Abstract
All eukaryotic cells possess an endoplasmic reticulum (ER), which is the site for synthesizing proteins that populate the cell surface or extracellular space. The environment of the ER is oxidizing, which supports the formation of intra- and interchain disulfide bonds that serve to stabilize the folding and assembly of nascent proteins. Recent experimental data reveal that the formation of disulfide bonds does not occur spontaneously but results from the enzymatic transfer of disulfide bonds through a number of intermediate proteins, with molecular oxygen serving as the terminal electron acceptor. Thus, each disulfide bond that forms during oxidative folding should produce a single reactive oxygen species (ROS). Dedicated secretory tissues like the pancreas and plasma cells have been estimated to form up to 3–6 million disulfide bonds per minute, which would be expected to result in the production of the same number of molecules of ROS. Although the methods used to deal with this amount of oxidative stress are not well understood, recent research suggests that different types of cells use distinct strategies and that the unfolded protein response (UPR) is a critical component of the defense. Antioxid. Redox Signal. 11, 2317–2331.
Introduction
The endoplasmic reticulum (ER) is the major site of synthesis of secretory and membrane proteins and forms a membranous network throughout the cell (Fig. 1). In many mammalian cells, approximately one third of the total proteins produced are synthesized in the ER, although this percentage can be much higher in specialized secretory cells like hepatocytes, pancreatic β islet cells, and plasma cells (116).
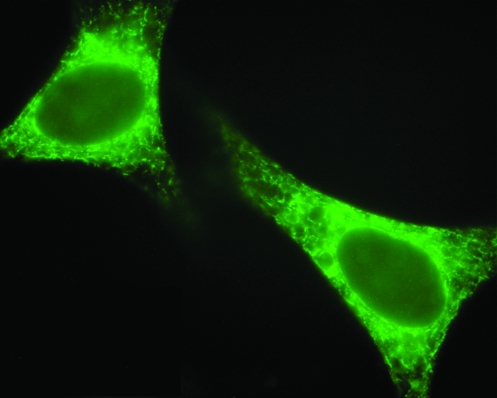
FIG. 1.
The ER is a membrane network that reaches throughout the cells. HeLa cells were transfected with a lymphoid-specific resident ER protein, pERpl, and visualized with a polyclonal pERp1 antiserum followed by FITC-conjugated anti-rabbit Ig antiserum. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
To prepare these nascent proteins properly for an extracellular fate, the ER lumen possesses a unique environment that is specialized for high-fidelity protein folding and assembly (33, 43). It contains high concentrations of molecular chaperones, folding enzymes, and ATP, which aid proper maturation of proteins. Unlike the cytosol, it also possesses an oxidizing environment, which favors intra- and intermolecular disulfide bond formation, and millimolar concentrations of Ca2+ that pose unusual complications for folding.
Only proteins that fulfill quality-control standards are allowed to exit the ER and travel farther along the secretory pathway toward their final destinations (25). However, if the amount of proteins to be folded exceeds the capacity of the folding machineries, unfolded proteins will accumulate in the ER. This upsets the normal ER homeostasis and induces a signaling pathway called the unfolded protein response (UPR), which serves to alleviate the stress, or alternatively, to eliminate the affected cells to protect the organism (60, 104). Although the ER exists in virtually all mammalian cells, the relative amounts of ER and the demands placed on this organelle are quite different between tissues. Given the particularly high demands placed on the ER in secretory tissues, it is easy to imagine that even a small fluctuation in the ER environment could dramatically affect cellular homeostasis.
Recent studies reveal that the UPR plays an important role to control this (39, 54). In addition, because reactive oxygen species (ROS) are produced as a side product of the oxidative folding in the ER (110, 125), secretory cells are likely to use certain defense strategies to protect themselves against oxidative stress.
In this review, we focus on protein folding in the ER, with particular emphasis on oxidative folding in several secretory tissues. We discuss how these cells use the UPR and other mechanisms to maintain ER homeostasis and other cellular functions in the face of high-level protein synthesis, and finally, we compare what is unique and what is similar between tissue types under physiologic and nonphysiologic conditions.
The ER as a Major Site of Protein Synthesis
Although the types and quantities of proteins synthesized in the ER vary dramatically between tissues, the basic mechanics of protein biosynthesis are largely the same (Fig. 2). Proteins that are destined for the secretory pathway are synthesized in the cytosol on ER-associated ribosomes. The presence of a hydrophobic signal sequence on the nascent polypeptide chain directs it to the ER membranes and plays a role in its insertion into the translocon, a proteinaceous channel that traverses the ER membrane (19, 46). This allows the protein to be translocated into the lumen of the ER, in many cases, as it is being synthesized. The elongating nascent chain first passes through a channel in the ribosome and then through the translocon (58, 135). It appears that the growing polypeptide chain remains largely unfolded during its transit through the ribosome and translocon, although recent studies indicate that some secondary structures can form during this process (20). After the chain enters the ER, N-terminal signal sequences are often removed by a signal peptidase that is positioned at the luminal side of the translocon.
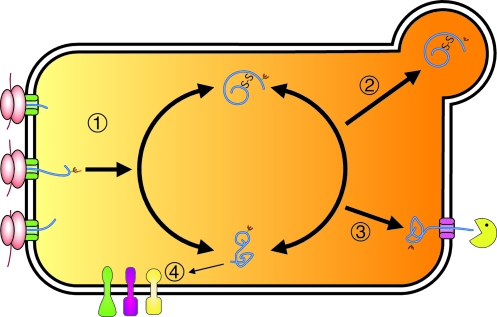
FIG. 2.
The ER is the site of secretory protein synthesis. (1) Proteins enter the ER co-translationally and begin to fold immediately. (2) Proteins that mature properly are transported to the Golgi, (3) whereas those that fail to fold are identified and retrotranslocated for degradation by the 26S proteasome. (4) In response to imbalances in the normal homeostasis of the ER, unfolded proteins accumulate and activate a signaling pathway known as the unfolded protein response (UPR). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Once the polypeptide chain emerges into the lumen, N-linked glycans are added co-translationally by the oligosaccharyl-transferase (OST) complex to asparagine residues that occur within the consensus N-X-S/T sequence (65), and the polypeptide chain often begins folding co-translationally (5, 17). However, for some proteins, folding occurs between regions that are very distal on the linear protein sequence, and thus are temporally separated in terms of protein synthesis (108).
The modification of secretory pathway proteins with N-linked glycans serves in part to limit the ways the nascent protein can fold. In addition, the ER environment itself causes further constraints on and benefits to protein folding and assembly. The calcium required for many signal-transduction pathways is stored here (83). Thus, proteins that are synthesized in this organelle have evolved to fold in a high-calcium environment, and changes in the ER calcium level can dramatically affect their folding (71, 75).
Like the extracellular space with which it is contiguous, the ER also possesses an oxidizing environment (53). This allows disulfide bonds to form between juxtaposed cysteine residues, which can serve to stabilize folded regions of the nascent chain (10). However, in the crowded environment of the ER lumen, where large numbers of unfolded polypeptide chains are being synthesized, the formation of disulfide bonds between the wrong regions of a protein or even between unrelated proteins could lead to misfolding or to the formation of large, insoluble aggregates (56). Surprisingly, this rarely occurs under normal conditions. Previously it was believed that the oxidizing environment of the ER was sufficient to promote disulfide bond formation directly (53). It is now clear that the oxidation of thiols or rearrangements of nonnative disulfide bonds is catalyzed by enzymes, which transfer oxidizing equivalents via a protein-relay system (28, 123) (Fig. 3). Disulfide bond formation is reversible, and the same catalytic proteins can often act to reduce disulfide bonds (109). The demands on this system can be particularly high in secretory cells. For example, plasma cells synthesize thousands of immunoglobulin M (IgM) pentamers per second (44). This requires the formation of ∼20 disulfide bonds per monomer or ∼100 disulfide bonds per pentamer, which means that ∼100,000 disulfide bonds are produced per second, and this does not include off-pathway products that must be reduced and reoxidized!
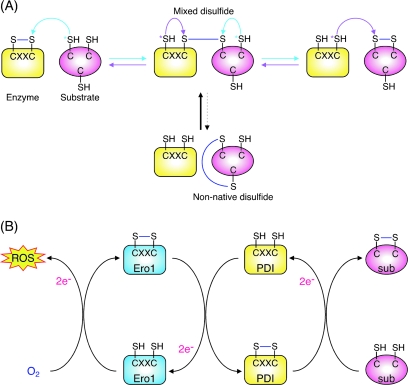
FIG. 3.
Mechanisms of disulfide bond formation and reduction. (A) Schematic of the intermediates formed during the transfer of disulfide bonds from an oxidoreductase to a substrate protein, as well as the mechanism for correcting off-pathway intermediates. (B) The pathway of disulfide bond transfer from Erol to PDI to a substrate and the flow of electrons to oxygen. This results in the production of one molecule of ROS per disulfide bond formed. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
The high fidelity of protein folding in the ER is due to a stringent quality-control apparatus (25). Newly synthesized proteins are carefully scrutinized by two major families of molecular chaperones, the Hsp70 cognate, BiP, and the resident lectin-like proteins, calnexin and calreticulin. BiP recognizes hydrophobic stretches of amino acids that are exposed on unfolded proteins, which disappear into the interior of the protein when folding is complete (7, 64). Calnexin and calreticulin interact with sugar residues on proteins that are characteristic of their folding status (21, 37) due to the interaction of UDP-glucosyltransferase with hydrophobic regions on the protein (15). The binding of these chaperones to unfolded proteins plays a critical role in preventing their aggregation, assisting in their correct folding, and finally retaining them in the ER (86). Only when all aspects of an unfolded state have disappeared do the chaperones cease to bind, and the newly synthesized protein is allowed to exit the ER for transport to the Golgi for further routing along the secretory pathway.
The regulated and carefully controlled assembly of subunit proteins into larger complexes also occurs in the ER and is monitored by the same quality-control apparatus. Proteins that ultimately fail to mature properly are targeted for intracellular degradation by the 26S proteasome (47, 81) (Fig. 2).
The Unfolded Protein Response
Although the resident ER molecular chaperones are expressed in all tissues and cell types, they are expressed highest in secretory tissues. This is achieved in part by sensing that the load of unfolded proteins in the ER is exceeding the capacity of ER chaperones to deal with them. The resulting accumulation of unfolded proteins triggers a signal-transduction cascade that has been termed the unfolded protein response (UPR). This response was first identified because of its activation by physiologic and pharmacologic conditions that impair normal protein folding in the ER (66, 71). The hallmark of the UPR is the upregulation of ER chaperones and folding enzymes, which are required to bind to unfolded proteins and prevent their aggregation (71). This component of the response is likely also to aid in the subsequent folding of these proteins if the stress conditions are alleviated.
A second part of the response is a transient attenuation of nascent protein synthesis (11, 49). Although it is not restricted to ER proteins, its effects appear to be greater in some cells for membrane-bound polysomes. Clearly this serves to limit the load of unfolded proteins under conditions that are not well suited to their proper maturation. The transient inhibition of protein synthesis also provides a window for the transcriptional upregulation of ER chaperones and folding enzymes (79), so the cell is better equipped with chaperones to accommodate the load of unfolded proteins when translation resumes. In addition, the degradative capacity of the cell is increased to aid in the turnover of misfolded proteins (70, 92), which also serves to limit the possibility of protein aggregation and destruction to the ER. These parts of the UPR are largely considered to be cytoprotective. However, if stress conditions are not resolved and normal homeostasis is not restored to the ER, then the UPR switches gears and guides the cell toward death to protect the organism (8, 88). How the balance between the cytoprotective and cytodestructive parts of the response is achieved is not well understood and varies greatly between cells, with some cell types being able to withstand ER stress for prolonged periods, and other cell types being extremely sensitive to stress.
Sensing stress in the ER
Three resident ER transmembrane sensors detect unfolded proteins in the ER to initiate three distinct UPR branches; inositol-requiring protein-1 (Ire1), activating transcription factor-6 (ATF6), and protein kinase RNA (PKR)-like ER kinase (PERK) (78). All the sensors have luminal domains that bind to the ER chaperone BiP under nonstress conditions. However, once unfolded proteins accumulate in the ER lumen, BiP is released from the UPR sensors to bind to the unfolded proteins, which triggers the activation of all three sensors simultaneously.
Ire1
Ire1 represents the most primitive of the UPR signal transducers and is conserved from yeast to humans (104). Two forms of Ire1 are present in mammalian cells, the ubiquitously expressed Ire1α (121) and Ire1β, which is expressed only in gut epithelium (134). When BiP is released from the luminal domain of Ire1 in response to ER stress, it dimerizes and autophosphorylates in trans (6) (Fig. 4). This activates an endoribonuclease activity encoded in the C-terminus of the cytoplasmic domain. The only target of the endonuclease activity of Ire1 to be identified thus far is the X-box binding protein-1 (XBP1) transcript, which, in the unspliced form, encodes a protein with a DNA-binding domain but no transactivation domain. The excision of 26 bases from the XBP1 transcript by activated Ire1 changes the reading frame of the C-terminus of XBP1, so that the spliced form of XBP1 now encodes both a DNA-binding domain and a transactivation domain (13, 141). The remodeled XBP1(S) regulates components of the ERAD pathway, like EDEM (142), cofactors of the ER chaperone BiP, including ERdj3 and ERdj4 (70), and components of lipid synthesis that play a role in the expansion of ER membranes during the differentiation of some secretory tissues (112, 117).
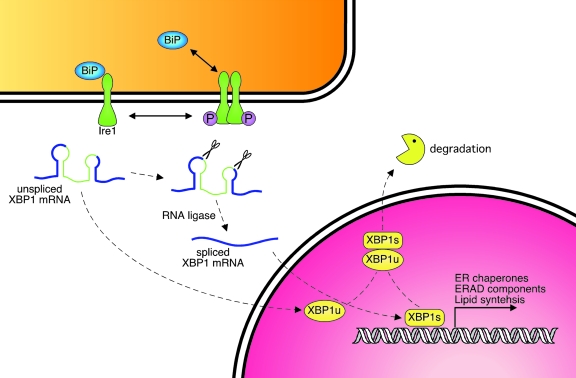
FIG. 4.
Ire1 signaling pathway. Ire1 exists as a BiP-bound monomer in the absence of ER stress. When unfolded proteins accumulate, BiP is released, and Ire1 forms homodimers that autophosphorylate and activate an endonuclease activity encoded in its C-terminus. Activated Ire1 removes 26 bases from the XBP1 transcript, which is relegated and now encodes a fully active transcription factor, XBP1(S). XBP1(S) transactivates a number of UPR targets and, later in the response, is negatively regulated via heterodimerization with the unspliced form of XBP1 [XBP1(U)]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Later in the ER stress response or when stress is relieved, unspliced XBP1 [XBP1(U)] acts as a negative regulator of XBP1(S) by forming an XBP1(U)-XBP1(S) heterodimer, which is transported to the cytosol for degradation (143). Ire1 activation was recently shown to target the degradation of some ER-attached mRNAs during ER stress in Drosophila (50), which would serve specifically to decrease protein synthesis in the ER. Recent studies demonstrate that both CD59 and insulin mRNAs are targeted by Ire1, suggesting that a similar destruction of ER-associated mRNAs may occur in mammalian cells during ER stress (74, 91).
Finally, in addition to its ribonuclease activity, Ire1 activates the c-Jun amino-terminal kinase (JNK) and caspase 12 via a direct interaction with TRAF2 (127, 140) and with proapoptotic Bcl-2 family members BAX and BAK (45). Thus, activated Ire1 is involved in both cytoprotective and cytodestructive aspects of the UPR. Its contribution to these opposing functions may vary temporally or between different tissues. Mice that are nullizygous for Ire1 (121) and XBP1 (101) have been produced, and both exhibit an embryonic lethal phenotype, which argues that these gene products are also likely to play a role in normal development.
PERK
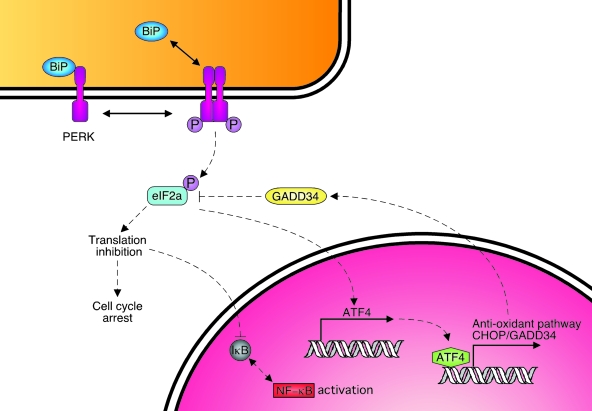
The second UPR transducer PERK is a member of the eIF2α kinase family (41, 115). Although PERK is not found in yeast, it is a component of the UPR signaling apparatus in all metazoans (114). Release of BiP in response to ER stress leads to PERK oligomerization and autophosphorylation in trans (Fig. 5). Activated PERK phosphorylates ser-51 of the α subunit of the translation-initiation factor-2 (eIF2α), which results in the inhibition of most cap-dependent translation (40). This is sufficient to affect 70–90% of the protein synthesis after UPR activation with conventional pharmacologic inducers like thapsigargin or DTT. Paradoxically, the phospho eIF2α-dependent translation arrest allows translation of the ATF4 transcription factor. ATF4 is constitutively transcribed, but the presence of a number of small open reading frames in the 5' untranslated region of the mRNA prevents translation initiation at the correct methionine (38). When general translation is dramatically reduced, ribosomes bind to the correct initiating methionine, and the ATF4 protein is synthesized. ATF4 in turn binds to the promoter of GADD34 (79), the regulatory subunit of the PP1 phosphatase, which serves to dephosphorylate eIF2α and restore cap-dependent translation (90).
FIG. 5.
PERK signaling pathway. Like Ire1, PERK exists as a BiP-bound monomer under normal physiologic conditions. On activation, PERK oligomerizes, and its kinase domain is activated. The target of this kinase is a component of the translation-initiation complex, EIF2α, which results in a transient inhibition of most cap-dependent translation. Downstream of the translational arrest is the activation of NF-κB due to loss of IκB, a G1 cell-cycle arrest, and paradoxically, the translation of ATF4, a transcription factor that upregulates a number of genes involved in ROS metabolism, as well as GADD34, which dephosphorylates eIF2α, allowing translation to resume. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Another target of ATF4 is the C/EBP homologous transcription factor CHOP (77). CHOP can heterodimerize with C/EBP family members, displacing them from their normal targets and allowing them to bind and transactivate new targets (103). In addition, CHOP homodimers have distinct targets of their own (126). Although CHOP induction occurs very early during UPR activation, it has been linked to apoptosis at later times in the stress response in some cell types and in various disease states (144).
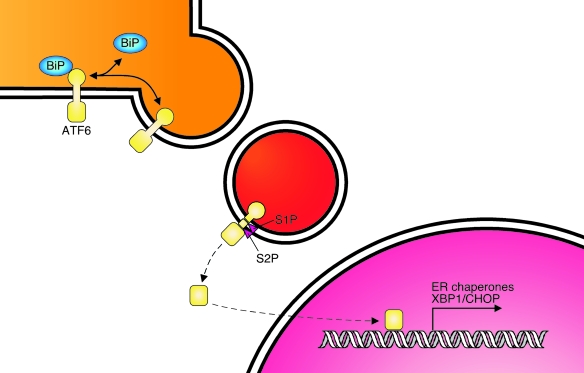
ATF6
The third ER stress sensor ATF6 is specific to metazoans and occurs in two isoforms in mammals (α and β), although ATF6α appears to play a larger transcriptional role in the UPR (1) (Fig. 6). ATF6 is retained in the ER during nonstress conditions via its association with BiP (113). Once BiP is released in response to the accumulation of unfolded proteins, ATF6 translocates to Golgi, which is regulated by two Golgi localization signals that are encoded within the luminal domains of these proteins (113). ATF6 then becomes a substrate of the site 1 and 2 (S1P and S2) proteases, which liberate the cytosolic domain that encodes the transcription factor (139). Cleaved ATF6 migrates to the nucleus to activate transcription of ER chaperones, a number of protein disulfide isomerases that catalyze the formation or reduction or both of disulfide bonds; Ero1β, which plays an important role in maintaining the oxidizing environment of the ER; several proteins involved in ER-associated degradation; and the XBP1 gene (1, 138). Studies using knockout MEFs revealed that ATF6α plays a major role in the induction of chaperones and folding enzymes, whereas ATF6β must heterodimerize with XBP1(S) to transactivate ERAD components. Interestingly, although mice that are null for either ATF6α or ATF6β develop normally, mice that are deficient in both ATF6 isoforms die in utero (1), arguing that functional redundancy exists between these two genes during development.
FIG. 6.
ATF6 signaling pathway. ATF6 is synthesized as a transmembrane protein with a luminal stress-sensing domain that binds to BiP and a cytosolically oriented transcription-factor domain. During ER stress, BiP is released, allowing it to be transported to the Golgi, where it is cleaved by S1P and S2P proteases. The liberated transcription factor trafficks to the nucleus and upregulates ER chaperones and the XBP1 mRNA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Cytoprotective versus cytodestructive aspects of the UPR
It appears that, in most cases, the UPR can be considered a cytoprotective response. In addition to the prosurvival aspects of the response outlined earlier, a number of antiapoptotic responses are initiated. For instance, the PERK-induced translational arrest leads to the loss of IκB, thereby activating NF-κB (57). This in turn upregulates antiapoptotic proteins like Bcl2 (82). Other studies suggest that a pool of BiP, which is upregulated by the ATF6 branch of the UPR, can relocalize to the cytosol and bind to caspase-7 and caspase-12, which prevents their activation (99, 100). Finally, glycogen synthase kinase-3β (GSK3β) is activated by NF-κB in response to ER stress and can contribute to cell survival by phosphorylating p53, which accelerates its degradation (97).
However, it is clear that if stress conditions persist for long periods or are particularly severe, the UPR also can lead to the activation of apoptotic cascades that kill the cell to protect the organ or organism. In addition to the three resident ER UPR transducers described earlier, a dedicated ER stress–inducible caspase has been identified. Procaspase-12 is localized to the cytosolic face of the ER membrane (87). Mice that are null for caspase-12 are more resistant to tunicamycin-induced apoptosis. Although the human caspase-12 gene contains several missense mutations and clearly cannot have a role in UPR-induced apoptosis, recent data indicate that caspase-4 in humans is homologous to murine caspase-12 and is activated in an ER-stress–specific manner, indicating that it might be the human caspase-12 orthologue (48).
Caspase-12/4 activation is a component of the intrinsic cell-death pathway and leads to the downstream cleavage of procaspase-9 and −3 (87). In addition to activation of the caspase cascade, several proapoptotic genes also are activated by the UPR pathway. CHOP, which is proapoptotic in a number of settings, downregulates antiapoptotic proteins like Bcl2 and increases free oxygen species, causing mitochondrial membrane damage and cytochrome c release, which induces apoptosis (82). The increases in cytosolic calcium that occur during UPR activation lead to upregulation of the proapoptotic protein BAD, leading to further cytochrome c release and activation of APAF1 (132).
The higher cytosolic calcium levels observed during ER stress also can induce calpain activation, which in turn leads to procaspase-12 cleavage (87) and activation of the caspase-9 cascade (85). Finally, during ER stress, Ire1 can recruit TRAF2 and induce procaspase-12 clustering and activation (140). Ask1 can be recruited to the Ire1/TRAF2 complex and activated, leading to JNK activation. JNK in turn induces the proapoptotic protein Bim (72, 96) and inhibits the antiapoptotic protein Bcl2 (137).
Oxidative Folding and ROS
To prepare proteins for ultimate expression outside the cell, which is an oxidizing environment, the ER of eukaryotic cells has evolved an appropriate environment and a host of proteins that can catalyze the formation of disulfide bonds between two thiol groups (-SH) on juxtaposed cysteine residues. Oxidative folding is dependent on the maintenance of an oxidizing environment in the ER, as disrupting this environment with agents like DTT inhibits the formation of disulfide bonds (9).
Recently, significant advances have occurred in our understanding of this process, which reveal that oxidative folding is an enzyme-driven reaction that involves a cascade of disulfide bond transfers from at least two enzymes to the substrate protein (109, 125) (Fig. 3). Enzymes that catalyze disulfide bond formation are referred to as oxidoreductases, because they usually have the ability both to initiate the formation of a disulfide bond during oxidative folding, and to reduce disulfide bonds. The reduction of disulfides is important for the isomerization of incorrect bonds that can form during folding (56, 69) and to prepare partially folded proteins for retrotranslocation and degradation (128). The activity of an oxidoreductase is dependent on the oxidative state of its active site, with the enzyme serving as an oxidase when the site is oxidized and as an isomerase or reductase when it is reduced (109, 125).
The disulfide-exchange reaction is initiated when a free thiol in the substrate is deprotonated, allowing it to interact with the disulfide bond on the oxidoreductase enzyme. This leads to an intermediate step in which the disulfide bond in the donor is reduced, permitting it to form a covalent bond with a cysteine in the acceptor protein. The resulting “mixed disulfide bond” is then resolved by a second exchange reaction in which the remaining thiolate anion of the substrate attacks the mixed-disulfide bond, allowing the bond to form within the substrate (109, 125). Once PDI donates its disulfide bond to a substrate, the resulting reduced active site must be reoxidized to catalyze another disulfide bond.
A combination of genetic and biochemical studies in yeast led to the discovery of an FAD-bound, ER oxidase, Ero1p (30, 123). Ero1p is tightly associated with the luminal face of the ER membrane and is essential to oxidative folding in yeast (29, 95) due to its ability to oxidize PDI, although it has no homology to any other redox enzymes. Ero1p is directly oxidized by molecular oxygen in a flavin-dependent reaction and is highly responsive to small changes in physiologic levels of free FAD, which explains the dependence of oxidative folding on cellular FAD levels (124). FAD is synthesized in the cytosol but can readily enter the ER lumen and promote Ero1p-catalyzed oxidation. Two Ero1p homologues exist in mammalian cells, Ero1α and β (12, 94), which are also flavo-enzymes that are oxidized by molecular oxygen (133). Consistent with its role in protein folding, both yeast Ero1p and human Ero1β are induced by the unfolded protein response. The disulfide reaction involves a flux of electrons from the substrate acceptor of the disulfide bond to the oxidoreductase enzyme to Ero1, which is consequently passed to molecular oxygen. This results in the production of stoichiometric amounts of reactive oxygen species (ROS) (36). In the case of isomerization or reductase activities, the flow of electrons and passage of disulfide bonds occurs in the opposite direction.
The best-characterized oxidoreductase enzyme is protein disulfide isomerase (PDI), which has four thioredoxin-like folds (31). Two of these contain CXXC catalytic sites, which must be oxidized to transfer this bond to the substrate, and at least one of the other thioredoxin-like folds is involved in substrate recognition (63). PDI exists largely in two semioxidized states in vivo (i.e., either the first or the second active site is oxidized), arguing that it can function as an oxidase or as a reductase/isomerase (2).
Nearly 20 PDI-like family members have been identified in human cells, which display a wide range of domain complements and active-site chemistries, and about half of them have been shown to have oxidoreductase activity in vitro (3). The large number of PDI-like proteins in the mammalian ER allows them to participate in specific functions. One of these, ERp57, interacts with the glycoprotein-specific ER chaperones calnexin and calreticulin and assists in the formation of disulfide bonds in glycosylated proteins (93). Another, ERdj5, acts as a reductase to promote protein unfolding to prepare misfolded proteins for retrotranslocation and degradation via the ERAD pathway (128). Although several of these (i.e., PDIp) have limited tissue expression, which may suggest a unique function or substrate, in most cases, specific roles for these oxidoreductases are not well understood. This is probably because of difficulty in determining their natural substrates, because the formation of mixed disulfides with substrates is usually very transient, and thus, it is very hard to detect endogenous enzyme–substrate complexes (2).
Integrative Stress Response
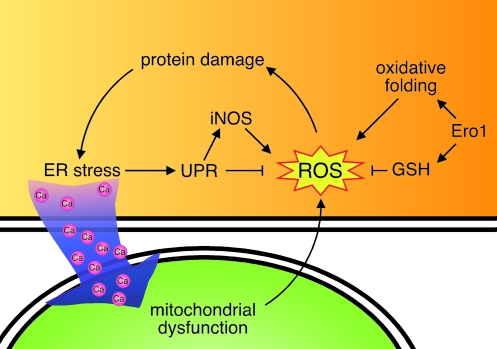
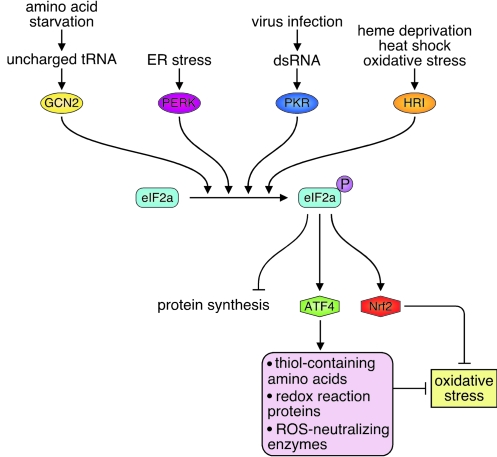
Because molecular oxygen serves as the electron acceptor for the disulfide reaction, it would appear that one molecule of hydrogen peroxide should be generated for each disulfide bond formed (36), suggesting that its activity must be regulated according to the folding load to protect the cell from the consequences of oxidative folding. In bacteria, this is controlled by coupling oxidative folding to the respiratory chain, which can reduce molecular oxygen to water (4). However, in eukaryotic cells, oxidative folding occurs in the ER, and respiration, in the mitochondria. Although the mechanism for dealing with the reactive oxygen species (ROS) generated in eukaryotic cells remains unclear, several recent studies shed some light on this problem (Fig. 7). Ero1 activity is regulated by reduction of its CXXC sites by reduced glutathione (GSH) (84; 111). Increased Ero1p activity, which occurs in secretory cells, leads to enhanced GSH synthesis (84), which in turn can inactivate ROS. Thus, it would appear that Ero1 is both a major contributor to the problem by catalyzing oxidative folding as well as part of the solution. A second level of controlling ROS levels is achieved through UPR induction, which can occur in response to oxidative damage to ER proteins (61, 68). The activation of PERK appears to play a critical role in controlling this damage, as studies in Caenorhabditis elegans revealed that stressing worms that lack PERK leads to significant accumulation of peroxide (42). This can be eliminated by reducing Ero1 levels, presumably because oxidative folding is dramatically decreased. The PERK-mediated phosphorylation of eIF2α leads to a translational arrest (40), which both reduces the number of proteins that would be undergoing oxidative folding and allows translation of ATF4. A number of different cellular stress conditions activate other eIF2α kinases and therefore also induce ATF4 expression (38) and its downstream targets, which has led to this branch of the UPR being referred to as the integrative stress response (42) (Fig. 8). Studies on ATF4-null cells revealed that ATF4 regulates a number of genes that are important in protection from oxidative stress. These include genes involved in the import and metabolism of thiol-containing amino acids that serve as precursors to glutathione, and proteins involved in redox reactions like heme oxygenase-1 (42), glutathione peroxidase (62), and peroxiredoxin-1 (62), which protect the cell from oxidative stress. Finally, the enhanced degradation that is a component of the UPR also appears to be important in controlling oxidative stress. Several studies have shown that inhibiting ERAD with proteasomal inhibitors leads to increased ROS accumulation in cells (52, 68).
FIG. 7.
The UPR and ROS. Ero1-catalyzed oxidative folding leads to the production of ROS, which in turn can damage proteins in the ER and activate the UPR, leading to the production of proteins that detoxify ROS. Conversely, UPR activation causes calcium release from the ER, which leads to mitochondrial dysfunction and the production of more ROS. Ero1 also leads to increases in GSH, which suppresses ROS, demonstrating the opposing functions of Ero1 and the UPR in regulating ROS. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
FIG. 8.
The integrative stress response. Four different eIF2α kinases have been identified in mammalian cells: GCN2, PERK, PKR, and HRI, which share the responses downstream of eIF2α phosphorylation, including ATF4 upregulation. ATF4 is responsible for much of the cellular response to combat ROS production in response to oxidative folding, including increased production of thiol-containing amino acids, which are precursors to GSH and proteins that regulate redox reactions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article at www.liebertonline.com/ars).
Although UPR activation can neutralize the affects of ROS production, it is clear that, in some cases, it can also contribute to the problem (Fig. 7). The leakage of Ca2+ from the ER in response to ER stress is taken up by the mitochondria, where it disrupts the electron-transport chain that produces ATP and generates ROS via multiple pathways (14, 23). Second, activation of the UPR in response to pharmacologic agents can lead to the upregulation of the gene for inducible nitric oxide synthase (iNOS), which generates nitric oxide and in turn produces ROS through both calcium-dependent and calcium-independent pathways (67). However, iNOS also regulates genes involved in thiol metabolism, which serves to limit the build-up of ROS (62), and pretreatment of a murine fibrosarcoma line with a UPR inducer diminished ROS production in response to TNF-α and reduced cell death (136). Together these data argue that, although the UPR can contribute to ROS production, it largely serves to protect cells by scavenging ROS and even repairing cellular damage (18).
Protein Folding in the Secretory Tissues
Secretory tissues like liver, pancreas, and plasma cells have a greatly expanded ER, which is populated with increased levels of molecular chaperones and folding enzymes to accommodate the large volume of proteins that are being synthesized for export. The vast majority of these nascent proteins undergo oxidative folding in the ER, which should lead to the production of large quantities of ROS. It has been estimated that Ero1-mediated folding could account for ∼25% of cellular ROS production during protein synthesis in the average cell, and this is likely to be even higher in secretory cells (125). Thus, it is reasonable to assume that these cells have developed specific ways to neutralize ROS and maintain themselves as protein factories, although at present, this has not been well studied. The unique properties of two very different types of secretory tissues and the distinct strategies they use that allow them to produce large quantities of secreted proteins that undergo oxidative folding provide insights into this important problem.
Pancreatic β cells
Pancreatic β islet cells are specialized in the production and secretion of insulin. In the absence of normal insulin synthesis and secretion, blood glucose levels increase and can result in diabetes. Conversely, excess insulin can lead to abnormally low blood glucose levels, termed hypoglycemia. Therefore, it is very important to maintain proper insulin levels. The pancreatic β cells produce insulin in response to increased blood glucose levels, and when blood glucose levels decrease, insulin synthesis is negatively regulated (105). Although a modest transcriptional upregulation of the insulin gene occurs when glucose levels increase or decrease, the majority of insulin regulation occurs posttranscriptionally at the level of both translation and regulated secretion. Insulin is synthesized as preproinsulin. Soon after translocation into the ER is initiated, the signal sequence is cleaved, and the soluble insulin precursor, proinsulin, begins to fold, which includes the formation of three intrachain disulfide bonds (35, 105). Once folding is complete, proinsulin is transported to the Golgi, where it undergoes further proteolyic processing by the PC1 and PC2 proteases, which remove an internal peptide. The mature insulin is then packaged into secretory granules and can be rapidly released as the need arises. Under hypoglycemic conditions, the pancreatic β−islet cells contain a large pool of insulin mRNA that can represent up to 20% of the total mRNA in these cells, which is transcriptionally quiescent (35, 130). This pool of insulin transcripts is rapidly recruited to polysomes in response to transient increases in the blood glucose level, which can lead to an ∼50-fold increase in insulin biosynthesis (35). Under these conditions, the insulin precursor can compose up to 50% of the total protein synthesized in the β cell (107, 130), and the rate of insulin translation in β cells can approach ∼1 million molecules per minute per cell (105). Given that three disulfide bonds are formed per molecule of insulin, this would conceivably result in the production of 3 million molecules of ROS being generated per minute! However, pancreatic β cells express relatively low levels of enzymes that neutralize ROS or are redox regulating (i.e., catalase, glutathione peroxidase, superoxide dismutase, and thioredoxin) (62, 89), arguing that they should be particularly sensitive to oxidative stress. In contrast, expression of genes encoding the rate-limiting enzyme for glutathione synthesis (i.e., γ-glutamylcysteine ligase) is similar in β-islet cells compared with other metabolic tissues, indicating that glutathione (GSH) may be important in suppressing ROS in these cells (122).
High-glucose treatment reduces ratio of GSH/GSSG, and this ratio was further decreased by inhibiting ERAD by proteasomal inhibitors (62). Under these conditions, γ-GCL, glutathione peroxidase, and peroxiredoin-1 were upregulated. However, inhibition of nitric oxide synthase weakened the effect, suggesting that, in some cases, nitric oxide may also play a protective role against ROS in β cells.
The balance between nitric oxide and ROS appears to be particularly important, as reaction of nitric acid with superoxide can generate peroxynitrite, which is a potent oxidant and can lead to apoptosis in multiple tissues, including the pancreas (24, 26). The accumulation of peroxynitrite can also induce the ER stress response and activate PERK, which is important in protecting against apoptosis (76). In other studies, the accumulation of ROS was shown to induce apoptosis in insulin-producing cells via c-Jun N-terminal protein kinase (JNK) and p38 MAPK activation (51). In addition, ROS suppresses the transcription of the insulin gene by decreasing the expression of pancreatic and duodenal homeobox factor-1 (PDX-1), which controls proinsulin mRNA levels and plays a major role in maintaining normal β-cell function (118).
Antioxidant treatment or overexpression of antioxidant enzymes in β-cells can restore the expression of insulin suppressed by high levels of glucose (119, 120), and using antioxidants to treat an animal model of diabetes preserved insulin expression and protected against decreases in pancreas size (59, 119). These data indicate that oxidative stress occurs in pancreatic cells and is tightly linked to the β-cell dysfunction and the onset of the diabetes.
It appears that a major mechanism for limiting oxidative stress in the pancreatic β cells is through controlling the translation of insulin. This largely occurs via changes in the phosphorylation status of eIF2α (105). The role of the UPR in regulating this was revealed in studies on the PERK-null mice, which develop diabetes (39). eIF2α is constitutively active at low levels in pancreatic cells under normal physiologic conditions. In the absence of PERK, insulin synthesis is initially much higher than in control littermates, which ultimately leads to increased cell death of the exocrine cells, and finally to hyperglycemia (39). These studies indicate that the ER stress-induced translational attenuation that occurs with glucose stimulation of the pancreatic β cell is critical for proper control of insulin production. However, mice that are heterozygous for mutation in the phosphorylation site of eIF2α (Ser 51 → A) also develop diabetes when fed a high-fat diet because of the reduced insulin secretion (106), underscoring that the translational control of insulin production must be carefully regulated.
Further investigation revealed that eIF2α is quickly dephosphorylated by protein phosphatase-1 (PP-1) when β cells are stimulated by glucose (131). Conversely, eIF2α is phosphorylated in response to decreases in glucose concentrations, which is also primarily mediated by the activation of PERK (34). How PP-1 and PERK are activated under hyper- or hypoglycemic conditions, respectively, is currently unclear, but it seems that both require novel upstream signaling molecules that are not used in the regular UPR pathway, and the balance of the activities of PP-1 versus PERK may be the largest factor that regulates the synthesis of insulin. Importantly, mutations in PERK were found to cause Wolcot-Rallison syndrome, a rare human autosomal recessive genetic disorder characterized by early infancy type I diabetes (22).
In addition to the PERK pathway, Ire1α is also constitutively activated in the pancreas (55). Ire1 phosphorylation is coupled to insulin biosynthesis in response to transient high-glucose treatment of pancreatic islet cells, and reducing Ire1 levels with shRNA interfered with insulin synthesis under these conditions (73). The importance of this pathway in insulin synthesis was further demonstrated by studies that revealed that Wolfram syndrome, another genetic form of juvenile diabetes, is caused by mutations in WFS1, a transmembrane ER protein that is regulated by Ire1 and plays an important role in ER homeostasis in pancreatic β cells (27). In contrast, when β cells are exposed to prolonged high glucose, Ire1α activation leads to the suppression of insulin gene expression (73). This was found to be due in part to an Ire1α-mediated degradation of insulin transcripts (74). This adaptation may play a critical role in maintaining homeostasis in pancreatic β cells and could explain why the β cells in type 2 diabetes patients with chronic hyperglycemia stop producing insulin, which occurs in the absence of apoptosis.
Plasma cells
Unlike pancreatic cells, most plasma cells are very short lived to limit the production of antibody once its specific antigen has been eliminated. These cells therefore encounter different types of problems and use different strategies to cope with them. Quiescent 8 lymphocytes are the precursor to antibody-secreting plasma cells. They have very little ER and are largely metabolically inactive (44). Once they are activated by antigens or mitogens, they undergo terminal differentiation to become plasma cells, which produce and secrete massive amounts of immunoglobulin (Ig) to protect the organism from microbial pathogens. Antibodies are assembled from two identical heavy chains (HCs) and two identical light chains (LCs), both of which are composed of multiple Ig domains. Each domain has one intrachain disulfide bond, and the HCs are linked by interchain disulfide bonds to form homodimers, to which each of the two LCs are added through an additional interchain disulfide bond. In the case of IgM and IgA, these monomers are further assembled into disulfide-bridged polymers (pentamers and dimers, respectively) with the aid of a J chain. In IgM-producing plasma cells that have been estimated to produce up to 1,000 antibodies per second (44), this entails the formation of ∼100,000 disulfide bonds per second, which should result in the formation of 100,000 molecules of ROS! However, it is important to note that no measurements of ROS accumulation in plasma cells have been performed. The activation of B cells by antigen is followed by dramatic changes in the intracellular environment, including explosive expansion of the ER and other organelles of the secretory pathway, upregulation of molecular chaperones and folding enzymes, and increased production of mitochondria to supply the energy required (112, 129). This process begins within a day of stimulation and lasts only a few days before the plasma cells undergo an apoptotic death.
The UPR plays a critical role in terminal plasma cell differentiation and contributes to the expansion of the secretory-pathway organelles, the increased production of mitochondria, and the upregulation of chaperones, folding enzymes, and proteins involved in amino acid synthesis, metabolism, and redox regulation of the ER (112). However, this does not appear to be a conventional UPR, as only some branches are activated, and the signal for activation is not clear, because UPR induction occurs before the massive increase in Ig synthesis in the ER (32, 54).
XBP1 was shown to be required for plasma cell differentiation (102) well before it was known that XBP1 must undergo Ire-1–mediated splicing to become a fully functional transcription factor. It was subsequently shown that only the spliced form of XBP1 supported differentiation (54). Studies on B cell lines and splenic B cells demonstrated that soon after LPS stimulation, XBP1 splicing could be detected, as well as the upregulation of its downstream targets. Microarray studies on differentiating plasma cells revealed that XBP1 is responsible for the vast majority of the architectural changes that take place, as well as the increases in proteins needed to support them (112). XBP1 also plays a role in increasing the degradative capacity of the cell and upregulating Ero1β. In addition, ATF6 cleavage occurs during differentiation, leading to increased transcription of molecular chaperones and enzymes that are essential to oxidative folding (138). However, unlike the conventional UPR, PERK does not appear to be activated, because PERK and eIF2α phosphorylation are not detected during plasma cell differentiation, nor is either GADD34 or CHOP induction observed (32). However, this is not because the PERK pathway is nonfunctional in B-lineage cells, because they can activate this branch in response to conventional UPR inducers like tunicamycin and thapsigargin.
Thus, whereas pancreatic cells use the PERK branch of the UPR to control protein synthesis (39), thus allowing them to limit oxidative stress and the demands on the system, plasma cells appear to use a “go for broke” strategy, which allows them to continue to produce extremely high levels of protein during this period. This underscores a dramatic difference between these two types of tissues. Whereas plasma cells are very short lived and therefore less sensitive to long-term effects of oxidative damage to DNA and other macromolecules, pancreatic cells must continuously deal with the consequences of the ROS produced in response to insulin synthesis. Plasma cells do use general methods for dealing with oxidative stress, including the upregulation of NADPH oxidases like NOXA2, Prx1, and a plasma cell–specific oxidoreductase, PC-TRP (80). In addition, Nrf-2, a redox-regulated transcription factor, is induced and mediates upregulation of glutathione S-transferase and glutamyl cysteine ligase, which are rate-limiting genes for GSH synthesis, as well as heme oxygenase-1, phase II detoxifying enzymes, and other antioxidant factors (80). Perhaps the upregulation of these enzymes allows them somewhat to limit oxidative damage during the relatively short lifetime of a plasma cell. Ultimately after only about 4 days of high-level protein synthesis, plasma cells succumb to an apoptotic death. It is likely that amino acid supplies and energy production become limiting to the plasma cell. The biosynthesis of any protein is likely to result in a certain number of misfolded proteins that must be identified, corrected, or targeted for degradation. It seems likely that the number of misfolded Ig proteins might increase as amino acid synthesis and ATP production are saturated. Studies have shown that the degradative capacity of the plasma cell is diminished at later stages, perhaps because the proteasomes are also saturated with misfolded proteins that must be disposed of (16). This correlates with stabilization of proteins like IκB, thus diminishing the protective role of NF-κB, as well as proapoptotic factors like Bim and Bax (16).
Finally, it is very likely that perturbations in the ER redox potential and accumulated ROS contribute to plasma cell death (80). Although the majority of plasma cells are short lived, a small proportion migrate to the bone marrow and become long-lived plasma cells, which continue to produce large quantities of Ig (98). How these cells are able to continue to do so without succumbing to apoptosis is not well understood. It is conceivable that they use strategies similar to those of pancreatic cells, or alternatively, that lineage-specific factors to deal with oxidative damage are induced.
In conclusion, the formation of disulfide bonds in secretory-pathway proteins is essential to their proper folding. However the byproduct of each bond formed is a molecule of ROS. This would seem to be an extremely high price for secretory tissues to pay to produce large quantities of secreted proteins. Recent studies have shed light on some of the strategies that these tissues use, although clearly, much more research is required to understand this very important problem of biology.
Abbreviations Used
- ATF6
activating transcription factor 6
- CHOP
C/EBP homologous protein
- eIF2α
eukaryotic initiation factor-2α
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- Ero1
ER oxidase 1
- GSH
reduced glutathione
- Ig
immunoglobulin
- Ire1
inositol-requiring protein 1
- OST
oligosaccharyl transferase
- PDI
protein disulfide isomerase
- PERK
PKR-like ER kinase
- PP-1
protein phosphatase-1
- ROS
reactive oxygen species
- S1P/S2P
site 1 protease, site 2 protease
- UPR
unfolded protein response
- XBP1
X box–binding protein-1
- XBP1(S)
spliced form of XBP1
- XBP1(U)
unspliced form of XBP1
Acknowledgments
This work is supported by NIH grant GM54068 (LMH), the Cancer Center CORE grant CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital.
References
- 1.Adachi Y. Yamamoto K. Okada T. Yoshida H. Harada A. Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33:75–89. doi: 10.1247/csf.07044. [DOI] [PubMed] [Google Scholar]
- 2.Appenzeller-Herzog C. Ellgaard L. In vivo reduction-oxidation state of protein disulfide isomerase: the two active sites independently occur in the reduced and oxidized forms. Antioxid Redox Signal. 2008;10:55–64. doi: 10.1089/ars.2007.1837. [DOI] [PubMed] [Google Scholar]
- 3.Appenzeller-Herzog C. Ellgaard L. The human PDI family: versatility packed into a single fold. Biochim Biophys Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Bader M. Muse W. Ballou DP. Gassner C. Bardwell JC. Oxidative protein folding is driven by the electron transport system. Cell. 1999;98:217–227. doi: 10.1016/s0092-8674(00)81016-8. [DOI] [PubMed] [Google Scholar]
- 5.Bergman LW. Kuehl WM. Co-translational modification of nascent immunoglobulin heavy and light chains. J Supramol Struct. 1979;11:9–24. doi: 10.1002/jss.400110103. [DOI] [PubMed] [Google Scholar]
- 6.Bertolotti A. Zhang Y. Hendershot LM. Harding HP. Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 7.Blond-Elguindi S. Cwirla SE. Dower WJ. Lipshutz RJ. Sprang SR. Sambrook JF. Gething MJ. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 8.Boyce M. Yuan J. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death Differ. 2006;13:363–373. doi: 10.1038/sj.cdd.4401817. [DOI] [PubMed] [Google Scholar]
- 9.Braakman I. Helenius J. Helenius A. Manipulating disulfide bond formation and protein folding in the endoplasmic reticulum. EMBO J. 1992;11:1717–1722. doi: 10.1002/j.1460-2075.1992.tb05223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Braakman I. Helenius J. Helenius A. Role of ATP and disulphide bonds during protein folding in the endoplasmic reticulum. Nature. 1992;356:260–262. doi: 10.1038/356260a0. [DOI] [PubMed] [Google Scholar]
- 11.Brostrom CO. Prostko CR. Kaufman RJ. Brostrom MA. Inhibition of translational initiation by activators of the glucose-regulated stress protein and heat shock protein stress response systems: role of the interferon-inducible double-stranded RNA-activated eukaryotic initiation factor 2alpha kinase. J Biol Chem. 1996;271:24995–25002. doi: 10.1074/jbc.271.40.24995. [DOI] [PubMed] [Google Scholar]
- 12.Cabibbo A. Pagani M. Fabbri M. Rocchi M. Farmery MR. Bulleid NJ. Sitia R. ERO1-L, a human protein that favors disulfide bond formation in the endoplasmic reticulum. J Biol Chem. 2000;275:4827–4833. doi: 10.1074/jbc.275.7.4827. [DOI] [PubMed] [Google Scholar]
- 13.Calfon M. Zeng H. Urano F. Till JH. Hubbard SR. Harding HP. Clask SG. Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 14.Camello-Almaraz C. Gomez-Pinilla PJ. Pozo MJ. Camello PJ. Age-related alterations in Ca2+ signals and mitochondrial membrane potential in exocrine cells are prevented by melatonin. J Pineal Res. 2008;45:191–198. doi: 10.1111/j.1600-079X.2008.00576.x. [DOI] [PubMed] [Google Scholar]
- 15.Caramelo JJ. Castro OA. de Prat-Gay G. Parodi AJ. The endoplasmic reticulum glucosyltransferase recognizes nearly native glycoprotein folding intermediates. J Biol Chem. 2004;279 (44 ):46280–46285. doi: 10.1074/jbc.M408404200. Epub 2004 Aug 279: 46280–46285, 2004. [DOI] [PubMed] [Google Scholar]
- 16.Cenci S. Mezghrani A. Cascio P. Bianchi G. Cerruti F. Fra A. Lelouard H. Masciarelli S. Mattioli L. Oliva L. Orsi A. Pasqualetto E. Pierre P. Ruffato E. Tagliavacca L. Sitia R. Progressively impaired proteasomal capacity during terminal plasma cell differentiation. EMBO J. 2006;25:1104–13. doi: 10.1038/sj.emboj.7601009. Epub 2006 Feb 23 25: 1104–1113, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W. Helenius J. Braakman I. Helenius A. Cotranslational folding and calnexin binding during glycoprotein synthesis. Proc Natl Acad Sci U S A. 1995;92:6229–6233. doi: 10.1073/pnas.92.14.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.costa-Alvear D. Zhou Y. Blais A. Tsikitis M. Lents NH. Arias C. Lennon CJ. Kluger Y. Dynlacht BD. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol Cell. 2007;27:53–66. doi: 10.1016/j.molcel.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 19.Crowley KS. Liao S. Worrell VE. Reinhart GD. Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- 20.Daniel CJ. Conti B. Johnson AE. Skach WR. Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J Biol Chem. 2008;283:20864–20873. doi: 10.1074/jbc.M803517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daniels R. Kurowski B. Johnson AE. Hebert DN. N-linked glycans direct the cotranslational folding pathway of influenza hemagglutinin. Mol Cell. 2003;11:79–90. doi: 10.1016/s1097-2765(02)00821-3. [DOI] [PubMed] [Google Scholar]
- 22.Delepine M. Nicolino M. Barrett T. Golamaully M. Lathrop GM. Julier C. EIF2AK3, encoding translation initiation factor 2-alpha kinase 3, is mutated in patients with Wolcott-Rallison syndrome. Nat Genet. 2000;25:406–409. doi: 10.1038/78085. [DOI] [PubMed] [Google Scholar]
- 23.Deniaud A. Sharaf el DO. Maillier E. Poncet D. Kroemer G. Lemaire C. Brenner C. Endoplasmic reticulum stress induces calcium-dependent permeability transition, mitochondrial outer membrane permeabilization and apoptosis. Oncogene. 2008;27:285–299. doi: 10.1038/sj.onc.1210638. [DOI] [PubMed] [Google Scholar]
- 24.Dickhout JG. Hossain GS. Pozza LM. Zhou J. Lhotak S. Austin RC. Peroxynitrite causes endoplasmic reticulum stress and apoptosis in human vascular endothelium: implications in atherogenesis. Arterioscler Thromb Vasc Biol. 2005;25:2623–2629. doi: 10.1161/01.ATV.0000189159.96900.d9. [DOI] [PubMed] [Google Scholar]
- 25.Ellgaard L. Helenius A. Quality control in the endoplasmic reticulum. Nat Rev Mol Cell Biol. 2003;4:181–191. doi: 10.1038/nrm1052. [DOI] [PubMed] [Google Scholar]
- 26.Emre Y. Hurtaud C. Karaca M. Nubel T. Zavala F. Ricquier D. Role of uncoupling protein UCP2 in cell-mediated immunity: how macrophage-mediated insulitis is accelerated in a model of autoimmune diabetes. Proc Natl Acad Sci U S A. 2007;104:19085–19090. doi: 10.1073/pnas.0709557104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fonseca SG. Fukuma M. Lipson KL. Nguyen LX. Allen JR. Oka Y. Urano F. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic beta-cells. J Biol Chem. 2005;280:39609–39615. doi: 10.1074/jbc.M507426200. [DOI] [PubMed] [Google Scholar]
- 28.Frand AR. Cuozzo JW. Kaiser CA. Pathways for protein disulphide bond formation. Trends Cell Biol. 2000;10:203–210. doi: 10.1016/s0962-8924(00)01745-1. [DOI] [PubMed] [Google Scholar]
- 29.Frand AR. Kaiser CA. The ERO1 gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell. 1998;1:161–170. doi: 10.1016/s1097-2765(00)80017-9. [DOI] [PubMed] [Google Scholar]
- 30.Frand AR. Kaiser CA. Ero1p oxidizes protein disulfide isomerase in a pathway for disulfide bond formation in the endoplasmic reticulum. Mol Cell. 1999;4:469–477. doi: 10.1016/s1097-2765(00)80198-7. [DOI] [PubMed] [Google Scholar]
- 31.Freedman RB. Klappa P. Ruddock LW. Protein disulfide isomerases exploit synergy between catalytic and specific binding domains. EMBO Rep. 2002;3:136–140. doi: 10.1093/embo-reports/kvf035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gass JN. Gifford NM. Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277:49047–49054. doi: 10.1074/jbc.M205011200. [DOI] [PubMed] [Google Scholar]
- 33.Gething MJ. Sambrook J. Protein folding in the cell. Nature. 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 34.Gomez E. Powell ML. Bevington A. Herbert TP. A decrease in cellular energy status stimulates PERK-dependent eIF2alpha phosphorylation and regulates protein synthesis in pancreatic beta-cells. Biochem J. 2008;410:485–493. doi: 10.1042/BJ20071367. [DOI] [PubMed] [Google Scholar]
- 35.Goodge KA. Hutton JC. Translational regulation of proinsulin biosynthesis and proinsulin conversion in the pancreatic beta-cell. Semin Cell Dev Biol. 2000;11:235–242. doi: 10.1006/scdb.2000.0172. [DOI] [PubMed] [Google Scholar]
- 36.Gross E. Sevier CS. Heldman N. Vitu E. Bentzur M. Kaiser CA. Thorpe C. Fass D. Generating disulfides enzymatically: reaction products and electron acceptors of the endoplasmic reticulum thiol oxidase Ero1p. Proc Natl Acad Sci U S A. 2006;103:299–304. doi: 10.1073/pnas.0506448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammond C. Braakman I. Helenius A. Role of N-linked oligosaccharide recognition, glucose trimming, and calnexin in glycoprotein folding and quality control. Proc Natl Acad Sci U S A. 1994;91:913–917. doi: 10.1073/pnas.91.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harding HP. Novoa I. Zhang Y. Zeng H. Wek R. Schapira M. Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 39.Harding HP. Zeng H. Zhang Y. Jungries R. Chung P. Plesken H. Sabatini DD. Ron D. Diabetes mellitus and exocrine pancreatic dysfunction in perk-/- mice reveals a role for translational control in secretory cell survival. Mol Cell. 2001;7:1153–1163. doi: 10.1016/s1097-2765(01)00264-7. [DOI] [PubMed] [Google Scholar]
- 40.Harding HP. Zhang Y. Bertolotti A. Zeng H. Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 41.Harding HP. Zhang Y. Ron D. Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature. 1999;397:271–274. doi: 10.1038/16729. [DOI] [PubMed] [Google Scholar]
- 42.Harding HP. Zhang Y. Zeng H. Novoa I. Lu PD. Calfon M. Sadri N. Yun C. Popko B. Paules R. Stojdl DF. Bell JC. Hettmann T. Leiden JM. Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 43.Hendershot LM. The ER chaperone BiP is a master regulator of ER function. Mt Sinai J Med. 2004;71:289–297. [PubMed] [Google Scholar]
- 44.Hendershot LM. Sitia R. Antibody synthesis and assembly. In: Alt FW, editor; Honjo T, editor; Neuberger MS, editor. Molecular biology of B cells. New York: Elsevier Science; 2004. pp. 261–273. [Google Scholar]
- 45.Hetz C. Bernasconi P. Fisher J. Lee AH. Bassik MC. Antonsson B. Brandt GS. Iwakoshi NN. Schinzel A. Glimcher LH. Korsmeyer SJ. Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1alpha. Science. 2006;312:572–576. doi: 10.1126/science.1123480. [DOI] [PubMed] [Google Scholar]
- 46.High S. Stirling CJ. Protein translocation across membranes: common themes in divergent organisms. Trends Cell Biol. 1993;3:335–339. doi: 10.1016/0962-8924(93)90103-8. [DOI] [PubMed] [Google Scholar]
- 47.Hiller MM. Finger A. Schweiger M. Wolf DH. ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science. 1996;273:1725–1728. doi: 10.1126/science.273.5282.1725. [DOI] [PubMed] [Google Scholar]
- 48.Hitomi J. Katayama T. Eguchi Y. Kudo T. Taniguchi M. Koyama Y. Manabe T. Yamagishi S. Bando Y. Imaizumi K. Tsujimoto Y. Tohyama M. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and A{beta}-induced cell death. J Cell Biol. 2004;165:347–356. doi: 10.1083/jcb.200310015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holcik M. Sonenberg N. Korneluk RG. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 2000;16:469–473. doi: 10.1016/s0168-9525(00)02106-5. [DOI] [PubMed] [Google Scholar]
- 50.Hollien J. Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 51.Hou N. Torii S. Saito N. Hosaka M. Takeuchi T. Reactive oxygen species-mediated pancreatic beta-cell death is regulated by interactions between stress-activated protein kinases, p38 and c-Jun N-terminal kinase, and mitogen-activated protein kinase phosphatases. Endocrinology. 2008;149:1654–1665. doi: 10.1210/en.2007-0988. [DOI] [PubMed] [Google Scholar]
- 52.Hsieh YH. Su IJ. Lei HY. Lai MD. Chang WW. Huang W. Differential endoplasmic reticulum stress signaling pathways mediated by iNOS. Biochem Biophys Res Commun. 2007;359:643–648. doi: 10.1016/j.bbrc.2007.05.154. [DOI] [PubMed] [Google Scholar]
- 53.Hwang C. Sinskey AJ. Lodish HF. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 54.Iwakoshi NN. Lee AH. Vallabhajosyula P. Otipoby KL. Rajewsky K. Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 55.Iwawaki T. Akai R. Kohno K. Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 56.Jansens A. van DE. Braakman I. Coordinated nonvectorial folding in a newly synthesized multidomain protein. Science. 2002;298:2401–2403. doi: 10.1126/science.1078376. [DOI] [PubMed] [Google Scholar]
- 57.Jiang HY. Wek SA. McGrath BC. Scheuner D. Kaufman RJ. Cavener DR. Wek RC. Phosphorylation of the alpha subunit of eukaryotic initiation factor 2 is required for activation of NF-kappaB in response to diverse cellular stresses. Mol Cell Biol. 2003;23:5651–5663. doi: 10.1128/MCB.23.16.5651-5663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Johnson AE. Haigh NG. The ER translocon and retrotranslocation: is the shift into reverse manual or automatic? Cell. 2000;102:709–712. doi: 10.1016/s0092-8674(00)00059-3. [DOI] [PubMed] [Google Scholar]
- 59.Kaneto H. Kajimoto Y. Miyagawa J. Matsuoka T. Fujitani Y. Umayahara Y. Hanafusa T. Matsuzawa Y. Yamasaki Y. Hori M. Beneficial effects of antioxidants in diabetes: possible protection of pancreatic beta-cells against glucose toxicity. Diabetes. 1999;48:2398–2406. doi: 10.2337/diabetes.48.12.2398. [DOI] [PubMed] [Google Scholar]
- 60.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 61.Kelsen SG. Duan X. Ji R. Perez O. Liu C. Merali S. Cigarette smoke induces an unfolded protein response in the human lung: a proteomic approach. Am J Respir Cell Mol Biol. 2008;38:541–550. doi: 10.1165/rcmb.2007-0221OC. [DOI] [PubMed] [Google Scholar]
- 62.Kitiphongspattana K. Khan TA. Ishii-Schrade K. Roe MW. Philipson LH. Gaskins HR. Protective role for nitric oxide during the endoplasmic reticulum stress response in pancreatic beta-cells. Am J Physiol Endocrinol Metab. 2007;292:E1543–E1554. doi: 10.1152/ajpendo.00620.2006. [DOI] [PubMed] [Google Scholar]
- 63.Klappa P. Ruddock LW. Darby NJ. Freedman RB. The b' domain provides the principal peptide-binding site of protein disulfide isomerase but all domains contribute to binding of misfolded proteins. EMBO J. 1998;17:927–935. doi: 10.1093/emboj/17.4.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knarr G. Gething MJ. Modrow S. Buchner J. BiP binding sequences in antibodies. J Biol Chem. 1995;270:27589–27594. doi: 10.1074/jbc.270.46.27589. [DOI] [PubMed] [Google Scholar]
- 65.Kornfeld R. Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 66.Kozutsumi Y. Segal M. Normington K. Gething MJ. Sambrook J. The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature. 1988;332:462–464. doi: 10.1038/332462a0. [DOI] [PubMed] [Google Scholar]
- 67.Kuang E. Wan Q. Li X. Xu H. Liu Q. Qi Y. ER Ca2+ depletion triggers apoptotic signals for endoplasmic reticulum (ER) overload response induced by overexpressed reticulon 3 (RTN3/HAP) J Cell Physiol. 2005;204:549–559. doi: 10.1002/jcp.20340. [DOI] [PubMed] [Google Scholar]
- 68.Lai WL. Wong NS. ROS mediates 4HPR-induced posttranscriptional expression of the Gadd153 gene. Free Radic Biol Med. 2005;38:1585–1593. doi: 10.1016/j.freeradbiomed.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 69.Land A. Zonneveld D. Braakman I. Folding of HIV-1 envelope glycoprotein involves extensive isomerization of disulfide bonds and conformation-dependent leader peptide cleavage. FASEB J. 2003;17:1058–1067. doi: 10.1096/fj.02-0811com. [DOI] [PubMed] [Google Scholar]
- 70.Lee AH. Iwakoshi NN. Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee AS. Coordinated regulation of a set of genes by glucose and calcium ionophores in mammalian cells. Trends Biochem Sci. 1987;12:20–23. [Google Scholar]
- 72.Lei K. Davis RJ. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc Natl Acad Sci U S A. 2003;100:2432–2437. doi: 10.1073/pnas.0438011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lipson KL. Fonseca SG. Ishigaki S. Nguyen LX. Foss E. Bortell R. Rossini AA. Urano F. Regulation of insulin biosynthesis in pancreatic beta cells by an endoplasmic reticulum-resident protein kinase IRE1. Cell Metab. 2006;4:245–254. doi: 10.1016/j.cmet.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 74.Lipson KL. Ghosh R. Urano F. The role of IRE1alpha in the degradation of insulin mRNA in pancreatic beta-cells. PLoS ONE. 2008;3:e1648. doi: 10.1371/journal.pone.0001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lodish HF. Kong N. Wikstrom L. Calcium is required for folding of newly made subunits of the asialoglycoprotein receptor within the endoplasmic reticulum. J Biol Chem. 1992;267:12753–12760. [PubMed] [Google Scholar]
- 76.Lu PD. Jousse C. Marciniak SJ. Zhang Y. Novoa I. Scheuner D. Kaufman RJ. Ron D. Harding HP. Cytoprotection by pre-emptive conditional phosphorylation of translation initiation factor 2. EMBO J. 2004;23:169–179. doi: 10.1038/sj.emboj.7600030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ma Y. Brewer JW. Diehl JA. Hendershot LM. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J Mol Biol. 2002;318:1351–1365. doi: 10.1016/s0022-2836(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 78.Ma Y. Hendershot LM. The unfolding tale of the unfolded protein response. Cell. 2001;107:827–830. doi: 10.1016/s0092-8674(01)00623-7. [DOI] [PubMed] [Google Scholar]
- 79.Ma Y. Hendershot LM. Delineation of the negative feedback regulatory loop that controls protein translation during ER stress. J Biol Chem. 2003;278:34864–34873. doi: 10.1074/jbc.M301107200. [DOI] [PubMed] [Google Scholar]
- 80.Masciarelli S. Sitia R. Building and operating an antibody factory: redox control during B to plasma cell terminal differentiation. Biochim Biophys Acta. 2008;1783:578–588. doi: 10.1016/j.bbamcr.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 81.McCracken AA. Brodsky JL. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J Cell Biol. 1996;132:291–298. doi: 10.1083/jcb.132.3.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCullough KD. Martindale JL. Klotz LO. Aw TY. Holbrook NJ. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21:1249–1259. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meldolesi J. Pozzan T. The endoplasmic reticulum Ca2+ store: a view from the lumen. Trends Biochem Sci. 1998;23:10–14. doi: 10.1016/s0968-0004(97)01143-2. [DOI] [PubMed] [Google Scholar]
- 84.Molteni SN. Fassio A. Ciriolo MR. Filomeni G. Pasqualetto E. Fagioli C. Sitia R. Glutathione limits Ero1-dependent oxidation in the endoplasmic reticulum. J Biol Chem. 2004;279:32667–32673. doi: 10.1074/jbc.M404992200. [DOI] [PubMed] [Google Scholar]
- 85.Morishima N. Nakanishi K. Takenouchi H. Shibata T. Yasuhiko Y. An endoplasmic reticulum stress-specific caspase cascade in apoptosis: cytochrome c-independent activation of caspase-9 by caspase-12. J Biol Chem. 2002;277:34287–34294. doi: 10.1074/jbc.M204973200. [DOI] [PubMed] [Google Scholar]
- 86.Munro S. Pelham HR. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 87.Nakagawa T. Yuan J. Cross-talk between two cysteine protease families: activation of caspase-12 by calpain in apoptosis. J Cell Biol. 2000;150:887–894. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakagawa T. Zhu H. Morishima N. Li E. Xu J. Yankner BA. Yuan J. Caspase-12 mediates endoplasmic-reticulum-specific apoptosis and cytotoxicity by amyloid-beta. Nature. 2000;403:98–103. doi: 10.1038/47513. [DOI] [PubMed] [Google Scholar]
- 89.Newsholme P. Haber EP. Hirabara SM. Rebelato EL. Procopio J. Morgan D. Oliveira-Emilio HC. Carpinelli AR. Curi R. Diabetes associated cell stress and dysfunction: role of mitochondrial and non-mitochondrial ROS production and activity. J Physiol. 2007;583:9–24. doi: 10.1113/jphysiol.2007.135871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Novoa I. Zeng H. Harding HP. Ron D. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2alpha. J Cell Biol. 2001;153:1011–1022. doi: 10.1083/jcb.153.5.1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Oikawa D. Tokuda M. Iwawaki T. Site-specific cleavage of CD59 mRNA by endoplasmic reticulum-localized ribonuclease, IRE1. Biochem Biophys Res Commun. 2007;360:122–127. doi: 10.1016/j.bbrc.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 92.Okuda-Shimizu Y. Hendershot LM. Characterization of an ERAD pathway for nonglycosylated BiP substrates, which requires Herp. Mol Cell. 2007;28:544–554. doi: 10.1016/j.molcel.2007.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oliver JD. van der Wal FJ. Bulleid NJ. High S. Interaction of the thiol-dependent reductase ERp57 with nascent glycoproteins. Science. 1997;275:86–88. doi: 10.1126/science.275.5296.86. [DOI] [PubMed] [Google Scholar]
- 94.Pagani M. Fabbri M. Benedetti C. Fassio A. Pilati S. Bulleid NJ. Cabibbo A. Sitia R. Endoplasmic reticulum oxidoreductin 1-lbeta (ERO1-Lbeta), a human gene induced in the course of the unfolded protein response. J Biol Chem. 2000;275:23685–23692. doi: 10.1074/jbc.M003061200. [DOI] [PubMed] [Google Scholar]
- 95.Pollard MG. Travers KJ. Weissman JS. Ero1p: a novel and ubiquitous protein with an essential role in oxidative protein folding in the endoplasmic reticulum. Mol Cell. 1998;1:171–182. doi: 10.1016/s1097-2765(00)80018-0. [DOI] [PubMed] [Google Scholar]
- 96.Putcha GV. Le S. Frank S. Besirli CG. Clark K. Chu B. Alix S. Youle RJ. LaMarche A. Maroney AC. Johnson EM., Jr. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron. 2003;38:899–914. doi: 10.1016/s0896-6273(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 97.Qu L. Huang S. Baltzis D. Rivas-Estilla AM. Pluquet O. Hatzoglou M. Koumenis C. Taya Y. Yoshimura A. Koromilas AE. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 2004;18:261–277. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Radbruch A. Muehlinghaus G. Luger EO. Inamine A. Smith KG. Dorner T. Hiepe F. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 99.Rao RV. Peel A. Logvinova A. Del Rio G. Hermel E. Yokota T. Goldsmith PC. Ellerby LM. Ellerby HM. Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 2002;514:122–128. doi: 10.1016/s0014-5793(02)02289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Reddy RK. Mao C. Baumeister P. Austin RC. Kaufman RJ. Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 101.Reimold AM. Etkin A. Clauss I. Perkins A. Friend DS. Zhang J. Horton HF. Scott A. Orkin SH. Byrne MC. Grusby MJ. Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 102.Reimold AM. Iwakoshi NN. Manis J. Vallabhajosyula P. Szomolanyi-Tsuda E. Gravallese EM. Friend D. Grusby MJ. Alt F. Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 103.Ron D. Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 104.Ron D. Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 105.Scheuner D. Kaufman RJ. The unfolded protein response: a pathway that links insulin demand with beta-cell failure and diabetes. Endocr Rev. 2008;29:317–333. doi: 10.1210/er.2007-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scheuner D. Mierde DV. Song B. Flamez D. Creemers JW. Tsukamoto K. Ribick M. Schuit FC. Kaufman RJ. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat Med. 2005;11:757–764. doi: 10.1038/nm1259. [DOI] [PubMed] [Google Scholar]
- 107.Schuit FC. Kiekens R. Pipeleers DG. Measuring the balance between insulin synthesis and insulin release. Biochem Biophys Res Commun. 1991;178:1182–1187. doi: 10.1016/0006-291x(91)91017-7. [DOI] [PubMed] [Google Scholar]
- 108.Segal MS. Bye JM. Sambrook JF. Gething MJ. Disulfide bond formation during the folding of influenza virus hemagglutinin. J Cell Biol. 1992;118:227–244. doi: 10.1083/jcb.118.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sevier CS. Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 110.Sevier CS. Kaiser CA. Ero1 and redox homeostasis in the endoplasmic reticulum. Biochim Biophys Acta. 2008;1783:549–556. doi: 10.1016/j.bbamcr.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 111.Sevier CS. Qu H. Heldman N. Gross E. Fass D. Kaiser CA. Modulation of cellular disulfide-bond formation and the ER redox environment by feedback regulation of Ero1. Cell. 2007;129:333–344. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 112.Shaffer AL. Shapiro-Shelef M. Iwakoshi NN. Lee AH. Qian SB. Zhao H. Yu X. Yang L. Tan BK. Rosenwald A. Hurt EM. Petroulakis E. Sonenberg N. Yewdell JW. Calame K. Glimcher LH. Staudt LM. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81–93. doi: 10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 113.Shen J. Chen X. Hendershot L. Prywes R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell. 2002;3:99–111. doi: 10.1016/s1534-5807(02)00203-4. [DOI] [PubMed] [Google Scholar]
- 114.Shen X. Ellis RE. Kurnit DM. Liu CY. Lee K. Solomon A. Morimoto RI. Yoshida H. Mori K. Kaufman RJ. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 115.Shi Y. Vattem KM. Sood R. An J. Liang J. Stramm L. Wek RC. Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol Cell Biol. 1998;18:7499–7509. doi: 10.1128/mcb.18.12.7499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shimizu Y. Hendershot LM. Organization of the functions and components of the endoplasmic reticulum. Adv Exp Med Biol. 2007;594:37–46. doi: 10.1007/978-0-387-39975-1_4. [DOI] [PubMed] [Google Scholar]
- 117.Sriburi R. Jackowski S. Mori K. Brewer JW. XBP1: a link between the unfolded protein response, lipid biosynthesis, and biogenesis of the endoplasmic reticulum. J Cell Biol. 2004;167:35–41. doi: 10.1083/jcb.200406136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stoffers DA. Thomas MK. Habener JF. Homeodomain protein IDX-1 A master regulator of pancreas development and insulin gene expression. Trends Endocrinol Metab. 1997;8:145–151. doi: 10.1016/s1043-2760(97)00008-8. [DOI] [PubMed] [Google Scholar]
- 119.Tanaka Y. Gleason CE. Tran PO. Harmon JS. Robertson RP. Prevention of glucose toxicity in HIT-T15 cells and Zucker diabetic fatty rats by antioxidants. Proc Natl Acad Sci U S A. 1999;96:10857–10862. doi: 10.1073/pnas.96.19.10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tanaka Y. Tran PO. Harmon J. Robertson RP. A role for glutathione peroxidase in protecting pancreatic beta cells against oxidative stress in a model of glucose toxicity. Proc Natl Acad Sci U S A. 2002;99:12363–12368. doi: 10.1073/pnas.192445199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tirasophon W. Welihinda AA. Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tran PO. Parker SM. LeRoy E. Franklin CC. Kavanagh TJ. Zhang T. Zhou H. Vliet P. Oseid E. Harmon JS. Robertson RP. Adenoviral overexpression of the glutamylcysteine ligase catalytic subunit protects pancreatic islets against oxidative stress. J Biol Chem. 2004;279:53988–53993. doi: 10.1074/jbc.M404809200. [DOI] [PubMed] [Google Scholar]
- 123.Tu BP. Ho-Schleyer SC. Travers KJ. Weissman JS. Biochemical basis of oxidative protein folding in the endoplasmic reticulum. Science. 2000;290:1571–1574. doi: 10.1126/science.290.5496.1571. [DOI] [PubMed] [Google Scholar]
- 124.Tu BP. Weissman JS. The FAD- and O(2)-dependent reaction cycle of Ero1-mediated oxidative protein folding in the endoplasmic reticulum. Mol Cell. 2002;10:983–994. doi: 10.1016/s1097-2765(02)00696-2. [DOI] [PubMed] [Google Scholar]
- 125.Tu BP. Weissman JS. Oxidative protein folding in eukaryotes: mechanisms and consequences. J Cell Biol. 2004;164:341–346. doi: 10.1083/jcb.200311055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ubeda M. Wang XZ. Zinszner H. Wu I. Habener JF. Ron D. Stress-induced binding of the transcriptional factor CHOP to a novel DNA control element. Mol Cell Biol. 1996;16:1479–1489. doi: 10.1128/mcb.16.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Urano F. Wang X. Bertolotti A. Zhang Y. Chung P. Harding HP. Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- 128.Ushioda R. Hoseki J. Araki K. Jansen G. Thomas DY. Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 129.van Anken E. Romijn EP. Maggioni C. Mezghrani A. Sitia R. Braakman I. Heck AJ. Sequential waves of functionally related proteins are expressed when B cells prepare for antibody secretion. Immunity. 2003;18:243–253. doi: 10.1016/s1074-7613(03)00024-4. [DOI] [PubMed] [Google Scholar]
- 130.Van LL. Janssens K. Quintens R. Tsukamoto K. Vander MD. Lemaire K. Denef C. Jonas JC. Martens G. Pipeleers D. Schuit FC. Probe-independent and direct quantification of insulin mRNA and growth hormone mRNA in enriched cell preparations. Diabetes. 2006;55:3214–3220. doi: 10.2337/db06-0774. [DOI] [PubMed] [Google Scholar]
- 131.Vander MD. Scheuner D. Quintens R. Patel R. Song B. Tsukamoto K. Beullens M. Kaufman RJ. Bollen M. Schuit FC. Glucose activates a protein phosphatase-1-mediated signaling pathway to enhance overall translation in pancreatic beta-cells. Endocrinology. 2007;148:609–617. doi: 10.1210/en.2006-1012. [DOI] [PubMed] [Google Scholar]
- 132.Wang HG. Pathan N. Ethell IM. Krajewski S. Yamaguchi Y. Shibasaki F. McKeon F. Bobo T. Franke TF. Reed JC. Ca2+-induced apoptosis through calcineurin dephosphorylation of BAD. Science. 1999;284:339–343. doi: 10.1126/science.284.5412.339. [DOI] [PubMed] [Google Scholar]
- 133.Wang L. Li SJ. Sidhu A. Zhu L. Liang Y. Freedman RB. Wang CC. Reconstitution of human Ero1-La/protein disulfide isomerase oxidative folding pathway in vitro: position-dependent differences in role between the A and A' domains of protein disulfide isomerase. J Biol Chem. 2009;284:199–206. doi: 10.1074/jbc.M806645200. [DOI] [PubMed] [Google Scholar]
- 134.Wang X-Z. Harding HP. Zhang Y. Jolicoeur EM. Kuroda M. Ron D. Cloning of mammalian Ire1 reveals diversity in the ER stress responses. EMBO J. 1998;17:5708–5717. doi: 10.1093/emboj/17.19.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.White SH. Von HG. The machinery of membrane protein assembly. Curr Opin Struct Biol. 2004;14:397–404. doi: 10.1016/j.sbi.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 136.Xue X. Piao JH. Nakajima A. Sakon-Komazawa S. Kojima Y. Mori K. Yagita H. Okumura K. Harding H. Nakano H. Tumor necrosis factor alpha (TNFalpha) induces the unfolded protein response (UPR) in a reactive oxygen species (ROS)-dependent fashion, and the UPR counteracts ROS accumulation by TNFalpha. J Biol Chem. 2005;280:33917–33925. doi: 10.1074/jbc.M505818200. [DOI] [PubMed] [Google Scholar]
- 137.Yamamoto K. Ichijo H. Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol. 1999;19:8469–8478. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yamamoto K. Sato T. Matsui T. Sato M. Okada T. Yoshida H. Harada A. Mori K. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 139.Ye J. Rawson RB. Komuro R. Chen X. Dave UP. Prywes R. Brown MS. Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 140.Yoneda T. Imaizumi K. Oono K. Yui D. Gomi F. Katayama T. Tohyama M. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem. 2001;276:13935–13940. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- 141.Yoshida H. Matsui T. Yamamoto A. Okada T. Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 142.Yoshida H. Matsui T. Hosokawa N. Kaufman RJ. Nagata K. Mori K. A time-dependent phase shift in the mammalian unfolded protein response. Dev Cell. 2003;4:265–271. doi: 10.1016/s1534-5807(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 143.Yoshida H. Oku M. Suzuki M. Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates UPR activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:562–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zinszner H. Kuroda M. Wang X. Batchvarova N. Lightfoot RT. Remotti H. Stevens JL. Ron D. CHOP is implicated in programmed cell death in response to impaired function of the endoplasmic reticulum. Genes Dev. 1998;12:982–995. doi: 10.1101/gad.12.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]