Abstract
Purpose
SAG (Sensitive to Apoptosis Gene, also known as RBX2 or ROC2) was originally cloned as a redox inducible antioxidant protein and was later characterized as a RING component of SCF E3 ubiquitin ligases. SAG overexpression inhibits apoptosis induced by many stimuli both in vitro and in vivo. SAG mRNA was over-expressed in human lung tumor tissues with the correlation to a poor patient survival. To investigate whether SAG serves as an anticancer target, we determined the effect of SAG silencing on cell proliferation, survival and radiosensitivity.
Experimental Design
SAG protein expression in human tumors was evaluated by immunohistochemistry staining using tumor tissue arrays. SAG expression in cancer cells was knocked down by siRNA silencing. The anticancer effects of SAG silencing were evaluated by in vitro assays for cell growth and survival, and by an in vivo orthotopic xenograft tumor model. Radiosensitization of SAG silencing in human cancer cells was determined by clonogenic survival assay. Apoptosis induction was evaluated by FACS analysis, caspase-3 activation assay, and western blotting of apoptosis-associated proteins.
Results
SAG was overexpressed in multiple human tumor tissues, compared to their normal counterparts. SAG silencing selectively inhibited cancer cell proliferation, suppressed in vivo tumor growth and sensitized radiation-resistant cancer cells to radiation. Mechanistically, SAG silencing induced apoptosis with accumulation of Noxa, while SAG over-expression reduced Noxa levels and shortened Noxa protein half-life.
Conclusions
The findings demonstrated that SAG E3 ubiquitin ligase plays an essential role in cancer cell proliferation and tumor growth, and may serve as a promising anticancer and radiosensitizing target.
Keywords: SAG/ROC2/Rbx2, SCF ubiquitin ligases, siRNA silencing, Apoptosis, Radiosensitization
Introduction
Ubiquitin–proteasome system (UPS) controls protein turnover and regulates a variety of signalling pathways and cellular processes, from cell proliferation, differenciation to death (1). Recent studies demonstrated that pharmacological inhibition of the UPS can be efficacious in the treatment of human cancers. Bortezomib (Velcade, formerly PS-341) represents the first FDA-approved proteasome inhibitor to treat multiple myeloma and haematologic as well as solid tumors (2). It mainly blocks activation of nuclear factor-kappa B (NF-kB) and induces pro-apoptotic protein NOXA, rendering cells to apoptosis (2, 3). The fast-track approval of Bortezomib has spurred a great wave of interest in the development of anticancer reagents against UPS.
The SCF (Skp1-Cullin1-F-box-protein) E3 ubiquitin ligases are multi-unit complexes and consist of scaffold protein cullins (cullin 1–7), ring-finger proteins (RBX1/ROC1 or RBX2/SAG), adaptor proteins (e.g. SKP1) and F-box proteins (e.g. SKP2 and FBW7) (4, 5). They mediate the transfer of ubiquitin molecules to substrates (substrate polyubiquitination) for subsequent recognition and degradation by proteasome. Since SCF E3 ubiquitin ligases control the degradation of a variety of protein substrates via UPS to regulate diverse cellular processes, SCF dysfunction could cause a variety of diseases including cancer. For example, oncogenic F-box proteins Skp2 and β-TrCP, which promote the degradation of tumor suppressor p27 or IκB, respectively, were overexpressed in a number of human cancers, which is associated with a higher degree of malignant and poor patient prognosis (6). On the other hand, mutation or deletion of tumor suppressor F-box protein Fbw7 caused the accumulation of a number of oncogenic protein substrates, such as c-Jun, c-Myc, cyclin E and mTOR, leading to an accelerated tumor cell proliferation and tumorigenesis (7, 8). Most recently, we found that RING-finger protein RBX1/ROC1 was overexpressed in diverse human primary tumors and downregulation of RBX1 by RNAi silencing inhibited cancer cell growth via activation of multiple cell-killing pathways, including cell cycle arrest, apoptosis and senescence (9). These findings suggested that the SCF E3 ubiquitin ligase complexes could function as an anticancer target.
SAG (Sensitive to Apoptosis Gene) was originally cloned in our laboratory as a redox inducible antioxidant protein and was later characterized as the second member of RBX/ROC RING component of SCF E3 ubiquitin ligases (10–12). As an antioxidant, SAG over-expression inhibits apoptosis induced by redox (10, 13), nitric oxide (14), ischemia/hypoxia (15), heat shock (16), neurotoxin and 1-methyl-4-phenylpyridinium (17) in vitro and in vivo. When complexed with other components of SCF E3 ubiquitin ligase, SAG has E3 ubiquitin ligase activity and promotes the stage-dependent degradation of c-Jun and IκBα, thus regulating carcinogenesis and tumor growth in a DMBA-TPA murine skin cancer model (18). In human lung cancers, SAG mRNA was significantly overexpressed in tumor tissues with the correlation to a poor patient survival (19). The findings suggest that SAG E3 ubiquitin ligase may be required for human carcinogenesis or progression and serve as an anticancer target.
Here we showed that SAG protein was overexpressed in multiple human primary tumor tissues, particularly in lung cancer. SAG siRNA silencing had no effect on normal cell growth, but inhibited the growth of cancer cells, both in vitro and in vivo by induction of apoptosis. Moreover, SAG silencing sensitized radio-resistant H1299 and U87 cells to ionizing radiation. Thus, SAG may serve as a target for anticancer therapy as well as radio-sensitization.
Materials and methods
Cell culture
H1299 human lung cancer cells, U87 human glioblastoma cells, PANC-1 human pancreatic carcinoma cells and MRC-5 lung fibroblast cells were purchased from ATCC and cultured in DMEM media with 10% FBS. Normal bronchial epithelial cells, NL20, were grown in Ham’s F12 medium with 4% FBS and essential supplements as described (20).
Immunohistochemistry (IHC) staining of human tumor tissue arrays
Multiple human tumor tissue arrays were provided and stained with purified monoclonal SAG antibody by the University of Michigan Comprehensive Cancer Tissue Core. Briefly, the tissue array sections in 5 microns were dehydrated and subject to peroxidase blocking. SAG monoclonal antibody [raised against the RING domain (AA44-113) was added at a dilution of 1:100 and incubated at room temperature for 30 minutes on the DAKO AutoStainer using the DakoCytomation EnVision+ System-HRP (DAB) detection kit. The slides were counterstained with hematoxylin (Surgipath). The stained slides were observed under microscope (OLYMPUS 1X71) and images were acquired using software DP controller (Ver. 3.1.1.267, OLYMPUS).
Lentivirus-based siRNA and lentivirus infection
Construction and preparation of lentivirus-based siRNA against SAG (LT-SAG) and lentivirus expressing scrambled control siRNA (LT-CONT) were described previously (21). The target sequences are as follows: LT-SAG02-0 1 5′-AACAAGAGGACTGTGTTGTGGTCTGGTTCAAGAGACCAGACCACAACACAGT CCTCTTGTTTTTTGT-3′; LT-SAG02-0 2 5′-CTAGACAAAAAACAAGAGGACT GTGTTGTGGTCTGGTCTCTTGAACCAGACCA CAACACAGTCCTCTTGTT-3′; LT-CONT-01 5′-ATTGTATGCGATCGCAGACTTTTCAAGAGAAAGTCTGCGATCGCA TACAATTTT TTGT-3′; and LT-CONT-02 5′-CTAGACAAAAAATTGTATGCGATCG CAGACTTTCTC TTGAAAA GTCTGCGATCGCATACAAT-3′.
ATPlite cell proliferation assay
Cells were infected with LT-SAG or LT-CONT for 96 hrs, then split and seeded into 96-well plates with 3000 cells per well in quadruplicate. At 24, 48, 72 and 96 hours post cell plating, cell proliferation assay using ATPlite 1step luminescence ATP detection assay system (PerkinElmer, USA) was performed according to the manusfacter’s instruction (22).
Clonogenic survival assay
Cells were infected with LT-SAG or LT-CONT for 96 hrs, then split and seeded into 6-well plates with 100 cells (H1299 and Panc-1) or 300 cells (U87) per well in triplicate, followed by incubation at 37 °C for 9 days. The colonies formed were fixed with 10% acidic acid in methanol, stained with 0.05% methylene blue and counted.
Soft agar assay
Ten thousand cells after lentivirus-based siRNA silencing were seeded in 0.33% agar containing 1× cell culture medium and 10% FBS in 60-mm petri dish, and grown at 37°C for 14 days. The cells were stained with p-iodonitrotetrazolium (1 mg/mL; Sigma) overnight and the colonies were counted (21).
Irradiation and radisensitization assay
Cells after lentivirus-based siRNA silencing were seeded in six-well plates at three different cell densities in duplicate. The next day, cells were exposed to different doses of radiation followed by incubation at 37°C for 9 days. The colonies formed were fixed and the surviving fraction was determined by the proportion of seeded cells following irradiation to form colonies relative to untreated cells as described (20).
FACS analysis
H1299 and U87 cells were infected with LT-SAG, along with LT-CONT for 96 hours, then split and cultured for 72 hours, followed by PI staining and FACS analysis for apoptosis detection. Briefly, infected cells were harvested and fixed in 70% EtOH at −20°C for 4 hours, stained with propidium iodide (18μg/ml) containing 400μg/mL RNaseA (Roche) with shaking for 1 hour, and analyzed by flow cytometry for apoptosis and cell cycle profile, as described previously (22). Apoptosis was measured by the percentage of cells in sub-G1 population.
Western blotting analysis
Whole cell lysates were prepared and subjected to IB analysis using antibodies against SAG (monoclonal antibody raised againt the RING domain (AA44-113), Bax, Bad, cIAP2, (Cell signaling), pRB, Puma, Mcl-1 (Santa Cruz Biotechnology), XIAP, Bcl-XL, p21, (BD Transduction Laboratories), β-Actin (Sigma), Bak (Upstate), Bim (Imgenex), Noxa (Oncogene Science), Bcl-2, (Dako), and survivin (Novus biologicals).
Orthotopic pancreatic tumor model
Panc-1 cells stably transfected with luciferase reporter were infected with LT-CONT or LT-SAG for 96 hrs and used to establish an orthotopic model of pancreatic tumor, as described (23). Briefly, cells (2×106 per mouse in a volume of 0.1ml) were injected orthotopically into the pancreas of nude mice as follows. Five mice in each group were anesthetized, a small left abdominal flank incision was made, and tumor cells were injected into the subcapsular region of the pancreas using a 30-gauge needle and a calibrated push button-controlled dispensing device. A cotton swab was held cautiously for 1 min over the site of injection to prevent leakage. The peritoneum and skin incision were closed sequentially with absorbable suture. After 5 weeks, tumors were bioluminescence imaged using a cryogenically cooled imaging system coupled to a data acquisition computer running Living Image software at the University of Michigan MRI core. Mice were then sacrificed and tumors were harvested and weighed.
Caspase-3 activation assay
Cells were infected with LT-SAG, along with LT-CONT for 96 hours, then split and seeded in 96-well plates at 1.5 × 104 cells per well for incubation at 37°C for 72h. The cells were then lysed and subjected to a fluorogenic caspase-3 assay with Ac-DEVD-AFC as a substrate (Biomol) (22). The results were expressed as fold change compared to control. Each treatment had three replicates.
Half-life analysis of Noxa
To evaluate the effect of SAG overexpression on the half life of NOXA, U87 cells cultured in six-well cell culture plates were transiently transfected with FLAG-NOXA in combination with FLAG-SAG or pcDNA3 with the total 2 μg DNA per well (1 μg FLAG-NOXA plus 1 μg FLAG-SAG or pcDNA3) for 24h. The transfected cells were then treated with CHX at 20 μg/ml. Protein samples were collected at 0h, 1h, 2h, 4h and 8h post CHX treatment, and subjected to Noxa detection using anti-FLAG antibody by Western blotting analysis with β-Actin as the loading control.
To determine the effect of SAG knockdown on the half life of endogenous Noxa, U87 cells were infected with LT-SAG, along with LT-CONT for 96 hours and then split and seeded into six-well cell culture plates. Twenty-four hrs later, cells were treated with CHX at 20 μg/ml for indicated periods of time, followed by Western blotting using antibody against endogenous Noxa with β-Actin as the loading control. The relative Noxa levels were quantified by densitometry analysis using ImageJ1.410 image processing software.
Statistical analysis
The statistical significance of differences between groups was assessed using GraphPad Prism4 software (version 4.03). The unpaired, two-tailed t-test was used for the comparison of parameters between groups. The level of significance was set at P < 0.05.
Results
SAG is overexpressed in human primary tumor tissues
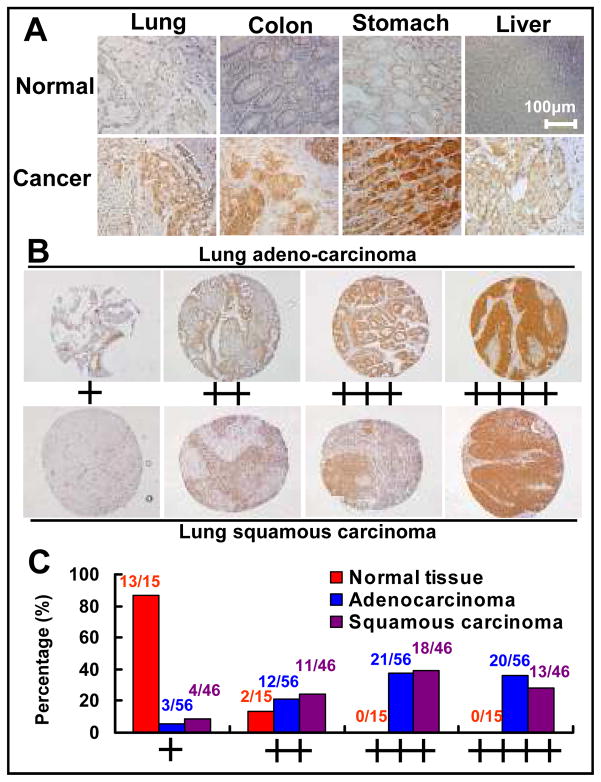
SAG over-expressed was previously shown in human lung cancer tissues by RT-PCR (19) and in a subset of colon cancer by Western blotting (24). These studies might underestimate SAG overexpression in cancer tissues due to normal tissue contamination and tumor stromal cell infiltration. To precisely determine the expression status of SAG in human tumor tissues, we performed immuno-staining analysis using a SAG monoclonal antibody (mAb), raised against purified human SAG RING domain (AA44-113) fused with GST (Creative Biolabs, Port Jefferson Station, NY). The antibody specificity against SAG was validated, via immuno-fluorescent staining, to detect SAG in wild type mouse embryonic stem (ES) cells, but not in SAG knockout ES cells (unpublished data). This specific mAb was then used to measure the SAG levels in human cancer tissue microarrays. As shown in Fig 1A, SAG was expressed weakly in several normal tissues (top panels), but over-expressed in a panel of human tumor tissues, including carcinomas of lung, colon, stomach and liver (bottom panel). To focus on lung cancer, we immuno-stained two sets of human lung cancer tissue arrays, consisting of 15 normal tissues and 102 tumor tissues of adenocarcinoma (N=56) and squamous carcinoma (N=46). Based upon the intensity of staining, we classified the samples into 4 groups with group 1 the least staining (+) and group 4 the highest staining (++++) (Figure 1B). We found that the SAG staining in normal tissues were either under group 1 (13 out of 15, 87%) or group 2 (2/15, 13%), whereas a majority of tumor tissues, regardless of tumor types, had a high SAG staining (mainly in cancer cells, but not in stromal cells) and were classified into Groups 3 and 4 (73% for adenocarcinoma and 67% for squamous carcinoma) (Figure 1C). Thus, SAG is overexpressed in a majority of human lung cancers. The overexpression of SAG in diverse primary human tumors suggests that SAG could play a role in carcinogenesis or in the maintenance of tumor cell phenotype.
Figure 1. The expression of SAG in human tumors and normal counterparts.
(A) SAG overexpression in multiple human primary tumor tissues. Tumor tissue arrays containing multiple normal and tumor tissues from different organs were stained with purified SAG monoclonal antibody on the DAKO AutoStainer using the DakoCytomation EnVision+ System-HRP (DAB) detection kit, and counterstained with hematoxylin (Surgipath). The stained slides were observed under microscope (OLYMPUS 1X71) and images were acquired using software DP controller. (B) SAG staining in lung tissues, normal vs. cancer. Lung tumor tissue arrays containing normal lung and tumor tissues were stained for SAG expression. Stained normal and tumor tissues were classified into four groups (+ to ++++) according to staining intensity of each tissue. (C) The percentage of normal or tumor tissues in each staining group. Tissue samples with different staining intensity were grouped and tabulated.
SAG silencing inhibits the growth of human cancer cells
We next determined, using siRNA knockdown approach, the role of SAG in regulation of cell proliferation of tumor vs. normal cells by an ATP-lite assay. As shown in Figure 2A, SAG silencing by a lentivirus-based SAG siRNA construct (LT-SAG) (21) caused reduction of SAG expression by over 90% in H1299 non-small cell lung carcinoma cells (panel 1), in U87 glioblastoma cells (panel 2), in NL20 normal bronchial epithelial cells (panel 3), and in MRC-5 lung fibroblast cells (panel 4). As a result, cell proliferation was inhibited significantly in H1299 and U87 cells (Figure 2B, panels 1&2). Interestingly, the normal cells were rather resistant to SAG knockdown with a very minor growth inhibition (B, panels 3&4), demonstrating a tumor cell selective growth suppression. We next determined the effect of SAG silencing on tumor cell survival by clonogenic assay. As shown in Figure 2C, SAG silencing caused an 80% or 85% inhibition in survival of H1299 or U87 cells, respectively. Furthermore, the ability of anchorage-independent growth of H1299 and U87 cells, as measured by soft agar assay, was inhibited up to 75% upon SAG silencing (Figure 2D). These findings indicated that SAG silencing significantly suppresses the growth and survival of human cancer cells, but is rather inactive on normal cell growth.
Figure 2. SAG silencing selectively inhibits the growth of human cancer cells.
H1299 human lung cancer cells, U87 human glioblastoma cells, NL20 normal bronchial epithelial cells and MRC-5 lung fibroblast cells were infected with LT-CONT and LT-SAG for 96 hrs, and then split for assays as follows. (A) SAG silencing effects, determined by immunobloting (IB) with β-actin as the loading control 96h post cells splitting. (B) ATPlite cell proliferation assay. Cells after lentivirus-based siRNA silencing were split and seeded into 96-well plates with 3000 cells per well in quadruplicates and subject to ATPlite cell proliferation assay over periods up to 96 hrs. *P<0.05; **P<0.01. (C) Clonogenic cell survival assay in H1299 (top panel) and U87 (bottom) cells. Cells after lentivirus-based siRNA silencing were split, seeded into 6-well plates with 100 cells (H1299) or 300 cells (U87) per well in triplicates, and incubated at 37 °C for 9 days, followed by 0.05% methylene blue staining and colony counting. (D) Soft agar anchorage-independent growth assay in H1299 and U87 cells. Ten thousand cells after lentivirus-based siRNA silencing were seeded in 0.33% agar containing 1× cell culture medium and 10% FBS in 60-mm petri dish, and grown at 37°C for 14 days, followed by staining with p-iodonitrotetrazolium overnight and colony counting.
SAG silencing sensitizes H1299 and U87 cells to radiation
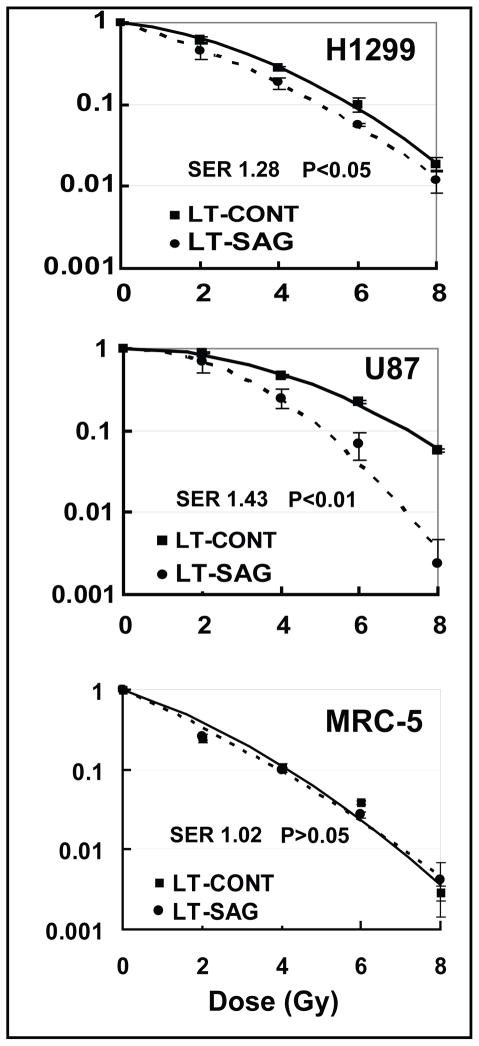
SAG was previously shown to have antioxidant activity and protected cells from redox compound-induced apoptosis (10, 12). Since a common mechanism by which radiation-induced cell killing is via production of reactive oxygen species (ROS) (25), we determined if SAG silencing would confer radiosensitivity of otherwise radioresistant cancer cells. Two radioresistant cell lines, H1299 and U87 (20, 26), were tested in a standard clonogenic assay. As shown in Figure 3, SAG silencing significantly sensitized them to radiation with a sensitizing enhancement ratio (SER) of 1.28 and 1.43, respectively (top and middle panel). To test whether normal cells are also sensitive to radiation upon SAG silencing, we performed a comparable assay in MRC-5 cells. As shown in Figure 3 (bottom panel), SAG silencing did not sensitize normal cells to radiation with a SER of 1.02. The findings suggest that SAG silencing selectively sensitizes cancer cells to radiation.
Figure 3. SAG silencing sensitizes cancer cells to radiation.
Cells after lentivirus-based siRNA silencing were seeded in six-well plates at three different cell densities in duplicates. The next day, cells were exposed to different doses of radiation followed by incubation at 37°C for 9 days for colony counting. The surviving fraction was calculated and plotted after comparison with 0 Gy corresponding controls. The sensitizing enhancement ratio (SER) was calculated as the ratio of the inactivation dose under scrambled siRNA control conditions divided by inactivation dose after SAG silencing. Shown is value ±SEM from three independent experiments.
SAG silencing induces apoptosis with an accumulation of pro-apoptosis protein NOXA
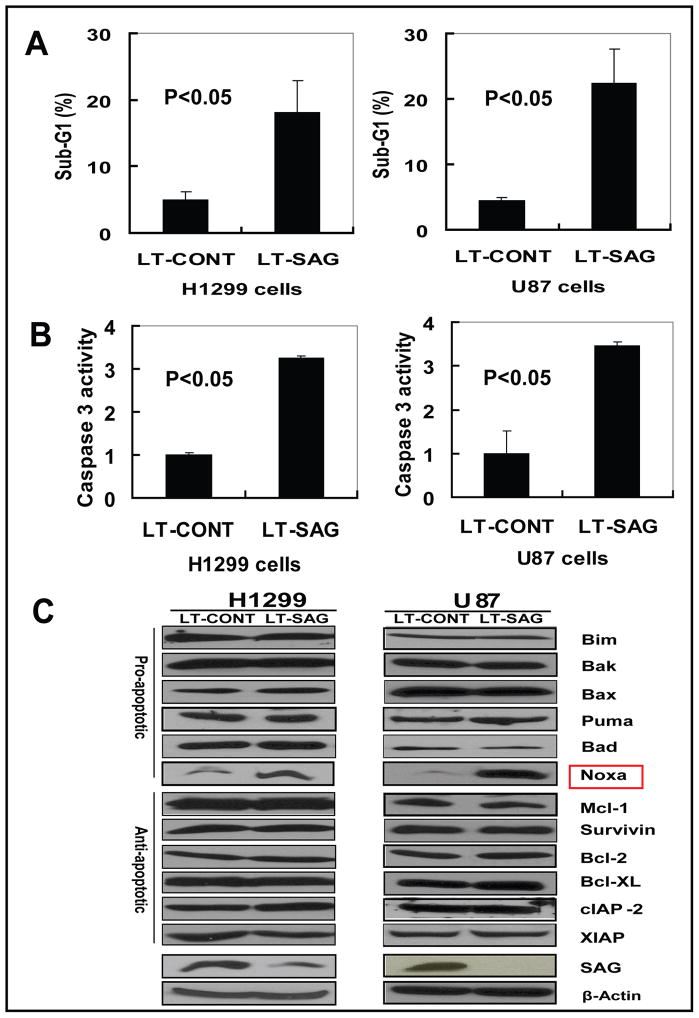
To investigate the potential mechanism of cell growth suppression induced by SAG silencing, we performed FACS analysis to profile the cell population at the sub-G1 (apoptotic population) or other phases of cell cycle in H1299 and U87 cells. As shown in Figure 4A, 20% of H1299 and U87 cell population underwent apoptosis upon SAG silencing, respectively, compared to less than 5% population seen in LT-CONT control cells. Apoptotic induction of these cells upon SAG silencing was further confirmed by the activation of caspase 3 (Figure 4B). No significant cell cycle disturbance was induced by SAG silencing in both cell lines (data not shown). Furthermore, only a small fraction of SAG silenced cells underwent senescence, as demonstrated by senescence-associated β-gal staining, but without a significant difference from the control cells (data not shown). These results clearly demonstrated that apoptosis induced by SAG silencing is the major mechanism for the inhibition of cell growth and survival.
Figure 4. SAG silencing induces apoptosis with Noxa accumulation.
H1299 and U87 cells were infected with LT-SAG, along with LT-CONT for 96 hours, then split and cultured for 72 hours, followed by PI staining and FACS analysis for apoptosis detection, caspase-3 activity assay, cell cycle profile and Western blotting for the levels of apoptosis-associated proteins. (A) Induction of apoptosis by SAG silencing. Apoptotic cells were determined by sub-G1 fraction in FACS analysis; (B) Caspase-3 activation upon SAG silencing. Caspase-3 activity in infected cells was determined by Caspase-3 activity assay. For A and B, shown is value ±SEM of three independent experiments. (C) The expression of apoptosis-associated proteins. The status of a panel of apoptosis-associated proteins including pro-apoptosis proteins and anti-apoptosis proteins were detected by Western blotting with β-Actin as the loading control.
To further understand how SAG silencing induced apoptosis, we analyzed the expression of a panel of pro-apoptotic proteins (Bax, Bak, Puma, Bim, Bad and Noxa), of anti-apoptotic proteins (Bcl-2, Mcl-1, Survivin, XIAP, Bcl-XL, and cIAP-2) and of known SCF E3 ligase substrates (β-catenin, cyclin D, cyclin E, IκB-α, p21 and pRB) (4, 5). As shown in Figure 4C, among apoptosis-regulatory proteins tested, pro-apoptotic Noxa was the only protein significantly accumulated in both H1299 and U87 cells upon SAG silencing. Other SCF E3 ligase substrates were either unchanged or undetectable except pRB, which was accumulated in U87 cells as well as A549 cells, but not H1299 cells (Fig S1). In addition, the expression of these proteins upon SAG silencing was also determined in another lung cancer cell line, A549. Consistently, upon SAG knockdown, Noxa was accumulated, whereas other proteins were unchanged (Fig S1).
SAG overexpression reduces Noxa levels and shortens Noxa protein half-life, whereas SAG knockdown extends its half-life
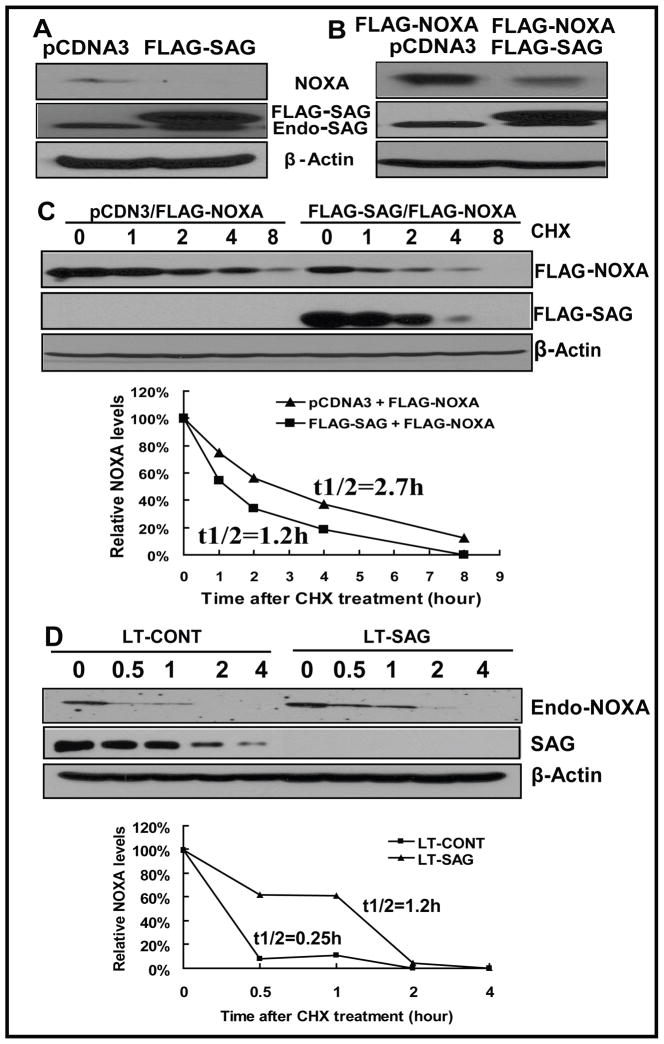
As a RING component of SCF E3 ubiquitin ligases, SAG knockdown by siRNA would disrupt the SCF complex and inactivate its ligase activity, leading to accumulation of its substrates, whereas SAG overexpression would likely promote the degradation of its substrates. We next characterized Noxa as a potential substrate of SAG SCF E3 ubiquitin ligases in U87 cells. Indeed, SAG overexpression eliminated endogenous Noxa (Fig 5A), as well as reduced the level of ectopically overexpressed Noxa (Fig 5B). Furthermore, SAG overexpression significantly shortened the protein half-life of Noxa from 2.7 hr to 1.2 hr (Fig 5C). Finally, we evaluated the effects of SAG knockdown on the half-life of endogenous Noxa and found that SAG silencing significantly extended the half-life of Noxa from about 0.25 hr to 1.2 hr (Fig 5D). Thus, Noxa appears to be a novel substrate of SAG E3 ligase.
Figure 5. SAG manipulation changes Noxa level and protein half-life.
(A) SAG overexpression eliminates endogenous Noxa. U87 cells were transiently transfected with pcDNA3-FLAG-SAG, along with pcDNA3 as a control. Cells were harvested 30 hr later and subjected to Western blotting using anti-Noxa antibody with β-Actin as the loading control. (B) SAG overexpression reduced the level of ectopically overexpressed Noxa. U87 cells were transiently cotransfected with FLAG -Noxa and FLAG-SAG or FLAG -Noxa and pcDNA3. Cells were harvested 30 hr later and subjected to Western blotting using anti-FLAG antibody (For FLAG-Noxa) and anti-SAG (for FLAG-SAG and endogenous SAG) with β-Actin as the loading control. (C) SAG overexpression shortened the protein half life of Noxa. U87 cells were transiently cotransfected with FLAG -Noxa and FLAG-SAG or FLAG -Noxa and pcDNA3. Twenty-four hrs later, cells were treated with CHX at 20 μg/ml for indicated periods of time, followed by Western blotting using antibodies against FLAG (for FLAG-Noxa), SAG (for FLAG-SAG) with β-Actin as the loading control. The relative Noxa levels were quantified by densitometry analysis using ImageJ1.410 image processing software. (D) SAG knockdown extended the half life of endogenous Noxa. U87 cells were infected with LT-SAG, along with LT-CONT for 96 hrs and then split. Twenty-four hrs later, cells were treated with CHX at 20 μg/ml for indicated periods of time, followed by Western blotting using antibody against endogenous Noxa with β-Actin as the loading control. The relative Noxa levels were quantified by densitometry analysis using ImageJ1.410 image processing software. Endo-Noxa: endogenous Noxa.
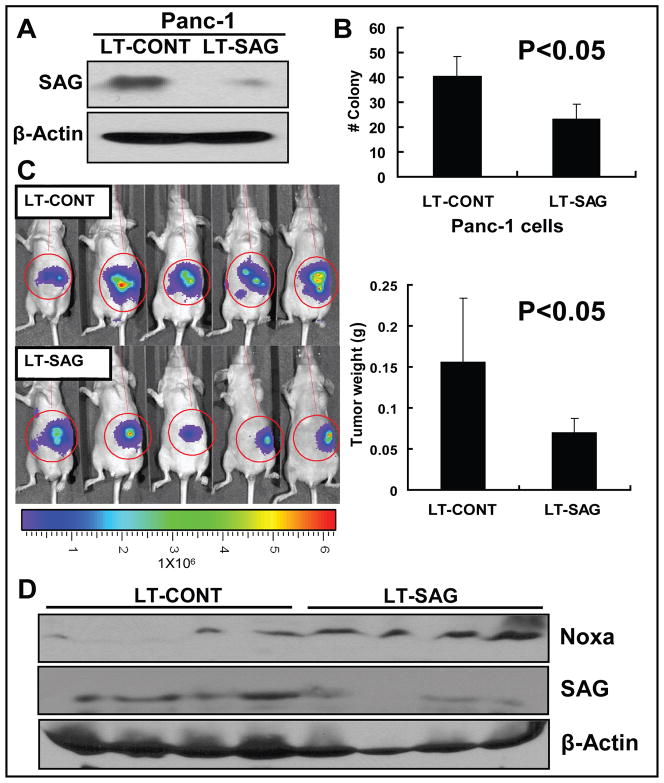
SAG silencing inhibits the in vivo tumor growth in an orthotopic mouse model of pancreatic cancer
We next assessed the potential effect of SAG silencing on in vivo tumor growth using an orthotopic pancreatic tumor xenograft model since it is a physiologically relevant and far superior to subcutaneously implanted tumor models. We first confirmed that SAG expression was indeed knocked down by LT-SAG in human pancreatic cancer line PANC-1 cells by ~80% (Fig 6A). Consistent to observations made in H1299 and U87 cells, SAG knock-down caused a 50% inhibition of PANC-1 cell survival in vitro (Fig 6B). The SAG silenced PANC-1 cells (LT-SAG), along with scrambled control cells (LT-CONT) were implanted into the pancreas of nude mouse (5 mice per group). After 5 weeks of in vivo growth, tumors were bioluminescent-imaged. As shown in Figure 6C (left panel), the strength of luminescent signal emitted from SAG-silenced tumors was on average much weaker than that from the control tumors. More precisely, each individual tumor was harvested and weighted. SAG silencing reduced overall tumor mass by 50% which is statistically significant (p < 0.05) (Fig 6C, right panel). Finally, we determined the level of Noxa in available tumor tissues harvested and found that NOXA was accumulated in SAG-silencing tumors (Fig 6D). Thus, the orthotopic in vivo growth of PANC-1 pancreatic cancer cells was significantly inhibited upon SAG silencing and NOXA accumulation may contribute to this process.
Figure 6. SAG silencing inhibits the growth of orthotopic pancreatic tumors.
PANC-1 human pancreatic carcinoma cells were infected with LT-CONT and LT-SAG, followed by the determination of SAG silencing effect (A), cell survival assay in vitro (B), tumor formation in vivo (C), and Western blotting (D). (A) SAG silencing effects. Panc-1 cells stably transfected with luciferase were infected with LT-CONT or LT-SAG, and the SAG levels were determined 96 hrs post infection by Western blotting with β-actin as the loading control. (B) Clonogenic cell survival assay. Cells after SAG silencing were split, seeded into 6-well plates with 100 cells per well in triplicates, and incubated at 37 °C for 9 days, followed by 0.05% methylene blue staining and colony counting. (C) Bioluminescence imaging of implanted tumors and measurement of tumor weight. Panc-1 cells stably transfected with luciferase were infected with LT-CONT or LT-SAG and implanted into pancreases of the mice (5 mice per group) for the evaluation of tumor growth in vivo. After 5 weeks, tumors were bioluminescence imaged using a cryogenically cooled imaging system coupled to a data acquisition computer running Living Image software at the University of Michigan Small Animal Imaging Core. Mice were then sacrificed, tumors harvested and weighed. (D) NOXA expression in tumors. The level of Noxa, SAG in tumor tissues was determined by Western blotting using antibodies against Noxa and SAG with β-Actin as the loading control.
Discussion
Ideal cancer targets should have the following features: a) they play an essential role in carcinogenesis, and/or are required for the maintenance of cancer cell phenotype, and/or are survival proteins to confer cancer cells resistance to apoptosis; b) they are over-expressed in cancer cells, which is associated with a poor prognosis of patient survival; c) inhibition of their expression or activity induces growth suppression and/or apoptosis in cancer cells, but not in normal cells, achieving a potential therapeutic window; d) it is “drugable”, that is, it is an enzyme (e.g. kinase) or a cell surface molecule (e.g. membrane bound receptor), which can be easily screened for small molecule inhibitors or being targeted by a specific antibody (27, 28). In this study, we validated SAG, a dual functional protein with antioxidant and ligase activities, as a potential anti-cancer target. We showed here that 1) SAG is over-expressed in a number of human cancers originated from different organs, particularly in lung cancer tissues; 2) while having no effect on normal cell growth, SAG silencing dramatically inhibits proliferation and survivals via apoptosis induction in multiple cancer cell models in in vitro and in a pancreatic cancer orthotopic xenograft model in vivo; 3) SAG silencing also sensitizes radioresistant cancer cells to radiation. In combination with our previous studies that SAG, when over-expressed, protects cells from apoptosis induced by a variety of stimuli (10, 11, 15, 29) and promotes in vivo tumor growth also by inhibiting apoptosis (18) and the fact that SAG is a “drugable” E3 ubiquitin ligase, SAG appears to be a promising anticancer target as well as the target for radio-sensitization.
Cancer cells tend to obtain apoptosis-escaping mechanisms by inactivating apoptosis signaling pathways via down-regulation of pro-apoptotic proteins or up-regulation of anti-apoptotic proteins. Thus, reactivation of cellular apoptotic signaling has become a major effort toward the development of anticancer therapies (30, 31). In this study, we found that SAG silencing induced apoptosis, which is associated with the accumulation of pro-apoptotic Noxa. Further characterization revealed that SAG overexpression promoted degradation of Noxa and shortened its protein half life, whereas SAG knockdown delayed the degradation of Noxa and extended its half-life. Consistently, others have recently reported that Noxa was accumulated or upregulated after treatment with proteasome inhibitor bortezomib (also known as Velcade, PS-341) in melanoma cells (32) and that Noxa with a protein half-life of less than 2 hrs in lymphoblastic leukemia cells was subjected to ubiquitin-proteasome-mediated degradation, which is inhibited by a proteasome inhibitor, MG132 (33). Taken together, our study suggested that Noxa may be a novel substrate of SAG-SCF E3 ligase and its accumulation, upon SAG silencing, could contribute to apoptosis induction. Future study is directed to identify the F-box protein that recognizes Noxa for further biochemical characterization of Noxa as a novel substrate of SAG-SCF E3 ligase. Finally, given the fact that SAG is the RING component of SCF E3 ligases, required for ubiquitination and subsequent degradation of a variety of protein substrates, one could anticipate that alterations of multiple protein substrates would contribute to the induction of apoptosis following SAG silencing. Mechanistic characterization of over 350 potential SCF E3 ligase substrates, identified through global protein stability profiling, which are closely involved in the regulation of apoptosis, cell cycle, and cell signaling (34, 35), would certainly broaden our understanding how SAG-SCF E3 ligases regulate cell proliferation and apoptosis.
Radioresistance is a major obstacle for effective cancer treatment. Particularly in cases of glioblastoma multiforme and non-small cell lung cancer (NSCLC), radiotherapy is very ineffective due to extreme radioresistance of cancer cells, contributing to poor patient survival rate (36, 37). Here we showed that SAG silencing sensitized H1299 NSCLC cells and U87 glioblastoma cells, two radioresistent lines, to radiation with the sensitizing enhancement ratio comparable to the silencing of TRAF2, a well-known cellular survival protein (20). Mechanistically, SAG silencing-mediated radiosensitization could be attributable to the loss of ROS-scavenging activity that blocks radiation-generated ROS (25). It has been previously demonstrated that SAG overexpression protects cells or tissues against damages induced by redox compound 1,10-phenanthroline or zinc ion (10, 13), nitric oxide (14), ischemia/hypoxia (15), neurotoxin and 1-methyl-4-phenylpyridinium (MPP) (17), heat shock (16), and UV-irradiation (38). On the other hand, radiosensitization upon SAG silencing could result from the accumulation of a panel of yet-to-be identified radiation-sensitizing substrates as a result of the inactivation of SAG SCF E3 ligase activity.
In summary, we validated SAG as a promising anticancer target as well as a target for radio-sensitizatiion on the basis of the following findings: (a) SAG is overexpressed in a number of human primary tumor tissues, particularly lung cancer; (b) SAG silencing selectively kills cancer cells but not normal cells in vitro and in vivo; (c) SAG silencing sensitizes cancer cells to anticancer radiation. Future challenge will be a) to identify specific inhibitors of SAG E3 ligases (27, 28) and to develop them as a novel class of anticancer agents and radiosensitizers; and b) to develop siRNA-based therapy against radio-resistant cancer using nano-particle packaged SAG siRNA (39, 40).
Supplementary Material
Figure S1: The effect of SAG silencing on the expression of SCF E3 ligase substrates in cancer cells. H1299, U87 and A549 cells were infected with LT-SAG, along with LT-CONT for 96 hrs, then split and cultured for 72 hrs. The levels of β-catenin, cyclin D, cyclin E, iκB-α, p21 and pRB, as known SCF substrates, were determined by Western blotting using specific antibodies with β-Actin as the loading control.
Acknowledgments
This work is supported by NCI grants (CA111554 and CA118762) to YS. We thank Drs. Dafydd Thomas and Thomas Giordano for providing us the primary human cancer tissue microarrays.
Footnotes
STATEMENT OF TRANSLATIONAL RELEVANCE
The interest in the development of anticancer drugs against ubiquitin–proteasome system (UPS) has been spurred by the fast-track FDA approval of proteasome inhibitor Bortezomib (also known as Velcade or PS-341) for the treatment of relapsed and refractory multiple myeloma. SCF (Skp1-Cullin1-F-box-protein) complexes, the largest multi-unit E3 ubiquitin ligases, target a variety of substrates for degradation via UPS and regulate carcinogenesis and cancer progression. We demonstrate here that SAG (Sensitive to Apoptosis Gene), the second member of RBX/ROC RING component of SCF E3 ubiquitin ligases, is a promising anticancer target, as follows: 1) SAG is over-expressed in a number of human primary cancers, particularly lung cancer; 2) SAG siRNA silencing selectively kills cancer in vitro and in vivo; and 3) SAG siRNA silencing selectively sensitizes radioresistant cancer cells to radiation. Thus, the development of small molecules or RNAi-based therapy targeting SAG holds a promise for future treatment of human cancer.
References
- 1.Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat Rev Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 2.Orlowski RZ. Proteasome inhibitors in cancer therapy. Methods Mol Biol. 2005;301:339–50. doi: 10.1385/1-59259-895-1:339. [DOI] [PubMed] [Google Scholar]
- 3.Nikiforov MA, Riblett M, Tang WH, et al. Tumor cell-selective regulation of NOXA by c-MYC in response to proteasome inhibition. Proc Natl Acad Sci U S A. 2007;104:19488–93. doi: 10.1073/pnas.0708380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6:369–81. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 5.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 6.Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat Rev Cancer. 2008;8:438–49. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 8.Mao JH, Kim IJ, Wu D, et al. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science. 2008;321:1499–502. doi: 10.1126/science.1162981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69:4974–82. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan H, Wang Y, Aviram M, et al. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol Cell Biol. 1999;19:3145–55. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid Redox Signal. 2001;3:635–50. doi: 10.1089/15230860152542989. [DOI] [PubMed] [Google Scholar]
- 12.Swaroop M, Bian J, Aviram M, et al. Expression, purification, and biochemical characterization of SAG, a RING finger redox sensitive protein. Free Radicals Biol Med. 1999;27:193–202. doi: 10.1016/s0891-5849(99)00078-7. [DOI] [PubMed] [Google Scholar]
- 13.Sun Y. Alteration of SAG mRNA in human cancer cell lines: Requirement for the RING finger domain for apoptosis protection. Carcinogenesis. 1999;20:1899–903. doi: 10.1093/carcin/20.10.1899. [DOI] [PubMed] [Google Scholar]
- 14.Yang ES, Park JW. Regulation of nitric oxide-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Res. 2006;40:279–84. doi: 10.1080/10715760500511500. [DOI] [PubMed] [Google Scholar]
- 15.Chanalaris A, Sun Y, Latchman DS, Stephanou A. SAG attenuates apoptotic cell death caused by simulated ischaemia/reoxygenation in rat cardiomyocytes. J Mol Cell Cardiol. 2003;35:257–64. doi: 10.1016/s0022-2828(03)00003-8. [DOI] [PubMed] [Google Scholar]
- 16.Lee SJ, Yang ES, Kim SY, Shin SW, Park JW. Regulation of heat shock-induced apoptosis by sensitive to apoptosis gene protein. Free Radic Biol Med. 2008;45:167–76. doi: 10.1016/j.freeradbiomed.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 17.Kim SY, Kim MY, Mo JS, Park JW, Park HS. SAG protects human neuroblastoma SH-SY5Y cells against 1-methyl-4-phenylpyridinium ion (MPP+)-induced cytotoxicity via the downregulation of ROS generation and JNK signaling. Neurosci Lett. 2007;413:132–6. doi: 10.1016/j.neulet.2006.11.074. [DOI] [PubMed] [Google Scholar]
- 18.Gu Q, Bowden TG, Normolle D, Sun Y. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage dependent targeting of c-Jun/AP1 and IkB/NF-kB. J Cell Biol. 2007;178:1009–23. doi: 10.1083/jcb.200612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sasaki H, Yukiue H, Kobayashi Y, et al. Expression of the sensitive to apoptosis gene, SAG, as a prognostic marker in nonsmall cell lung cancer. Int J Cancer. 2001;95:375–7. doi: 10.1002/1097-0215(20011120)95:6<375::aid-ijc1066>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 20.Zheng M, Morgan-Lappe SE, Yang J, et al. Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 2008;68:7570–8. doi: 10.1158/0008-5472.CAN-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007;67:3616–25. doi: 10.1158/0008-5472.CAN-06-4020. [DOI] [PubMed] [Google Scholar]
- 22.Bockbrader KM, Tan M, Sun Y. A small molecule Smac-mimic compound induces apoptosis and sensitizes TRAIL- and etoposide-induced apoptosis in breast cancer cells. Oncogene. 2005;24:7381–8. doi: 10.1038/sj.onc.1208888. [DOI] [PubMed] [Google Scholar]
- 23.Khanbolooki S, Nawrocki ST, Arumugam T, et al. Nuclear factor-kappaB maintains TRAIL resistance in human pancreatic cancer cells. Mol Cancer Ther. 2006;5:2251–60. doi: 10.1158/1535-7163.MCT-06-0075. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Duan H, Sun Y. Elevated expression of SAG/ROC2/Rbx2/Hrt2 in human colon carcinomas: SAG does not induce neoplastic transformation, but its antisense transfection inhibits tumor cell growth. Mol Carcinog. 2001;30:62–70. [PubMed] [Google Scholar]
- 25.Cook JA, Gius D, Wink DA, Krishna MC, Russo A, Mitchell JB. Oxidative stress, redox, and the tumor microenvironment. Semin Radiat Oncol. 2004;14:259–66. doi: 10.1016/j.semradonc.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 26.Wang CC, Liao YP, Mischel PS, Iwamoto KS, Cacalano NA, McBride WH. HDJ-2 as a target for radiosensitization of glioblastoma multiforme cells by the farnesyltransferase inhibitor R115777 and the role of the p53/p21 pathway. Cancer Res. 2006;66:6756–62. doi: 10.1158/0008-5472.CAN-06-0185. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y. Targeting E3 ubiquitin ligases for cancer therapy. Cancer Biol Therapy. 2003;2:623–9. [PubMed] [Google Scholar]
- 28.Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–54. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang GY, Pang L, Ge HL, et al. Attenuation of ischemia-induced mouse brain injury by SAG, a redox- inducible antioxidant protein. J Cereb Blood Flow Metab. 2001;21:722–33. doi: 10.1097/00004647-200106000-00010. [DOI] [PubMed] [Google Scholar]
- 30.Fulda S. Inhibitor of apoptosis proteins as targets for anticancer therapy. Expert Rev Anticancer Ther. 2007;7:1255–64. doi: 10.1586/14737140.7.9.1255. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson DW. From bench to clinic with apoptosis-based therapeutic agents. Nature. 2000;407:810–6. doi: 10.1038/35037747. [DOI] [PubMed] [Google Scholar]
- 32.Fernandez Y, Verhaegen M, Miller TP, et al. Differential regulation of noxa in normal melanocytes and melanoma cells by proteasome inhibition: therapeutic implications. Cancer Res. 2005;65:6294–304. doi: 10.1158/0008-5472.CAN-05-0686. [DOI] [PubMed] [Google Scholar]
- 33.Ploner C, Rainer J, Lobenwein S, Geley S, Kofler R. Repression of the BH3-only molecule PMAIP1/Noxa impairs glucocorticoid sensitivity of acute lymphoblastic leukemia cells. Apoptosis. 2009;14:821–8. doi: 10.1007/s10495-009-0355-5. [DOI] [PubMed] [Google Scholar]
- 34.Yen HC, Elledge SJ. Identification of SCF ubiquitin ligase substrates by global protein stability profiling. Science. 2008;322:923–9. doi: 10.1126/science.1160462. [DOI] [PubMed] [Google Scholar]
- 35.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–23. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 36.Short SC. External beam and conformal radiotherapy in the management of gliomas. Acta Neurochir Suppl. 2003;88:37–43. doi: 10.1007/978-3-7091-6090-9_7. [DOI] [PubMed] [Google Scholar]
- 37.Spiro SG, Silvestri GA. One hundred years of lung cancer. Am J Respir Crit Care Med. 2005;172:523–9. doi: 10.1164/rccm.200504-531OE. [DOI] [PubMed] [Google Scholar]
- 38.He H, Gu Q, Zheng M, Normolle D, Sun Y. SAG/ROC2/RBX2 E3 ligase promotes UVB-induced skin hyperplasia, but not skin tumors, by simultaneously targeting c-Jun/AP-1 and p27. Carcinogenesis. 2008;29:858–65. doi: 10.1093/carcin/bgn021. [DOI] [PubMed] [Google Scholar]
- 39.de Fougerolles AR. Delivery vehicles for small interfering RNA in vivo. Hum Gene Ther. 2008;19:125–32. doi: 10.1089/hum.2008.928. [DOI] [PubMed] [Google Scholar]
- 40.Howard KA, Kjems J. Polycation-based nanoparticle delivery for improved RNA interference therapeutics. Expert Opin Biol Ther. 2007;7:1811–22. doi: 10.1517/14712598.7.12.1811. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: The effect of SAG silencing on the expression of SCF E3 ligase substrates in cancer cells. H1299, U87 and A549 cells were infected with LT-SAG, along with LT-CONT for 96 hrs, then split and cultured for 72 hrs. The levels of β-catenin, cyclin D, cyclin E, iκB-α, p21 and pRB, as known SCF substrates, were determined by Western blotting using specific antibodies with β-Actin as the loading control.