Abstract
Prolyl-4-hydroxylase α(I) (P4Hα(I)) is the rate-limiting subunit forP4Henzyme activity, which is essential for procollagen hydroxylation and secretion. In the current study, we have characterized the human P4Hα(I) promoter for transcription factors and DNA elements regulating P4Hα(I) expression. Using a progressive deletion cloning approach, we have constructed pGL3-P4Hα(I) recombinant plasmids. We have identified a positive regulatory region at the positions of bp −184 to −97 responsible for ~80% of the P4Hα(I) promoter efficiency. Three E-boxes were located within this region, and the E-box at position bp −135 explains most of the regulatory capacity. Upstream stimulatory factors (USF1/USF2) were shown to bind on the E-box using chromatin immunoprecipitation assay. Suppression of USF1 and/or USF2 using specific short interference RNA resulted in a significant reduction in P4Hα(I) promoter activity, and overexpressed USF1 or USF2 increased P4Hα(I) promoter activity significantly. Although transforming growth factor β1 increased the USF1/USF2-E-box binding and P4Hα(I) promoter activity, this up-regulatory effect can be largely prevented by USF1/USF2-specific short interference RNA. On the other hand, cigarette smoking extracts, which have been shown to suppress P4Hα(I) expression, inhibited the binding between the USF1/USF2 and E-box, resulting in a reduced P4Hα(I) promoter activity. Furthermore, the E-box on the P4Hα(I) promoter appeared to indiscriminately bind with either USF1 or USF2, with a similar outcome on the promoter efficiency. In conclusion, our study shows that USF1/USF2 plays a critical role in basal P4Hα(I) expression, and both positive (transforming growth factor β1) and negative (cigarette smoking extract) regulators appear to influence the USF-E-box interaction and affect P4Hα(I) expression.
Extracellular matrix (ECM),2 the structural framework of all tissues, plays a key role in pathogenesis. In arterial wall, ECM holds various types of cells, including endothelial cells, vascular smooth muscle cells (VSMC), and fibroblasts, in their functional places. Dysregulated ECM is frequently associated with atherogenesis, vulnerable plaques, and aneurysm (1–5). ECM is maintained by a balanced synthesis and degradation of mainly collagens and elastins, which are in turn regulated by other molecules, e.g. fibrillin. Collagens form a multigene family with at least 39 members, the genes of which are known to be dispersed throughout at least 15 chromosomes. Collagen molecules consist of three identical or dissimilar polypeptide chains (called α chains) and contain at least one triple helical collagenous domain with repeating (GXY)n sequences, i.e. a glycine residue at every third amino acid, with the frequent presence of proline and 4-hydroxyproline in the X and Y positions, respectively. Most collagen molecules form supramolecular assemblies, and the superfamily can be divided into nine different families based on their polymeric structures or other features, such as collagens that form fibrils (types I–III, V, and XI). In normal arteries, fibrillar collagens I and III are the major constituents of the intima, media, and adventitia, whereas types I, III, IV, and V are in endothelial and VSMC basement membranes.
The biosynthesis of the precursors of fibrillar collagens (called procollagens) involves a number of post-translational modifications. The intracellular modifications require five specific enzymes, three collagen hydroxylases (6, 7) and two collagen glycosyltransferases (8, 9). Among these hydroxylases, prolyl 4-hydroxylase (P4H), a α2β2 heterotetramer, plays a key role in collagen metabolism. The β subunit is identical to the enzyme protein disulfide isomerase (6, 10) and is produced in excess of the β subunit, which contains a major portion of the catalytic sites of the enzyme, and the abundance of the α subunit restricts the enzymatic activity (6). P4H is an intracellular enzyme required for the synthesis and formation of all known types of collagen. It catalyzes the formation of hydroxyproline from proline residues located in repeating Xaa-Pro-Gly triplets in the procollagens during post-translational processing and is the process essential for folding the procollagen polypeptide chains into stable triple helical molecules (6). Inhibition of the P4H enzymes generates unstable collagen that is degraded inside the cell and is not secreted, which is associated with reduced collagen production (5). On the other hand, overexpression of the P4Hα(I) gene is associated with excess collagen production (11). P4H isoenzymes are expressed in most tissues, although there may be tissue specificity with regard to type I or II isoforms for the α subunit (12–14). Although relatively little data is available for factors affecting P4H expression and activity, P4H may be regulated by nitric oxide, tumor necrosis factor α, transforming growth factor β (TGFβ), insulin-like growth factor 1, β fibroblast growth factor, certain cytokines, and cigarette smoking (15–22). Environmental stimuli, such as hypoxia, can induce P4Hα(I) expression at both transcriptional and post-transcriptional levels, which results in excess accumulation of collagen, a process relevant to fibrosis (23, 24).
The promoter of rat P4Hα(I) has been described by Takahashi et al. (24). However, little is known with regard to the functional features of human P4Hα(I) promoter (AL731563). In the present study, we characterized the elements in the promoter of human P4Hα(I). We found that the E-box-like sequence (CACGGG), which has been shown to bind upstream stimulatory factors (USF) (25–27), in the position of bp −135 is vital for P4Hα(I) transcription. Upstream stimulatory factor (USF1/USF2) appears to be the transcription factor that controls the basal P4Hα(I) expression. Factors including TGFβ1 and cigarette smoking extract (CSE) up-regulated and down-regulated the USF-E-box interaction and affected P4Hα(I) expression.
MATERIALS AND METHODS
Plasmid Construction
pGL3-basic (Promega, Madison WI) was used to construct pGL-P4Hα(I) reporter vector to evaluate transcription efficiency. We used a PCR-based method to clone serially deleted P4Hα(I) promoter fragments into the pGL3-basic vector at the KpnI and HindIII sites. Genomic DNA from HeLa cells was used as the template for the cloning. The inserts were confirmed for the correct sequence and orientation by direct sequencing. We named each recombinant pGL3 vector according to the size of the inserted P4Hα(I) fragment (Table 1). The same antisense oligonucleotide primer that corresponded to the intron 1 region IVS1 + 76–IVS1 + 96 (5′-CCAAAGCTTACCACCACAGCGGGAAGGAAT- 3′) was used for the generation of all of the clones. A KpnI linker was incorporated into the 5′-end of the sense primers, and a HindIII linker was incorporated into the 5′-end of the antisense primers. The digested PCR products were subcloned into pGL3-basic at KpnI and HindIII sites. The mutated construct pGL- 145M was obtained by site-directed mutagenesis with a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions using primers specifically introducing a mutation at the E-box site of the P4Hα(I) pGL-145 vector. The primers used for site-directed mutagenesis are as follows (underlines refer to mutated nucleotides): sense 5′ AGC AGG GAG GCA CTC TCT TGG CGC CTG C-3; antisense 5′-GCA GGC GCC AAG AGA GTG CCT CCC TGC T-3′. The E-box mutation was confirmed by the direct sequencing. The USF1-pSG5 and USF2-pSG5 expression vectors were generous gifts from the laboratory of Dr. M. Sawadogo at the University of Texas MD Anderson Cancer Center, Houston, TX. Details of the vectors were described previously (28).
TABLE 1. Primers for the generation of serially deleted P4Hα(I) promoter fragments.
The same antisense oligonucleotide primer was used for the generation of all of the clones shown. A KpnI linker (CTGGTACC), shown in bold, was incorporated into the 5′-end of the sense primers, and a HindIII linker (CCAAAGCTT) was incorporated into the 5′-end of the antisense primers. The IVS1 indicates the location within the first intron counting from the end of exon 1 (Fig. 1A). The insertion sites of the cloned DNA fragments are at the multiple cloning region of the +5 (KpnI) and +53 (HindIII) upper stream of the luciferase gene.
| Inserts | Primer positions | Sense primer locations and sequences |
|---|---|---|
| pGL−580 | −580 to −560 | 5′-CCTGGTACCGAGATAACCTGACCCTGGAGC-3′ |
| pGL−480 | −480 to −457 | 5′-CCTGGTACCGAGGAACAAGACAGCTGAGATACC-3′ |
| pGL−417 | −417 to −394 | 5′-CCTGGTACCTGCTTTATTTTCTACACTCCCACG-3′ |
| pGL−320 | −320 to −298 | 5′-CCTGGTACCAAGGAGGCAAACTGAACAGGAGC-3′ |
| pGL−271 | −271 to −251 | 5′-CCTGGTACCAACGGTGGCGGGGCGAGAACC-3′ |
| pGL−184 | −184 to −164 | 5′-CCTGGTACCCTCGGCCTCAGACTCCACGGG-3′ |
| pGL−145 | −145 to −125 | 5′-CCTGGTACCAGCAGGGAGGCACGGGCTTGG-3′ |
| pGL−97 | −97 to −76 | 5′-CCTGGTACCGTGATCGAGCTCACGTAGCGAG-3′ |
| pGL−32 | −32 to −8 | 5′-CCTGGTACCGGTTATAAAAGGGCTAACGGGCTCC-3′ |
| pGL+18 | +18 to +38 | 5′-CCTGGTACCGCGGGCTGAGGGTAGGAAGTA-3′ |
| Antisense primer locations and sequence | ||
| IVS1+96−IVS1+76 | 5′-CCAAAGCTTACCACCACAGCGGGAAGGAAT-3′ | |
Cell Culture, Transfection, and Luciferase Assays
HeLa cells purchased from the American Type Culture Collection (Masassas, VA) were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum at 37 °C with 5% CO2. Human aortic VSMCs were purchased from Cambrex BioScience, Inc. (Walkersville, MD). VSMCs were cultured in smooth muscle basal medium containing 10% fetal bovine serum (Cambrex BioScience, Inc.), and the cells in passages 6–8 were used for the experiments. Plasmid DNA was prepared using a plasmid extraction kit (Qiagen Inc., Valencia, CA). One microgram of pGL- P4Hα(I) hybrid DNA and/or 1 µg of siRNA or USF-pSG5 were transfected into the cells cultured in a 6-well plate with Lipofectamine 2000 (Invitrogen), and 0.25 µg of pSVβgal (Promega), which expresses β-galactosidase, was co-transfected to allow normalization of the transfection efficiency. After 4 h, the cells were treated with or without 10 ng/ml TGFβ1 (Leinco Technologies, Inc., St. Louis, MO) for 24 h in the same medium but containing 0.2% fetal bovine serum. We also treated the transfected cells with CSE as described previously (29). Briefly, the smoke of a research cigarette (2R4F from Tobacco Health Research, University of Kentucky; nicotine, 0.85 mg/cigarette; tar, 9.70 mg/cigarette) was bubbled into a flask containing 20 ml of prewarmed F12K complete medium. The ignited cigarette was completely consumed in puffs by 10-ml syringe suction at one end of the tube within 5 min. The aqueous smoke extract was filtered through 0.2-µm syringe filter, and the pH of the CSE was adjusted to 7.4. The concentration of CSE was calculated in arbitrary units as cigarette equivalents/ml of the medium. CSE was used to treat the cells within 30 min of extraction. After 24 h, the cells were harvested and lysed in reporter lysis buffer (Promega), and cell lysates were assayed for luciferase and β-galactosidase activities. Luciferase activity was measured with an FB12 luminometer (Berthold Detection Systems, Pforzheim, Germany) using luciferin (Promega) as the substrate.β-Galactosidase was assayed in an assay buffer (Promega), and the activity was measured at 420 nm in a spectrophotometer. The luciferase activity, which reflects the promoter activity, was normalized to the β-galactosidase to account for the variation in transfection efficiency.
For siRNA transfection, 100 nm siRNA was transfected into the cells cultured in a 6-well plate with Lipofectamine 2000. The siRNA duplex (Integrated DNA Technologies, Houston, TX) sequences for USF1 were: sense, 5′-GAC CCA ACC AGU GUG GCU ATT-3′ and antisense, 5′-UAG CCA CAC UGG UUG GGU CTT-3′. The siRNA duplex sequences for USF2 were: sense, 5′-CCU CCA CUU GGA AAC GGU ATT-3′, and antisense, 5′-UAC CGU UUC CAA GUG GAG GTT-3′. As the negative control siRNA, we used scrambled siRNA designed and tested by Ambion, Inc. (catalog number 4611G, Austin, Texas), which has no significant sequence similarities to mouse, rat, or human gene sequences.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was performed using the histone H3 ChIP assay kit according to the manufacturer’s protocol (Upstate Biotechnology, Charlottesville, VA). In brief, ~3 × 107 cells were used per ChIP assay. The cells were cross-linked with 1% formaldehyde at 37 °C for 10 min and rinsed twice with ice-cold phosphate-buffered saline. The cells were harvested by a brief centrifugation. Cell pellets were resuspended in SDS lysis buffer (50 mmol/liter Tris-HCl, pH 8.1, 10 mmol/liter EDTA, 1% SDS, and protease inhibitors). Sonication was performed on ice using a Sonifier II 450 (Branson, Danbury, CT) with a 3-mm tip set at duty cycle 20 and an output level 2 to achieve chromatin fragments ranging between 200 and 1000 bp in size, followed by centrifugation at 15,000 × g for 10 min at 4 °C. Supernatants were collected and diluted 10-fold in ChIP dilution buffer (a 20-µl aliquot was removed to serve as an input sample) followed with preimmunoprecipitation clearing with 80 µl of a mixture of salmon sperm DNA/protein A/protein G at 4 °C with rotation for 30 min. Immunoprecipitation was carried out with 2 µg of anti-USF1 or -USF2 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) at 4 °C overnight with rotation. After immunoprecipitation, 60 µl of a mixture of salmon sperm DNA/protein A/protein G was added and incubated at 4 °C with rotation for 30 min and followed by brief centrifugation. The precipitates were washed twice (5 min each at 4 °C) with low salt buffer, once with high salt buffer, and once with LiCl buffer. Then the precipitates were washed again with TE buffer (10 mm Tris-Cl 1 mm EDTA) twice for 5 min each. The immune complexes were extracted three times with 200 µl of elution buffer. The eluates and the input were heated at 65 °C for at least 4 h to reverse the cross-link by the addition of 20 µl of 5 mol/liter NaCl. After proteinase K treatment, DNA was extracted by phenol/chloroform solution and precipitated with ethanol. The recovered DNA was resuspended in 30µl of H2O, and 4µl was used for PCR. PCR was performed using sense (5′-AGC AGG GAG GCA CGG GCT TGG-3′) and antisense (5′-TAC TTC CTA CCC TCA GCC CGC-3′) primers. The PCR product (bp 183), which contained the USF binding E-box cis-element, was analyzed by 2% agarose gel electrophoresis. Anti-FLAG antibody (Sigma) served as the negative control in the assay.
Real Time Quantitative Reverse Transcription (RT)-PCR
Primers were designed with Beacon Designer version 2.0 software. The primers for the human P4Hα(I) mRNA were: sense, 5′-CGT CTC CAG GAT ACC TAC AA-3′ and antisense, 5′-AAG CAG TCC TCA GCC GTT AG-3′. The primers for the housekeeping gene human β-actin were: sense, 5′-CTG GAA CGG TGA AGG TGA CA-3′ and antisense, 5′-AAG GGA CTT CCT GTA ACA ATG CA-3′. Total RNA was extracted from the cells with TRIzol (Invitrogen) according to the manufacturer’s protocol. The RNA solution was treated with 1 unit of RNase-free DNase I (1 unit/µl; Promega) in a final volume of 20 µl at 37 °C for 15 min to remove trace amounts of genomic DNA contamination. The DNase I was inactivated by incubation at 75 °C for 10 min. One microgram of total RNA was used for reverse transcription. The mRNAs were reverse-transcribed into cDNAs with an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA) containing a mixture of oligo(dT) and random primers. Real time PCR was performed using an iCycler iQ real time PCR detection system with a SYBR Green I Supermix kit (Bio-Rad) with the program running for 40 cycles at 95 °C for 20 s and 60 °C for 1 min. The PCR efficiency for the primers was examined by serially diluting the cDNA templates. The melting curve analysis was performed over the range 55–95 °C by monitoring SYBR Green fluorescence at increasing temperatures (0.5 °C increment changes at 10-s intervals). PCR-specific products were determined as a clear single peak, at the melting curves, of >80 °C. The specificity of primers was also confirmed as a single band with the correctly amplified fragment size through agarose gel electrophoresis of the real time RT-PCR products. Real time PCR was duplicated for each cDNA sample. Each genemRNA level was acquired from the value of the threshold cycle (Ct) of the real time PCR as related to that of β-actin through the formula 2ΔCt (ΔCt = β-actin Ct − gene of interest Ct).
RESULTS
Identification of a Positive Regulatory Element in the P4Hα(I) 5′-Flanking Sequence
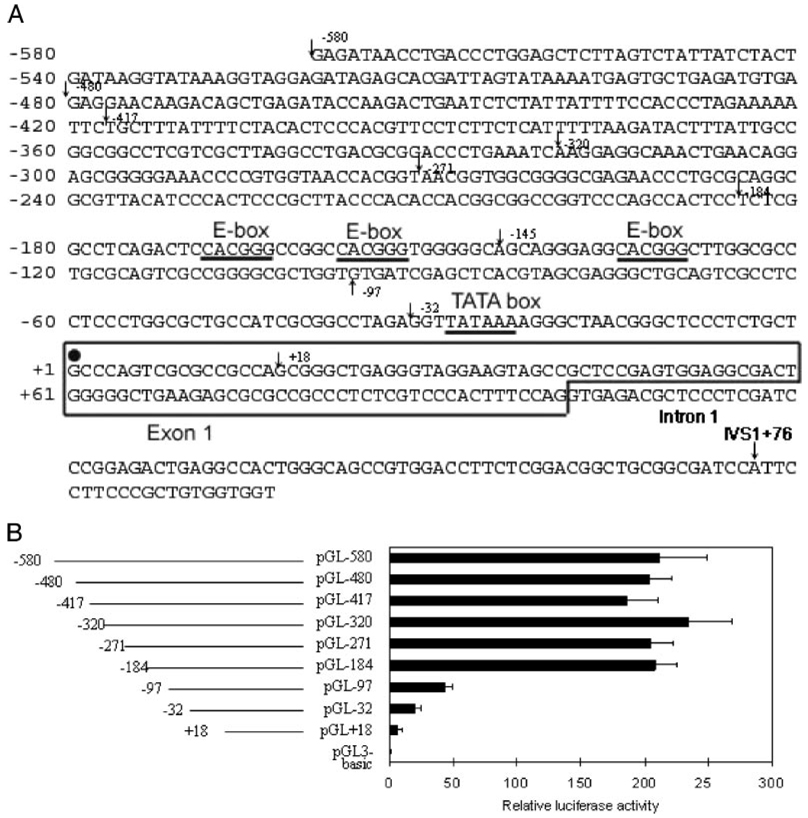
To clarify the regions responsible for the basal transcriptional regulation of the P4Hα(I) gene, we prepared several reporter constructs by cloning the 5′-flanking sequence of the P4Hα(I) gene (Fig. 1A) into the upstream of the luciferase reporter. We then transfected HeLa cells with these constructs containing the progressively deleted 5′-flanking regions of the P4Hα(I) gene (Fig. 1B) and measured the luciferase activity of the resulting cell extracts. The deletion of the 5′-flanking sequence from bp −580 to −184 did not affect the transcription efficiency (Fig. 1B). However, a deletion up to bp −97 resulted in an ~80% decrease in the transcription activity. These results suggest that the basal transcription of the P4Hα(I) gene was positively regulated by a region located between bp −184 and −97. One or more specific factors interacting with a cis-element(s) located between bp −184 and −97 may regulate the P4Hα(I) promoter efficiency.
FIGURE 1. Structure of P4Hα(I) transcriptional regulatory region.
A, the nucleotide sequence of the promoter region of P4Hα(I) is shown. The putative binding sites for transcriptional factors are underlined. The major transcriptional initiation site (solid circle) is designated +1. The starting positions for primers used in the serial cloning are indicated in the figure. B, schematic representation of the 5′-deleted P4Hα(I) promoter-luciferase constructs and their transcriptional activities in HeLa cells. HeLa cells were transfected with the 5′-deletion P4Hα(I) promoter-luciferase constructs. The relative luciferase activities were assessed. Results are expressed as the ratio of luciferase activity in each extract relative to that in the extract from HeLa cells transfected with pGL3-basic. The data were normalized to the activity of the co-transfected pSVβgal plasmid and presented as the means±S.D. of three independent experiments.
Effects of Mutation in the −135 E-box
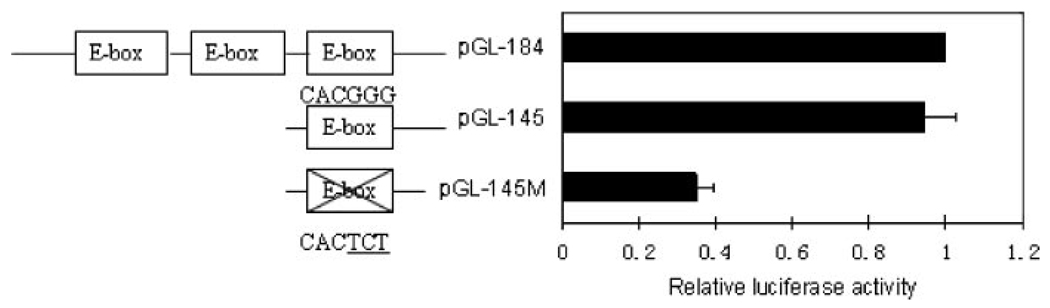
Through a bioinformatics search, we have located three consensus E-box-like elements (CACGGG) located between bp −184 and −97. They are at bp −169, −158, and −135 (Fig. 1A). To confirm the importance of the E-boxes in P4Hα(I) gene transcription, we generated a wild-type pGL-145 (Fig. 2) and a mutated reporter gene construct pGL-145M in which the −135 E-box-like sequence (CACGGG) was mutated to CACTCT. We then transfected HeLa cells with the pGL-184, pGL-145, and pGL-145M vectors (Fig. 2). The deletion of the 5′-flanking sequence from −184 to −145, which removed the −169 and −158 E-boxes, did not affect the transcription efficiency. The mutation of the −135 E-box (pGL-145M), however, resulted in an ~60% reduction in the activity. These results suggest that the basal transcription of the P4Hα(I) gene was positively regulated by the −135 E-box.
FIGURE 2. Mutating the E-box reduces P4Hα(I) promoter activity.
Plasmids pGL-184, pGL-145, and the mutated construct pGL-145M were transfected into HeLa cells. The relative luciferase activities were assessed. Results are expressed as the ratio of luciferase activity in each extract relative to that in the extract from HeLa cells transfected with pGL3-basic. The data were normalized to the activity of the co-transfected pSVβgal plasmid and presented as the means±S.D. of three independent experiments.
Involvement of USF1/USF2 in the P4Hα(I) Promoter Activity
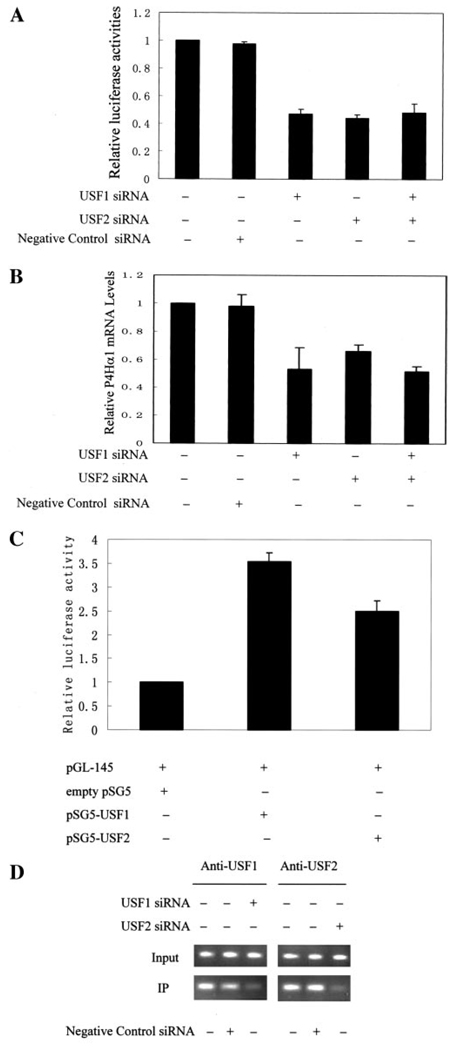
The USF is one of the main families of transcription factors binding to the E-box (30, 31). To investigate the involvement of USFs in transactivating the P4Hα(I) promoter, we first examined the effects of silencing USF1 and USF2 by siRNAs on the activity of the P4Hα(I) promoter. Transfection of USF1 siRNA and/or USF2 siRNA into HeLa cells showed a significantly decreased pGL-145 promoter activity (Fig. 3A). Using HeLa cells, we further examined whether USF1/USF2 silencing could also suppress P4Hα(I) expression. Indeed, the mRNA levels of the P4Hα(I) gene measured by the quantitative real time RT-PCR were significantly reduced when USF1/USF2 were silenced (Fig. 3B). In the meantime, negative control siRNA had no effect on P4Hα(I) expression. Our experiments further demonstrated that the degree of inhibition by the USF1 or USF2 siRNA appeared to be similar, and treatment with both siRNAs did not induce any additive effect comparable to the reduction by a single siRNA. These results suggest that the transcription of the P4Hα(I) gene was positively regulated by the USF1/USF2, either one of which would be efficient for P4Hα(I) expression.
FIGURE 3. Silencing of USF1/USF2 by short interference RNA (siRNA) decreased the activity of the P4Hα(I) promoter and overexpressed USF1/USF2 increased the P4Hα(I) promoter activity.
A, 1 µg of pGL-145 hybrids was transfected with (+) or without (−) 100 nm USF1 siRNA and/or USF2 siRNA into HeLa cells cultured in a 6-well plate with Lipofectamine 2000. We used scrambled siRNA designed and tested by Ambion, which has no significant sequence similarities to mouse, rat, or human gene sequences, as the negative control siRNA. 0.25 µg of pSVβgal was co-transfected to allow normalization of transfection efficiency. Results are expressed as the ratio of luciferase activity in each cell extract relative to that in the extract from cells transfected with pGL-145 but without siRNA treatment. B, HeLa cells were transfected with (+) or without (−) 100 nm USF1 siRNA and/or USF2 siRNA. After 24 h, the P4Hα(I) mRNA level was quantitated by real time RT-PCR and normalized to the housekeeping gene β-actin. Results are expressed as the ratio relative to HeLa cells without siRNA treatment. C, humanaortic smooth muscle cells were co-transfected with 1µg of pGL-145 hybrids and 1µg of empty expression vector (empty pSG5) or expression constructs for USF1 or USF2 using Lipofectamine 2000. 0.25µg of pSVβgal was co-transfected to allow normalization of transfection efficiency. After 28 h, the cells were harvested for luciferase assay. Results in each extract are expressed as the ratio relative to that in the extract from human aortic smooth muscle cells co-transfected with pGL-145 and empty pSG5. D, ChIP assays were performed with anti-USF1 or anti-USF2 antibody for the HeLa cells treated with (+) or without (−) 100 nm USF1 siRNA or USF2 siRNA for 24 h. Primers for the P4Hα(I) 5′-flanking region from −145 to −38 were used to amplify the DNA isolated from the ChIP assay. All data are presented as the means±S.D. of three independent experiments. IP, immunoprecipitation.
We then overexpressed USF1 or USF2 in HeLa cells and human aortic VSMCs by co-transfecting pGL-145 vectors with USF1- or USF2-pSG5 vectors. As shown in Fig. 3C, USF1 and USF2 significantly up-regulated P4Hα(I) promoter activities as measured by luciferase activity in VSMCs. The same effect was also observed in HeLa cells (data not shown). In addition, USF1 appeared to induce a higher P4Hα(I) promoter activity than did USF2 (Fig. 3C). Such differential effect was not elucidated by the siRNA-mediated USF1 and/or USF2 suppression.
Direct Binding of USF1/USF2 to the P4Hα(I) Promoter
To further assess the involvement of USF1/USF2 in P4Hα(I) transcription, we examined whether USF transcription factors could bind to the E-box in the P4Hα(I) promoter in vivo. The binding of E-box-binding proteins USF1/USF2 to the P4Hα(I) promoter was illustrated by the ChIP assay (Fig. 3D). We then examined the effect of USF1/USF2 silencing on the binding activity of the USF1/USF2 using the ChIP assay. We found a decreased binding activity of both USF1 and USF2 to the E-box (Fig. 3D). Meanwhile, the negative control siRNA had a minimal effect on the binding.
Effects of TGFβ1 on P4Hα(I) Promoter Activity
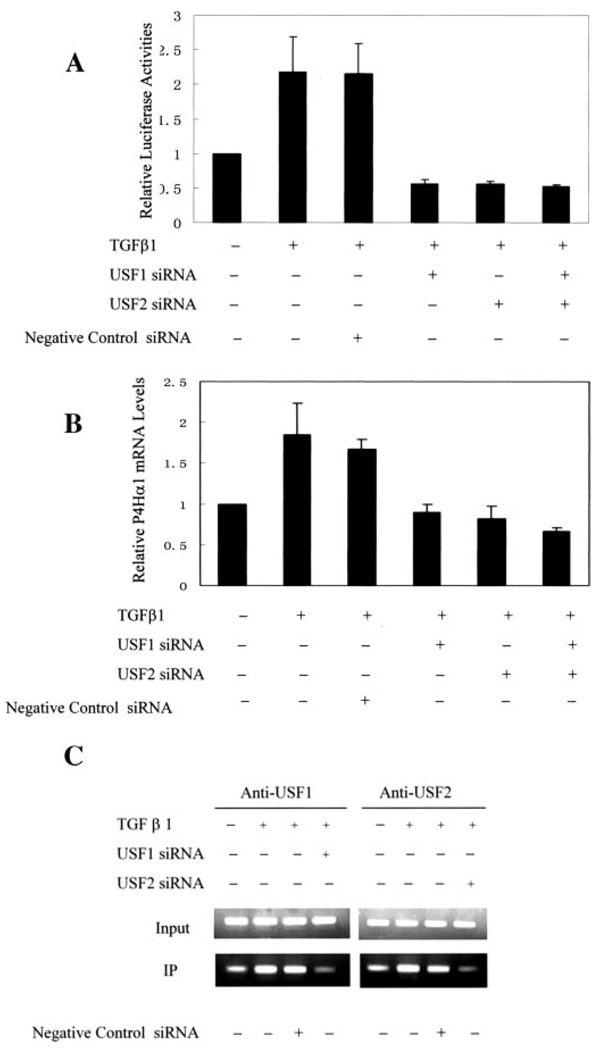
P4Hα(I) has a central role in the biosynthesis of collagens. TGFβ1 can induce up-regulated collagen synthesis. To evaluate whether and how the P4Hα(I) promoter would respond to TGFβ1 stimulation, pGL-145 luciferase plasmids were transfected into HeLa cells. The normalized luciferase activity was calculated after TGFβ1 treatment. We found a 2-fold increase in the luciferase activity after TGFβ1 treatment (Fig. 4A). This effect was blocked by USF1/USF1 silencing using siRNAs (Fig. 4A), whereas negative siRNA had no effect. We further demonstrated the effect of TGFβ1 treatment on the P4Hα(I) mRNA levels in HeLa cells (Fig. 4B). We found a 1.8-fold increase in P4Hα(I) mRNA levels, and this effect was blocked by USF1/USF2 silencing using siRNA (Fig. 4B). The same effect was also shown in VSMCs (data not shown). ChIP assay showed TGFβ1 increased in the band intensity for the binding between the USF1/USF2 and the E-box in the P4Hα(I) promoter (Fig. 4C). Furthermore, negative control siRNA had no effect on the binding activity. Taken together, these results suggest that the transcription of the P4Hα(I) gene was positively regulated by TGFβ1 through the USF1/USF2 and E-box interaction.
FIGURE 4. Effects of TGFβ1 on the P4Hα(I) promoter activity.
A, one microgram of pGL-145 hybrids was transfected with (+) or without (−) 100 nm USF1 siRNA and/or USF2 siRNA into HeLa cells with Lipofectamine 2000. We used scrambled siRNA designed and tested by Ambion, which has no significant sequence similarities to mouse, rat, orhuman gene sequences, as the negative control siRNA. 0.25µgof pSVβgal was co-transfected to allow normalization of transfection efficiency. After 4 h, the cells were treated with or without 10 ng/ml TGFβ1 in the same medium but containing 0.2% fetal bovine serum for 24 h. Results are expressed as the ratio of luciferase activity in each cell extract relative to that in the extract from cells transfected with pGL-145 but without TGFβ1 and siRNA treatment. B, HeLa cells were transfected with or without 100 nmUSF1 siRNA and/or USF2 siRNA and then treated with or without 10 ng/ml TGFβ1 for 24 h. After 24 h, the P4Hα(I) mRNA level was quantitated by real time RT-PCR and normalized to the housekeeping gene β-actin. Results are expressed as the ratio relative to HeLa cells without TGFβ1 and siRNA treatment. C, ChIP assays were performed with anti-USF1 or anti-USF2 antibody for HeLa cells transfected with or without 100 nm USF1 siRNA or USF2 siRNA and then treated with or without 10 ng/ml TGFβ1 for 24 h. Primers for the P4Hα(I) 5′-flanking region from bp mRNA level was quantitated by real time RT-PCR 145 to mRNA level was quantitated by real time RT-PCR 38 were used to amplify the DNA isolated from the ChIP assay. All data are presented as the means±S.D. of three independent experiments. IP, immunoprecipitation.
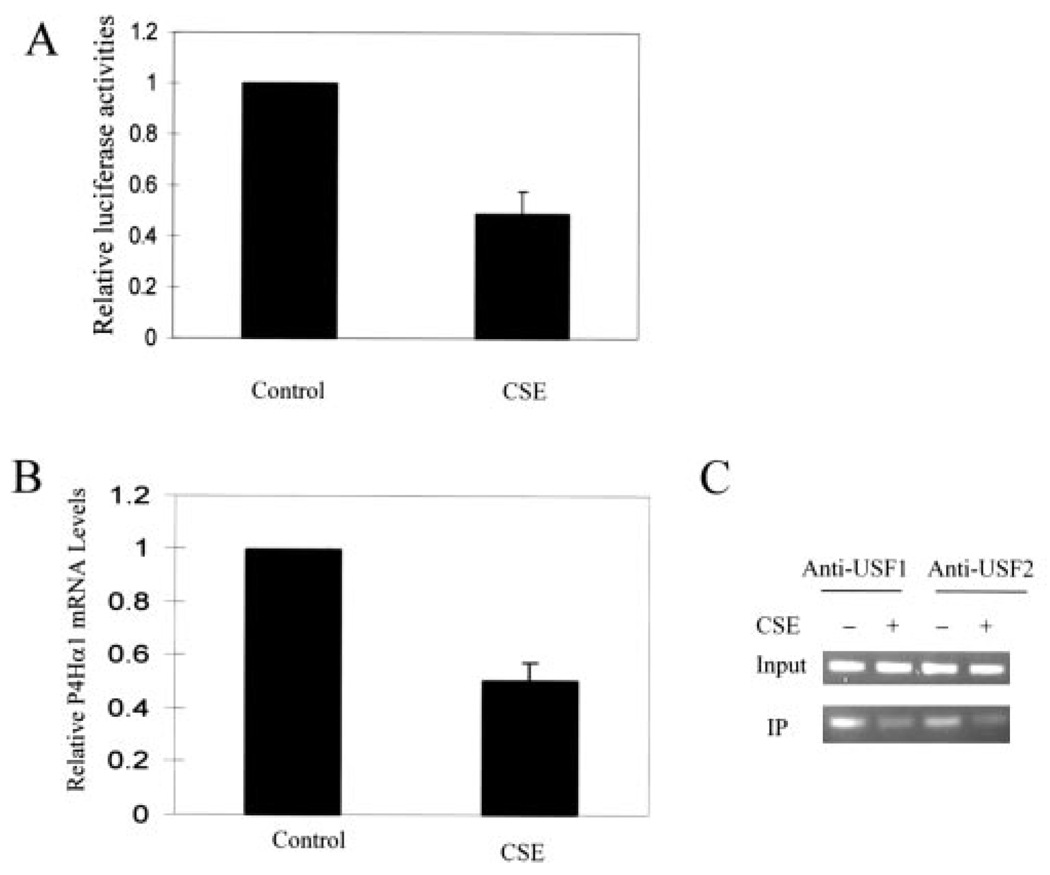
Effects of CSE on P4Hα(I) Promoter Activity
Previously, we have shown (15) that CSE could down-regulate P4Hα(I) expression in endothelial cells and VSMCs. To evaluate the responsiveness of the P4Hα(I) promoter to CSE treatment, pGL-145 luciferase plasmids were transfected into HeLa cells, and the normalized luciferase activity was calculated after CSE treatment. We found an ~50% reduction in the luciferase activity by the CSE treatment (Fig. 5A). We further showed that CSE also decreased the P4Hα(I) mRNA levels in HeLa cells (Fig. 5B), which was consistent with our previous report (15) on endothelial cells and VSMCs. ChIP assay demonstrated that CSE decreased the band intensity for the binding between the USF1/USF2 transcription factor and the E-box in the P4Hα(I) promoter (Fig. 5C). These results suggest that the transcription of the P4Hα(I) gene was negatively regulated by CSE through USF1/USF2-E-box interaction.
FIGURE 5. Effect of CSE treatment on the P4Hα(I) promoter activity.
A, one microgram of pGL-145 hybrid DNA was transfected into HeLa cells with Lipofectamine 2000. 0.25µg of pSVβgal was co-transfected to allow normalization of transfection efficiency. After 4 h, the cells were then treated with or without CSE (0.01 unit) for 24 h. Results are expressed as the ratio of luciferase activity in each cell extract relative to that in the extract from cells transfected with pGL-145 but without CSE treatment. B, HeLa cells were treated with or without CSE (0.01 unit) for 24 h. After 24 h, the P4Hα(I) mRNA level was quantitated by real time RT-PCR and normalized to the housekeeping gene β-actin. Results are expressed as the ratio relative to HeLa cells without CSE treatment. C, ChIP assays were performed with anti-USF1 or anti-USF2 antibody for HeLa cells treated with or without CSE (0.01 unit) for 24 h. Primers for the P4Hα(I) 5′-flanking region from bp −145 to +38 were used to amplify the DNA isolated from the ChIP assay. All data are presented as the means±S.D. of three independent experiments. IP, immunoprecipitation.
DISCUSSION
Because collagen is the major structural protein in the ECM, which provides the framework for all tissues including arteries, there is little doubt that factors regulating collagen synthesis, process, secretion, and degradation will play a significant role in health and disease. It is therefore important to understand the molecular mechanisms regulating P4Hα(I) expression, a rate-limiting subunit for the P4H enzyme, which is critical for procollagen modification and secretion (32, 33). Our study unequivocally shows that the E-box-like sequence located in the region of bp −184 to −97 upstream of the P4Hα(I) promoter is the positive regulator and is responsible for nearly 80% of the basal promoter activity. As far as we are aware, this is the first characterization of the human P4Hα(I) promoter (12, 34).
Although there is a significant sequence homology between human and rodent P4Hα(I), the regulatory elements appear to be different between them. Takahashi and colleagues showed that the positive regulatory region is located at positions bp −246 to −165 in the rat P4Hα(I) promoter (24). We showed that this positive regulator is located at bp −184 to −97, particularly the E-box at bp −135 in the human P4Hα(I) promoter. Although their study did not show experimental evidence for any cis-acting element in P4Hα(I) regulation, our experiments have shown convincing evidence for the involvement of the USF family and E-box interaction in regulating P4Hα(I) expression. This conclusion is supported by the siRNA-specific suppression of the USF1/USF2, USF1/USF2 overexpression and the E-box mutagenesis. It should be noted, however, our study only demonstrates the regulatory elements for basal expression. DNA elements and corresponding transcription factors regulating the response to stimulation are likely to be different from those governing the basal expression.
USF1 and USF2 are ubiquitous proteins characterized by highly conserved C-terminal basic helix-loop-helix and leucine zipper domains responsible for dimerization and DNA binding activities (28, 35, 36). The E-box, the consensus sequence of which is CANNTG, is known as the binding site for USF and regulates the transcription of several genes (37, 38). They appear to be involved in the regulation of developmental and tissue-specific or gene-specific expression of a number of genes (39, 40), including those for surfactant protein A (41, 42), steroidogenic factor 1 (43), and carboxyl ester lipase (44). Our study adds a collagen regulator P4Hα(I) as another USF target in regulating ECM metabolism in addition to its suppressing effect on cathepsin B expression (45). Together with a long list of genes including those involved in glucose metabolism, cholesterol metabolism, cell proliferation, detoxification, and hypoxia that can be potentially regulated by the USF (46–54), it appears clear that USF may have a role in vascular disease more than we have appreciated so far.
The major USF species present in most tissues and cell lines is the heterodimer between USF1 and USF2, which interact with E-box (36). USF1 homodimers are less abundant, and USF2 homodimers are scarce (55, 56), which may have different effects (57, 58). In our study, however, we have shown that USF1 and USF2 appear to be interchangeable in P4Hα(I) promoter activity. Suppressing either one will result in a similar degree of inhibition in P4Hα(I) promoter activity; suppressing both does not induce an additive suppressing effect, which is consistent with the recent reports on TGFβ1 (59, 60).
Our experiments have further shown that the positive effect of TGF±1 on collagen production is at least partly mediated by the up-regulated P4Hα(I) expression, which is regulated through the USF-E-box interaction at the bp −135 of the P4Hα(I) promoter. Likewise, the same promoter mechanism also appears to be negatively regulated by cigarette smoking, which has been shown to down-regulate P4Hα(I) expression and collagen production (29). Clearly both TGFβ1 and cigarette smoking would regulate ECM metabolism through more than one target. Our study shows the USF-E-box interaction on the P4Hα(I) promoter is one of the targets. The same mechanism may apply to other target genes of TGFβ1 and cigarette smoking if they have the same cis-acting element. However, we should be aware that the promoter elements responsible for basal expression may differ from those responding to stimulation. Although our experiments show that TGFβ1 may regulate USF-E-box interaction, studies of others show that USF is also a positive transcription regulator for the TGFβ1 gene via the glucose response element (59, 60). These findings suggest that, in addition to directly regulating P4Hα(I) expression, USF proteins may regulate collagen or ECM metabolism through TGFβ1 pathways. These effects could be cell type-specific. Together with their effects on growth factors, e.g. insulin-like growth factor-binding proteins (25), it appears that USF may have a significant role in ECM metabolism.
In summary, our study has characterized that USF plays an important role in P4Hα(I) expression. The USF-E-box interaction appears to be the site wherein the positive (TGFβ1) and negative (cigarette smoking) regulators of collagen production and P4Hα(I) expression will act. Further studies on how TGFβ1 or cigarette smoking regulates USF-E-box interaction will be of significant value in understanding vascular diseases, such as vulnerable atherosclerotic plaque or aortic aneurysm that involves dysregulated ECM metabolism.
Footnotes
This work was supported by American Heart Association Established Investigator Award AHA 0440001N (to X. L. W.) and National Institutes of Health Grants HL066053 and HL71608. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: ECM, extracellular matrix; P4H, prolyl-4-hydroxylase; USF, upstream stimulatory factor; ChIP, chromatin immunoprecipitation; siRNA, short interference RNA; VSMC, vascular smooth muscle cell; TGF, transforming growth factor; RT, reverse transcription; Ct, threshold cycle.
REFERENCES
- 1.Stary HC, Chandler AB, Glagov S, Guyton JR, Insull W, Jr., Rosenfeld ME, Schaffer SA, Schwartz CJ, Wagner WD, Wissler RW. Circulation. 1994;89:2462–2478. doi: 10.1161/01.cir.89.5.2462. [DOI] [PubMed] [Google Scholar]
- 2.Stary HC, Chandler AB, Dinsmore RE, Fuster V, Glagov S, Insull W, Jr., Rosenfeld ME, Schwartz CJ, Wagner WD, Wissler RW. Arterioscler. Thromb. Vasc. Biol. 1995;15:1512–1531. doi: 10.1161/01.atv.15.9.1512. [DOI] [PubMed] [Google Scholar]
- 3.Fabunmi RP, Sukhova GK, Sugiyama S, Libby P. Circ. Res. 1998;83:270–278. doi: 10.1161/01.res.83.3.270. [DOI] [PubMed] [Google Scholar]
- 4.Libby P. Circulation. 1995;91:2844–2850. doi: 10.1161/01.cir.91.11.2844. [DOI] [PubMed] [Google Scholar]
- 5.Rocnik EF, Chan BM, Pickering JG. J. Clin. Investig. 1998;101:1889–1898. doi: 10.1172/JCI1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kivirikko KI, Pihlajaniemi T. Adv. Enzymol. Relat. Areas Mol. Biol. 1998;72:325–398. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 7.Kivirikko KI, Myllyharju J. Matrix Biol. 1998;16:357–368. doi: 10.1016/s0945-053x(98)90009-9. [DOI] [PubMed] [Google Scholar]
- 8.Kivirikko KI, Myllyla R. Methods Enzymol. 1982;82:245–304. doi: 10.1016/0076-6879(82)82067-3. [DOI] [PubMed] [Google Scholar]
- 9.Prockop DJ, Kivirikko KI. Annu. Rev. Biochem. 1995;64:403–434. doi: 10.1146/annurev.bi.64.070195.002155. [DOI] [PubMed] [Google Scholar]
- 10.Kivirikko KI, Myllyla R, Pihlajaniemi T. FASEB J. 1989;3:1609–1617. [PubMed] [Google Scholar]
- 11.John DC, Watson R, Kind AJ, Scott AR, Kadler KE, Bulleid NJ. Nat. Biotechnol. 1999;17:385–389. doi: 10.1038/7945. [DOI] [PubMed] [Google Scholar]
- 12.Annunen P, Helaakoski T, Myllyharju J, Veijola J, Pihlajaniemi T, Kivirikko KI. J. Biol. Chem. 1997;272:17342–17348. doi: 10.1074/jbc.272.28.17342. [DOI] [PubMed] [Google Scholar]
- 13.Annunen P, Autio-Harmainen H, Kivirikko KI. J. Biol. Chem. 1998;273:5989–5992. doi: 10.1074/jbc.273.11.5989. [DOI] [PubMed] [Google Scholar]
- 14.Nissi R, Autio-Harmainen H, Marttila P, Sormunen R, Kivirikko KI. J. Histochem. Cytochem. 2001;49:1143–1153. doi: 10.1177/002215540104900908. [DOI] [PubMed] [Google Scholar]
- 15.Raveendran M, Senthil D, Utama B, Shen Y, Dudley D, Wang J, Zhang Y, Wang XL. Biochem. Biophys. Res. Commun. 2004;323:592–598. doi: 10.1016/j.bbrc.2004.08.129. [DOI] [PubMed] [Google Scholar]
- 16.Hiramatsu M, Kumegawa M, Hatakeyama K, Yajima T, Minami N, Kodama H. Endocrinology. 1982;111:1810–1816. doi: 10.1210/endo-111-6-1810. [DOI] [PubMed] [Google Scholar]
- 17.Mizutani K, Ikeda K, Yamori Y. Biochem. Biophys. Res. Commun. 2000;274:61–67. doi: 10.1006/bbrc.2000.3097. [DOI] [PubMed] [Google Scholar]
- 18.Kato Y, Inoue H, Fujiyama Y, Bamba T. J. Gastroenterol. 1996;31:565–571. doi: 10.1007/BF02355058. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki T. J. Dermatol. 1992;19:664–666. doi: 10.1111/j.1346-8138.1992.tb03755.x. [DOI] [PubMed] [Google Scholar]
- 20.Kawaguchi Y, Kitani A, Hara M, Harigai M, Hirose T, Suzuki K, Kawakami M, Hidaka T, Ishizuka T, Kawagoe M. J. Rheumatol. 1992;19:1195–1201. [PubMed] [Google Scholar]
- 21.Muguerza B, Castilla-Cortazar I, Garcia M, Quiroga J, Santidrian S, Prieto J. Biochim. Biophys. Acta. 2001;1536:185–195. doi: 10.1016/s0925-4439(01)00045-x. [DOI] [PubMed] [Google Scholar]
- 22.Cao M, Westerhausen-Larson A, Niyibizi C, Kavalkovich K, Georgescu HI, Rizzo CF, Hebda PA, Stefanovic-Racic M, Evans CH. Biochem. J. 1997;324:305–310. doi: 10.1042/bj3240305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi Y, Takahashi S, Shiga Y, Yoshimi T, Miura T. J. Biol. Chem. 2000;275:14139–14146. doi: 10.1074/jbc.275.19.14139. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi S, Dohi N, Takahashi Y, Miura T. Biochim. Biophys. Acta. 2002;1574:354–358. doi: 10.1016/s0167-4781(01)00360-8. [DOI] [PubMed] [Google Scholar]
- 25.Matsukawa T, Inoue Y, Oishi Y, Kato H, Noguchi T. Endocrinology. 2001;142:4643–4651. doi: 10.1210/endo.142.11.8513. [DOI] [PubMed] [Google Scholar]
- 26.Crissey MA, Leu JI, De Angelis RA, Greenbaum LE, Scearce LM, Kovalovich K, Taub R. Hepatology. 1999;30:1187–1197. doi: 10.1002/hep.510300520. [DOI] [PubMed] [Google Scholar]
- 27.Coulson JM, Edgson JL, Marshall-Jones ZV, Mulgrew R, Quinn JP, Woll PJ. Biochem. J. 2003;369:549–561. doi: 10.1042/BJ20021176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo X, Sawadogo M. Mol. Cell. Biol. 1996;16:1367–1375. doi: 10.1128/mcb.16.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raveendran M, Wang J, Senthil D, Wang J, Utama B, Shen Y, Dudley D, Zhang Y, Wang XL. FEBS Lett. 2005;579:733–740. doi: 10.1016/j.febslet.2004.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kyriacou CP, Rosato E. J. Biol. Rhythms. 2000;15:483–490. doi: 10.1177/074873040001500605. [DOI] [PubMed] [Google Scholar]
- 31.Calomme C, Nguyen TL, de Launoit Y, Kiermer V, Droogmans L, Burny A, Van Lint C. J. Biol. Chem. 2002;277:8775–8789. doi: 10.1074/jbc.M107441200. [DOI] [PubMed] [Google Scholar]
- 32.Kivirikko KI, Helaakoski T, Tasanen K, Vuori K, Myllyla R, Parkkonen T, Pihlajaniemi T. Ann. N. Y. Acad. Sci. 1990;580:132–142. doi: 10.1111/j.1749-6632.1990.tb17925.x. [DOI] [PubMed] [Google Scholar]
- 33.Myllyharju J. Matrix Biol. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 34.Kukkola L, Hieta R, Kivirikko KI, Myllyharju J. J. Biol. Chem. 2003;278:47685–47693. doi: 10.1074/jbc.M306806200. [DOI] [PubMed] [Google Scholar]
- 35.Gregor PD, Sawadogo M, Roeder RG. Genes Dev. 1990;4:1730–1740. doi: 10.1101/gad.4.10.1730. [DOI] [PubMed] [Google Scholar]
- 36.Sirito M, Walker S, Lin Q, Kozlowski MT, Klein WH, Sawadogo M. Gene Expr. 1992;2:231–240. [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang B, Mendelson CR. Mol. Cell. Biol. 2003;23:6117–6128. doi: 10.1128/MCB.23.17.6117-6128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang B, Mendelson CR. Mol. Cell. Biol. 2005;25:8824–8833. doi: 10.1128/MCB.25.20.8824-8833.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qyang Y, Luo X, Lu T, Ismail PM, Krylov D, Vinson C, Sawadogo M. Mol. Cell. Biol. 1999;19:1508–1517. doi: 10.1128/mcb.19.2.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawadogo M. J. Biol. Chem. 1988;263:11994–12001. [PubMed] [Google Scholar]
- 41.Gao E, Wang Y, Alcorn JL, Mendelson CR. J. Biol. Chem. 1997;272:23398–23406. doi: 10.1074/jbc.272.37.23398. [DOI] [PubMed] [Google Scholar]
- 42.Gao E, Wang Y, Alcorn JL, Mendelson CR. Am. J. Physiol. 2003;284:L1027–L1036. doi: 10.1152/ajplung.00219.2002. [DOI] [PubMed] [Google Scholar]
- 43.Harris AN, Mellon PL. Mol. Endocrinol. 1998;12:714–726. doi: 10.1210/mend.12.5.0100. [DOI] [PubMed] [Google Scholar]
- 44.Bengtsson SH, Madeyski-Bengtson K, Nilsson J, Bjursell G. Biochem. J. 2002;365:481–488. doi: 10.1042/BJ20020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan S, Sloane BF. Gene. 2004;337:199–206. doi: 10.1016/j.gene.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 46.Griffin MJ, Sul HS. IUBMB Life. 2004;56:595–600. doi: 10.1080/15216540400022474. [DOI] [PubMed] [Google Scholar]
- 47.Shoulders CC, Naoumova RP. Trends Mol. Med. 2004;10:362–365. doi: 10.1016/j.molmed.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 48.Schweizer M, Roder K, Zhang L, Wolf SS. Biochem. Soc. Trans. 2002;30:1070–1072. doi: 10.1042/bst0301070. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi Y, Kamataki T. Drug Metab. Rev. 2001;33:37–47. doi: 10.1081/dmr-100000139. [DOI] [PubMed] [Google Scholar]
- 50.Girard J, Ferre P, Foufelle F. Annu. Rev. Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 51.Rousseau GG. Horm. Res. (Basel) 1992;37 Suppl. 3:88–92. doi: 10.1159/000182407. [DOI] [PubMed] [Google Scholar]
- 52.Sellak H, Choi C, Browner N, Lincoln TM. J. Biol. Chem. 2005;280:18425–18433. doi: 10.1074/jbc.M500775200. [DOI] [PubMed] [Google Scholar]
- 53.Pajukanta P, Lilja HE, Sinsheimer JS, Cantor RM, Lusis AJ, Gentile M, Duan XJ, Soro-Paavonen A, Naukkarinen J, Saarela J, Laakso M, Ehnholm C, Taskinen MR, Peltonen L. Nat. Genet. 2004;36:371–376. doi: 10.1038/ng1320. [DOI] [PubMed] [Google Scholar]
- 54.Shoulders CC. Nat. Genet. 2004;36:322–323. doi: 10.1038/ng0404-322. [DOI] [PubMed] [Google Scholar]
- 55.Sirito M, Lin Q, Maity T, Sawadogo M. Nucleic Acids Res. 1994;22:427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sirito M, Lin Q, Deng JM, Behringer RR, Sawadogo M. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3758–3763. doi: 10.1073/pnas.95.7.3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rodriguez CI, Girones N, Fresno M. J. Biol. Chem. 2003;278:43135–43145. doi: 10.1074/jbc.M300053200. [DOI] [PubMed] [Google Scholar]
- 58.Vallet VS, Casado M, Henrion AA, Bucchini D, Raymondjean M, Kahn A, Vaulont S. J. Biol. Chem. 1998;273:20175–20179. doi: 10.1074/jbc.273.32.20175. [DOI] [PubMed] [Google Scholar]
- 59.Weigert C, Brodbeck K, Sawadogo M, Haring HU, Schleicher ED. J. Biol. Chem. 2004;279:15908–15915. doi: 10.1074/jbc.M313524200. [DOI] [PubMed] [Google Scholar]
- 60.Zhu Y, Casado M, Vaulont S, Sharma K. Diabetes. 2005;54:1976–1984. doi: 10.2337/diabetes.54.7.1976. [DOI] [PubMed] [Google Scholar]