Abstract
In the last decade, thermal melt/thermal shift assays have become a common tool for identifying ligands and other factors that stabilize specific proteins. Increased stability is indicated by an increase in the protein's melting temperature (Tm). In optimizing the assays for subsequent screening of compound libraries, it is important to minimize the variability of Tm measurements so as to maximize the assay's ability to detect potential ligands. Here we present an investigation of Tm variability in recombinant proteins from Plasmodium parasites. Ligands of Plasmodium proteins are particularly interesting as potential starting points for drugs for malaria, and new drugs are urgently needed. A single standard buffer (100 mM HEPES, pH 7.5, 150 mM NaCl) permitted estimation of Tm for 58 of 61 Plasmodium proteins tested. However, with several proteins, Tm could not be measured with a consistency suitable for high-throughput screening unless alternative protein-specific buffers were employed. We conclude that buffer optimization to minimize variability in Tm measurements increases the success of thermal melt screens involving proteins for which a standard buffer is suboptimal.
Keywords: thermal shift assays, protein unfolding, protein stabilization, superoxide dismutase
INTRODUCTION
Determining small-molecule ligands of proteins is important in the drug discovery process, as some ligands alter protein function and thus are potential leads for new drugs. A number of new techniques for ligand detection have been developed, including surface Plasmon resonance,1 frontal affinity chromatography,2 ultrafiltration followed by LC-MS,3 and thermal melt/thermal shift techniques.4 Thermal melt assays have the advantage of being amenable to 96- and 384-well plate formats, and thus can be used to scan lots of compounds.4 A disadvantage of this approach is that it consumes multimilligram quantities of pure proteins and thus is primarily suitable for recombinantly expressed proteins.5
Thermal melt assays gradually heat a protein in order to determine its melting temperature (Tm), at which it unfolds from its natural three-dimensional configuration.6 In fluorescence-based versions of these assays, Tm is determined with a dye that fluoresces upon binding to exposed hydrophobic regions of the protein.4 This technique – also known as Differential Scanning Fluorimetry (DSF)5 and ThermoFluor7 – can be used to find small-molecule ligands that bind to and stabilize proteins, as indicated by increases in Tm.4,8–11 It can also identify other conditions (such as variations in buffer, pH, salt and glycerol concentration, types of cations, etc.) that stabilize proteins and thus make them more amenable to purification and crystallization.12–15 It is applicable to a wide variety of eukaryotic and prokaryotic proteins and yields results comparable to static light scattering (SLS) measurements of protein aggregation.4,11,16
Previous studies of factors affecting proteins' melting curves have focused almost exclusively on identifying factors that increase Tm.12–15 However, in setting up a thermal melt-based high-throughput screen (HTS) designed to identify inhibitors or other ligands of a particular protein, the absolute value of the Tm is less important than the variability of Tm measurements. A baseline Tm with a large standard deviation will make it difficult to distinguish true hits (which raise Tm above its baseline) from false positives and thus will limit the robustness of the assay (as quantified by statistics such as the Z-factor17).
We are currently using thermal melt assays to screen for ligands of Plasmodium proteins, which may lead to new anti-malaria drugs. There is a critical need for such drugs, as Plasmodium parasites have become resistant to essentially all known anti-malaria drugs.18,19 As part of the process of thermal melt screen optimization, we have investigated the variability of Tm determinations for proteins from Plasmodium parasites and the extent to which this variability can be reduced via buffer optimization. Our results indicate that, for some proteins, buffer optimization is necessary to reduce Tm variability to levels acceptable for HTS.
MATERIALS AND METHODS
Chemical compounds were purchased from Sigma-Aldrich (St. Louis, MO) with the following exceptions: glycerol, sodium acetate, sodium chloride, and sodium citrate were obtained from Fisher Scientific (Fair Lawn, NJ); iron (II) chloride, iron (III) chloride, potassium phosphate (monobasic and dibasic), and sodium phosphate (monobasic and dibasic) were obtained from Mallinckrodt Baker (Phillipsburg, NJ). Pig heart citrate synthase was from Sigma. SYPRO Orange dye was from Invitrogen (Carlsbad, CA). 96-well hard-shell PCR plates and plate seals were from Bio-Rad (Hercules, CA).
Protein cloning, purification, and purity analysis
Numerous recombinant proteins from Plasmodium berghei, P. falciparum, P. knowlesi, P. vivax, and P. yoelii were expressed and purified essentially as described previously.20,21 Some plasmids or proteins were obtained from the sources listed under Acknowledgments. In brief, proteins were produced in E. coli using a modification of the Studier system22 and purified by immobilized metal affinity chromatography (IMAC) followed by size exclusion chromatography (SEC).21 When IMAC-purified proteins were judged to be over 95% pure by SDS-PAGE and coomassie-blue staining, the SEC step was omitted. Proteins were dialyzed overnight into dialysis buffer (25 mM HEPES, pH 7.0–7.25, 5% glycerol, 500 mM NaCl, 1–2 mM DTT), concentrated to a range of 1.5 to 33 mg/mL, and flash-frozen in 20-µL aliquots as previously described.21
Estimates of protein purity were made with Quantity One software (Bio-Rad). Images of SDS-PAGE gels were analyzed to determine the fraction of a lane's total signal contained in the band representing the protein of interest. Although the software assigned an exact percentage to each band in an automated fashion, the gels were not optimized for the purpose of quantifying protein purity, so our estimates of purity should be considered semiquantitative.
Sample preparation
Samples were generally prepared in a standard buffer similar to those used in previous work11,23,24: 100 mM HEPES, pH 7.5, with 150 mM NaCl. However, during the "buffer optimization" part of the present study, numerous other buffers were also tested. These included 100 mM Bis-Tris, Bis-Tris propane, cacodylate, CHES, citrate, glycine, MES, MOPS, potassium phosphate, sodium phosphate, and Tris, with pH's ranging from 3.0 to 9.5, and including NaCl concentrations of either 0.1 or 0.5 M and glycerol concentrations of either 0% or 10%. In other experiments, various metal chlorides were added to the standard buffer, with final concentrations of 1 mM (CoCl2, FeCl2, FeCl3, and MnCl2) or 10 µM (ZnCl2).
Samples were pipetted by hand into 96-well plates (final volume: 20 µL per well). The final concentration of Plasmodium protein in all experiments was 100 µg/mL. The fluorescent dye SYPRO Orange, used for tracking protein denaturation, is sold as a stock solution of "5000X" and was added to a final concentration of "3.2X." In some experiments with adenosine deaminase from P. vivax, diethylstilbestrol was added to a final concentration of 100 µM. In control wells, the final concentration of citrate synthase was 176 µg/uL and that of oxaloacetic acid (when present) was 1 mM. 96-well plates were centrifuged for 1 minute at ~4000g just prior to each thermal melt assay to ensure that samples were at the bottom of their wells.
Fluorescence measurements and analysis
96-well plates were heated in 0.2-degree increments from 20 to 90 °C over a 59-minute period by a DNA Engine Opticon 2 (manufactured by MJ Research, now part of Bio-Rad). Fluorescence readings for each well (excitation window, 470–505 nm; absorption window, 540-700 nm) were taken after each 0.2-degree increase.
A Tm for each well was determined with the First Derivative method, which used Opticon Monitor software (Bio-Rad) to smooth the raw fluorescence data (converting each data point into a moving average of itself and adjacent points) and then finding the temperature at which the upward slope of the fluorescence-vs.-temperature curve was the steepest, i.e., the temperature at which the first derivative of this curve was maximal (Fig. 1). In some cases, a second, independent method was also used to measure Tm. The Multiparameter Fit method, implemented in Microsoft Excel, used a nonlinear least-squares fitting algorithm to estimate Tm and four other parameters from the raw fluorescence data, as previously described.4,6,10
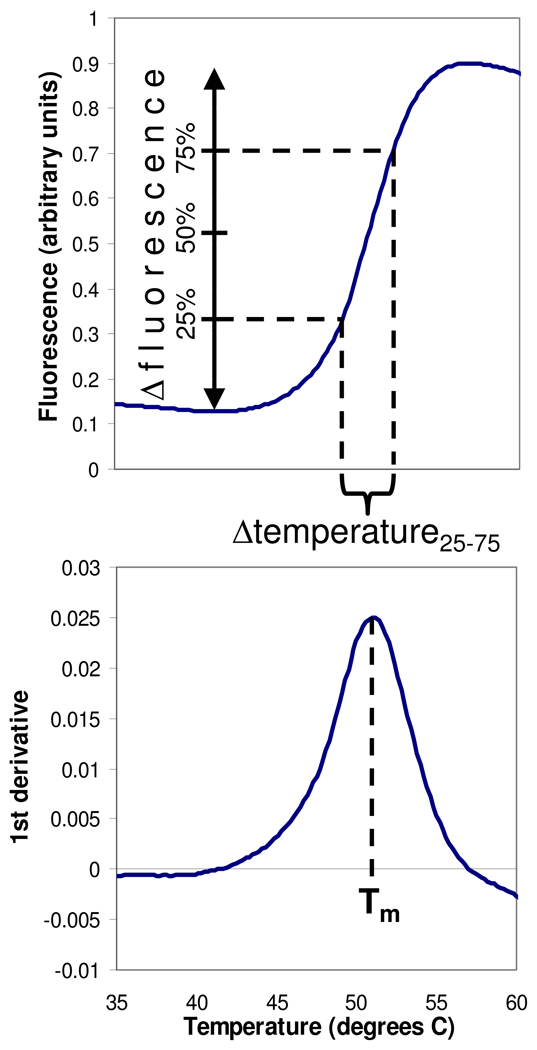
FIG. 1.
Quantitation of protein melting curves. Top panel: raw fluorescence data as a function of temperature. Bottom panel: first derivative of the raw data curve. Melting temperature (Tm) was defined as the temperature where the first derivative is maximal. Δfluorescence was defined as the difference between the minimal fluorescent signal prior to unfolding and the maximal signal. Δtemperature25–75 was defined as the temperature range spanning the middle 50% of the increase in fluorescence.
Raw fluorescence data were also used to determine the magnitude of the change in fluorescent signal due to unfolding (Δfluorescence) and the temperature range over which the unfolding occurred (Δtemperature25–75). These quantities were defined as shown in Figure 1.
For plates containing positive control wells, the Z'-factor (an indicator of assay robustness) was calculated as previously described.17
RESULTS
Preliminary testing of Plasmodium proteins
Initial thermal melt assays on 61 distinct Plasmodium proteins showed that a Tm could be estimated for 58 of them (Table 1). However, inspection of the melting curves and standard deviations suggested that the quality of the data varied greatly from protein to protein. Among the 14 most-studied proteins (5–6 plates apiece), the mean intra-plate standard deviation of the Tm exhibited a range of about 50-fold, from a low of 0.02 °C (aspartate carbamoyltransferase) to a high of 1.1 °C (6-pyruvoyltetrahydropterin synthase from P. vivax).
Table 1.
Results of thermal melt assays on 61 Plasmodium proteins. Proteins are from P. falciparum unless otherwise noted. The second column refers to identifiers assigned by PlasmoDB.org.28 The third colum uses the abbreviation of "SGC" to refer to the Structural Genomics Consortium of the University of Toronto and "UW" to refer to the University of Washington. The fourth column indicates the mean Tm, the mean intra-plate standard deviation (based on 3 wells per plate), and the number of plates run for each protein. Tm's and standard deviations are in °C. "NA" means Not Available (i.e., Tm could not be estimated from the melting curves).
| Protein | PlasmoDB ID | Source | Tm in standard buffer |
|---|---|---|---|
| 2C-methyl-D-erythritol 2,4- cyclodiphosphate synthase (P. vivax) |
PVX_003920 | SGC | 45.0; 0.2 (N=6) |
| 6-phosphogluconolactonase | PF14_0511 | UW | 50.6; 0.1 (N=4) |
| 6-pyruvoyltetrahydropterin synthase |
PFF1360w | UW | 58.2; 1.0 (N=6) |
| 6-pyruvoyltetrahydropterin synthase (P. vivax) |
PVX_114505 | UW | 51.0; 1.1 (N=5) |
| Adenosine deaminase | PF10_0289 | UW | 51.1; 0.6 (N=4) |
| Adenosine deaminase (P. vivax) |
PVX_111245 | UW | 59.8; 0.5 (N=3) |
| Adenylosuccinate lyase (P. vivax) |
PVX_114710 | SGC | 44.9; 0.1 (N=3) |
| Adenylosuccinate synthetase | PF13_0287 | UW | 51.5; 0.1 (N=3) |
| Aspartate carbamoyltransferase (P. vivax) |
PVX_083135 | UW | 70.9; 0.0 (N=5) |
| CDK-related protein kinase 6 | MAL13P1.185 | Glasgow | 44.9; 0.4 (N=4) |
| Choline kinase | PF14_0020 | UW | 45.5; 0.1 (N=4) |
| Cyclophilin | PFE1430c | UW | 52.1; 0.2 (N=4) |
| D-ribulose-5-phosphate 3- epimerase |
PFL0960w | UW | 47.5; 0.6 (N=5) |
| Dihydroorotate dehydrogenase |
PFF0160c | Harvard | 47.9; 0.4 (N=1) |
| dUTPase | PF11_0282 | UW | 48.9; 0.3 (N=2) |
| Dynein light chain 1 | PFL0660w | SGC | 64.2; 0.2 (N=3) |
| Eukaryotic translation initiation factor 5A |
PFL0210c | UW | 53.1; 0.5 (N=4) |
| Farnesyl pyrophosphate synthase (P. vivax) |
PVX_092040 | SGC | 46.5; 0.3 (N=4) |
| Glutamate dehydrogenase, NADP-specific |
PF14_0164 | UW | 69.1; 0.1 (N=3) |
| Glyceraldehyde-3-phosphate dehydrogenase |
PF14_0598 | UW | 49.9; 0.3 (N=3) |
| Glycerol-3-phosphate dehydrogenase |
PFL0780w | UW | 53.9; 0.6 (N=5) |
| Glycogen synthase kinase 3 | PFC0525c | UW | 47.5; 0.2 (N=3) |
| Glycophorin binding protein | PF14_0010 | UW | NA (N=2) |
| Guanylate kinase (P. vivax) | PVX_099895 | UW | 47.2; 0.1 (N=5) |
| HSP90 | PF11_0188 | Harvard | NA (N=2) |
| Hypothetical protein, conserved |
MAL13P1.257 | UW | 54.2; 0.1 (N=2) |
| Hypothetical protein, conserved |
PFF0880c | UW | 44.6; 0.2 (N=3) |
| Methionine adenosyltransferase |
PFI1090w | UW | 44.7; 0.4 (N=4) |
| Methionine aminopeptidase 1 | PF10_0150 | UW | 43.3; 0.1 (N=5) |
| Methionine aminopeptidase 2 | PF14_0327 | UW | NA (N=1) |
| N-myristoyltransferase | PF14_0127 | UW | 59.3; 0.2 (N=5) |
| Nucleoside diphosphate kinase B |
PF13_0349 | UW | 61.8; 0.2 (N=3) |
| Nucleosome assembly protein 1 |
PKH_130240 | UW | 55.3; 0.1 (N=5) |
| Ornithine aminotransferase (P. yoelii) |
PY00104 | SGC | 47.0; 0.2 (N=3) |
| Orotidine 5' monophosphate decarboxylase |
PF10_0225 | UW | 65.6; 0.1 (N=3) |
| Peptidase | PF14_0517 | UW | 49.8; 0.5 (N=3) |
| Phosphatidylethanolamine- binding protein (P. vivax) |
PVX_123630 | UW | 42.3; 0.4 (N=6) |
| Phosphoethanolamine N- methyltransferase |
MAL13P1.214 | UW | 52.4; 0.6 (N=3) |
| Phosphoglycerate kinase | PFI1105w | UW | 45.1; 0.1 (N=3) |
| Phosphoglycerate mutase | PF11_0208 | UW | 57.0; 0.0 (N=3) |
| Phosphomethylpyrimidine kinase |
PFE1030c | UW | 62.6; 0.2 (N=3) |
| Protein-L-isoaspartate O- methyltransferase |
PF14_0309 | SGC | 48.8; 0.2 (N=3) |
| Rab11a GTPase | PF13_0119 | SGC | 41.5; 0.5 (N=4) |
| Rab18 GTPase | PF08_0110 | UW | 44.0; 0.2 (N=4) |
| Ribonucleotide reductase, small subunit (P. vivax) |
PVX_086155 | SGC | 46.3; 0.8 (N=3) |
| Ribose 5-phosphate isomerase |
PFE0730c | UW | 55.5; 0.4 (N=5) |
| Ribosomal RNA methyltransferase |
PF13_0052 | SGC | 38.1; 0.2 (N=4) |
| S-adenosyl-homocysteine hydrolase |
PFE1050w | UW | 43.8; 0.3 (N=4) |
| Spermidine synthase | PF11_0301 | SGC | 44.3; 0.1 (N=4) |
| Superoxide dismutase (P. berghei) |
PB000490.02.0 | UW | 58.6; 0.3 (N=5) |
| Superoxide dismutase (P. knowlesi) |
PKH_142350 | SGC | 60.4; 0.5 (N=3) |
| Thioredoxin | PF14_0545 | UW | 74.9; 0.4 (N=3) |
| Thioredoxin reductase | PFI1170c | UW | 50.2; 0.0 (N=3) |
| Tryptophan tRNA ligase | PF13_0205 | UW | 46.6; 0.1 (N=4) |
| Ubiquitin carboxyl-terminal hydrolase |
PF14_0576 | UW | 49.4; 1.1 (N=2) |
| Ubiquitin conjugating enzyme | PF10_0330 | UW | 52.0; 1.0 (N=5) |
| Ubiquitin conjugating enzyme | PFE1350c | SGC | 53.1; 0.1 (N=3) |
| Ubiquitin conjugating enzyme | PF13_0301 | UW | 55.7; 0.3 (N=2) |
| Ubiquitin conjugating enzyme | PF08_0085 | UW | 52.4; 0.2 (N=2) |
| Ubiquitin conjugating enzyme | PFC0255c | SGC | 51.0; 0.1 (N=4) |
| Uridine phosphorylase | PFE0660c | UW | 57.2; 0.1 (N=4) |
Why do Plasmodium proteins differ in Tm variability?
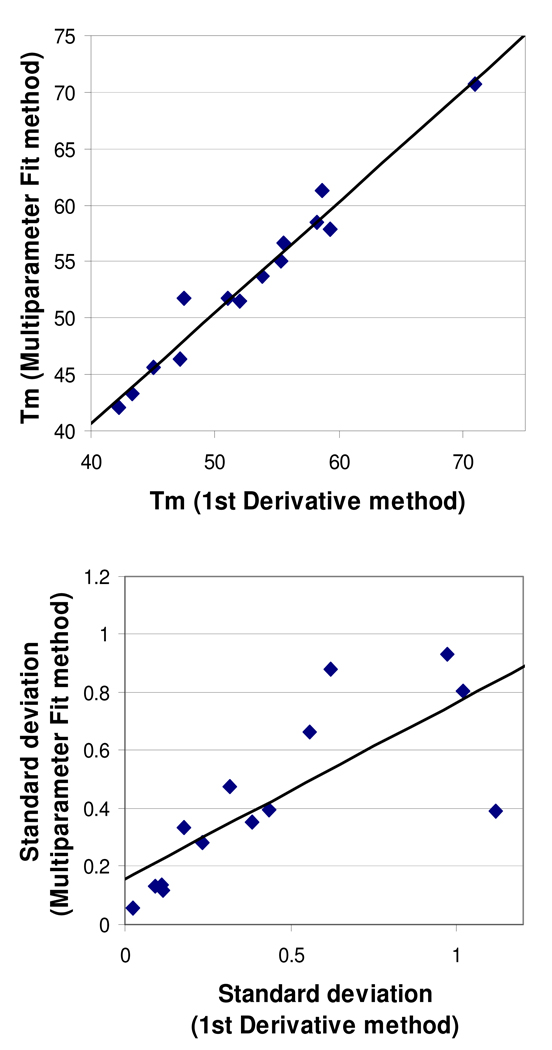
The large range in Tm variability could, in theory, reflect the limitations of our First Derivative method for measuring Tm, which differs from the Multiparameter Fit method used by most other investigators. However, when Tm's and standard deviations were determined by both methods and compared, the agreement of the two methods was quite respectable (Fig. 2).
FIG. 2.
Comparison of the First Derivative and Multiparameter Fit methods of determining Tm. Each data point represents one of the 14 most-studied proteins (5–6 plates per protein). Top panel: mean Tm values (in °C) estimated by each method. Equation: y = 0.98x + 1.40; R2 = 0.97. Bottom panel: mean standard deviations in Tm (in °C) estimated by each method. Equation: y = 0.61x + 0.16; R2 = 0.60.
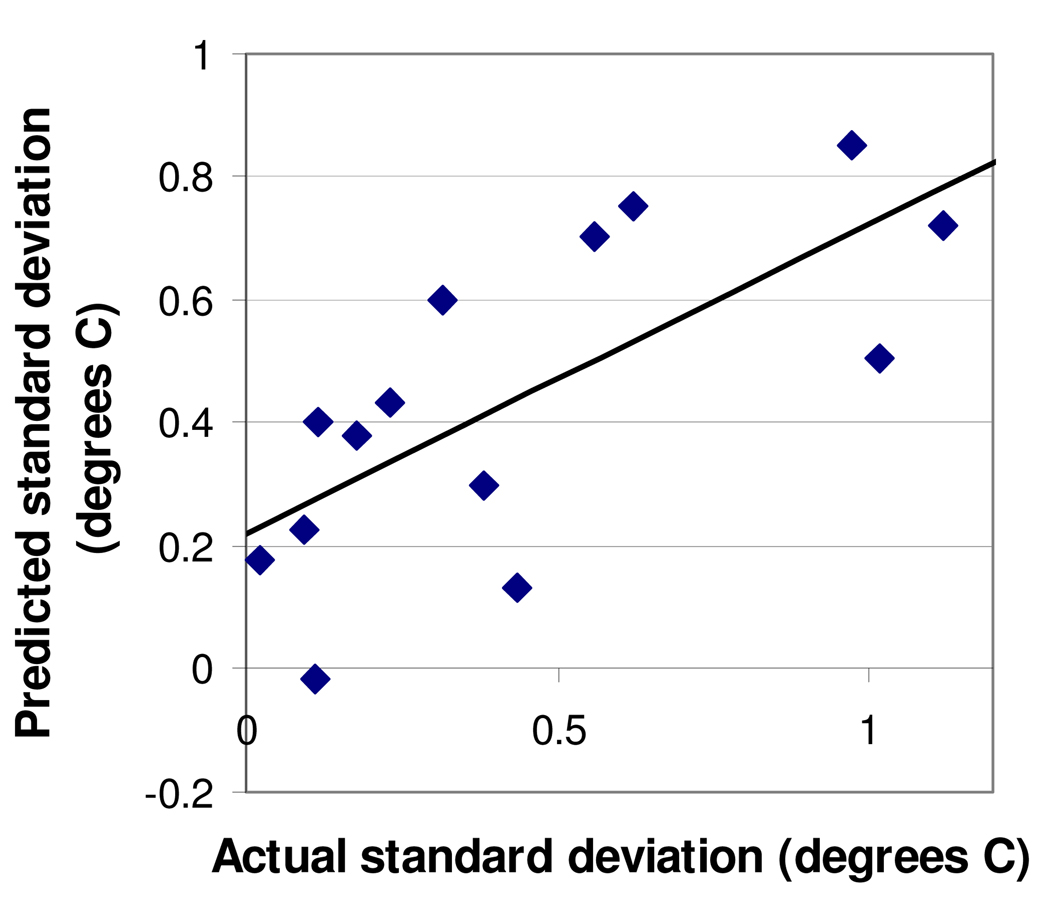
The proteins' differences in Tm variability might also simply represent occasional inconsistency on the part of the experimenter and/or the DNA Engine Opticon 2. However, two lines of evidence argue against this interpretation. First, control wells of pig heart citrate synthase + oxaloacetic acid, included on the plates of all 14 of the most-studied Plasmodium proteins, had a mean standard deviation of less than 0.45 °C for all 14, suggesting that standard deviations well above 0.45 °C are due to the specific protein being tested rather than experimenter or machine error. Second, the standard deviations in Tm were not random; rather, they were related to the proteins' Δfluorescence and Δtemperature25–75 (Fig. 3), with high standard deviations tending to coincide with low Δfluorescence and/or high Δtemperature25–75 values. This too suggests that some proteins are inherently more variable than others.
FIG. 3.
Prediction of proteins' standard deviations in Tm based upon Δfluorescence and Δtemperature. Predicted standard deviation (the Y axis) was calculated as 0.046(Δtemperature) minus 0.40(Δfluorescence) plus 0.60, a "best-fit" equation that minimized the difference between predicted and actual standard deviations. The R2 value for the predicted-vs.-actual graph is 0.50. Each data point represents one of the 14 most-studied proteins (5–6 plates per protein).
Why then do some proteins reproducibly show higher-than-normal variability in unfolding? One possibility is that unfolding relates to protein size; perhaps larger, multidomain, and/or multimeric proteins melt in a less uniform manner. However, there was no significant correlation between standard deviations in Tm and proteins' molecular weights (data not shown). Moreover, for proteins for which a crystal structure (of the protein itself or a close homolog) is available in the Protein Data Bank25, there was no correlation between the standard deviation in Tm and the number of domains or number of polypeptides in the protein (data not shown).
It is also possible that Tm variability relates to the hydrophobicity of the proteins, since the thermal melt method depends on the binding of dye to hydrophobic regions. However, proteins' Grand Average of Hydropathy (GRAVY)26 did not correlate with their standard deviations in Tm (data not shown).
Yet another possibility is that Tm variability reflects protein purity; perhaps less pure proteins unfold in a less consistent manner. When purity was estimated by SDS-PAGE and Coomassie staining, there was no obvious relationship between percent purity and standard deviation in Tm (data not shown).
Buffer optimization can improve Tm variability
Still another potential source of variability might be variations in the stability of the proteins in our standard buffer (100 mM HEPES, pH 7.5, 150 mM NaCl), which is similar to that used in many protein-heating studies.11,23,24 Therefore we asked whether Tm variability could be improved by the use of alternative buffers. A first step in this direction was to add specific metal cations to the standard buffer for proteins thought to bind these cations. Although most additions did not reduce Tm variability, the presence of 1 mM MnCl2 decreased the Δtemperature25–75 and Tm standard deviation of the P. berghei and P. knowlesi superoxide dismutases by ≥50% (Table 2).
Table 2.
Improvements in Tm variability made through buffer optimization. Proteins are from P. falciparum unless otherwise noted. Results in the fourth column are based on comparisons of the standard buffer (100 mM HEPES, pH 7.5, 150 mM NaCl) and optimal buffers side-by-side on the same plate (8–16 wells per buffer treatment per plate). "N" in the fourth column is the number of plates run for that protein.
| Protein | PlasmoDB ID | Optimal buffer | Improvement vs. standard buffer |
|---|---|---|---|
| 6-pyruvoyltetrahydropterin synthase |
PFF1360w | 100 mM citric acid, pH 5.5, 100 mM NaCl |
Decreased mean standard deviation in Tm from 2.7 to 0.4 °C (N=6). |
| Adenosine deaminase | PF10_0289 | 100 mM Bis-Tris, pH 6.0, 100 mM NaCl, 10% Glycerol |
Increased mean Δfluorescence from 0.05 to 1.73 (arbitrary units), permitting Tm's to be measured in all wells (N=3). Some wells were uninterpretable when using standard buffer. |
| Adenosine deaminase (P. vivax) |
PVX_111245 | 100 mM potassium phosphate, pH 7.0, 500 mM NaCl, 10% Glycerol |
Decreased mean Δtemperature25–75 from 8.9 to 3.9 °C and decreased mean standard deviation in Tm from 1.1 to 0.3 degrees (N=9). Increased mean Z'-factor from −0.10 to 0.47 (N=6). |
| Phosphoethanolamine N-methyltransferase |
MAL13P1.214 | 100 mM MOPS, pH 7.0, 100 mM NaCl |
Increased mean Δfluorescence from 0.07 to 0.23 (arbitrary units), permitting Tm's to be measured in all wells (N=3). Some wells were uninterpretable when using standard buffer.. |
| Superoxide dismutase (P. berghei) |
PB000490.02.0 | Standard buffer + 1 mM MnCl2 |
Decreased mean Δtemperature25–75 from 8.8 to 4.6 °C and decreased mean standard deviation in Tm from 1.0 to 0.3 °C (N=10). Increased mean Z'-factor from 0.39 to 0.75 (N=6). |
| Superoxide dismutase (P. knowlesi) |
PKH_142350 | Standard buffer + 1 mM MnCl2 |
Decreased mean Δtemperature25–75 from 11.4 to 5.5 °C and decreased mean standard deviation in Tm from 3.1 to 0.4 °C (N=7). Increased mean Z'-factor from −0.61 to 0.68 (N=6). |
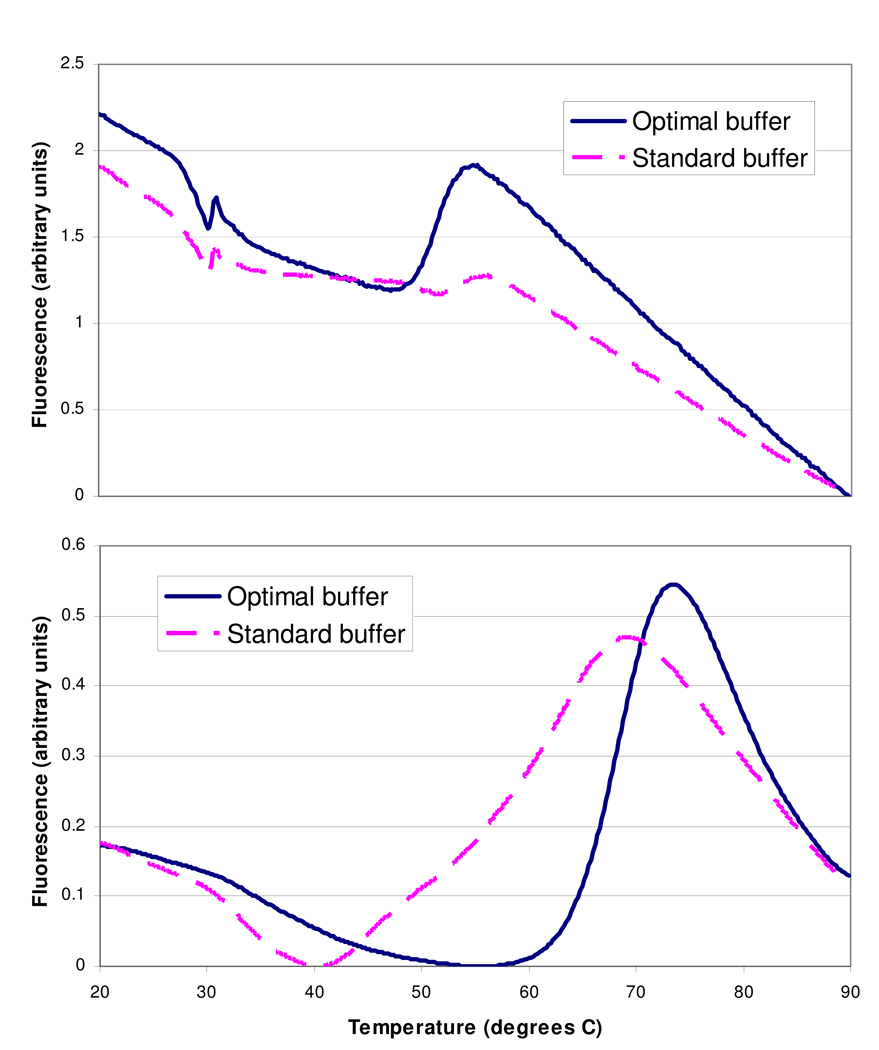
As a second step of buffer optimization, selected proteins with relatively high Tm variability were tested in a variety of buffer solutions differing in pH and NaCl and glycerol concentration. For a few of these proteins, switching buffers led to substantial improvements in melting curves (Table 2 and Fig. 4).
FIG. 4.
Examples of improvements in melting curves made through buffer optimization. Data shown are unsmoothed fluorescence data from representative wells. Top panel: phosphoethanolamine N-methyltransferase. Note that the optimal buffer increases the Δfluorescence at around 50–55 degrees from an almost-uninterpretable blip to a much clearer indication of the protein unfolding. Bottom panel: adenosine deaminase from P. vivax. Note the extremely broad temperature range over which the protein unfolds in the standard buffer, making it difficult to assign a precise Tm.
Buffer optimization can improve Z'-factors
Decreases in Tm variability, while presumably desirable, do not in and of themselves prove that one buffer is superior to another when conducting high-throughput screens for ligands of proteins. A buffer is useful for screening purposes only if, in addition to minimizing Tm variability, it allows "hit" compounds to raise Tm above the baseline. Therefore, as a final test of whether buffer optimization is useful for thermal melt screening, we ran buffer comparison experiments that included positive controls (i.e., Tm-raising hits). These experiments consisted of 96-well plates divided into wells with the standard buffer and wells with the optimal buffer; each of these sets of wells was further subdivided into wells with and without positive controls (i.e., Tm-raising hits).
For adenosine deaminase from P. vivax, the ligand diethylstilbestrol was used as a Tm-raising positive control. In six separate experiments over five days, a mean Z'-factor of 0.47 was achieved with the optimal buffer (100 mM potassium phosphate, pH 7.0, 500 mM NaCl, 10% glycerol). This is consistent with NIH guidelines for assay validation27, which state in part that Z'-factors should consistently exceed 0.4 over several days. In contrast, the mean Z'-factor with the standard buffer was −0.10.
In setting up similar experiments with superoxide dismutase (SOD), no inexpensive Tm-raising ligands could be found, so the condition of a lower pH (6.5) was used as a positive control. This raises the Tm several degrees above what it is at pH 7.5. A series of six experiments on the P. knowlesi SOD over four days yielded a mean Z'-factor of 0.68 with the optimal buffer – acceptable according to NIH guidelines – whereas the mean was -0.61 with the standard buffer. A similar but less dramatic difference was seen when the same two buffers were applied to the P. berghei SOD: the mean Z'-factors were 0.75 with the optimal buffer and 0.39 with the standard buffer.
DISCUSSION
This study constitutes, to our knowledge, the most comprehensive look to date at the applicability of thermal melt assays to a large number of proteins from a single taxonomic genus. For 58 of 61 Plasmodium proteins, we could detect an increase in fluorescence sufficient for estimation of a melting temperature (Tm). This ~95% success rate compares favorably to the data of Vedadi et al.11, who reported successful determination of Tm's for 44 of 61 human and parasite proteins studied, including 16 of 26 Plasmodium proteins. At least four proteins were common to both studies. The control protein of citrate synthase from pig heart has a Tm of 52.4–53.0 °C according to Vedadi et al. and 53.0 °C according to the present study, constituting excellent agreement. Dynein light chain 1 (PFL0660w) has a Tm of 65.8 °C according to the previous study and 64.2 °C according to the present one, also indicating acceptable consistency. The small subunit of ribonucleotide reductase (PVX_086155) was flagged by Vedadi et al. as having a high initial fluorescence in thermal melt assays, so no Tm was reported, but these researchers used static light scattering (SLS) to measure a temperature of aggregation (Tagg). Tagg data generally correlate well with Tm data, both for baseline measurements and for ligand-induced shifts in temperature;11 in this case, our estimated Tm (46.3 °C) is within the usual ±4-degree window of the previously reported Tagg (42.7 °C). Finally, Vedadi et al. did not report a Tm or Tagg for the P. knowlesi superoxide dismutase (PKH_142350) but noted that it was stabilized (as indicated by SLS) by MnCl2. Likewise, we observed a MnCl2-induced rise in the Tm of this enzyme (data not shown).
The primary question of this study was whether buffers other than the standard buffer (100 mM HEPES, pH 7.5, 150 mM NaCl) could be used to improve the quality of thermal melt data. We found that, for several proteins, an alternative buffer increased the magnitude of the fluorescent signal due to unfolding (Δfluorescence) and/or decreased the temperature range over which the unfolding occurred (Δtemperature25–75), and that these improvements tended to be associated with reduced Tm variability. Reduced Tm variability, in turn, improved assay robustness (as quantified with the Z'-factor), suggesting that, for certain proteins, selection of an appropriate protein-specific buffer will improve the success of thermal melt screens.
One example of improving melting curves through buffer optimization was the addition of Mn2+ to the standard buffer, which reduced Tm variability for superoxide dismutases (SODs) from P. berghei (PB000490.02.0) and P. knowlesi (PKH_142350). This effect was not seen with Fe2+ (data not shown) – a somewhat unexpected finding, since both proteins are annotated as Fesuperoxide dismutases rather than Mn-superoxide dismutases28 and are 90–92% identical to a P. falciparum SOD previously found to be iron-dependent.29 Furthermore, all three Plasmodium SODs are predicted by the SODa webtool30 to use iron, not manganese, as a cofactor. On the other hand, the P. berghei SOD has been reported as binding manganese rather than iron (structure 2A03 in the Protein Data Bank25). Thus the physiological cofactor of these SODs has not yet been established with 100% certainty.
While Mn2+ improved Tm variability in both SODs studied, other pairs of Plasmodium proteins showed divergent responses to buffer optimization. The adenosine deaminases from P. falciparum and P. vivax had different buffer preferences (Table 2); in addition, Tm variability of the P. falciparum 6-pyruvoyltetrahydropterin synthase was reduced by a buffer (100 mM citric acid, pH 5.5, 100 mM NaCl) that did not have a comparable effect on the P. vivax 6-pyruvoyltetrahydropterin synthase (data not shown). These differences, along with the 8–9 °C differences in Tm between the pairs (Table 1), underscore the limitations of extrapolating from one protein to an ortholog within the same genus.
Although Tm variability was found to be buffer-dependent, it did not appear closely related to protein traits such as purity, molecular weight, number of domains and polypeptides, and overall hydrophobicity. Future research may provide a more complete explanation of why some proteins yield better melting curves than others.
Another interesting issue not directly addressed by the present study is the influence of buffer selection on proteins' ligand-binding behavior. If a given buffer is optimal for minimizing Tm variability because it maintains a protein in a state close to its natural configuration in vivo, the buffer might also cause the protein's affinities for its ligands to more closely approximate affinities in vivo. Such speculation awaits further experimentation.
ACKNOWLEDGMENTS
We thank Vishal Patel and Jon Clardy (Harvard University) for supplying the dihydroorotate dehydrogenase and HSP90 used in this study, and Sylvain Eschenlauer and Christian Doerig (University of Glasgow) for supplying the CDK-related protein kinase 6. We also acknowledge Angela Kelley, Eric Larson, Ethan Merritt, Kayode K. Ojo, and Frank Zucker (University of Washington) and Martin Kramer (Genzyme) for helpful ideas and advice. Funding for this project came from the Medicines for Malaria Venture (MVV), the Medical Structural Genomics for Pathogenic Protozoa (MSGPP) program project (P01AI067921 from NIAID), the NIH (GM64655 to WCVV), and the UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases (05–00508).
Contributor Information
Alberto J. Napuli, Email: napuli@u.washington.edu.
Andrew P. Thomas, Email: androothomas@gmail.com.
Diana J. Chung, Email: diana.j.chung@gmail.com.
Kuzma V. Kovzun, Email: kuzmak@u.washington.edu.
David J. Leibly, Email: leiblydj@u.washington.edu.
Lisa J. Castaneda, Email: ljj@u.washington.edu.
Janhavi Bhandari, Email: janhavi@u.washington.edu.
Christopher J. Damman, Email: cjdamman@u.washington.edu.
Raymond Hui, Email: raymond.hui@utoronto.ca.
Wim G. J. Hol, Email: wghol@u.washington.edu.
Frederick S. Buckner, Email: fbuckner@u.washington.edu.
Christophe L. M. J. Verlinde, Email: verlinde@u.washington.edu.
Zhongsheng Zhang, Email: zhzhang@u.washington.edu.
Erkang Fan, Email: erkang@u.washington.edu.
REFERENCES
- 1.Neumann T, Junker HD, Schmidt K, Sekul R. SPR-based fragment screening: advantages and applications. Curr Top Med Chem. 2007;7:1630–1642. doi: 10.2174/156802607782341073. [DOI] [PubMed] [Google Scholar]
- 2.Slon-Usakiewicz JJ, Ng W, Dai JR, Pasternak A, Redden PR. Frontal affinity chromatography with MS detection (FAC-MS) in drug discovery. Drug Discov Today. 2005;10:409–416. doi: 10.1016/S1359-6446(04)03360-4. [DOI] [PubMed] [Google Scholar]
- 3.Kamchonwongpaisan S, Vanichtanankul J, Tarnchompoo B, Yuvaniyama J, Taweechai S, Yuthavong Y. Stoichiometric selection of tight-binding inhibitors by wild-type and mutant forms of malarial (Plasmodium falciparum) dihydrofolate reductase. Anal Chem. 2005;77:1222–1227. doi: 10.1021/ac0487597. [DOI] [PubMed] [Google Scholar]
- 4.Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. doi: 10.1177/108705710100600609. [DOI] [PubMed] [Google Scholar]
- 5.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 6.Brandts JF, Lin LN. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry. 1990;29:6927–6940. doi: 10.1021/bi00481a024. [DOI] [PubMed] [Google Scholar]
- 7.Cummings MD, Farnum MA, Nelen MI. Universal screening methods and applications of ThermoFluor. J Biomol Screen. 2006;11:854–863. doi: 10.1177/1087057106292746. [DOI] [PubMed] [Google Scholar]
- 8.Carver TE, Bordeau B, Cummings MD, Petrella EC, Pucci MJ, Zawadzke LE, et al. Decrypting the biochemical function of an essential gene from Streptococcus pneumoniae using ThermoFluor technology. J Biol Chem. 2005;280:11704–11712. doi: 10.1074/jbc.M413278200. [DOI] [PubMed] [Google Scholar]
- 9.Giuliani SE, Frank AM, Collart FR. Functional Assignment of Solute-Binding Proteins of ABC Transporters Using a Fluorescence-Based Thermal Shift Assay. Biochemistry. 2008;47:13974–13984. doi: 10.1021/bi801648r. [DOI] [PubMed] [Google Scholar]
- 10.Lo MC, Aulabaugh A, Jin G, Cowling R, Bard J, Malamas M, et al. Evaluation of fluorescence-based thermal shift assays for hit identification in drug discovery. Anal Biochem. 2004;332:153–159. doi: 10.1016/j.ab.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 11.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Jr, Wasney GA, et al. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci U S A. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 13.Kean J, Cleverley RM, O'Ryan L, Ford RC, Prince SM, Derrick JP. Characterization of a CorA Mg2+ transport channel from Methanococcus jannaschii using a Thermofluor-based stability assay. Mol Membr Biol. 2008;25:653–663. doi: 10.1080/09687680802541169. [DOI] [PubMed] [Google Scholar]
- 14.Mezzasalma TM, Kranz JK, Chan W, Struble GT, Schalk-Hihi C, Deckman IC, et al. Enhancing recombinant protein quality and yield by protein stability profiling. J Biomol Screen. 2007;12:418–428. doi: 10.1177/1087057106297984. [DOI] [PubMed] [Google Scholar]
- 15.Richard AJ, Liu CC, Klinger AL, Todd MJ, Mezzasalma TM, LiCata VJ. Thermal stability landscape for Klenow DNA polymerase as a function of pH and salt concentration. Biochim Biophys Acta. 2006;1764:1546–1552. doi: 10.1016/j.bbapap.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 16.Senisterra GA, Markin E, Yamazaki K, Hui R, Vedadi M, Awrey DE. Screening for ligands using a generic and high-throughput light-scattering-based assay. J Biomol Screen. 2006;11:940–948. doi: 10.1177/1087057106294699. [DOI] [PubMed] [Google Scholar]
- 17.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 18.Plowe CV. The evolution of drug-resistant malaria. Trans R Soc Trop Med Hyg. 2008 doi: 10.1016/j.trstmh.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D. Fukuda MM: Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 20.Vedadi M, Lew J, Artz J, Amani M, Zhao Y, Dong A, et al. Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms. Mol Biochem Parasitol. 2007;151:100–110. doi: 10.1016/j.molbiopara.2006.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Mehlin C, Boni E, Buckner FS, Engel L, Feist T, Gelb MH, et al. Heterologous expression of proteins from Plasmodium falciparum: results from 1000 genes. Mol Biochem Parasitol. 2006;148:144–160. doi: 10.1016/j.molbiopara.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 22.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Senisterra GA, Soo Hong B, Park HW, Vedadi M. Application of high-throughput isothermal denaturation to assess protein stability and screen for ligands. J Biomol Screen. 2008;13:337–342. doi: 10.1177/1087057108317825. [DOI] [PubMed] [Google Scholar]
- 24.Lorca GL, Ezersky A, Lunin VV, Walker JR, Altamentova S, Evdokimova E, et al. Glyoxylate and pyruvate are antagonistic effectors of the Escherichia coli IclR transcriptional regulator. J Biol Chem. 2007;282:16476–16491. doi: 10.1074/jbc.M610838200. [DOI] [PubMed] [Google Scholar]
- 25.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 27.Eli Lilly and Company and NIH Chemical Genomics Center. [last accessed February 17, 2009];Assay Guidance Manual Version 5.0. 2008 Available online at: http://www.ncgc.nih.gov/guidance/manual_toc.html.
- 28.Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, et al. PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 2009;37:D539–D543. doi: 10.1093/nar/gkn814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gratepanche S, Menage S, Touati D, Wintjens R, Delplace P, Fontecave M, et al. Biochemical and electron paramagnetic resonance study of the iron superoxide dismutase from Plasmodium falciparum. Mol Biochem Parasitol. 2002;120:237–246. doi: 10.1016/s0166-6851(02)00004-x. [DOI] [PubMed] [Google Scholar]
- 30.Kwasigroch JM, Wintjens R, Gilis D, Rooman M. SODa: an Mn/Fe superoxide dismutase prediction and design server. BMC Bioinformatics. 2008;9:257. doi: 10.1186/1471-2105-9-257. [DOI] [PMC free article] [PubMed] [Google Scholar]