Marine-derived actinomycete bacteria are emerging as a valuable resource for bioactive natural products encompassing a variety of unique structural classes.[1] In our hands, early detection of cell growth inhibitors using in vitro cytotoxicity assays against the colon carcinoma cancer cell line HCT-116, followed by extensive mechanism of action studies, has proven to be an effective approach. As such, the HCT-116 assay has been instrumental in the identification of potentially important anticancer agents.[2]
In the course of our continued studies, Streptomyces strain CNR-698[3] was isolated from bottom sediments collected at a depth of 1618 meters in the Bahamas Islands in 2003. Cytotoxicity-guided (HCT-116) fractionation by C18 flash chromatography and RP-HPLC of crude extract led to the isolation of ammosamides A (1) and B (2) as blue and red solids, respectively (3 and 4 mg L−1). Structure assignments for 1 and 2 proved to be particularly difficult due to their inherent insolubility (soluble only in dimethyl sulfoxide (DMSO)) and a lack of descriptive NMR signals, ultimately requiring the integration of NMR spectral analysis, mass spectrometry data, and single crystal X-ray diffraction studies.
High-resolution (ESI) mass spectrometric analysis of ammosamide A (1) indicated a molecular formula C12H1035ClN5OS (m/z [M+H]+: 308.0303]. The molecular weight of ammosamide B (2) was found to be 16 amu lower (m/z [M+H]+: 292.0604) consistent with the molecular formula C12H1035ClN5O2. The UV/Vis spectrum of 1 was indicative of an unusually highly conjugated structure with absorptions at λmax=580, 430, 350, and 290 nm.
Inspection of the 1H NMR spectrum of 1 in [D6]DMSO revealed six singlets between δ=6.0 and 9.0 ppm and one methyl singlet at δ=4.03 ppm, while the 13C NMR spectra revealed the presence of eleven sp2 hybridized carbon atoms and a single sp3 hybridized carbon atom at δc=33.3 ppm (Table 1). The addition of D2O (20 μL) to the sample in [D6]DMSO resulted in the immediate disappearance of 1H NMR signals at δ=7.16 (1H), 6.63 (1H), 6.89 ppm (2H) and the slower disappearance of singlets at δ=8.92 (1H), and 7.68 ppm (1H) (less than 10 min). The exchangeable protons at δ=7.16, 6.63 and 6.89 ppm were assigned as aromatic amines at C-6 and C-8 (based on HMBC correlations), while the slowly exchanging protons at δ=8.92 and 7.68 ppm were assigned to a primary amide on the basis of COSY and HMBC correlations. The only non-exchangeable hydrogen atoms were the methyl singlet resonance at δ=4.03 ppm and a one-proton singlet at δ=8.47 ppm. The 13C NMR spectrum of 1 indicated the presence of two carbonyl groups (δc=177.2 and 166.0 ppm), as well as two upfield sp2 carbon atoms (δc=103.1 and 110.5 ppm). HMBC correlations between the downfield carbonyl (δc=177.2 ppm) and the proton methyl singlet at δ=4.03 ppm, we thought, defined an N-methyl amide, although a carbon chemical shift so far downfield would not be expected. In addition to correlations from the aromatic δ=8.47 ppm singlet, the only other HMBC correlations were from the exchangeable protons at δ=7.16/6.63 ppm to C-7 (δc=103.1 ppm) and from δ=6.89 ppm to C-7 and C-8a (δc=110.5 ppm).
Table 1.
NMR spectral data for 1 and 2 ([D6]DMSO).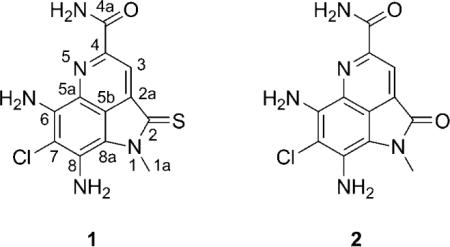
| Ammosamide A (1) | Ammosamide B (2) | ||||
|---|---|---|---|---|---|
| Position | δC[a,c] | δH[d] | HMBC | δC[b,c] | δH[d] |
| 1a | 33.3 | 4.03 (s) | C-2, C-8a | 28.5 | 3.59 (s) |
| 2 | 177.2 | 164.0 | |||
| 3 | 116.5 | 8.47 (s) | C-2, C-4 | 115.3 | 8.34 (s) |
| C-5b, C-4a | |||||
| 4 | 144.6 | 144.7 | |||
| 4a | 166.0 | 166.2 | |||
| 5b | 119.7 | 119.0 | |||
| 7 | 103.1 | 104.5 | |||
| 8a | 110.5 | 106.3 | |||
| CONH2 | 7.68 (brs) | C-4 | 7.66 (brs) | ||
| 8.92 (brs) | C-4a | 8.91 (brs) | |||
| NH2 (C-6) | 7.16 (brs)[e] | C-7 | 6.73 (brs)[f] | ||
| 6.63 (s)[e] | |||||
| NH2 (C-8) | 6.89 (s) | C-7, C-8a | 6.18 (brs)[f] | ||
75 MHz.
125 MHz.
C-2a, C-5a, C-6, and C-8 (1: δC = 132.6, 134.7, 136.8, 142.7 ppm; 2: δC = 130.6, 130.8, 132.4, 140.5 ppm) could not be unambiguously assigned.
600 MHz.
At certain concentrations, these two one-proton signals coalesce to a two-proton signal at δ = 7.09 ppm.
Assigned by analogy to 1.
The spectral data for 1 suggested a highly unsaturated azaaromatic metabolite possessing three rings. However, the lack of definitive NMR assignments that could be used to link these features forced us to concentrate efforts toward obtaining an X-ray crystal structure. We were fortunate to obtain small crystals of 1 by the slow diffusion of H2O into a saturated solution in DMSO.[4] The X-ray assignment of ammosamide A (1) is shown in Figure 1. Once X-ray data became clear, the spectral data for 1 could be assigned.
Figure 1.
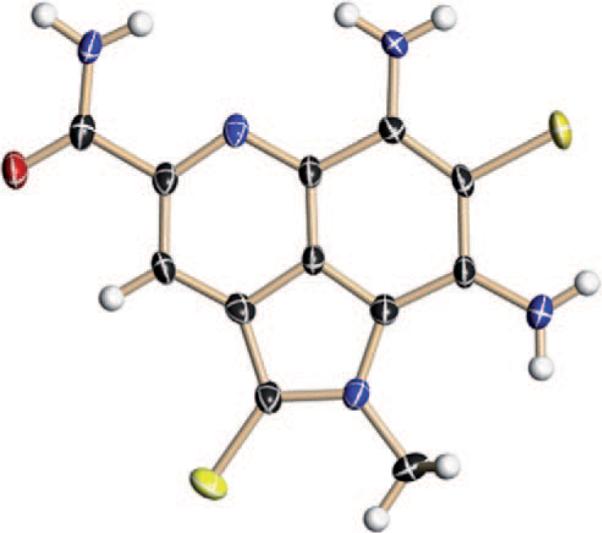
X-ray crystal structure of ammosamide A (1). Red O, blue N, yellow Cl, black C, white H.
The structure assignment of ammosamide B (2) followed from analysis of spectral data and chemical interconversion. Comparison of the C-2 carbonyl chemical shifts in 1 (δC=177.2 ppm) and 2 (δC=164.0 ppm) revealed a difference of 13 ppm, consistent with the typical 13C chemical shift difference between a carbonyl and a thiocarbonyl (ca. 20 ppm).[5] In order to chemically confirm the presence of the thiolactam functionality, we used Lawesson's reagent [2,4-bis(p-methoxyphenyl)-1,3-dithiadiphosphetane-2,4-disulfide] to convert lactam 2 into thiolactam 1.[6] The low yield of this reaction is likely attributable the nucleophilic amines in 2.[7] Exposed to air during storage, 1 was gradually converted to ammosamide B (2). Notably, the transformation could also be accomplished in 10 min, upon treatment of 1 with hydrogen peroxide in aqueous methanol.[8] This reactivity has been previously observed in other thioamide-containing compounds.[9]
The structural similarities between the ammosamides and the microbial product lymphostin (3) are clear,[10] as is the relationship of the ammosamides to several sponge-derived pyrroloiminoquinone natural products, including batzelline A (4),[11a] isobatzelline D (5),[11b] and makaluvamine A (6)[11c] (Scheme 1). The sponge metabolites 4–6 possess different patterns of Cl and NH2 substitution and assume p-iminoquinone and o-quinone structures. The presence of an amino group at C-8 in the ammosamides results in a fundamentally different structure type in which the quinoline tautomer predominates. The pyrrole moiety in 3–6 is uniquely oxidized to the pyrrolidinone in ammosamide B (2). Finally, though methyl sulfides are present in 4 and 5, ammosamide A (1) is the first natural product to contain a thio-γ-lactam functionality.[12]
Scheme 1.
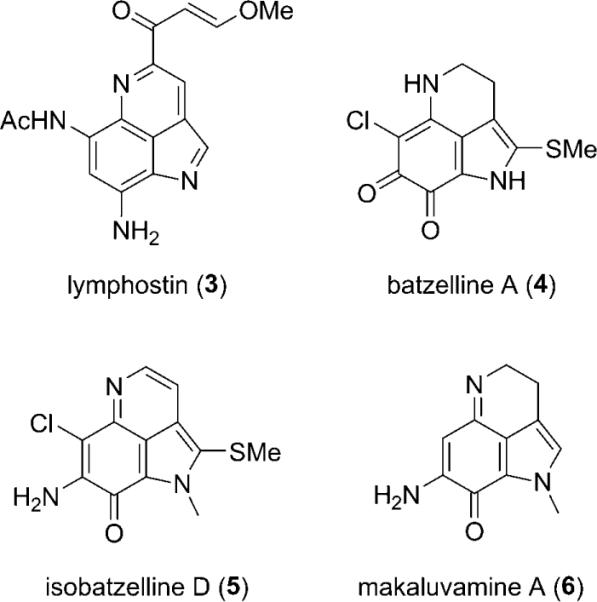
Related metabolites from bacteria and sponges.
The fact that the ammosamides are highly colored (1: λmax=580 nm; 2: λmax=530 nm), yet lack quinone or iminoquinone functionalities, leads to speculation about the electronic character and reactivity of these metabolites. The intense colors of these compounds could reflect a strong charge separation between the two six-membered aromatic rings due to the effects of electron-donating groups on the chlorine-containing ring and electron-withdrawing substituents on the pyridine ring. It is, conceptually, also explained by the potential for ammosamide A to exist in an equilibrium with its bis-iminoquinone tautomer (Scheme 2). Furthermore, in 1 and 2 the chlorine atom at C-7 is poised to engage in nucleophilic aromatic substitution with a suitable nucleophile, particularly when the molecule exists as its bis-iminoquinone tautomer.[13] This reactivity may be relevant to the molecule's interaction with its protein target.[14]
Scheme 2.
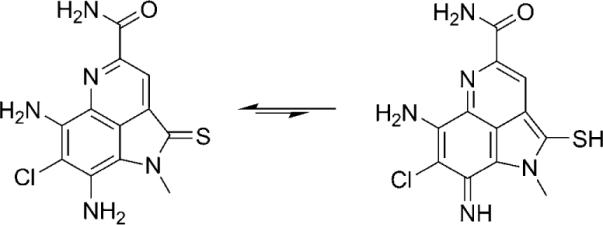
Possible tautomeric form of ammosamide A (1).
Ammosamides A (1) and B (2) exhibited significant in vitro cytotoxicity against HCT-116 colon carcinoma, each with IC50=320 nM. These compounds also demonstrated pronounced selectivity in a diversity of cancer cell lines with values ranging from 20 nM to 1 μM, indicating a specific target mechanism of action. To explore the intracellular target of the ammosamides, ammosamide B (2) was converted to a highly fluorescent molecule by conjugation.[14] Treatment of HCT-116 colon carcinoma or HeLa cells with this fluorescent molecule produced immediate and irreversible labeling of a specific protein in the cellular cytosol. Using a cell and molecular biology approach, the target of the ammosamides was identified as a member of the myosin family, important cellular proteins that are involved in numerous cell processes, including cell cycle regulation, cytokinesis, and cell migration.
Supplementary Material
Acknowledgments
This work was supported by a grant from the US National Cancer Institute, NIH under grant CA44848 (to W.F.). S.P.G. is grateful to the Fundação para a Ciência e Tecnologia, Portugal, for a postdoc fellowship. The authors thank Arnold Rheingold (UCSD) for X-ray diffraction data, Lisa Zeigler and Wolf Wrasidlo (UCSD Cancer Center) for in vitro cytotoxicity data, and Michelle Leibrand for help with purification.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/anie.200804890.
References
- [1].a) Fenical W, Jensen PR. Nat. Chem. Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]; b) Koehn FE, Carter GT. Nat. Rev. Drug Discovery. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- [2].a) Feling RH, Buchanan GO, Mincer TJ, Kauffman CA, Jensen PR, Fenical W. Angew. Chem. 2003;115:369–371. doi: 10.1002/anie.200390115. [DOI] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2003;42:355–357. [Google Scholar]; b) Cho JY, Kwon HC, Williams PG, Jensen PR, Fenical W. Org. Lett. 2006;8:2471–2474. doi: 10.1021/ol060630r. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kwon HC, Kauffman CA, Jensen PR, Fenical W. J. Am. Chem. Soc. 2006;128:1622–1632. doi: 10.1021/ja0558948. [DOI] [PubMed] [Google Scholar]
- [3].Strain CNR-698 was isolated from a sediment sample collected at Little San Salvador, Bahamas. The strain does not require seawater for growth. Its partial 16S rRNA sequence (Genbank #EU863183) is 100% identical to another bacterium of the family Streptomycetaceae, also isolated from marine sediments (20 m) (#EU099411).
- [4].X-ray measurements were made on a SMART CCD area detector with graphite monochromated MoKα radiation (λ=1.54178 Å). 1 (C12H10ClN5OS): crystal dimensions 0.30×0.20×0.10 mm, monoclinic, space group P21/c, a=9.3766(8), b=18.2930(12), c=7.0462(6) Å, V=1208.55(17) Å3, Z=4, ρcalcd=1.691 mg m−3, μ=4.456 mm−1, T=100(2) K, 2θmax=136.3°, 7060 measured reflections, 1872 independent reflections (Rint=0.0399), 183 parameters refined, R=0.0541 (for 4205 reflections with I>2.00σ(I)), Rw=0.1523, max/min residual peaks in the final difference map 0.495/−0.817 e Å−3. CCDC 694293 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via www.ccdc.cam.ac.uk/data_request/cif.
- [5].Pretsch EP, Seibl J, Clerc T, Simon W, editors. Tables of Spectral Data for Structure Determination of Organic Compounds. 2nd ed. Springer; Berlin: 1989. [Google Scholar]
- [6].Pedersen BS, Scheibye S, Nilsson NH, Lawesson S-O. Bull. Soc. Chim. Belg. 1978;87:223. [Google Scholar]
- [7].Amines (and alcohols) react with Lawesson's reagent to afford a variety of N-phosphorus heterocycles: Jesberger M, Davis TP, Barner L. Synthesis. 2003:1929–1953.; Jesberger M, Davis TP, Barner L. Synthesis. 2003:1943–1948..
- [8].Conversion to the amide occurs via the thioamide S oxide: Hillhouse JH, Blair IA, Field L. Phosphorus Sulfur Relat. Elem. 1986;26:169–184..
- [9].a) Magnus P, Mendoza JS, Stamford A, Ladlow M, Willis P. J. Am. Chem. Soc. 1992;114:10232–10245. [Google Scholar]; b) Nowaczyk S, Alayrac C, Reboul V, Metzner P, Averbuch-Pouchot M-T. J. Org. Chem. 2001;66:7841–7848. doi: 10.1021/jo015956m. [DOI] [PubMed] [Google Scholar]; c) Sośnicki JG. Synlett. 2003:1673–1677. [Google Scholar]
- [10].Aotani Y, Nagata H, Yoshida M. J. Antibiot. 1997;50:543–545. doi: 10.7164/antibiotics.50.543. [DOI] [PubMed] [Google Scholar]
- [11].a) Sakemi S, Sun HH. Tetrahedron Lett. 1989;30:2517–2520. [Google Scholar]; b) Sun HH, Sakemi S, Burres N, McCarthy P. J. Org. Chem. 1990;55:4964–4966. [Google Scholar]; c) Radisky DC, Radisky ES, Barrows LR, Copp BR, Kramer RA, Ireland CM. J. Am. Chem. Soc. 1993;115:1632–1638. [Google Scholar]; d) Antunes EM, Copp BR, Davies-Coleman MT, Samaai T. Nat. Prod. Rep. 2005;22:62–72. doi: 10.1039/b407299p. For a review, see: [DOI] [PubMed] [Google Scholar]
- [12].For a review of sulfur-containing metabolites from marine organisms, see: Christophersen C, Anthoni U. Sulfur Rep. 1986;4:365–442..
- [13].A common strategy in the total synthesis of C-7-N containing natural products, like makaluvamine A (6), is the displacement of a C-7 leaving group with a primary amine. See: Iwao M, Motoi O, Fukuda T, Ishibashi F. Tetrahedron. 1998;54:8999–9010..
- [14].Hughes CC, MacMillan JB, Gaudêncio SP, Fenical W. J. J. La Clair, Angew. Chem. 2009;121:742. doi: 10.1002/anie.200804107. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew. Chem. Int. Ed. 2009;48:728. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


