Abstract
Hypoxia inducible factors (HIFs) are heterodimeric transcription factors responsible for the metazoan hypoxia response and are required for tumor growth, metastasis and resistance to cancer treatment. The C-terminal PAS domain of HIF2α (HIF2α PAS-B) contains a preformed solvent-inaccessible cavity that binds artificial ligands that allosterically perturb the formation of the HIF heterodimer. To better understand how small molecules bind within this domain, we examined the structures, equilibrium and transition state thermodynamics of HIF2α PAS-B with several artificial ligands using ITC, NMR exchange spectroscopy and X-ray crystallography. Rapid association rates reveal that ligand binding is not dependent upon a slow conformational change in the protein to permit ligand access, despite the closed conformation observed in NMR and crystal structures. Compensating enthalpic and entropic contributions to the thermodynamic barrier for ligand binding suggest a binding-competent transition state characterized by increased structural disorder. Finally, molecular dynamics simulations reveal conversion between open and closed conformations of the protein and pathways of ligand entry into the binding pocket.
Keywords: PAS domain, ligand binding
Introduction
A cell's ability to sense and respond to environmental stimuli is critical for survival. In metazoans, oxygen concentration is monitored by the hypoxic response pathway, where hypoxia inducible factors (HIF) regulate numerous genes in response to oxygen.1 HIF proteins are heterodimeric basic-helix-loop-helix Per-ARNT-Sim (bHLH-PAS) transcription factors2 whose activity is regulated in an oxygen-dependent manner.3 With adequate oxygen levels (normoxia), oxygen-dependent hydroxylations of the HIFα subunit block interaction with CBP/p300 family coactivator proteins4 and also trigger its ubiquitin-mediated degradation.5 Under low oxygen conditions (hypoxia), unmodified HIFα subunits bind the aryl hydrocarbon receptor nuclear translocator (ARNT, also known as HIFβ) protein, forming the transcriptionally active heterodimer. Thus, cells translate environmental oxygen concentration into altered gene expression. HIF proteins are of considerable interest due to their role in promoting solid tumor growth6,7 and their role in chemotherapy resistance.8
Both HIFα and ARNT contain an N-terminal basic-helix-loop-helix DNA binding domain and two adjacent PAS domains, referred to as PAS-A and PAS-B. PAS domains are structural modules found in proteins from all kingdoms of life that have significant structural homology despite little conservation in amino acid sequence. PAS domains often serve as protein-protein interaction components, as in the case of HIFα and ARNT where they are needed for assembly of the HIF2 PAS-B heterodimer via their β-sheet surfaces.9–11 For many PAS domains, these protein/protein interactions can be environmentally-regulated by internally-bound small molecules or cofactors,12 including FAD, FMN,13 heme,14 or 4-hydroxycinnamic acid.15 For example, flavin-binding PAS domains serve as blue light photoreceptors in bacteria and plants16 and assess energy content in bacteria,17 while heme-based PAS domains monitor dissolved gases.18
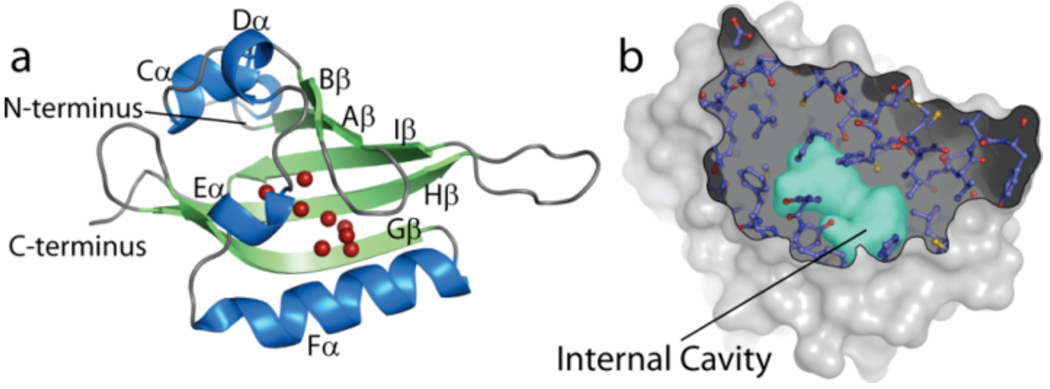
The sensory roles of many PAS domains, including those from the HIF proteins, remain an open question as they have often been isolated without cofactors. Notably, recent structural work on the HIF2α PAS-B domain shows that it contains a 290 Å3 water-filled cavity at its core, isolated from bulk solvent10 (Figs. 1A,B). Cavities of this size are quite rare in proteins and suggest a missing cofactor or ligand binding site.10 While any natural HIF2α ligands remain unknown, we have discovered a number of artificial ligands that bind within this site with low micromolar affinity using an NMR-based small molecule screen.19 Several of these compounds modulate the interaction between HIF2α and ARNT PAS-B domains in vitro, suggesting a linkage between internal ligand and external protein binding on opposite faces of the central β-sheet of the protein, consistent with PAS domains in other proteins.10,20–23 This raises the question as to how these ligands gain access to the internal cavity of HIF2α PAS-B, which is completely inaccessible to solvent in our structures.
Figure 1. Crystal structures of HIF2α PAS-B reveal and internal solvent-filled cavity which binds artificial ligands.
(a). Crystal structure of the PAS B domain of HIF2α in the apo form showing the eight bound solvent atoms within the core of the domain (red spheres). (b). Cutaway of the HIF-2α surface revealing the 290 Å3 internal cavity, depicted as a surface (cyan).
To address this point and better understand the nature of HIF/ligand interactions in general, we determined the crystal structures of complexes of HIF2α PAS-B with two artificial ligands bound at this internal site. These structural data complement thermodynamic studies of HIF2α PAS-B with these and three additional ligands by isothermal titration calorimetry (ITC). Using NMR exchange spectroscopy, we determined the association and dissociation rate constants for each of these compounds, and through their temperature dependence, also the transition-state thermodynamics of ligand binding. These parameters reflect the degree of protein conformational distortion required to reach a ligand-accessible transition state. Finally, we combined solution NMR studies and molecular dynamics (MD) simulations to identify ligand entry/exit pathways into the pocket of HIF2α PAS-B. The NMR-derived results suggest that HIF2α PAS-B ligands bind rapidly to a partially disordered, more open conformation of HIF2α PAS-B that must rapidly interconvert with a ligand-inaccessible conformation illustrated by crystal and solution NMR structures. In the MD simulations, the HIF2α PAS-B protein populates both ‘open’ and ‘closed’ conformers, with the closed form being preferred. We identified ligand entry/exit pathways by observing routes taken by solvent water transferring to and from the ligand binding pocket. This transfer takes place primarily in an open conformation of the protein, although there is a minor route in the closed form. Taken together, these data provide structural and energetic characterizations of a protein that is apparently poised to bind natural or artificial regulatory ligands and cofactors.
Experimental Methods
Sample preparation
15N-labeled HIF2α PAS-B was purified as described.10 Briefly, an expression construct encoding HIF2α PAS-B residues 240–350 was generated through PCR amplification from cDNA ligated into the pHis6x-GB1 expression plasmid.20 For ITC studies, protein was expressed in Luria broth while NMR studies utilized 15N-labeled protein as produced by expression in M9 media containing 1 g/L 15NH4Cl as the sole nitrogen source. Protein was purified by nickel chromatography and, after cleavage of the N-terminal affinity tag by TEV protease,24 a second round of nickel chromatography followed by Superdex 75 size exclusion chromatography. For crystallization, PAS-B* mutants of the HIF2α PAS-B (R247E) and ARNT PAS-B (E362R) domains were produced as described.10
Crystallization, structure determination and refinement
Crystals of HIF2α PAS-B* were grown in a high-affinity heterodimer complex with the ARNT PAS-B* domain. Liganded co-crystals were grown by vapor diffusion of 340 µM PAS-B* heterodimer, 440 µM compound solution against 100 mM Bis-Tris, pH 6.0, 17–25% PEG-3350. Crystals were subsequently treated with 25% PEG-3350 and 10% PEG-400 as a cryoprotectant prior to freezing in liquid nitrogen. X-ray diffraction data were collected at the 19-BM beamline of the Structural Biology Center at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Data were reduced using HKL200025 and refined using REFMAC 526 and COOT.27 Data scaling and refinement statistics are summarized in Table 1. THS-017 co-crystals grew in the C2 space group reported previously10 and were amenable to Fourier synthesis methods using the apo-protein structure (3F1P) as an initial model. The THS-020 co-crystals adopted the P21 space group and the diffraction data were phased by molecular replacement using the program PHASER.28 Experimental electron density for each of the structures is shown in Supporting Information Figure 1.
Table 1.
X-ray Crystallography Data Processing and Refinement Statistics (molecular replacement)
| THS-017 | THS-020 | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 73.60, 82.67, 41.10 | 40.89, 70.80, 42.36 |
| β (°) | 106.39 | 108.69 |
| Resolution (Å) | 20.7 to 1.65 | 26.4 to 1.5 |
| (1.68 to 1.65) | (1.53 to 1.50) | |
| Rsym or Rmerge | 5.1 (46.2) | 3.7 (23.8) |
| I / σI | 25.0 (2.1) | 21.8 (2.5) |
| Observed reflections | 105,492 | 101,923 |
| Unique reflections | 28,092 | 34,380 |
| Completeness % (highest shell) | 97.6 (88.5) | 94.6 (68.8) |
| Refinement | ||
| Resolution (Å) | 22.5 to 1.65 | 26.3 to 1.5 |
| (1.68 to 1.65) | (1.53 to 1.50) | |
| Rwork / Rfree | 20.2 / 23.8 | 19.6 / 23.4 |
| No. atoms | ||
| Protein | 1857 | 1933 |
| Ligand/ion | 20 | 20 |
| Water | 182 | 215 |
| B-factors | ||
| Protein | 19.6 | 17.2 |
| Ligand/ion | 30.5 | 18.7 |
| Water | 31.4 | 28.1 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.013 | 0.019 |
| Bond angles (°) | 1.48 | 1.83 |
Values in parentheses are for highest-resolution shell.
Determination of ligand binding affinities
Ligand binding affinities were determined using a MicroCal VP-ITC calorimeter (Northampton, MA). Protein was extensively dialyzed against buffer (25 mM Tris, pH 7.5, 17 mM NaCl, 5 mM β-mercaptoethanol), which was used to prepare compound solutions from 50 mM stocks (in D6-DMSO). Solutions containing 200–330 µM HIF2α PAS-B were titrated from the syringe of this instrument into a cell containing 10–20 µM compound. Compound and protein solutions all contained 0.02% DMSO, except for KG2-023, which was supplemented with 5% DMSO to facilitate compound solubility (minimal effects on thermodynamic parameters were observed in 5% DMSO controls with THS-017 and THS-020). Heats of dilution were experimentally determined from control titrations of HIF2α PAS-B into compound-free buffer, which were subsequently subtracted from the corresponding ligand titration prior to fitting the data to a single-site binding model (Origin v7.0). Reported equilibrium thermodynamic parameters are average values derived from three independent measurements (apart from KG2-023, at two measurements).
NMR spectroscopy
Experiments were conducted using a 600 MHz Varian Inova spectrometer equipped with a cryogenically cooled probe and a Z-axis pulsed-field gradient. To determine association and dissociation rate constants for ligand binding, we employed a ZZ-exchange modified 15N/1H HSQC experiment which gives rise to time-dependent exchange peaks via heteronuclear longitudinal magnetization transfer between the bound and unbound states.29 Samples were prepared for NMR spectroscopy as follows: 133 µM ligand (from 50 mM D6-DMSO stocks) was added to 266 µM HIF2α PAS-B in 17 mM NaCl, 50 mM Tris pH 7.4, 5 mM DTT, 10% D2O. Total DMSO concentration in the prepared sample was 0.26%. Ligand association and dissociation rates were extracted from a simultaneous fit of the measured crosspeak and autopeak intensities to the McConnell equations.29,30 Data processing and analysis were performed with NMRpipe31 and NMRviewJ32 respectively, while curve fitting was performed using Berkeley Madonna software.33 Data were collected at 25°C for determination of binding and dissociation rate constants and further at 10, 15, 20, 25, and 30°C for determination of transition state thermodynamic parameters, which were obtained by linear least-squares fit to experimental values for koff and kon by Eyring analysis.34 Backbone 15N-relaxation measurements29 were collected from 300 µM 15N-HIF2α PAS-B in the presence of and absence of 400 µM THS-044. Relaxation rate constants were estimated from serially collected spectra with time delays of 10, 30, 70, 130, 250, 370, 490, 630, 750, 850 and 1110 ms for R1 and 10, 30, 50, 70, 90, 110, 130, 150 and 190 ms for R2. 15N{1H} NOE values were determined from spectra collected with and without a 3 s saturation period.
Molecular dynamics simulations
All computations were performed using the in lucem molecular mechanics (ilmm) program.35 These simulations are part of the Daggett laboratory’s Dynameomics effort,36–38 which is an effort to create a repository of molecular dynamics simulations and the corresponding metadata.
The starting model for simulations was the first conformer of the NMR structure of the C-terminal PAS domain of HIF2α (PDB entry 1P97).9 The structure was minimized in vacuo for 1000 steps of steepest descent minimization. Atomic partial charges and the potential energy function were taken from Levitt et al.39 The minimized structures were then solvated in a rectangular box of flexible three-center (F3C) waters40 with walls located ≥ 10 Å from any protein atom. The solvent density of the box was pre-equilibrated to 0.997 g/mL, the experimental water density for 298 K and 1 atm pressure.41 The solvent was minimized for 1000 steps. This solvent minimization was followed by 1 ps of dynamics of the solvent only, an additional 500 steps of solvent minimization and 500 steps of minimization of the entire system. After completion of this solvation process, the whole system was heated to 298 K.
Three separate 35 ns MD simulations were performed at 298 K and neutral pH in the microcanonical (NVE, constant volume, total energy, and number of particles) ensemble. Protocols and the potential energy function have been described previously.39,42 A force-shifted nonbonded cutoff of 10 Å was used,39,42,43 and the nonbonded interaction pair list was updated every three steps. A time step of 2 fs was applied in all simulations, and structures were saved every 1 ps for analysis. Analysis of MD simulations was performed using ilmm.
Results
Thermodynamics of HIF2α PAS-B ligand binding
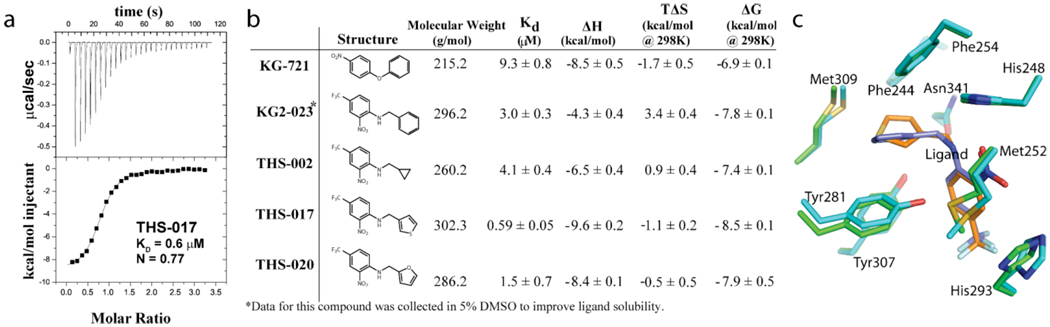
In-house NMR-based small molecule screening efforts identified a number of artificial ligands for HIF2α PAS-B,10 most of which are composed of a two substituted ring structure connected by a one or two atom linker. To investigate the structural dependence of the thermodynamics of interactions between HIF2α PAS-B and small molecules, we compared calorimetric data collected on complexes of HIF2α PAS-B with five related bicyclic ligands (Fig. 2). Four of the five compounds used in this study share a common scaffold consisting of a functionalized benzyl ring with trifluoromethyl and nitro groups located para- and ortho-to the linker, respectively. The fifth compound, KG-721, lacks a trifluoromethyl group, and contains a nitro group para- to its single atom ether linkage. To determine the binding affinities and thermodynamic parameters for these compounds to the isolated HIF2α PAS-B domain, we used ITC to monitor heat evolved upon complex formation. We found that ligand binding is enthalpically driven with minor entropic penalties for compounds KG-721, THS-017, and THS-020, while THS-002 and KG2-023 had favorable entropic contributions to binding. The enthalpies of KG-721, THS-017, and THS-020 are quite similar, notable in light of the reduced functionality of the KG-721 ligand relative to the other two compounds. Evidently, the additional functional groups are making equivalent enthalpic contributions in THS-017 and THS-020 to the single para-nitro group of KG-721. THS-002 and KG2-023 show favorable entropic contributions to binding, both of which are poorly soluble in aqueous solution and likely draw an entropic benefit upon binding protein due to desolvation.
Figure 2. HIF2α PAS-B equilibrium ligand-binding thermodynamics.
(a). Typical isothermal titration calorimetry results from compound THS-017 binding to HIF2α PAS-B. (b). Compounds evaluated in this study, shown in wireframe with binding and thermodynamic parameters derived from ITC. (c). The ligand binding pocket of HIF2α PAS-B of the THS-017 (green) and THS-020 (cyan) bound structures. All panels are shown in the same orientation and the ARNT PAS-B* domain heterodimer has been omitted for clarity.
Crystal structures of the THS-017 and THS-020/PAS B* heterodimer complexes
To provide a structural context for the varied thermodynamics we observed, we determined two additional X-ray crystal structures of HIF2α PAS-B ligand complexes. A readily crystallized variant of the HIF2α PAS-B heterodimer (see Methods), referred to as HIF2α PAS-B*, yielded high-resolution protein/ligand complex structures with compounds, THS-017 (1.65 Å resolution) and THS-020 (1.5 Å) (Table 1). As previously reported for another compound, THS-044,10 THS-017 and THS-020 occupy the internal HIF2α PAS-B* cavity in nearly identical conformations (Fig. 2C). Both ligands within these complexes, like THS-044, are a minimum of 6.6 Å from bulk solvent, completely isolating them within the protein core. Based on the three available HIF2α PAS-B/ligand cocomplex structures (THS-017, THS-020 and our previously-reported complex with THS-04410) and the chemical similarity of the compounds evaluated here, we believe that the remaining bicyclic compounds evaluated in this study likely bind analogously as previously observed.
In general, our structural data suggests that HIF2α PAS-B preorganizes the ligand binding cavity, using shape complementarity with few specific protein/ligand interactions to guide binding. This observation is supported by comparisons between structures of three protein/ligand complexes and one unliganded form. Ligand binding is associated with minimal distortion relative to the water-filled unliganded form, with few side chain rearrangements (His 248, Met 252). Around the variable ring of the ligands (“ring B” in SI Fig. 2), we observe a predominantly hydrophobic environment built with Phe244, Phe254, Phe280, Tyr307, Met309 and Leu319 side chains. This site within the HIF2α PAS-B cavity contains a single low-occupancy water molecule in the unliganded crystal structure, consistent with its hydrophobic composition. As previously noted, we observe few direct hydrogen bonds or electrostatic interactions between the protein and the bound ligands, most of which involve the “A ring” that is common to four of our five ligands. Ligand-protein interactions consist primarily of van der Waals contacts, a dipole-π interaction between the Tyr281 side chain and the ligand aromatic ring and hydrogen bonds between the His248 side chain and the ligand nitro and secondary amine groups. THS-020 has an additional hydrogen bond between the furan oxygen atom of THS-020 and the hydroxyl group of Tyr307. The preferred orientation of the ligands we observe could also help stabilize a hydrogen bond inside the ligand, between the secondary amine linker and the nitro group on the “A ring.” Notably, the scarcity of hydrogen bonds in the liganded structures starkly contrasts with our structure of unliganded HIF2α PAS-B that contains 8 solvent-inaccessible water molecules within the protein core. These water molecules make up an extensive internal hydrogen bonding network10 with a number of the side chains lining the cavity (Supp. Figure 2). Thus, a number of protein hydrogen bond partners remain unpaired when ligand is bound, leaving us to suggest that much of the energetics favoring ligand binding stem from other forces (e.g. hydrophobic interactions with the “B ring”).
HIF2α PAS-B ligand-binding kinetics reveal characteristics of an open conformation
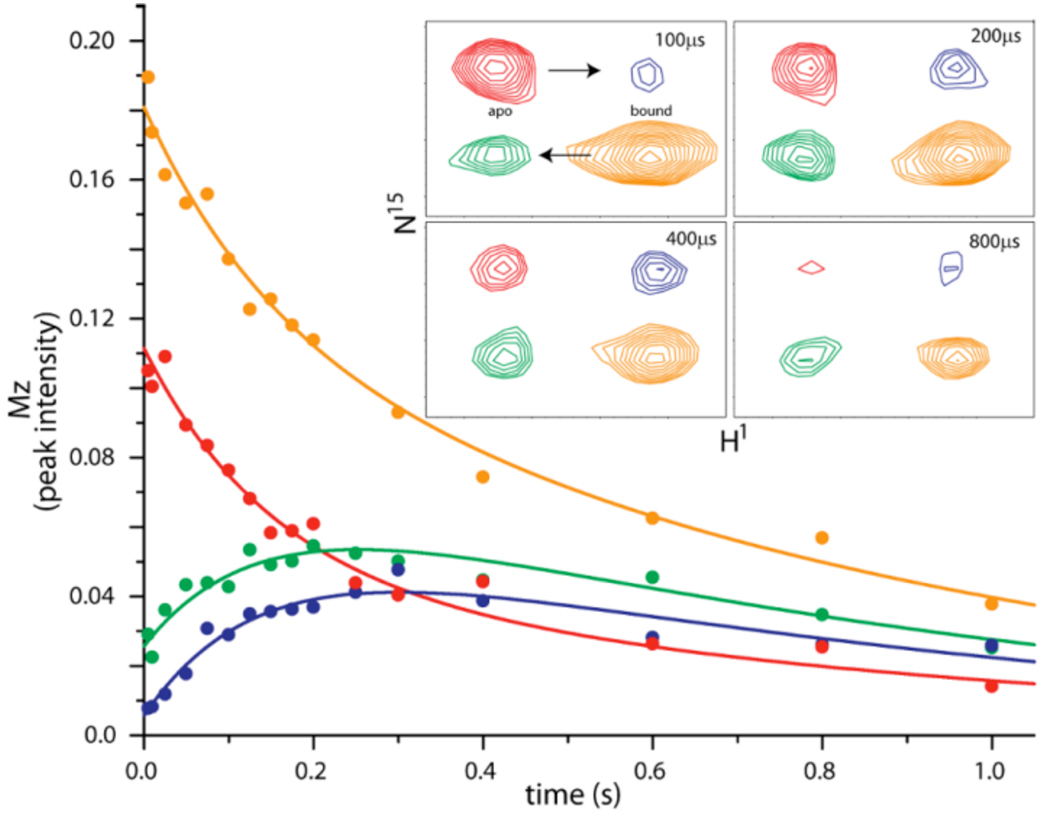
To complement these structural and thermodynamic data, we determined ligand association and dissociation rate constants under equilibrium conditions using NMR ZZ-exchange spectroscopy (Fig. 3). These methods have been used to measure such exchange rates in a range of macromolecular systems undergoing slow (millisecond to second timescale) dynamic equilibria.29,30,44 The time-dependence of cross- and auto-peak intensities from these spectra provide exchange rates (kex), from which a first order dissociation rate constant is obtained. The association rate constant can then be calculated using the equilibrium constant derived from ITC measurements. For complexes of HIF2α PAS-B and these compounds, ligand dissociation rate constants are approximately proportional to the affinity determined by ITC; thus, dissociation is the primary determinant of binding affinity for these compounds. Despite the completely buried nature of the HIF2α PAS-B pocket, we detected rapid ligand association kinetics with kon values greater than 106 M−1 s−1 for THS-017 and THS-020 at 25°C (Table 2). Compounds KG2-023 and THS-002 are somewhat slower, likely due to solvation effects due to their poor solubility. The narrow range of association rate constants suggests that the compounds traverse a similar barrier to binding, which we interpret as the conformational rearrangement necessary for HIF2α PAS-B to convert from a closed structure into a binding-competent ‘open’-state. Given the considerable size of these compounds relative to the scale of the HIF2α PAS-B domain,10 it seems apparent that a substantial conformational change is necessary to permit entry. Yet, the rapid association rates we observe are more typical of solvent accessible ligand binding sites than internal cavities. Thus, the open conformation of HIF2α PAS-B must either be present in significant concentration at equilibrium or rapidly interconvert with the ‘closed’ native state.
Figure 3. NMR ZZ-exchange data from HIF-2α PAS B and compound.
Shown are fits to NMR peak intensities as a function of exchange mixing time, with a subset corresponding crosspeaks shown in inset. Arrows indicating the directionality of autopeak to crosspeak are shown in the first inset.
Table 2.
Association and dissociation rate constants at 25°C
| Compound | kon × 105 (M−1 s−1) | koff (s−1) | No. of probes (amide cross/autopeaks) |
|---|---|---|---|
| KG-721 | 9.2 ± 1.1 | 9.2 ± 1.0 | 18 |
| KG2-023 | 4.5 ± 0.4 | 1.6 ± 0.1 | 5 |
| THS-002 | 6.9 ± 0.6 | 2.8 ± 0.6 | 8 |
| THS-017 | 13.9 ± 2.2 | 1.4 ± 0.3 | 6 |
| THS-020 | 13.0 ± 1.0 | 2.5 ± 0.1 | 12 |
The transition state barrier exhibits compensating enthalpic and entropic contributions
To better characterize the ligand-binding transition state, we measured the temperature dependence of the kon and koff rate constants for three HIF2α PAS-B complexes, THS-002, THS-020, and KG-721. Eyring analyses (Fig. 4) demonstrated a primarily enthalpic barrier to ligand binding (Table 3). The transition-state enthalpy reflects breaking of favorable interactions within the protein to reach an accessible, open transition state. We observed the largest enthalpic penalty in the transition state with compound THS-020, while binding of THS-002 had the lowest transition state enthalpic barrier. For each compound, this distortion of the protein structure is partially compensated by an increase in activation entropy. Such compensation suggests that ligand accommodation entails a transition state with increased entropy from protein flexibility, release of ordered solvent or a combination of these effects.
Figure 4. Eyring plots for HIF-2α PAS B ligands KG-721, THS-002 and THS-020.
(a). Temperature dependence of koff. (b). Temperature dependence of kon.
Table 3.
Transition state thermodynamics determined using NMR ZZ-exchange measurements at 10 to 25°C
| kon | ΔH‡ (kcal/mol) |
TΔS‡ (kcal/mol@25C) |
ΔS‡ (cal/mol K) |
ΔG‡ (kcal/mol@25C) |
|---|---|---|---|---|
| KG-721 | 16.8 ± 0.6 | 7.4 ± 0.2 | 25.0 ± 0.7 | 9.3 ± 0.6 |
| THS-002 | 13.5 ± 0.6 | 4.0 ±0 .1 | 13.4 ± 0.4 | 9.5 ± 0.6 |
| THS-020 | 22.9 ± 0.8 | 13.9 ± 0.4 | 46.5 ± 1.4 | 9.1 ± 0.9 |
| koff | ΔH‡ (kcal/mol) |
TAS‡ (kcal/mol@25C) |
ΔS‡ (cal/mol K) |
ΔG‡ (kcal/mol@25C) |
|---|---|---|---|---|
| KG-721 | 20.4 ± 1.1 | 4.5 ± 0.3 | 15.0 ± 0.9 | 15.9 ± 1.1 |
| THS-002 | 16.7 ± 0.5 | 0.0 ± 0.1 | 0.0 ± 1.8 | 16.7.± 0.5 |
| THS-020 | 23.5 ± 1.1 | 6.7 ± 0.3 | 22.5 ± 1.1 | 16.8 ± 1.1 |
Molecular dynamics simulations reveal conversion between open and closed conformations and pathways of ligand entry
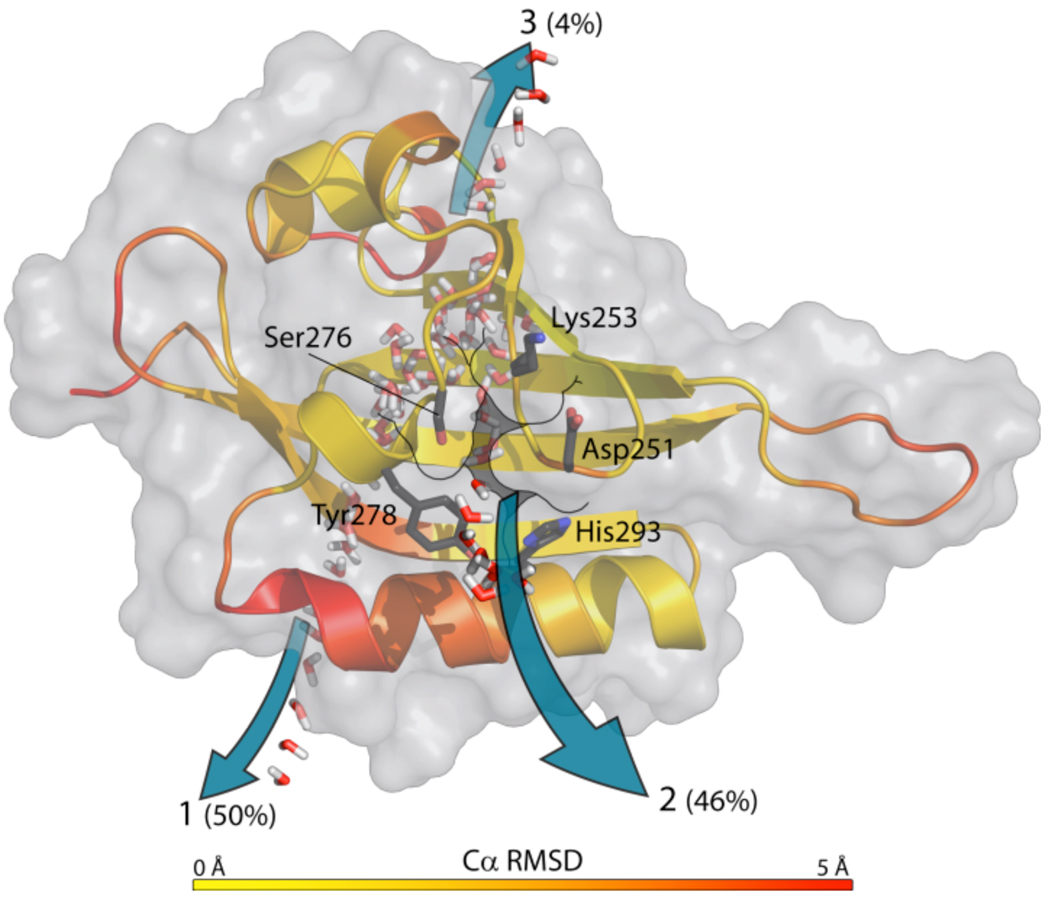
To identify possible pathways used by ligands to enter/exit the cavity within HIF2α PAS-B, we conducted multiple MD simulations beginning from an NMR-derived structure of apo-HIF2α PAS-B (PDB: 1P97).9 Despite the significantly smaller cavity (ca. 120 Å3) found in this starting structure compared with the crystal structure, we observed water entering and exiting the protein core via two primary routes in each of three independent 35 ns MD trajectories. The most common entry/exit pathway (pathway 1) is between the Fα helix and the Gβ strand, with 50% of solvent entry/exit events proceeding via this route (Fig. 5). The other major pathway (46% of solvent entry/exit events) is between the short Eα helix, the Fα helix and the AB loop (pathway 2, Fig. 5). In pathway 2, motion in the AB loop and Eα relative to Fα displaces the Met252 and Tyr278 side chains, opening a passageway for water entry and egress. An additional minor pathway (pathway 3) involved one water molecule entering and another escaping between Cys257 and Asp259 (between Bα and Cα), but this was only observed early in a single simulation. Consequently, we focus on pathways 1 and 2, which were observed multiple times and in multiple simulations (Supp. Table 1).
Figure 5. Molecular dynamics suggest routes of ligand entry and egress.
Heat map of HIF2α PAS-B derived from Cα-RMSD values over the course of 35 ns molecular dynamics simulations. Water passage pathways are superimposed on the crystal structure and noted with pathway numbers and percentages of total observed water entry/exit events.
To determine whether distinct ‘open’ and ‘closed’ protein conformers were populated in these simulations, we performed conformational clustering via an all-versus-all comparison of structures sampled during MD, which was projected into three-dimensional space via multidimensional scaling (Supp. Fig. 3A). Two dominant conformations were evident in all three simulations, corresponding to ‘open’ and ‘closed’ conformers (Supp. Fig. 3B). Overall, HIF2α PAS-B spent 57% of the time in the closed conformer, 41% in the open state and 2% in neither. There were 3 transitions from the closed to the open form and 2 transitions from the open form back to the closed form detected in the combined 105 ns of simulation time. Thus, these structures were not continuously converting on the MD timescale. Overall there were 0.05 transitions/ns, which unfortunately falls in a time regime slower than the rotational correlation time of this domain (τc = 9.1 ns), and into a timescale that is difficult to experimentally examine with NMR relaxation methods.
The open and closed conformations both contain all of the secondary structure elements found in the HIF2α PAS-B NMR9 and crystal10 structures, but differ mainly in their tertiary packing of the distorted, one-turn Dα and Eα helices and the AB loop on the β-sheet (Supp. Fig. 3B). The movement of Fα also contributes to the open conformation and is correlated with the changes in the positions of the Dα and Eα helices. Fα moves away from the protein core, pivoting on its C-terminus such that the N-terminus of the helix shifts by 20–30°. These dynamics are the primary factor enabling transport of water along pathway 1 (between the Fα helix and the Gβ strand). The Dα and Eα helices move by 5–8 Å away from the AB loop in the open conformation observed in MD simulations, opening the front of the molecule (Supp. Fig. 3). Interestingly, the C-terminus of the Dα helix also extends by two residues in the open form (Supp. Fig. 3C). The Dα and Eα helices and neighboring AB loop are highly dynamic, but the neighboring Aβ-Bβ hairpin becomes more ordered when the short helices move away. The hairpin maintains its contacts with the Iβ strand while Aβ adds residues at its C-terminus and Bβ gains structure and forms hydrogen bonds with Aβ. The combined result of these motions is that a channel is created from the solvent into the protein core (Supp. Fig. 3d) that serves as the route for water passage via pathway 2 (Fig. 5). The average Cα RMSD between the open and closed forms range from 2.2–3.6 Å in the three simulations.
The majority of the water entry and exit events (85%) occurred while the protein was in the open conformation (Supp. Table 1). Transport via pathway 1 was only observed in the open conformation; in contrast, most transport via pathway 2 was in the open conformation although five cases were observed from the closed state. In these latter cases, the entry and exit of water via pathway 2 was not controlled solely by large backbone movements. Instead, the side chain of residue Met 289 exists in different rotamers, acting as a gate for water passage. In one rotamer, Met 289 interacts with Met 252 and Tyr 278, effectively blocking the channel (Supp. Fig. 4), while an alternate rotamer swings out of the way and permits water passage (Supp. Table 1). Our simulations indicate that both pathways 1 and 2 are sufficiently wide (~8 Å) in the open conformation to allow entry of the ligands in this study, all of which have dimensions of approximately 8 × 12 Å in their shortest and longest dimensions.
Discussion
Ligand binding by proteins forms the basis for numerous biological processes, including the specificity of enzymes, hormonal control, environmental sensing, and cell-cell recognition. Biological systems have tuned their respective ligand affinities for these purposes, utilizing high-affinity interactions for transport and relatively low affinity for rapid response to stimuli. Thus, understanding the thermodynamics of a protein/ligand interaction is key to understanding biological function. Ligand binding also provides an attractive avenue for exogenous control of biological systems, as exploited by pharmaceuticals that modulate natural function through specific binding with molecular targets.
In several cases where proteins bind ligands with high affinity, evolution has optimized the association rate and achieved the diffusion limit for ligand binding,45,46 where each molecular collision between protein and ligand produces a successful binding event. For the majority of proteins, however, reduced accessibility of a ligand-binding site provides a barrier to ligand binding that is reflected in the ligand association rate constant relative to this diffusion limit. This principle is well illustrated by the work of Scott and colleagues47 in myoglobin where the bulk of distal residue side chains represent a ‘conformational gate’ to diffusional ligand access which is evident in O2 association rate constants.
Given the high HIF2α PAS-B ligand association rate constants, we conclude that the conformational barrier to ligand entry is small. This was unexpected considering the inaccessibility of the crystallographically determined protein binding pocket, the size of bound ligands, and the required expulsion of eight water molecules from the binding site. Given this low barrier, one might suppose that a proportion of HIF2α PAS-B exists in a distinct open conformation in equilibrium with a closed state in a manner similar to periplasmic carbohydrate binding proteins, such as maltose binding protein (MBP).48 While MBP and HIF2α PAS-B both exhibit low micromolar affinity and rapid ligand association rates for similar sized ligands,45 much of our experimental data cannot independently confirm that HIF2α PAS-B exists to any significant degree in a discrete open state. A likely cause of this is the rapid timescale of interconversion between the ‘open’ and ‘closed’ states observed in MD simulations (on average 21 ns between interconversion events), which we anticipate leading to time averaging many NMR parameters that could be different between the two states. Our data are consistent with this, as we did not observe distinct chemical shifts for two conformations (which would require millisecond or slower interconversion), nor significant 15N relaxation dispersion (sensitive to high microsecond to low millisecond events). Further, our crystallographic data reveal no substantial differences between structures of the free and liganded states, even at the high resolutions under consideration (1.2 to 1.6 Å), but this is to be expected with crystallization trapping a single, lowest-energy conformation. From these data, unliganded HIF2α PAS-B appears to be in a single, closed conformation.
However, the combination of other NMR methods – sensitive to motions on different timescales – ligand association rates, and MD simulations suggest that the protein also populates a more open conformation. Furthermore, the combination of NMR and MD provides clues to the mechanism of ligand entry into the core of HIF2α PAS-B. NMR-based 2H exchange measurements are particularly useful in this context. For much of the central core of HIF2α PAS-B, binding of the THS-044 ligand significantly (>100x) stabilizes many of amides near the ligand, particularly in the Eα helix and the central Aβ, Hβ and Iβ strands of the β-sheet10 (shown schematically in Supp. Fig 5). This is consistent with a ligand-dependent shift of an “open”/”closed” conformational equilibrium in the apo protein being shifted towards a “closed” conformation (although it is likely that a component of this stabilization comes from the expulsion of water from the core of the domain). Further, sites with high r.m.s.d. values in the MD simulations show no significant 2H exchange protection in the apo-protein (Fig. 5, Supp. Fig 5). This is particularly conspicuous for the long Fα helix10, despite this section having backbone chemical shift and 1H-1H NOE characteristics of helical secondary structure.9 Notably, complex formation with THS-044 significantly broadens NMR signals from many AB-loop, Eα, and Fα sites with high r.m.s.d. values in the MD simulations, suggesting another link between experiment and simulation indicative of enhanced protein conformational dynamics induced by ligand binding or a more complicated conformational exchange between both components of the complex (Supp. Fig 5 in ref 10). Many of these same sites show elevated 15N R2/R1 ratios in the apo protein (Supp. Fig. 6), suggesting that the motions required for entry exist in the absence of ligand. Taken together, these observations strongly suggest the Fα helix has significant conformational heterogeneity, likely including components of an apo-HIF2α PAS-B conformation that facilitates ligand binding just as it enables water entry via both pathways 1 and 2 in the MD simulations.
The existence of an open, binding-competent conformation of HIF2α PAS-B is also supported by the enthalpy-entropy compensation observed in our transition state thermodynamics. Enthalpy-entropy compensation of this nature is characteristic of a loss of protein structure49 in the transition state that permits ligand access to the binding site. Mulder and coworkers have characterized a similar interconversion between a sterically inaccessible closed state and a partially unstructured open state of the T4 lysozyme L99A mutant50 that also binds small ligands with an association rate of 106 M−1 s−1.51 The L99A mutation in T4 lysozyme creates a 150 Å3 cavity (significantly smaller than the 290 Å3 HIF2α PAS-B cavity) that binds small substituted benzene compounds.52 Though HIF2α PAS-B ligands are substantially larger and were observed in a natural protein rather than a point mutant variant, the principle here appears to be the same, i.e. a conformational equilibrium exists between a highly populated ligand-inaccessible ground state and a binding competent state characterized by increased dynamics and altered structure.
A notable difference between T4 lysozyme and HIF2α PAS-B is the time and spatial scale of conformational motions necessary to permit access to the ligand-binding site. T4 lysozyme requires increased millisecond timescale dynamics within the Eα, Fα and Iα helices and their adjacent loops that permit ligand access.50 The HIF2α PAS-B dynamics appear to be more rapid than these motions, suggesting ligand entry requires nanosecond timescale motions, likely side chain rotamerization, loop displacement, and helical motions, as observed in the MD simulations. While the experimental studies cannot flesh out the details of the PAS-B conformational substates, these states were both detectable and characterizable by MD. Future studies will involve using the MD-generated models to devise specific experiments to check the predictions.
We close by asserting that a ligand accessible state of HIF2α PAS-B exists in equilibrium with the native state, although this open state is characterized by increased dynamics and altered structure. As such, the open state conformation is better described by the open state model of L99A T4 lysozyme than the discrete open structure of MBP. Efforts to quantitate the relative populations of native and open conformations of HIF2α PAS-B are challenging due to the rapid exchange involved, but they have important implications for both the binding of a natural ligand and for the development of targeted ligands aimed at disrupting the functionally important HIF2α PAS-B / ARNT PAS-B interaction within the intact HIF2 transcription factor.
Supplementary Material
Acknowledgements
We would like to acknowledge Paul Erbel and Rick Bruick for ongoing discussions through various aspects of this work, Doug Frantz and the UT Southwestern Medicinal Chemistry facility for synthesis of the THS series compounds used in this study and Carlos Amezcua for development of the initial small compound library and assistance with NMR spectroscopy. This work was supported by grants from the NIH (P01 CA95471 to KHG, R01 GM50789 to VD and T15 LM07442 to PCA) and the American Cancer Society (High Plains Division-North Texas Postdoctoral Fellowship PF-06-270-01-GMC to THS).
Abbreviations
- HIF
hypoxia inducible factor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- PAS
Per-ARNT-Sim domain
- NMR
nuclear magnetic resonance
- ITC
isothermal titration calorimetry
- LOV
Light-Oxygen-Voltage domain
- MBP
maltose binding protein
- MD
molecular dynamics
Footnotes
Accession codes. Protein Data Bank: The coordinates and structure factors of the PAS B* heterodimer bound to THS-017 and THS-020 have been deposited with the Protein Data Bank under accession codes 3H7W and 3H82, respectively.
Supporting Information
Experimental electron density maps of ligand-bound structures, protein/ligand interactions within the core of HIF2α PAS-B, details of fitting NMR exchange data, conformational changes revealed in molecular dynamics simulations, deuterium exchange protection factors mapped onto the structure of HIF2α PAS-B, 15N relaxation measurements from HIF2α PAS-B in the absence of presence of an artificial ligand, and a table of water entry/exit events.
References
- 1.Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology (Bethesda) 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- 2.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294(5545):1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 4.Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci U S A. 2002;99(8):5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaelin WG., Jr. The von Hippel-Lindau protein, HIF hydroxylation, and oxygen sensing. Biochem Biophys Res Commun. 2005;338(1):627–638. doi: 10.1016/j.bbrc.2005.08.165. [DOI] [PubMed] [Google Scholar]
- 6.Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, Semenza GL, van Diest PJ, van der Wall E. Levels of hypoxia-inducible factor-1alpha independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer. 2003;97(6):1573–1581. doi: 10.1002/cncr.11246. [DOI] [PubMed] [Google Scholar]
- 7.Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. Expression of hypoxia-inducible factor 1alpha in brain tumors: association with angiogenesis, invasion, and progression. Cancer. 2000;88(11):2606–2618. [PubMed] [Google Scholar]
- 8.Sullivan R, Pare GC, Frederiksen LJ, Semenza GL, Graham CH. Hypoxia-induced resistance to anticancer drugs is associated with decreased senescence and requires hypoxia-inducible factor-1 activity. Mol Cancer Ther. 2008;7(7):1961–1973. doi: 10.1158/1535-7163.MCT-08-0198. [DOI] [PubMed] [Google Scholar]
- 9.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor. Proc Natl Acad Sci U S A. 2003;100(26):15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheuermann TH, Tomchick DR, Machius M, Guo Y, Bruick RK, Gardner KH. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proc Natl Acad Sci U S A. 2009;106(2):450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, Zhang L, Erbel PJ, Gardner KH, Ding K, Garcia JA, Bruick RK. Functions of the Per/ARNT/Sim domains of the hypoxia-inducible factor. J Biol Chem. 2005;280(43):36047–36054. doi: 10.1074/jbc.M501755200. [DOI] [PubMed] [Google Scholar]
- 12.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 13.Crosson S, Rajagopal S, Moffat K. The LOV domain family: photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42(1):2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 14.Gilles-Gonzalez MA, Gonzalez G. Heme-based sensors: defining characteristics, recent developments, and regulatory hypotheses. J Inorg Biochem. 2005;99(1):1–22. doi: 10.1016/j.jinorgbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Hoff WD, Dux P, Hard K, Devreese B, Nugteren-Roodzant IM, Crielaard W, Boelens R, Kaptein R, van Beeumen J, Hellingwerf KJ. Thiol ester-linked p-coumaric acid as a new photoactive prosthetic group in a protein with rhodopsin-like photochemistry. Biochemistry. 1994;33(47):13959–13962. doi: 10.1021/bi00251a001. [DOI] [PubMed] [Google Scholar]
- 16.Briggs WR. The LOV domain: a chromophore module servicing multiple photoreceptors. J Biomed Sci. 2007;14(4):499–504. doi: 10.1007/s11373-007-9162-6. [DOI] [PubMed] [Google Scholar]
- 17.Taylor BL. Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox and energy. Mol Microbiol. 2007;65(6):1415–1424. doi: 10.1111/j.1365-2958.2007.05889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilles-Gonzalez MA, Gonzalez G. Regulation of the kinase activity of heme protein FixL from the two-component system FixL/FixJ of Rhizobium meliloti. J Biol Chem. 1993;268(22):16293–16297. [PubMed] [Google Scholar]
- 19.Amezcua CA, Harper SM, Rutter J, Gardner KH. Structure and interactions of PAS kinase N-terminal PAS domain: model for intramolecular kinase regulation. Structure. 2002;10(10):1349–1361. doi: 10.1016/s0969-2126(02)00857-2. [DOI] [PubMed] [Google Scholar]
- 20.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301(5639):1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 21.Craven CJ, Derix NM, Hendriks J, Boelens R, Hellingwerf KJ, Kaptein R. Probing the nature of the blue-shifted intermediate of photoactive yellow protein in solution by NMR: hydrogen-deuterium exchange data and pH studies. Biochemistry. 2000;39(47):14392–14399. doi: 10.1021/bi001628p. [DOI] [PubMed] [Google Scholar]
- 22.Moglich A, Moffat K. Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol. 2007;373(1):112–126. doi: 10.1016/j.jmb.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong W, Hao B, Chan MK. New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL. Biochemistry. 2000;39(14):3955–3962. doi: 10.1021/bi992346w. [DOI] [PubMed] [Google Scholar]
- 24.Blommel PG, Fox BG. A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif. 2007;55(1):53–68. doi: 10.1016/j.pep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. A heteronuclear correlation experiment for simultaneous determination of 15N longitudinal decay and chemical exchange rates of systems in slow equilibrium. J Biomol NMR. 1994;4(5):727–734. doi: 10.1007/BF00404280. [DOI] [PubMed] [Google Scholar]
- 30.Iwahara J, Clore GM. Direct observation of enhanced translocation of a homeodomain between DNA cognate sites by NMR exchange spectroscopy. J Am Chem Soc. 2006;128(2):404–405. doi: 10.1021/ja056786o. [DOI] [PubMed] [Google Scholar]
- 31.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 32.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 33.Macey R, Oster G, Zahnley T. Berkeley Madonna User’s Guide. 2000 [Google Scholar]
- 34.Eyring H. The activated complex in chemical reactions. J. Chem. Phys. 1935;3:107–115. [Google Scholar]
- 35.Beck DAC, Alonso DOV, Daggett V. ilmm, in lucem Molecular Mechanics. Seattle, WA: University of Washington; pp. 2000–2008. [Google Scholar]
- 36.Beck DA, Jonsson AL, Schaeffer RD, Scott KA, Day R, Toofanny RD, Alonso DO, Daggett V. Dynameomics: mass annotation of protein dynamics and unfolding in water by high-throughput atomistic molecular dynamics simulations. Protein Eng Des Sel. 2008;21(6):353–368. doi: 10.1093/protein/gzn011. [DOI] [PubMed] [Google Scholar]
- 37.Kehl C, Simms AM, Toofanny RD, Daggett V. Dynameomics: a multi-dimensional analysis-optimized database for dynamic protein data. Protein Eng. Des. Sel. 2008;21:379–386. doi: 10.1093/protein/gzn015. [DOI] [PubMed] [Google Scholar]
- 38.Simms AM, Toofanny RD, Kehl C, Benson NC, Daggett V. Dynameomics: design of a computational lab workflow and scientific data repository for protein simulations. Protein Eng. Des. Sel. 2008;21:369–377. doi: 10.1093/protein/gzn012. [DOI] [PubMed] [Google Scholar]
- 39.Levitt M, Hirshberg M, Sharon R, Daggett V. Potential-Energy Function and Parameters for Simulations of the Molecular-Dynamics of Proteins and Nucleic-Acids in Solution. Computer Physics Communications. 1995;91(1–3):215–231. [Google Scholar]
- 40.Levitt M, Hirshberg M, Sharon R, Laidig KE, Daggett V. Calibration and testing of a water model for simulation of the molecular dynamics of proteins and nucleic acids in solution. Journal of Physical Chemistry B. 1997;101(25):5051–5061. [Google Scholar]
- 41.Kell GS. Precise Representation of Volume Properties of Water at One Atmosphere. Current Contents/Engineering Technology & Applied Sciences. 1979;(31):16–16. [Google Scholar]
- 42.Beck DA, Daggett V. Methods for molecular dynamics simulations of protein folding/unfolding in solution. Methods. 2004;34(1):112–120. doi: 10.1016/j.ymeth.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 43.Beck DA, Armen RS, Daggett V. Cutoff size need not strongly influence molecular dynamics results for solvated polypeptides. Biochemistry. 2005;44(2):609–616. doi: 10.1021/bi0486381. [DOI] [PubMed] [Google Scholar]
- 44.Rubinstenn G, Vuister GW, Mulder FA, Dux PE, Boelens R, Hellingwerf KJ, Kaptein R. Structural and dynamic changes of photoactive yellow protein during its photocycle in solution. Nat Struct Biol. 1998;5(7):568–570. doi: 10.1038/823. [DOI] [PubMed] [Google Scholar]
- 45.Miller DM, 3rd, Olson JS, Pflugrath JW, Quiocho FA. Rates of ligand binding to periplasmic proteins involved in bacterial transport and chemotaxis. J Biol Chem. 1983;258(22):13665–13672. [PubMed] [Google Scholar]
- 46.Thorsteinsson MV, Bevan DR, Potts M, Dou Y, Eich RF, Hargrove MS, Gibson QH, Olson JS. A cyanobacterial hemoglobin with unusual ligand binding kinetics and stability properties. Biochemistry. 1999;38(7):2117–2126. doi: 10.1021/bi9819172. [DOI] [PubMed] [Google Scholar]
- 47.Scott EE, Gibson QH, Olson JS. Mapping the pathways for O2 entry into and exit from myoglobin. J Biol Chem. 2001;276(7):5177–5188. doi: 10.1074/jbc.M008282200. [DOI] [PubMed] [Google Scholar]
- 48.Tang C, Schwieters CD, Clore GM. Open-to-closed transition in apo maltose-binding protein observed by paramagnetic NMR. Nature. 2007;449(7165):1078–1082. doi: 10.1038/nature06232. [DOI] [PubMed] [Google Scholar]
- 49.Dunitz JD. Win some, lose some: enthalpy-entropy compensation in weak intermolecular interactions. Chem Biol. 1995;2(11):709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 50.Mulder FA, Mittermaier A, Hon B, Dahlquist FW, Kay LE. Studying excited states of proteins by NMR spectroscopy. Nat Struct Biol. 2001;8(11):932–935. doi: 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- 51.Feher VA, Baldwin EP, Dahlquist FW. Access of ligands to cavities within the core of a protein is rapid. Nat Struct Biol. 1996;3(6):516–521. doi: 10.1038/nsb0696-516. [DOI] [PubMed] [Google Scholar]
- 52.Eriksson AE, Baase WA, Zhang XJ, Heinz DW, Blaber M, Baldwin EP, Matthews BW. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255(5041):178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.