining standards for the Phase 3 studies. Phase 3 confirms the utility of the biomarker in a large, diverse cohort. The clinical utility of a given marker assay, its performance and interpretation is established in this phase. Lastly, Phase 4 involves the validation and technology transfer associated with the biomarker [13].
Table 1.
Sensitivity and specificity of urine-based biomarkers.
| Marker | Sensitivity | Range | Specificity | Range |
|---|---|---|---|---|
| Cytology | 35 | 13–75 | 94 | 85–100 |
| NMP 22 | 71 | 47–100 | 73 | 55–98 |
| BTAstat™ | 58 | 29–74 | 73 | 56–86 |
| BTA-TRAK™ | 71 | 60–83 | 66 | 60–79 |
| UroVysion™ | 79 | 70–86 | 70 | 66–93 |
| ImmunoCyt® | 67 | 52–100 | 75 | 62–82 |
| Telomerase | 39 | 29–66 | N/A | N/A |
| Microsatellite | 82 | 75–92 | 89 | 79–100 |
| Hyaluronic acid | 91 | N/A | 84 | N/A |
BTA: Bladder tumor antigen; N/A: Not available.
Modified from [16].
The concept that the presence or absence of one molecular marker will aid diagnostic or prognostic evaluation has not proved to be the case, which makes sense when one analyses the complex interactions between various molecules within a single-pathway, the cross-talk between molecular pathways, the redundancy of some pathways and the oligoclonality of many tumors. There needs to be a paradigm shift from single-marker/single-pathway research to a more global assessment of bladder cancer. To look for such a profile in bladder cancer requires high-throughput technology, such as genomics or proteomics.
Genomics
The advent of high-throughput microarray gene-expression technology has greatly enabled the search for clinically important disease biomarkers. Numerous exploratory studies have demonstrated the potential value of gene expression signatures in tumor classification [43], diagnosis [44] and in assessing the risk of postsurgical disease recurrence [45–47] in many tissue types, including bladder cancer [14–22]. To date, gene-expression profiling studies of urological samples have focused on the analysis of excised solid tumor tissue [48–56]. The number of published studies of bladder cancer using microarray expression profiling has been rather limited and only a few publications have involved clinical material. The first report assessed the expression patterns of noninvasive and invasive tumor cell suspensions prepared from 36 normal and 29 bladder tumor biopsies using oligonucleotide microarrays containing approximately 5000 genes. The study analyzed pools of cells made from normal urothelium, as well as pools of tumors of different stages [44]. Hierarchical clustering of gene-expression data grouped bladder cancer specimens based on tumor stage and separated noninvasive tumors from invasive tumors [44]. The same group then reported further advances in tumor classification and prediction of disease outcome in bladder cancer patients using the same oligonucleotide microarrays [45]. Based on the initial expression data findings, a 32-gene tumor-stage classifier was constructed through the profiling of 40 tumor tissue biopsies. Another 31 tumor tissues were profiled for delineation of recurrence-specific expression patterns, and a 26 gene-expression signature for prediction of recurrence was generated. This recurrence predictor identified 75% of the samples correctly by cross-validation; however, the prediction performance using independent test samples was not tested. The most recent study from the same group aimed at identifying a CIS-associated expression signature [46]. Hierarchical cluster analysis of profiles representing 41-biopsy specimens (13 CIS patients) was able to separate transitional cell carcinomas according to the presence or absence of CIS in the surrounding urothelium. In recent studies by one other research group, Sanchez-Carbayo and colleagues used cDNA microarrays containing 17,842 gene targets to profile tumor biopsies from 15 patients [47]. The application of hierarchical clustering and multidimensional analyses segregated invasive from noninvasive transitional carcinomas. It is worth noting that the authors found some early-stage tumors to harbor expression profiles similar to invasive carcinomas, however, no progression of the disease during the follow-up period was recorded. Profiles gleaned from solid tissue specimens are confounded, to some extent, by cellular heterogeneity, and it is not clear whether candidate biomarkers present in solid tissue will necessarily translate to utility in noninvasively obtained urine specimens. Thus, solid tissue profiling data are perhaps more likely to enhance histological evaluation of excised tissue for tumor subtype classification, treatment options and prognostication.
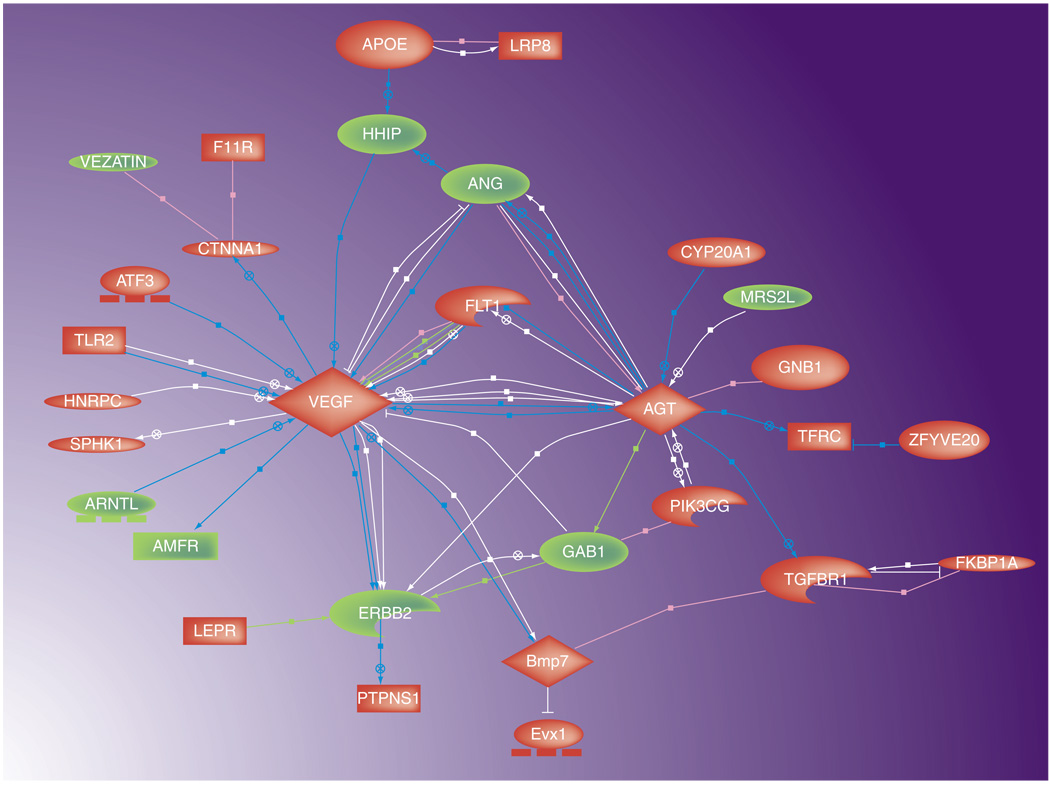
In order to progress towards the development of novel molecular assays for noninvasively obtained material, the more clinically appropriate material for profiling is the urine, and/or the surface transitional urothelia that are naturally shed into the urine. We recently reported the feasibility of evaluating the urine from healthy patients and patients with bladder cancer to demonstrate a unique gene-expression profile. After comparing the two cohorts with high-throughput transcriptome profiling on microarrays, the analysis revealed significant differences in gene expression. Specifically, 319 genes were found to have different expression levels between the two cohorts. After focusing in on the top 45 gene-expression differences, the analysis revealed connectivity between many of the genes associated with bladder disease status. Mapping these relationships showed that this connectivity is mediated through a few key factors that act as signaling hubs (Figure 1). The two major hubs, VEGF and angiotensinogen, were both upregulated in tumor cases, and are linked directly biologically, and indirectly through three minor hubs FLT1, ANG and ERBB2. Gene-expression differences between tumor and normal urothelial cell samples may implicate specific genes in malignant processes and, thus, reveal insights into tumor biology. Utilizing a selection/classification algorithm, we aimed to identify the gene signature that could most accurately diagnose the 46 cases with respect to the presence of bladder cancer. With this modeling classification approach, a 14-gene model achieved 76% overall accuracy in predicting class label during leave-one-out cross-validation (Table 2). This level of accuracy supports the feasibility that gene-expression differences can potentially be used to classify urothelial cell samples from patients of unknown clinical status [57]. The 76% accuracy of the genomic profile is comparable to other currently available assays [13,15]. Currently, this profile is being externally validated. The ultimate goal is not to use genomics as the screening/detection modality, but to acquire a better profile that can then be applied with conventional technology (i.e., ELISA or PCR). By translating the profile into a common laboratory assay, its applicability and appeal is greatly strengthened.
Figure 1. Genes with a significant change in expression taken from the urines of tumor-bearing versus urines from nontumor-bearing subjects.
Using gene-association files from the Kyoto Encyclopedia of Genes and Genomes Consortium, genes significantly regulated by urothelial carcinoma are depicted in a functional map annotator pathway profiler created by Pathway Studio. Upregulated genes are depicted in red, downregulated genes are depicted in green. Reproduced with permission from [57].
Table 2.
Genes comprising a 14-gene diagnostic model derived from the genomic profiling of urothelia. Expression patterns are described as either upregulated or downregulated in bladder cancer samples.
| Gene symbol | Function |
|---|---|
| Upregulated | |
| COX1 | Oxidation/reduction, aerobic respiration |
| AGT | Serpin peptidase inhibitor |
| WBSCR27 | Methyltransferase |
| B3GNT3 | Glycosyltransferase |
| PTPN23 | Protein tyrosine phosphatase |
| ND4 | NADH dehydrogenase (ubiquinone) activity |
| GRIPAP1 | Guanine nucleotide exchange factor for the Ras family |
| KIAA0841 | Hypothetical protein |
| LOC727916 | N/A |
| LOC100131581 | N/A |
| 243525_at* | N/A |
| Downregulated | |
| DMBT1 | Scavenger receptor, immune response |
| 237668_at* | Transmembrane glycoprotein |
| 1559057_at* | N/A |
Proteomics
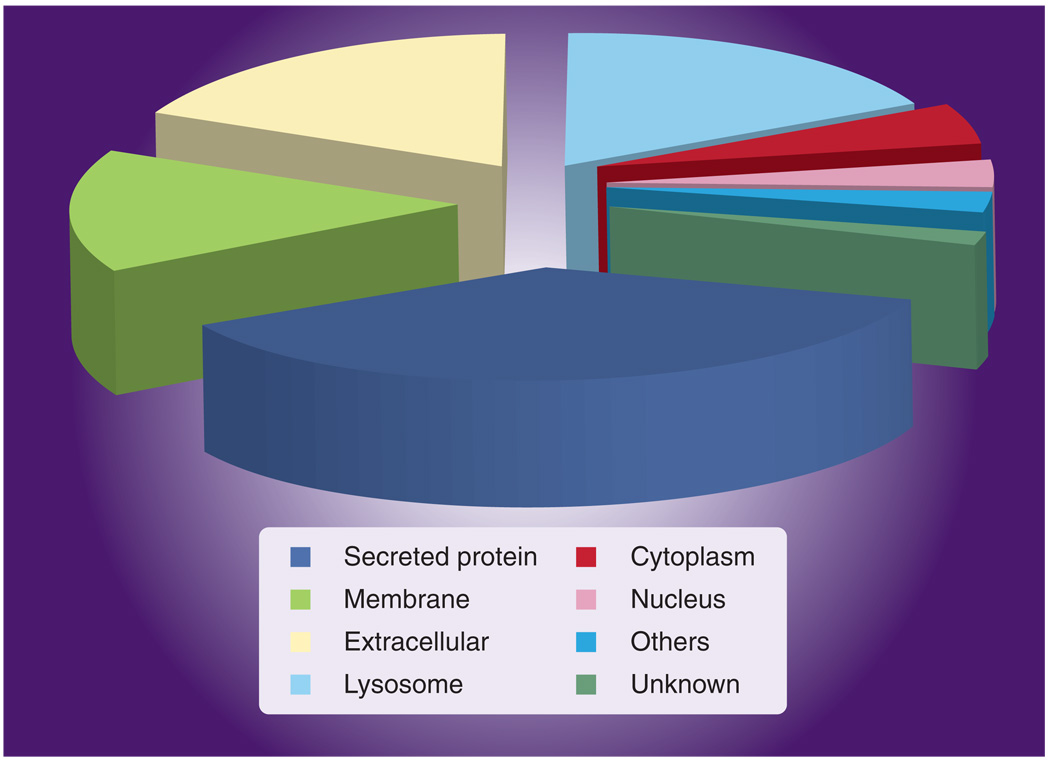
Newly developed high-throughput technology (i.e., time-of-flight mass spectroscopy, sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] with matrix-assisted laser desorption/ionization ion trap mass spectroscopy, and liquid phase 2D separation techniques) can greatly facilitate the analyses of proteins in biological specimens [58], however, only a limited number of studies, which have utilized these newer technologies, have been conducted in the analysis of urologic cancer to date. To this end, we have recently applied a novel proteomic strategy to monitor the glycoprotein component of naturally voided urine [59]. Voided urine samples taken from subjects with and without bladder cancer were subjected to concanavalin A affinity chromatography coupled to nanoflow liquid chromatography to separate a peptide mixture that was subjected to mass spectroscopy analysis. By utilizing concanavalin A, only the glycoproteins in the urine samples were investigated. A total of 186 urinary proteins were identified with approximately 40–65 proteins detected in each urine sample. The majority of glycoproteins had molecular masses within the range of 30,000–80,000 kDa, but glycoproteins as large as 300,000 were detected. Of the 186 proteins, 128 (69%) were glycoproteins as annotated by the Swiss-Prot database. The majority of identified proteins were secreted (40%), membrane-bound (18%) and extracellular (14%) (Figure 2). Zinc α2-glycoprotein and α1-microglobulin (AMBP protein) were the most common proteins in all urine samples. Table 3 ranks the identified proteins that were most discriminatory between samples of urine from subjects with and without bladder cancer. It is worth noting that α1B-glycoprotein was detected in all urine samples from subjects with bladder cancer, but was not detected in urine samples from subjects without bladder cancer [59].
Figure 2. Subcellular location of proteins identified in naturally micturated urine samples.
Reproduced with permission from [59].
Table 3.
Urinary glycoproteins detected in a bladder cancer proteomic study.
| Swiss-Prot ID | Protein name | Subcellular location | Protein class |
|---|---|---|---|
| KLK1_HUMAN‡ | Kallikrein-1 | Secreted protein | Protease |
| ATRN_HUMAN‡ | Attractin | Secreted protein | Immune system |
| ARSA_HUMAN | Arylsulfatase A | Lysosome | Enzyme |
| LAMP2_HUMAN | LAMP-2 | Membrane | Adhesion |
| LYAG_HUMAN | Lysosomal α-glucosidase | Lysosome | Glycosidase |
| UROM_HUMAN | Uromodulin | Secreted protein | Unknown |
| ITIH4_HUMAN | Inter-α-inhibitor heavy chain 4 | Extracellular | Protease inhibitor |
| CD44_HUMAN | CD44 antigen | Membrane | Adhesion |
| KNG1_HUMAN | Kininogen-1 | Secreted protein | Protease inhibitor |
| KV2A_HUMAN | Igk chain V–II region Cum | Extracellular | Immune system |
| PIGR_HUMAN | Polymeric-immunoglobulin receptor | Secreted protein | Carrier/transport protein |
| CD59_HUMAN | CD59 glycoprotein | Membrane | Inhibitor |
| KAC_HUMAN | Igκ chain C region | Extracellular | Immune system |
| LAC_HUMAN | Igλ chain C regions | Extracellular | Immune system |
| PTGDS_HUMAN | Prostaglandin-H2 D-isomerase | Secreted protein | Enzyme |
| ZA2G_HUMAN | Zinc-α-2-glycoprotein | Secreted protein | Unknown |
| ALBU_HUMAN | Serum albumin | Secreted protein | Carrier/transport protein |
| IGHA1_HUMAN | Igα-1 chain C region | Secreted protein | Immune system |
| AMBP_HUMAN | α-1-microglobulin | Secreted protein | Protease inhibitor |
| KV3A_HUMAN | Igκ chain V–III region B6 | Extracellular | Immune system |
| KV3B_HUMAN | Igκ chain V–III region SIE | Extracellular | Immune system |
| CADH1_HUMAN | Epithelial-cadherin | Membrane | Adhesion |
| APOA_HUMAN | ApoA | Extracellular | Protease |
| A1AT_HUMAN* | α-1-antitrypsin | Secreted protein | Protease inhibitor |
| TRFE_HUMAN* | Serotransferrin | Secreted protein | Carrier/transport protein |
| HPT_HUMAN* | Haptoglobin | Secreted protein | Carrier/transport protein |
| A1BG_HUMAN* | α-1B-glycoprotein | Secreted protein | Unknown |
Proteins associated with tumor-bearing cases.
Proteins associated with nontumoring-bearing cases.
Data taken from [59].
Larger, confirmatory studies are needed to further assess and develop clinically applicable tests. However, the data presented suggest that it may be possible to detect and characterize bladder cancer based upon gene-expression analysis of urothelia or proteomic profile of voided urine. Such strategies would allow for the reduction of invasive procedures, improve surveillance and provide asymptomatic screening of high-risk populations.
Future perspective
Detecting bladder cancer using diagnostic markers still remains a challenge. The need for novel markers for the detection of bladder cancer will continue in this prevalent and deadly disease. Thus, the field of urine assays for detecting bladder tumors is an expanding field. The emergence of high-throughput technology possesses the ability to usher in a new era in bladder cancer diagnosis with the detection of early, more treatable tumors causing a major paradigm shift in the management of this disease. It is hoped that these emerging marker technologies will have a profound impact on bladder cancer in the same way that serum prostate-specific antigen determination has had on prostate cancer in the last decade. However, caution must be exercised when evaluating high-throughput data since they are not validated. Although, to date, none of the noninvasive assays have replaced cystoscopy, perhaps with more robust assays (higher sensitivity, specificity, positive predictive value and negative predictive value), the frequency of cystoscopy may be reduced or even eliminated [60].
As urinary biomarkers advance, the ultimate goal is to find a noninvasive test that is equal or superior to cystoscopy. Yossepowitch et al. surveyed 200 patients undergoing surveillance cystoscopy with regard to the use of urine markers in order to detect recurrence of the disease. Their results indicated that patients would not compromise diagnostic accuracy in favor of a less invasive test or a rigorous schedule [61]. The lowest accepted sensitivity for a urine marker was 90%. At this time, there is no FDA approved urine marker that can surpass that threshold.
Executive summary
Clinical need for biomarkers in bladder cancer
As a result of its high recurrence rates, bladder cancer has one of the highest prevalence rates in the USA.
Currently, bladder cancer is detected via an invasive procedure known as cystoscopy and bladder biopsy.
Urine cytology: the gold standard of noninvasive bladder tumor marker
Urinary cytology is not a laboratory test, but a pathologic interpretation.
Sensitivity of urinary cytology is dismal.
Existing biomarkers for the detection of bladder cancer
Point-of-care assays (NMP 22 and BTAstat™) allow the diagnosis of bladder cancer to be made immediately in the physician’s office.
NMP 22, BTAstat and UroVysion™ have superior sensitivity than urinary cytology and are quickly gaining acceptance.
International consensus panel on bladder tumor markers assay development
Phase 1: feasibility studies.
Phase 2: evaluation studies of clinical utility.
Phase 3: confirmation studies.
Phase 4: validation and technology transfer as application studies.
Novel biomarkers of bladder cancer using genomics & proteomics
Genomics and proteomics promise to provide new bladder cancer markers.
Further clinical testing is required to identify and validate effective biomarkers.
Footnotes
Financial & competing interests disclosure
Flight Attendant Medical Research Institute (FAMRI) and National Cancer Institute (NCI) financed the genomics and proteomics research reported in this review. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics 2008. Cancer J. Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Millan-Rodriguez F, Chechile-Toniolo G, Salvador-Bayarri J, Palou J, Algaba F, Vicente-Rodriguez J. Primary superficial bladder cancer risk groups according to progression, mortality and recurrence. J. Urol. 2000;164:680–684. doi: 10.1016/s0022-5347(05)67280-1. [DOI] [PubMed] [Google Scholar]
- 3.Hall MC, Chang SS, Dalbagni G, et al. Guidelines for the management on nonmuscle invasive bladder cancer (stages Ta, t1, and Tis): 2007 update. J. Urol. 2007;178:2314–2330. doi: 10.1016/j.juro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 4.Montie JE, Abrahams NA, Bahnson RR, et al. Bladder cancer. Clinical guidelines in oncology. J. Natl Compr. Canc. Netw. 2006;4:984–1014. doi: 10.6004/jnccn.2006.0083. [DOI] [PubMed] [Google Scholar]
- 5.Almallah YZ, Rennie CD, Stone J, Lancashire MJ. Urinary tract infection and patient satisfaction after flexible cystoscopy and urodynamic evaluation. Urology. 2000;56:37–39. doi: 10.1016/s0090-4295(00)00555-0. [DOI] [PubMed] [Google Scholar]
- 6.Botteman MF, Pashos CL, Redaelli A, Laskin B, Hauser R. The health economics of bladder cancer. Pharmoeconomics. 2003;21:1315–1330. doi: 10.1007/BF03262330. [DOI] [PubMed] [Google Scholar]
- 7.Greene KL, Konety B. Urinary markers for bladder cancer. AUA Update. 2007;26:326–335. [Google Scholar]
- 8.Denzinger S, Burger M, Walter B, et al. Clinically relevant reduction in risk of recurrence of superficial bladder cancer using 5-aminolevulinic acid-induced fluorescence diagnosis. Urology. 2007;69:675. doi: 10.1016/j.urology.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 9.Schrag D, Hsieh LJ, Rabbani F, Bach PB, Herr H, Begg CB. Adherence to surveillance among patients with superficial bladder cancer. J. Natl Cancer Inst. 2003;95:588–597. doi: 10.1093/jnci/95.8.588. [DOI] [PubMed] [Google Scholar]
- 10.Wiener HG, Vooijs GP, Hof-Grootenboer BV. Accuracy of urinary cytology in the diagnosis of primary and recurrent bladder cancer. Acta Cytol. 1993;37:163–169. [PubMed] [Google Scholar]
- 11.Gregoire M, Fradet Y, Meyer F, et al. diagnostic accuracy of urinary cytology and deoxyribonucleic acid flow cytometry and cytology on bladder washings during follow up for bladder tumors. J. Urol. 1997;157:1660–1664. [PubMed] [Google Scholar]
- 12.Rife CC, Farrow GM, Utz DC. Urine cytology of transitional cell neoplasms. Urol. Clin. North Am. 1979;6:599–612. [PubMed] [Google Scholar]
- 13. Lokeshwar VB, Habuchi T, Grossman HB, et al. Bladder tumor marker beyond cytology: International Consensus Panel on bladder tumor markers. Urology. 2005;66:35–63. doi: 10.1016/j.urology.2005.08.064. • Comprehensive review of bladder tumor markers.
- 14.Glas AS, Roos D, Deutekom M, et al. Tumor markers in the diagnosis of primary bladder cancer: a systematic review. J. Urol. 2003;169:1975–1982. doi: 10.1097/01.ju.0000067461.30468.6d. [DOI] [PubMed] [Google Scholar]
- 15. van Rhijn BWG, van der Poel HG, van der Kwast TH. Urine markers for bladder cancer surveillance: a systematic review. Eur. Urol. 2005;47:736–748. doi: 10.1016/j.eururo.2005.03.014. • Comprehensive review of bladder tumor markers.
- 16.Cohen RA, Brown RS. Microscopic hematuria. N. Engl. J. Med. 2003;348:2330–2338. doi: 10.1056/NEJMcp012694. [DOI] [PubMed] [Google Scholar]
- 17.Murphy WM. Current status of urinary cytology in the evaluation of bladder neoplasms. Hum. Pathol. 1990;21:886–896. doi: 10.1016/0046-8177(90)90171-z. [DOI] [PubMed] [Google Scholar]
- 18.Karakiewecz PI, Benayoun S, Zippe C, et al. Institutional variability in the accuracy of urinary cytology for predicting recurrence of transitional cell carcinoma of the bladder. BJU Int. 2006;97:997. doi: 10.1111/j.1464-410X.2006.06036.x. [DOI] [PubMed] [Google Scholar]
- 19.Talwar R, Sinha T, Karan S, et al. Voided urine cytology in bladder cancer: is it time to review the indications? Urology. 2007;70:267–271. doi: 10.1016/j.urology.2007.03.070. [DOI] [PubMed] [Google Scholar]
- 20.Berezney R, Covey DS. Identification of a nuclear protein matrix. Biochem. Biophys. Res. Commun. 1974;60(4):1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 21.Ponsky LE, Sharma S, Pandrangi L, et al. Screening and monitoring for bladder cancer: refining the use of NMP22. J. Urol. 2001;166(1):75–78. [PubMed] [Google Scholar]
- 22. Grossman HB, Messing E, Soloway M, et al. Detection of bladder cancer using a point-of-care proteomic assay. JAMA. 2005;293(7):810–816. doi: 10.1001/jama.293.7.810. •• Large clinical trial illustrating the utility of NMP 22 in the detection of bladder cancer.
- 23. Grossman HB, Soloway M, Messing E, et al. Surveillance for recurrent bladder cancer using point-of-care proteomic assay. JAMA. 2006;295:299. doi: 10.1001/jama.295.3.299. •• Large clinical trial illustrating the utility of NMP 22 in the surveillance of bladder cancer.
- 24.Malkowicz SB. The application of human complement factor H-related protein (BTA TRAK) in monitoring patients with bladder cancer. Urol. Clin. North Am. 2000;27:63. doi: 10.1016/s0094-0143(05)70235-4. [DOI] [PubMed] [Google Scholar]
- 25.Heicappell R, Muller M, Fimmers R, et al. Qualitataive determination of urinary human complement factor H-related protein (hcfHrp) in patients with bladder cancer, healthy controls and patients with benign urologic disease. Urol. Int. 2000;65:181–184. doi: 10.1159/000064872. [DOI] [PubMed] [Google Scholar]
- 26.Sarosdy MF, Hudson ML, Ellis WJ, et al. Improved detection of recurrent bladder cancer using the Bard BTA stat test. Urology. 1997;50:349. doi: 10.1016/s0090-4295(97)00292-6. [DOI] [PubMed] [Google Scholar]
- 27.Nasuti JF, Gomella LG, Ismial M, et al. Utility of the BTAstat test kit for bladder cancer screening. Diagn. Cytopathol. 1999;21:27–29. doi: 10.1002/(sici)1097-0339(199907)21:1<27::aid-dc8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 28.Oge O, Atsu N, Sahin A, et al. Comparison of BTA stat and NMP 22 tests in the detection of bladder cancer. Scand. J. Urol. Nephrol. 2000;34:349. doi: 10.1080/003655900455404. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerman RL, Bagley D, Hawthorne C, et al. Utility of the Bard BTA test in detecting upper tract transitional cell carcinoma. Urology. 1998;51:956. doi: 10.1016/s0090-4295(98)00115-0. [DOI] [PubMed] [Google Scholar]
- 30.Landman J, Chang Y, Kavaler E, et al. Sensitivity and specificity of NMP-22, telomerase, and BTA in the detection of human bladder cancer. Urology. 1998;52:398. doi: 10.1016/s0090-4295(98)00219-2. [DOI] [PubMed] [Google Scholar]
- 31.Johnston B, Morales A, Emerson L, et al. Rapid detection of bladder cancer: a comparative study of point of care tests. J. Urol. 1997;158:2098. doi: 10.1016/s0022-5347(01)68166-7. [DOI] [PubMed] [Google Scholar]
- 32.Junker K, Boerner D, Schulze W, et al. Analysis of genetic alterations in normal bladder urothelium. Urology. 2003;62:1134–1138. doi: 10.1016/s0090-4295(03)00692-7. [DOI] [PubMed] [Google Scholar]
- 33.Sokolova IA, Halling KC, Jenkins RB, et al. The development of a multitarge, multicolor fluorsecnce in situ hybridization assay for the detection of urothelial cacinoma in urine. J. Mol. Diagn. 2000;2:116–123. doi: 10.1016/S1525-1578(10)60625-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halling KC, King W, Sokolova IA, et al. A comparison of cytology and fluorsecnce in situ hybridization for the detection of urothelial carcinoma. J. Urol. 2000;164:1768–1775. [PubMed] [Google Scholar]
- 35.Sarosdy MF, Schellenhammer P, Bokinsky G, et al. Clinical evaluation of multi-target fluorescent in situ hybridization assay for detection of bladder cancer. J. Urol. 2002;168:1950–1954. doi: 10.1016/S0022-5347(05)64270-X. [DOI] [PubMed] [Google Scholar]
- 36.Kipp BR, Karnes RJ, Brankley SM, et al. Monitoring intravesical therapy for superficial bladder cancer using fluorescence in situ hybridization. J. Urol. 2005;173:401. doi: 10.1097/01.ju.0000149825.83180.a4. [DOI] [PubMed] [Google Scholar]
- 37.Whitson J, Berry A, Carroll P, Konety B. A multicolor fluorescence in situ hybridization test predicts recurrence in patients with high-risk superficial bladder tumors undergoing intravesical therapy. BJU Int. 2009 doi: 10.1111/j.1464-410X.2009.08375.x. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 38.Knudson W. Tumor-associated hyaluronan. Providing an extracellular matrix that facilitates invasion. Am. J. Pathol. 1996;148:1721. [PMC free article] [PubMed] [Google Scholar]
- 39.Lokeshwar VB, Obek C, Soloway MS, et al. Tumor-associated hyaluronic acid: a new sensitive and specific urine marker for bladder cancer. Cancer Res. 1997;57:773. [PubMed] [Google Scholar]
- 40.Lokeshwar VB, Block NL. HA-HAase urine test. A sensitive and specific method for detecting bladder cancer and evaluating its grade. Urol. Clin. North Am. 2000;27:53. doi: 10.1016/s0094-0143(05)70234-2. [DOI] [PubMed] [Google Scholar]
- 41.Pham HT, Block NL, Lokeshwar VB. Tumor-derived hyaluronidase: a diagnostic urine marker for high-grade bladder cancer. Cancer Res. 1997;57:778. [PubMed] [Google Scholar]
- 42.Hautmann SH, Schroeder GL, Civantos F, et al. Hyaluronic acid and hyaluronidase. Two new bladder carcinoma markers. Urologe A. 2001;40:121. doi: 10.1007/s001200050449. [DOI] [PubMed] [Google Scholar]
- 43. Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. •• Illustrates gene-expression profiling that is necessary for gene discovery.
- 44. Stuart RO, Wachsman W, Berry CC, et al. In silico dissection of cell-type-associated patterns of gene expression in prostate cancer. Proc. Natl Acad. Sci. USA. 2004;101:615–620. doi: 10.1073/pnas.2536479100. •• Illustrates gene-expression profiling that is necessary for gene discovery in clinical samples.
- 45.LaTulippe E, Satagopan J, Smith A, et al. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]
- 46.Sun Y, Todorovic S, Goodison S. A feature selection algorithm capable of handling extremely large data dimensionality; Proceedings of the 8th SIAM International Conference on Data Mining; 24–26 April 2008; Atlanta, GA, USA. [Google Scholar]
- 47.Sun Y, Goodison S, Li J, Liu L, Farmerie W. Improved breast cancer prognosis through the combination of clinical and genetic markers. Bioinformatics. 2007;23:30–37. doi: 10.1093/bioinformatics/btl543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blaveri E, Brewer JL, Roydasgupta R, et al. Bladder cancer stage and outcome by array-based comparative genomic hybridization. Clin. Cancer Res. 2005;11:7012–7022. doi: 10.1158/1078-0432.CCR-05-0177. [DOI] [PubMed] [Google Scholar]
- 49.Blaveri E, Simko JP, Korkola JE, et al. Bladder cancer outcome and subtype classification by gene expression. Clin. Cancer Res. 2005;11:4044–4055. doi: 10.1158/1078-0432.CCR-04-2409. [DOI] [PubMed] [Google Scholar]
- 50.Dyrskjot L, Kruhoffer M, Thykjaer T, et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 51.Dyrskjot L, Thykjaer T, Kruhoffer M, et al. Identifying distinct classes of bladder carcinoma using microarrays. Nat. Genet. 2003;33:90–96. doi: 10.1038/ng1061. [DOI] [PubMed] [Google Scholar]
- 52.Dyrskjot L, Zieger K, Kruhoffer M, et al. A molecular signature in superficial bladder carcinoma predicts clinical outcome. Clin. Cancer Res. 2005;11:4029–4036. doi: 10.1158/1078-0432.CCR-04-2095. [DOI] [PubMed] [Google Scholar]
- 53.Dyrskjot L, Zieger K, Real FX, et al. Gene expression signatures predict outcome in non-muscle-invasive bladder carcinoma: a multicenter validation study. Clin. Cancer Res. 2007;13:3545–3551. doi: 10.1158/1078-0432.CCR-06-2940. [DOI] [PubMed] [Google Scholar]
- 54.Frohlich C, Albrechtsen R, Dyrskjot L, Rudkjaer L, Orntoft TF, Wewer UM. Molecular profiling of ADAM12 in human bladder cancer. Clin. Cancer Res. 2006;12:7359–7368. doi: 10.1158/1078-0432.CCR-06-1066. [DOI] [PubMed] [Google Scholar]
- 55.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J. Clin. Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez-Carbayo M, Socci ND, Lozano JJ, Haab BB, Cordon-Cardo C. Profiling bladder cancer using targeted antibody arrays. Am. J. Pathol. 2006;168:93–103. doi: 10.2353/ajpath.2006.050601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Rosser CJ, Liu L, Sun Y, et al. Bladder cancer-associated gene expression signatures identified by profiling of exfoliated urothelia. Cancer Epidemiol. Biomarkers Prev. 2009;18:444–453. doi: 10.1158/1055-9965.EPI-08-1002. •• First manuscript illustrating the ability to perform genomic profiling on human-voided urine samples.
- 58.Kreunin P, Yoo C, Urquidi V, Lubman DM, Goodison S. Differential expression of ribosomal proteins in a human metastasis model identified by coupling 2-D liquid chromatography and mass spectrometry. Cancer Genomics Proteomics. 2007;4:329–339. [PMC free article] [PubMed] [Google Scholar]
- 59. Kreunin P, Zhao J, Rosser CJ, Urquidi V, Lubman DM, Goodison S. Bladder cancer asociated glycoprotein signatures revealed by urinary proteomic profiling. J. Proteome Res. 2007;6:2631–2639. doi: 10.1021/pr0700807. •• First manuscript illustrating the ability to perform glycoprotein proteomic profiling on human-voided urine samples.
- 60.Lotan Y, Roehrborn CG. Cost–effectiveness of a modified care protocol substituting bladder tumor markers for cystoscopy for the follow-up of patients with transitional cell carcinoma of the bladder: a decision analytical approach. J. Urol. 2002;167:75–79. [PubMed] [Google Scholar]
- 61.Yossepowitch O, Herr HW, Donat SM. Use of urinary biomarkers for bladder cancer surveillance: patient perspectives. J. Urol. 2007:1277–1282. doi: 10.1016/j.juro.2006.11.066. [DOI] [PubMed] [Google Scholar]
Website
- 101.Surveillance Epidemiology and End Results Program (SEER) National Cancer Institute, cancer incidence public-use database. CD-ROM. 2001 http://seer.cancer.gov.