Summary
The orderly deposition of histones onto DNA is mediated by conserved assembly complexes, including Chromatin Assembly Factor-1 (CAF-1) and the Hir proteins [1–4]. CAF-1 and the Hir proteins operate in distinct but functionally overlapping histone deposition pathways in vivo [5, 6]. The Hir proteins and CAF-1 share a common partner, the highly conserved histone H3/H4-binding protein Asf1, which binds the middle subunit of CAF-1 as well as to Hir proteins [7–11]. Asf1 binds to newly synthesized histones H3/H4 [12] and this complex stimulates histone deposition by CAF-1 [7, 12, 13]. In yeast, Asf1 is required for the contribution of the Hir proteins to gene silencing [7, 14]. Here, we demonstrate that Hir1, Hir2, Hir3 and Hpc2 comprise the HIR complex, which co-purifies with histone deposition protein Asf1. Together, the HIR complex and Asf1 deposit histones onto DNA in a replication-independent manner. Histone deposition by the HIR complex and Asf1 is impaired by a mutation in Asf1 that inhibits HIR binding. These data indicate that the HIR complex and Asf1 proteins function together as a conserved eukaryotic pathway for histone replacement throughout the cell cycle.
Keywords: chromatin assembly, HIR, Asf1, histones
Results and Discussion
Asf1 and the Hir proteins exist as a complex in vivo
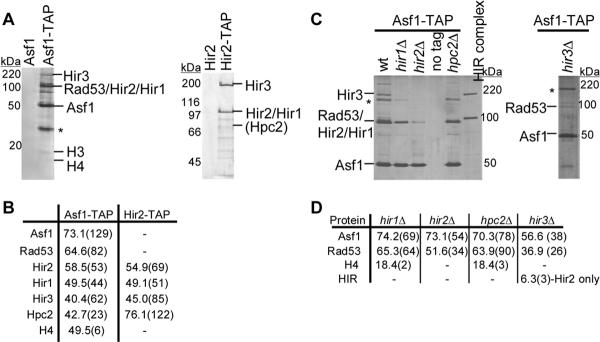
We first examined Asf1-containing protein complexes. A double affinity “TAP” tag containing the S peptide from RNase A [15] flanked by a TEV protease site and a minimal Protein A binding domain (ZZ tag) [16, 17] was fused to the C-terminus of Asf1 by genomic integration at the endogenous locus [18]. This ASF1-STAP allele was functional in vivo because it did not cause sensitivity to hydroxyurea or cause growth or silencing defects in the absence of a CAF-1 subunit (data not shown). Asf1-STAP complexes were purified using buffers containing 100 mM KCl [17]. After the second affinity step, the eluted protein complex was analyzed by SDS-PAGE and silver staining (Figure 1A, left panel), which revealed a predominant band at the approximate molecular weight of Asf1-STAP and multiple co-purifying species. The eluted material was also directly analyzed by mass spectrometry (Figure 1B), which identified checkpoint kinase Rad53, the four Hir proteins, and histones H3 and H4. The copurifying proteins were absent when the purification was performed at higher ionic strength (300 mM KCl; Figure S1A), suggesting that their recovery resulted from electrostatic interactions with Asf1 and not because of insolubility. Furthermore, ethidium bromide, which intercalates between DNA bases, did not disrupt any of the observed interactions (Figure S1B), suggesting that these protein-protein interactions are not bridged by DNA [19]. These data confirmed the known interactions between Asf1 and Rad53 [20, 21], and between Asf1 and histones H3/H4 [12]. This is the first evidence that Asf1 interacts with all four Hir proteins as a group.
Figure 1.
The four Hir proteins exist as a complex in vivo and co-purify with Asf1. (A) TAP purifications were performed using wild-type (PKY028) and ASF1-STAP (PKY3121) cells (left panel) and from wildtype (PKY028) and HIR2-STAP cells (PKY3062) (right panel). The Asf1-STAP purification was performed at 100 mM KCl and the Hir2-STAP purification was performed at 300 mM KCl. Proteins eluted from the second TAP affinity resin were analyzed on a SDS-PAGE gel and detected by silver staining. Polypeptides identified by mass spectrometry, immunoblotting or molecular weight (histone H3) are indicated. *indicates an unidentified polypeptide. (B) Summary of Asf1-STAP and Hir2-STAP mass spectrometry data. The percent sequence coverage is indicated in the table, with the number of unique peptides shown in parentheses. Although histone H3 peptides were not detected in all Asf1-STAP preparations by mass spectrometry, H3 was always apparent on the silver-stained protein gels. (C) Efficient association between Asf1 and the HIR complex is dependent upon all four Hir proteins. Asf1-TAP complexes were purified from wildtype (PKY3121), hir1Δ (PKY3906), hir2Δ (PKY3908), hpc2Δ (PKY3912) and hir3Δ (PKY3928) cells, analyzed on SDS-PAGE gels and detected by silver staining. Purified Hir2-STAP complex is shown for comparison. The Hir2-STAP subunit migrates more slowly than Rad53, which is still present in the complexes from hir mutant cells. *indicates an unidentified polypeptide. (D) A summary of Asf1-TAP mass spectrometry data. The percent sequence coverage of each protein is shown, with the number of unique peptides in parentheses.
These data suggested that the Hir proteins represent a macromolecular complex. To further examine Hir protein partners, we generated a HIR2-STAP allele. The HIR2-STAP allele was functional in vivo because it did not cause synergistic growth or silencing defects in cells lacking a CAF-1 subunit (Figure S2). The Hir2-STAP purification was performed as described for Asf1-STAP, with the exception that the ionic strength was maintained at 300 mM KCl. In the final protein complex (Figure 1A, right panel), two predominant bands were observed at the predicted molecular weights of Hir2-STAP (101 kD) and Hir3 (191 kD). Mass spectrometry confirmed that Hir1, Hir3 and Hpc2 co-purified with Hir2 (Figure 1B). We note that Hir1 (93 kD) and Hir2-STAP (101 kD) appear to co-migrate, and that Hpc2 does not stain well with silver. All other species identified were common contaminants of TAP tag purifications (data not shown). Moreover, Hir1, Hir3 and Hpc2 co-purified with Hir2 in the presence of ethidium bromide (data not shown). These data confirmed that the four Hir proteins interact in vivo, forming what we term the HIR complex.
Asf1-HIR complex architecture
To further define the protein interaction relationships, Asf1-STAP-containing complexes were purified from yeast strains lacking individual HIR genes. In the absence of any one of the HIR complex members, the remaining Hir proteins were generally not detected in the mass spectroscopic analysis of the Asf1-STAP complexes (Figure 1C). The one exception was in the Asf1-STAP complexes from a hir3Δ strain, in which a much reduced number of Hir2 peptides (three) were recovered. Western blot analyses of whole cell extracts confirmed that protein levels of Asf1-STAP did not change in any of the hir mutants (data not shown). Likewise, steady-state levels of Hir2-STAP were unaltered in hir and asf1 mutant cells (Figure S3A). We conclude that the interaction between Asf1 and the HIR complex is greatly reduced in the absence of any of the four Hir subunits. The minor recovery of Hir2 in hir3Δ cells suggests Hir3 may be a more peripheral subunit of the HIR complex.
These data also suggested that the HIR complex may not form in the absence of any subunit. To test this idea, the Hir2-STAP purification was performed from yeast strains lacking individual HIR genes (Figure S3B). In the absence of either Hir1 or Hir3, the remaining subunits still associated with Hir2. However, Hir3 was not detected by mass spectroscopy in Hir2-STAP complexes from hpc2Δ cells, suggesting that Hir3 associates with Hir1 and Hir2 via an interaction with Hpc2, or becomes less stable in the absence of Hpc2. Therefore, although the HIR complex is not completely dissociated when individual components are missing, all Hir subunits are required for normal interaction with Asf1.
To assess the functionality of HIR complexes lacking individual subunits in vivo, interaction of Hir2 with the HTA1-HTB1 promoter was investigated using chromatin immunoprecipitation. The Hir proteins are negative regulators of the HTA1-HTB1 locus [1, 2], repressing promoter activity outside of S phase [22, 23]. We observed that the Hir2-TAP protein was enriched 2.9-fold at the HTA1-HTB1 promoter relative to a non-specific locus, ACT1 (Figure S3C), confirming that Hir2-STAP binds the promoter in vivo [23]. However, in the absence of any of the other three HIR genes, Hir2-TAP no longer specifically associated with the HTA1-HTB1 promoter. In contrast, Hir2-TAP was enriched at the promoter 2.3-fold relative to ACT1 in asf1Δ cells. We conclude that association of the HIR complex with the HTA1-HTB1 promoter in vivo is dependent on Hir1, Hir3 and Hpc2 but not Asf1. In accordance with these data, previous studies have shown that transcriptional repression of HTA1 by Hir2 is fully dependent on Hir1 and partially dependent on Hir3 [24]. However, because HIR complex binding to the promoter is independent of Asf1, the deregulation of histone gene expression in asf1Δ cells [9] is not due to the inability of the HIR complex to interact with promoter DNA. Instead, increased histone gene expression in asf1Δ cells may be due to altered chromatin structure at the promoter, and/or may result from indirect defects on cell cycle progression [25].
Replication-independent chromatin assembly by the HIR complex and Asf1
Genetic and biochemical experiments had suggested that the yeast HIR complex would function as a histone deposition factor in conjunction with Asf1 [6, 7, 26]. To test this idea, we purified native HIR complexes using gentle elution conditions via an EGTA-elutable TAP tag (CBP-TEV-ZZ, [16]; Experimental Procedures). We used an electrophoretic mobility shift assay (EMSA) to detect formation of nucleoprotein complexes. When a radiolabeled 250 bp DNA probe was incubated with histones H3/H4 (Figure 2A, lane 2), a small amount of two slower migrating species were observed in the gel (arrows), suggesting inefficient formation of histone-DNA complexes. The addition of the HIR complex slightly increased the amounts of these complexes (Figure 2A, lane 3), suggesting a mild stimulation of histone deposition. When Asf1/H3/H4 complexes were incubated with the DNA, more of the two shifted DNA species appeared (Figure 2A, lane 5), with the predominant shifted band being the faster migrating of the two species. Addition of increasing amounts of the HIR complex to Asf1/H3/H4 resulted in greater amounts of probe shifted (Figure 2A, lanes 6–8), and a higher proportion of the slower migrating species. The observed shifts were dependent on histones H3/H4, because Asf1 or the HIR complex did not shift the DNA in the absence of histones (Figure 2A, lanes 10, 11, 13). We therefore hypothesized that the two shifted species represent histone H3/H4/DNA complexes. Upon addition of anti-H3 antibodies to these reactions, a novel band of slower migration was formed specifically in reactions containing both the HIR complex and Asf1/H3/H4 (Figure 2B, lane 10). These data confirm that the HIR and Asf1/H3/H4 complexes together promote histone deposition.
Figure 2.
Histone deposition by HIR and Asf1/H3/H4 complexes. (A) 30 fmol of a 250 bp radiolabeled DNA probe was incubated with 100 fmol of histones H3/H4 (lanes 2–4), 100 fmol of pre-formed Asf1/H3/H4 complex (lanes 5–9), 100 fmol of Asf1 (lanes 10–12) and either 1.75 μL (lane 6), 3.5 μL (lane 7), or 7 μL (lanes 3, 8, 11, 13) of HIR complex or HIR dialysis buffer (7 μL; lanes 4, 9, 12, 14). Reaction products were resolved on a native 4% polyacrylamide gel and detected by autoradiography. The upper and lower arrows indicate two distinct shifted species. (B) The shifted species contain histone H3. The EMSA was performed as described above, except that 5 μL of HIR complex or HIR dialysis buffer was used. Anti-histone H3 (0.125 μg, Abcam) was added to lanes 6–10. The arrow indicates the supershifted species. (C) Plasmid DNA pre-relaxed with human topoisomerase I was incubated with CBP-TAP purified HIR complex (20 μL) pre-incubated with 0.4 (lanes 2 and 4) and 0.8 (lanes 3 and 5) pmol of histones H3/H4. DNA was analyzed by agarose gel electrophoresis and visualized by SYBR Gold (Molecular Probes) staining.
To determine whether Asf1 was essential for histone deposition by the HIR complex, histones H3/H4 without Asf1 were incubated with plasmid DNA in the presence or absence of HIR complex (Figure 2C). To complete nucleosome formation, H2A/H2B were also added. Topoisomerase I was present to relax all unrestrained supercoils, so that any supercoiling of the DNA could be attributed to nucleosome formation. We observed that the HIR complex stimulated supercoiling under these conditions. These data indicate that the HIR complex alone is capable of replication-independent histone deposition.
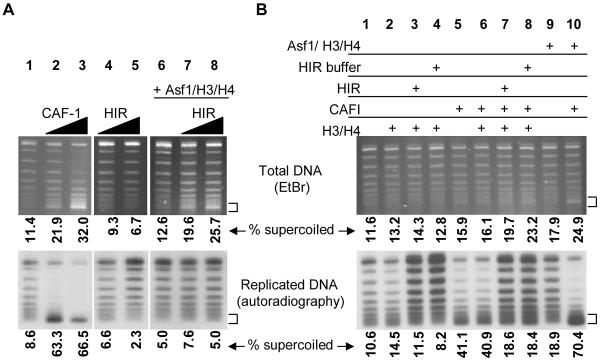
We next determined whether histone deposition by Hir/Asf1 proteins would occur during DNA replication. Nucleosome formation was monitored by the extent of supercoiling of plasmid DNA replicated in a human cell extract. A radioactive deoxyribonucleotide was incorporated into replicated DNA molecules, distinguishing them from non-replicated molecules. The addition of CAF-1 to this reaction results in preferential supercoiling of the radioactive, replicated molecules ([27–29]); Figure 3A, lanes 1–3), indicative of nucleosome formation on the replicated DNA templates. Hir protein complexes did not alter supercoiling of either the replicated or total DNA populations (Figure 3A, lanes 4–5). However, when preformed Asf1/H3/H4 complexes were added to reactions containing HIR complex, supercoiling increased almost two-fold in the total DNA visualized by ethidium bromide (lanes 7–8). However, comparison of the replicated molecules (lower panel) to the total DNA (upper panel) showed that there was no preferential supercoiling of replicated molecules in the presence of the Asf1/H3/H4 and HIR complexes. No supercoiled DNA was produced in the presence of either an equivalent amount of Asf1/H3/H4 alone (Figure 3A, lane 6), or in the presence of the HIR complex with an equivalent amount of histones in the absence of Asf1 (Figure 3B, lane 3). These data confirm that Asf1/H3/H4 and the HIR complex together promote replication-independent histone deposition. Notably, recent experiments [30] demonstrate that both Asf1 and the HIR complex are important for replication-independent re-deposition of histones at the yeast PHO5 promoter, consistent with our biochemical observations.
Figure 3.
The HIR complex and Asf1 assemble chromatin independently of DNA replication or CAF-1. (A) SV40 replication reactions containing α-32P-dATP. CAF-1 (10 ng, lane 2 or 20 ng, lane 3) or CBP-TAP purified HIR complex (2.25 μL, lanes 4 and 6 or 4.5 μL, lanes 5 and 7) were performed as indicated. Asf1/H3/H4 complexes (0.9 pmol) were added in lanes 6–8. Chromatin assembly was detected by the appearance of supercoiled DNA (indicated by arrow) on an agarose gel. Total DNA was visualized by staining with ethidium bromide (upper panel). Replicated molecules that incorporated α-32P-dATP were detected by autoradiography (lower panel). The values shown indicate the percentage of supercoiled DNA in each lane for both the total DNA and the replicated DNA. (B) The HIR complex does not stimulate replication dependent, CAF-1-mediated chromatin assembly. SV40 replication reactions were performed as in (A) in the presence of CAF-1 (7.5 ng, lanes 5–8, 10), histones H3/H4 (5 ng, lanes 2–4 and 6–8), HIR complex (4 μL, lanes 3, 7), HIR dialysis buffer (4 μL, lanes 4, 8), and pre-formed Asf1/H3/H4 complex (3.6 pmol, lanes 9, 10).
Asf1/H3/H4 stimulates replication-linked histone deposition by CAF-1 ([7, 10, 13, 31] and Figure 3B, lanes 5, 9–10). Because mutations in either the HIR or ASF1 genes cause synthetic growth and silencing defects in the absence of CAF-1 subunits [7, 14], we hypothesized that the HIR complex might affect CAF-1 activity. This idea was tested in replication-coupled chromatin assembly assays containing a substoichiometric amount of CAF-1. However, the addition of HIR complex pre-incubated with histones H3 and H4 did not significantly increase the amount of supercoiled DNA (Figure 3B, compare lanes 6, 7 and 8). Although the percentage of supercoiled replicated DNA appeared to decrease with the addition of the HIR complex to CAF-1, this was likely due to stimulation of DNA replication in the presence HIR buffer components (lanes 7 and 8). In contrast, this same amount of HIR complex triggered replication-independent histone deposition in the presence of Asf1/H3/H4 (Figure 3A). Therefore, the HIR/Asf1 proteins are sufficient for replication-independent chromatin assembly and operate independently of CAF-1. Furthermore, these data provide a biochemical distinction between the activities of the HIR complex and Asf1 despite their similar genetic phenotypes and physical association.
HIR-binding mutations in Asf1 inhibit histone deposition
Physical interaction between human Asf1a and the Hir protein homolog HIRA is disrupted by mutation of a small region of conserved residues on the surface of Asf1a [32]. Mutation of the homologous residues in budding yeast blocks the ability of Asf1 to contribute to telomeric gene silencing, suggesting that direct interaction between Asf1 and the HIR complex is important for silencing [33]. We therefore predicted that histone deposition by Asf1/HIR complexes would be impaired by the H36A,D37A mutations that affect the Asf1-HIR interaction. To test this idea, we purified recombinant Asf1-H36AD37A and tested its replication-independent chromatin assembly activity. As observed previously (Figures 2 and 3), wildtype Asf1 promoted DNA supercoiling by the HIR complex (Figure 4, lanes 12 and 13), indicating increased histone deposition. In contrast, the Asf1-H36AD37A mutant stimulated histone deposition by the HIR complex (lanes 14 and 15) less efficiently. However, Asf1-H36AD37A promoted histone deposition in the absence of the HIR complex at a level comparable to wildtype Asf1 (Figure S4). Therefore, the Asf1-H36AD37A mutant was not intrinsically defective for histone deposition, but could not be stimulated by HIR complexes as well as wildtype Asf1. These data indicate that a direct physical interaction between Asf1 and the HIR complex is important for their combined histone deposition activity.
Figure 4.
Asf1-H36AD37A impairs histone deposition by the HIR/Asf1 complex. Histone deposition was assayed as in Figure 2C. Plasmid DNA pre-relaxed with human topoisomerase I was incubated with 0.4 and 0.8 pmol histones H3 and H4 (lanes 2–3), 0.8 and 1.6 pmol Asf1 (lanes 4–5) or Asf1-H36AD37A (lanes 7–8) pre-incubated with 0.4 and 0.8 pmol histones H3/H4. CBP-TAP-purified HIR complex (15 μL) was added to reactions in lanes 10–16.
Conclusions
Hir1, Hir2, Hir3 and Hpc2 together comprise the HIR complex, a novel macromolecular chromatin assembly factor that co-purifies with chromatin assembly factor Asf1. Asf1 and the HIR complex together promote replication-independent histone deposition in a manner stimulated by their interaction. The Asf1/Hir proteins thus form a conserved histone deposition pathway [6, 7, 26, 34] implicated in histone replacement during RNA polymerase movement [35] and reassembly of promoters after histone eviction [30]. Because all components of the yeast HIR complex have been identified here, this study provides the groundwork for mechanistic analysis of replication-independent chromatin assembly in a genetically tractable organism.
Studies from the Workman laboratory describe an additional function of the HIR complex, formation of a SNF/SWI-resistant nucleoprotein structure, consistent with the role of Hir proteins in histone gene repression (Prochasson et al., in press). We note that our preparations of the HIR complex display the same polypeptide composition.
Experimental Procedures
All methods are described in the Supplemental Materials.
Supplementary Material
Acknowledgements
We thank Philippe Prochasson and Jerry Workman for generous communication of results prior to publication; Mary Ann Osley, Karsten Weis and Bertrand Séraphin for reagents; and members of the Kaufman Lab for support and helpful discussions. This work was supported by NIH grant GM55712 and NSF grant MCB-0234014. J.R.Y. is supported by NIH grant RR11823-09.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Osley MA, Lycan D. Trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 1987;7:4204–4210. doi: 10.1128/mcb.7.12.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H, Kim UJ, Schuster T, Grunstein M. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992;12:5249–5259. doi: 10.1128/mcb.12.11.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamour V, Lecluse Y, Desmaze C, Spector M, Bodescot M, Aurias A, Osley MA, Lipinski M. A human homolog of the S. cerevisiae HIR1 and HIR2 transcriptional repressors cloned from the DiGeorge syndrome critical region. Hum. Mol. Genet. 1995;4:791–799. doi: 10.1093/hmg/4.5.791. [DOI] [PubMed] [Google Scholar]
- 4.Franco AA, Kaufman PD. Histone deposition proteins: links between the DNA replication machinery and epigenetic gene silencing. Cold Spring Harbor Symposia on Quantitative Biology. 2004;69:1–8. doi: 10.1101/sqb.2004.69.201. doi:10.1101/sqb.2004.1169.1017. [DOI] [PubMed] [Google Scholar]
- 5.Sharp JA, Franco AA, Osley MA, Kaufman PD. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 2002;16:85–100. doi: 10.1101/gad.925302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 7.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 8.Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sutton A, Bucaria J, Osley MA, Sternglanz R. Yeast ASF1 Protein is Required for Cell-Cycle Regulation of Histone Gene Transcription. Genetics. 2001;158:587–596. doi: 10.1093/genetics/158.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krawitz DC, Kama T, Kaufman PD. Chromatin Assembly Factor-I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol. Cell. Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 13.Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G. Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep. 2002;3:329–334. doi: 10.1093/embo-reports/kvf068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J-S, Raines RT. Ribonuclease S-peptide as a carrier in fusion proteins. Protein Sci. 1993;2:348–356. doi: 10.1002/pro.5560020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigaut GA, Shevchenko B, Rutz M, Wilm M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 17.Cheeseman IM, Brew C, Wolyniak M, Desai A, Anderson S, Muster N, Yates JR, 3rd, Huffaker TC, Drubin DG, Barnes G. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J Cell Biol. 2001;155:1137–1145. doi: 10.1083/jcb.200109063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 19.Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci U S A. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emili A, Schieltz DM, Yates JR, Hartwell L. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 21.Hu F, Alcasabas AA, Elledge SJ. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dimova D, Nackerdien A, Furgeson S, Eguschi S, Osley MA. Role for transcriptional repressors in targeting the yeast Swi/Snf complex. Mol. Cell. 1999;4:75–83. doi: 10.1016/s1097-2765(00)80189-6. [DOI] [PubMed] [Google Scholar]
- 23.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, Zeitlinger J, Jennings EG, Murray HL, Gordon DB, Ren B, Wyrick JJ, Tagne JB, Volkert TL, Fraenkel E, Gifford DK, Young RA. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 24.Spector M, Raff A, DeSilva H, Lee K, Osley MA. Hir1p and Hir2p function as transcriptional corepressors to regulate histone gene transcription in the Saccharomyces cerevisiae cell cycle. Mol. Cell. Biol. 1997;17:545–552. doi: 10.1128/mcb.17.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zabaronick SR, Tyler JK. The histone chaperone anti-silencing function 1 is a global regulator of transcription independent of passage through S phase. Mol Cell Biol. 2005;25:652–660. doi: 10.1128/MCB.25.2.652-660.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray-Gallet D, Quivy JP, Scamps C, Martini EM, Lipinski M, Almouzni G. HIRA Is Critical for a Nucleosome Assembly Pathway Independent of DNA Synthesis. Mol Cell. 2002;9:1091–1100. doi: 10.1016/s1097-2765(02)00526-9. [DOI] [PubMed] [Google Scholar]
- 27.Stillman B. Chromatin assembly during SV40 DNA replication in vitro. Cell. 1986;45:555–565. doi: 10.1016/0092-8674(86)90287-4. [DOI] [PubMed] [Google Scholar]
- 28.Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- 29.Kaufman PD, Kobayashi R, Stillman B. Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev. 1997;11:345–357. doi: 10.1101/gad.11.3.345. [DOI] [PubMed] [Google Scholar]
- 30.Schermer UJ, Korber P, Horz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol Cell. 2005;19:279–285. doi: 10.1016/j.molcel.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast Histone Deposition Protein Asf1p Requires Hir Proteins and PCNA for Heterochromatic Silencing. Current Biology. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 32.Zhang R, Poustovoitov MV, Ye X, Santos HA, Chen W, Daganzo SM, Erzberger JP, Serebriiskii IG, Canutescu AA, Dunbrack RL, Pehrson JR, Berger JM, Kaufman PD, Adams PD. Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev Cell. 2005;8:19–30. doi: 10.1016/j.devcel.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Daganzo SM, Erzberger JP, Lam WM, Skordalakes E, Zhang R, Franco AA, Brill SJ, Adams PD, Berger JM, Kaufman PD. Structure and function of the conserved core of histone deposition protein Asf1. Curr Biol. 2003;13:2148–2158. doi: 10.1016/j.cub.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 34.Blackwell C, Martin KA, Greenall A, Pidoux A, Allshire RC, Whitehall SK. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol. 2004;24:4309–4320. doi: 10.1128/MCB.24.10.4309-4320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades A, Kaufman PD, Stillman DJ. Defects in SPT16 or POB3 (yFACT) Cause Dependence on the Hir/Hpc Pathway: Accessing DNA May Degrade Chromatin Structure. Genetics. 2002;162:1557–1571. doi: 10.1093/genetics/162.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.