SUMMARY
3′ end processing of histone pre-mRNA requires U7 snRNP, which binds downstream of the cleavage site and recruits the endonuclease CPSF-73. U7 snRNP contains a unique Sm ring in which the canonical SmD2 protein is replaced by Lsm11. We used the yeast two-hybrid system to identify binding partners of Lsm11 and selected the pro-apoptotic protein FLASH. Human FLASH interacts with Lsm11 in vitro and stimulates 3′ end processing of histone pre-mRNA in mammalian nuclear extracts. We also identified the FLASH ortholog in Drosophila and demonstrate that it interacts with Lsm11 in vitro and in vivo. Drosophila FLASH localizes to histone locus bodies and its depletion in fly cells inhibits U7-dependent processing resulting in polyadenylation of histone mRNAs. These results demonstrate that FLASH is an essential factor required for 3′ end maturation of histone mRNAs in both vertebrates and invertebrates and suggest a potential link between this process and apoptosis.
Keywords: histone pre-mRNA, 3′ end processing, U7 snRNP, Lsm11, FLASH, apoptosis
INTRODUCTION
3′ end processing of mRNA precursors (pre-mRNAs) is an essential step in generating mature mRNAs and plays an important role in regulation of gene expression at the post-transcriptional level. For the vast majority of pre-mRNAs, 3′ end processing occurs through cleavage/polyadenylation, a two-step mechanism in which the initial endonucleolytic cleavage downstream of the AAUAAA hexamer is followed by the addition of a poly(A) tail to the newly generated 3′ end (Zhao et al., 1999; Mandel et al., 2008). The only exceptions to this rule are animal replication-dependent histone pre-mRNAs, which are processed at the 3′ end by an endonucleolytic cleavage that is not followed by polyadenylation (Dominski and Marzluff, 2007). Cleavage of histone pre-mRNAs occurs within approximately 50–75 nucleotides 3′ of the stop codon at a site located between a highly conserved stem-loop structure and a purine-rich histone downstream element (HDE). The resultant histone mRNA ends with the stem-loop followed by either a 4- (Drosophila) or 5-nucleotide (mammals) single-stranded tail (Dominski and Marzluff, 2007).
The stem-loop structure located upstream of the cleavage site forms a tight complex with the stem-loop binding protein (SLBP), whereas the HDE interacts with the U7 snRNP. The U7 snRNP is composed of an approximately 60-nucleotide U7 snRNA and a heptameric Sm ring, which differs from the canonical Sm ring found in the spliceosomal snRNPs by the presence of Lsm10 and Lsm11 instead of SmD1 and SmD2 (Schumperli and Pillai, 2004). The U7 snRNP binds histone pre-mRNA primarily through a base-pairing interaction between the 5′ end of U7 snRNA and the HDE, and the binding is additionally stabilized by an interaction between the U7 snRNP and the SLBP/stem-loop complex. In mammalian cells, this interaction is likely mediated by a 100 kDa zinc finger protein (ZFP100), which was shown to interact with both the U7 snRNP and the SLBP/stem-loop complex (Dominski and Marzluff, 2007). A role in histone pre-mRNA 3′ end processing in vivo has also been postulated for the U2 snRNP, which interacts with a conserved region present in the majority of histone mRNAs (Friend et al., 2007), and for negative elongation factor (NELF), which forms a complex with SLBP (Narita et al., 2007).
Cleavage of histone pre-mRNAs is catalyzed by CPSF-73 (Dominski et al., 2005a), a member of the metallo-β-lactamase family of zinc-dependent hydrolytic enzymes (Dominski, 2007). The reaction also requires symplekin, a heat-sensitive protein of unknown function (Kolev and Steitz, 2005), and CPSF-100, a close homolog and an interacting partner of CPSF-73 (Dominski et al., 2005b; Wagner et al., 2007). Interestingly, CPSF-73, CPSF-100 and symplekin were first identified as essential components of the conventional 3′ end processing mechanism that generates polyadenylated mRNAs (Mandel et al., 2008; Gilmartin, 2005).
It is not known how CPSF-73 is recruited to histone pre-mRNA. One component of the processing complex that likely directly or indirectly participates in this task is Lsm11. Lsm11 is characterized by an unusually long N-terminal portion that is indispensable for 3′ end cleavage of histone pre-mRNAs in Xenopus oocytes (Schumperli and Pillai, 2004). We used the yeast two-hybrid system to screen a HeLa cDNA library with human Lsm11 and identified FLASH as a protein that interacts with the N-terminal region of Lsm11. FLASH was initially described as a pro-apoptotic protein required for activation of caspase-8 via the extrinsic pathway linked to the Fas receptor (Imai et al., 1999). Subsequently, FLASH has been implicated in modulating transcriptional activity of steroid hormone receptors and activating transcription of both NF-κB (Krieghoff et al., 2007) and the c-Myb oncogene (Alm-Kristiansen et al., 2008). Importantly, FLASH has also been shown to localize in the vicinity of histone gene loci and is required for S phase progression, suggesting a role in expression of histone genes (Barcaroli et al., 2006). Here, we demonstrate that FLASH fulfills this role primarily, if not exclusively, as an essential factor for 3′ end processing of histone pre-mRNAs. We also show that this role is conserved between vertebrates and invertebrates. FLASH likely occupies a central place in the processing complex by recruiting and/or activating CPSF-73 and may also play a critical role in integrating expression of histone genes with other cellular events, including cell cycle progression and apoptosis.
RESULTS
Human Lsm11 as bait in the yeast two-hybrid system
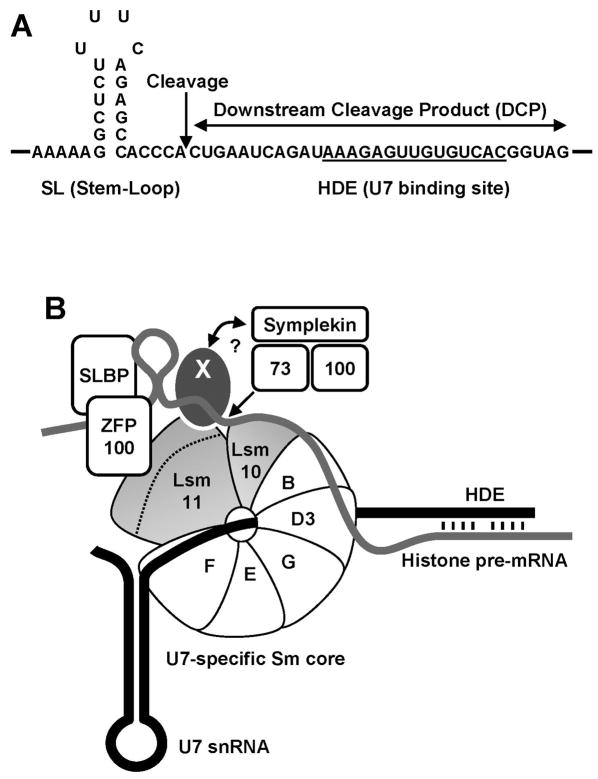
In the processing complex, binding of the U7 snRNP to the HDE in histone pre-mRNA is predicted to position Lsm11 close to the cleavage site (Yang et al., 2009) (Fig. 1). Human Lsm11 is a 360 amino acid protein that can be divided in two halves (Fig. 2A). The N-terminal half of 170 amino acids is unusually long for proteins of the Sm family and contains no recognizable domains (Schumperli and Pillai, 2004). The C-terminal half corresponds to a typical Sm protein and contains two conserved motifs, Sm1 and Sm2, followed by a relatively short C-terminus.
Figure 1. Sequences and proteins involved in 3′ end processing of histone pre-mRNA.
A. Sequence elements in histone pre-mRNA required for the cleavage reaction. The Histone Downstream Element (HDE) that base pairs with the 5′ end of the U7 snRNA is underlined and the site of cleavage located 5 nucleotides after the stem-loop is indicated with an arrow. B. A tentative configuration of trans-acting factors involved in 3′ end processing of histone pre-mRNA (thick grey line). The U7 snRNA is represented by a thick black line. The arrow indicates the site where CPSF-73 cleaves histone pre-mRNA. The X symbolizes an unknown protein that interacts with the N-terminal region of Lsm11 (indicated with a dashed line) and provides a link (question mark) to other components of the processing machinery, including the endonuclease CPSF-73.
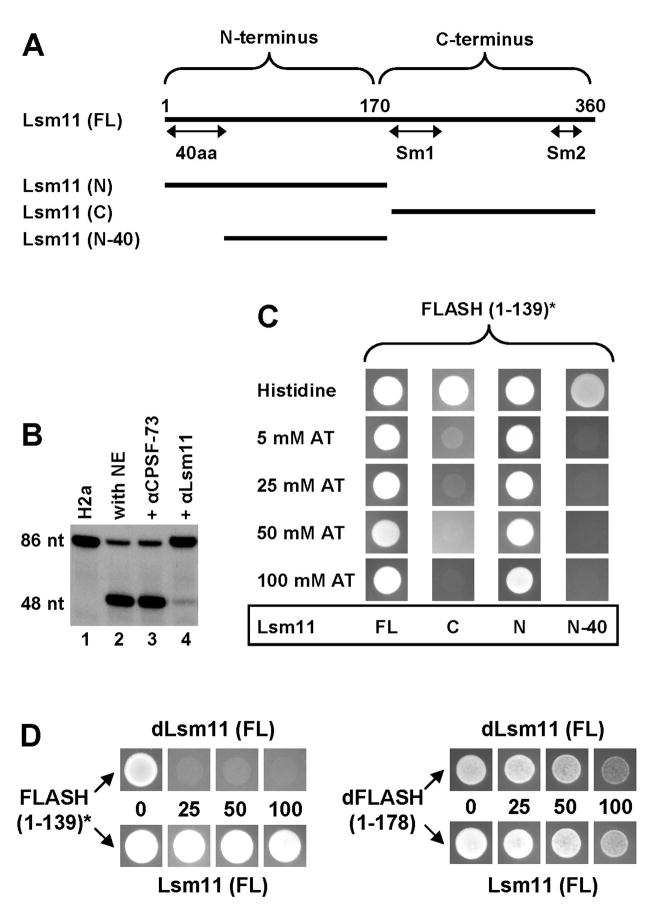
Figure 2. FLASH interacts with the N-terminal domain of Lsm11.
A. Domain organization of Lsm11 and schematic representation of deletion mutants used in the directed yeast two-hybrid system to map the region of interaction with FLASH. B. An antibody directed against the N-terminus of Lsm11 inhibits cleavage of the H2a pre-mRNA in a mouse nuclear extract (NE). The substrate alone is shown in lane 1. An anti-CPSF-73 antibody that does not recognize CPSF-73 in nuclear extracts was used as a negative control (lane 3). C. The directed yeast two-hybrid system was used to analyze the interaction of various fragments of Lsm11 fused to the GAL4 DNA-BD with the N-terminal fragment of FLASH fused to the GAL4 AD. The asterisk indicates that the N-terminal fragment of FLASH is placed out of frame with the GAL4 AD. Suspensions of yeast cells expressing Lsm11 and FLASH were spotted on a control plate containing histidine or on selective plates lacking histidine and supplemented with the indicated concentrations of 3-aminotriazole (3-AT). The plates were analyzed after 3 days of incubation at 27°C. D. The directed yeast two-hybrid system was used to analyze the interaction between the N-terminus of human FLASH (left) and the N-terminus of Drosophila dFLASH (right) with either Drosophila or human Lsm11 (top and bottom columns, respectively).
We generated an antibody against the N-terminal 170 amino acids of human Lsm11 that is active for Western blot analysis (Yang et al., 2009) and immunostaining (Ghule et al., 2008). Importantly, the same antibody when added to a mouse nuclear extract significantly reduced the efficiency of the cleavage reaction (Fig. 2B, lane 4). Thus, this portion of Lsm11 must perform an important function in 3′ end processing of histone pre-mRNAs in mammalian cells. An antibody against the N-terminus of CPSF-73 that does not recognize native CPSF-73 in a nuclear extract (Dominski et al., 2005a) had no effect on processing (Fig. 2B, lane 3).
Lsm11 was previously shown to interact in vitro with ZFP100 (Fig. 1B). However, this interaction does not involve highly conserved amino acids of Lsm11 that are required for 3′ end processing (Azzouz et al., 2005). Thus, there are likely other proteins that make essential contacts with the N-terminal region of Lsm11. To look for these proteins we screened a HeLa cDNA library against the full length human Lsm11 fused to the GAL4 DNA Binding Domain (GAL4 DNA-BD). Of nearly 20 million diploids screened, fewer than 200 showed substantial growth on selective medium, suggesting that they may express a protein interacting with Lsm11.
The most frequently obtained clones (total 7) contained various cDNA fragments encoding the N-terminal portion of the protein FLASH (for “Flice Associated Huge” protein), also known as CASP8AP2 (for “caspase-8 associated protein 2”) (Imai et al., 1999). Strikingly, in all of these clones the FLASH cDNA was fused out of frame with the upstream region encoding the GAL4 Activation Domain (AD), or contained a portion of the 5′UTR with an in-frame stop codon that should have precluded normal translation of the FLASH coding region. To determine whether the selected FLASH clones represented false positives, we subcloned the shortest selected cDNA encoding the first 139 amino acid of FLASH in frame with the GAL4 AD and reevaluated the interaction of this fusion protein with Lsm11 using the yeast two-hybrid system. Surprisingly, yeast cells expressing the in-frame fusion protein grew very slowly making it virtually impossible to test the interaction with Lsm11. This result suggests that high level expression of the N-terminal portion of FLASH is toxic to yeast cells and that the presence of a stop codon or a frame shift effectively reduced the expression of the fusion protein thus minimizing its toxic effect (Fig. S2A).
Mapping the region of human Lsm11 that interacts with the N-terminus of FLASH
Yeast cells expressing the out-of-frame N-terminal FLASH fragment (amino acids 1–139) and the full length Lsm11 displayed robust growth in the presence of 100 mM 3-aminotriazole (3-AT), indicative of a strong interaction (Fig. 2C, column 1). We carried out directed two-hybrid experiments to map the region of Lsm11 that is involved in the interaction with the N-terminal part of FLASH. The two halves of Lsm11 were separately cloned in frame with the GAL4 DNA-BD and tested for the interaction with the N-terminus of FLASH fused out-of-frame to the GAL4 AD. Yeast cells expressing the first 170 amino acids of Lsm11 retained the ability to robustly grow in the presence of 100 mM 3-AT, suggesting that this region is sufficient for a strong interaction with FLASH (Fig. 2C, column 3). Almost no growth at the lowest tested concentration of 3-AT (5 mM) was observed for yeast cells expressing the C-terminal half of Lsm11 (Fig. 2C, column 2). Western blot analysis demonstrated that all three Lsm11 clones were detectably expressed in yeast cells, with the expression level of the C-terminal portion exceeding that for the full length protein (not shown). Based on these results, we conclude that the N-terminal half of Lsm11 is sufficient for the interaction with FLASH.
Deletion of the first 40 amino acids from the N-terminal half of Lsm11 completely abolished the growth of yeast cells, demonstrating that residues 1–40 are essential for the interaction with FLASH (Fig. 2C, column 4). This N-terminal region contains a cluster of amino acids (positions 24–40) that have been highly conserved between vertebrates and Drosophila and likely play an important role in 3′ end processing of histone pre-mRNAs (Fig. S1C).
Identification of a Drosophila homolog of FLASH
To determine whether the interaction between Lsm11 and FLASH has been conserved during evolution, we used BLAST to search for a FLASH homolog in the Drosophila NCBI database. Only one Drosophila protein, designated CG4616, displayed a weak sequence similarity to mammalian FLASH, with the region of homology confined to the first ~180 amino acids of a total of 844 amino acids (Fig. S1A and B). Based on the functional data below we refer to this protein as Drosophila FLASH (dFLASH). We cloned the first 178 amino acids of dFLASH in frame with the GAL4 AD and tested its interaction in the yeast two-hybrid system with the full length Drosophila Lsm11 fused to the GAL4 DNA-BD (Fig. 2D). To control for the specificity of the interaction, we also co-expressed each protein in heterologous combinations: the N-terminal portion of dFLASH with human Lsm11, and Drosophila Lsm11 with the out-of-frame N-terminal human FLASH. Surprisingly, yeast cells expressing the N-terminal portion of dFLASH also grew at a slower rate and placing the insert out of frame with the GAL4 AD restored the normal growth rate (Fig. S2B). This result suggests that the N-terminal portion of FLASH and dFLASH is toxic to yeast cells. Since the inhibitory effect of dFLASH was not as detrimental as that caused by FLASH, we were able to carry out directed two-hybrid assays using the N-terminal fragment of Drosophila protein cloned in frame with the GAL4 AD.
The ability of yeast cells to grow in the presence of 100 mM 3-AT suggests that dFLASH tightly interacts with Drosophila Lsm11 (Fig. 2D, upper right corner). Surprisingly, an equally strong interaction was observed between the heterologous pair of dFLASH and human Lsm11 (Fig. 2D, bottom right corner), although no growth in the presence of 25 mM 3-AT was observed for cells co-expressing the N-terminal region of human FLASH and Drosophila Lsm11 (Fig. 2D, top left corner).
In vitro interaction between Lsm11 and the N-terminus of FLASH
We next analyzed the interaction between human Lsm11 and FLASH using the glutathione S-transferase (GST) pull-down assay. The N-terminal portion of FLASH (amino acids 1–139) was expressed in E. coli as a fusion protein with GST and tested for its ability to bind and pull down various portions of human Lsm11 labeled with 35S methionine.
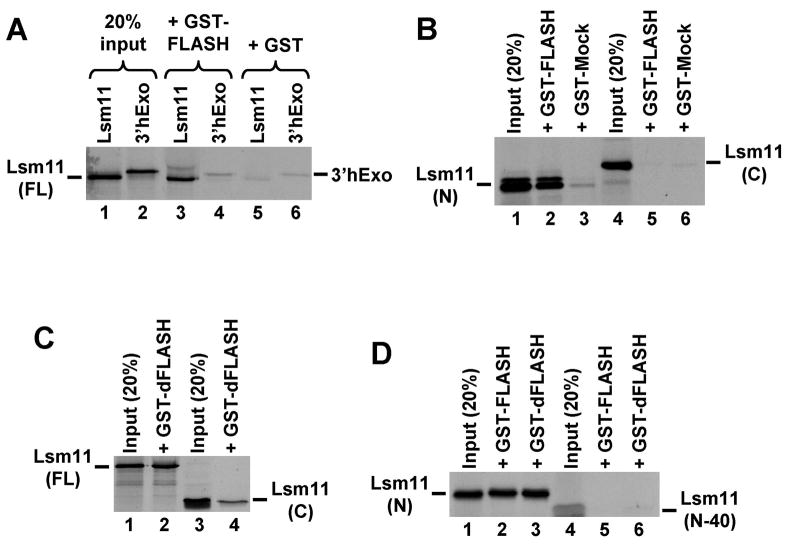
GST alone did not bind 35S-labeled human Lsm11 or 3′hExo, a protein of a similar size to Lsm11 that was used as a negative control (Fig. 3A, lanes 5 and 6). In contrast, GST-FLASH fusion protein bound up to 20% of the input full length Lsm11 (Fig. 3A, lane 3) but did not bind 3′hExo (Fig. 3A, lane 4). These results demonstrate that in vitro the N-terminal fragment of human FLASH specifically interacts with full length human Lsm11. The GST-FLASH fusion protein reproducibly pulled down 10–20% of the N-terminal half of Lsm11 (Fig. 3B, lane 2 and Fig. 3D, lane 2) and only a background amount of the C-terminal half (Fig. 3B, lane 5). Only a background amount of each Lsm11 fragment was pulled downed by a control fusion protein, GST-Mock (Fig. 3B, lanes 3 and 6). Importantly, the deletion of the first 40 amino acids of the N-terminal half of Lsm11 abolished the interaction with FLASH in vitro (Fig. 3D, lane 5). These data are in agreement with the data from the yeast directed two-hybrid assay and demonstrate that FLASH interacts with the N-terminal half of Lsm11, and that the interaction requires the first 40 amino acids of Lsm11.
Figure 3. Human and Drosophila FLASH interact with human Lsm11 in vitro.
A. Full length human Lsm11 and 3′hExo were synthesized and labeled by in vitro translation with 35S-methionine and incubated with a fusion protein consisting of GST and the N-terminal portion of FLASH (lanes 3 and 4), or with GST alone (lanes 5 and 6). Lanes 1 and 2 contain 20% of each radioactive protein used in the pull-down assay. B. The ability of GST-FLASH and GST-Mock fusion proteins to interact with the 35S-labeled Lsm11 N-terminal half (lanes 2 and 3), or C-terminal half (lanes 5 and 6). Lanes 1 and 4 contain 20% of each radioactive protein used in the pull-down assay. C. The ability of GST fused to the N-terminal region of Drosophila FLASH (GST-dFLASH) to interact with the 35S-labeled full length (lane 2) or C-terminal half of human Lsm11 (lane 4). Lanes 1 and 3 contain 20% of each radioactive protein used in the pull-down assay. D. The ability of GST-FLASH and GST-dFLASH fusion proteins to interact with the 35S-labeled N-terminal half (lanes 2 and 3) or the N-terminal half lacking the first 40 amino acids of human Lsm11 (lanes 5 and 6). Lanes 1 and 4 contain 20% of each radioactive protein used in the pull-down assay.
The fusion protein of GST and the N-terminal part of dFLASH bound more than 20% of the full length 35S-labeled human Lsm11 and only a background amount of the C-terminal fragment (Fig. 3C, lanes 2 and 4, respectively). The same fusion protein also bound the N-terminal part of human Lsm11 and the deletion of the first 40 amino acids from Lsm11 completely eliminated the binding (Fig. 3D, lanes 3 and 6, respectively). We conclude that FLASH and dFLASH bind the same region in the N-terminal half of human Lsm11 and make no essential contacts with the C-terminal half.
Depletion of dFLASH activates polyadenylation of a reporter gene in Drosophila cells
In Drosophila, depletion of any factor involved in U7-dependent 3′ end processing of histone pre-mRNAs by either RNAi or mutation results in a switch to the conventional mechanism of 3′ end processing and accumulation of polyadenylated histone mRNAs (Wagner et al., 2007). Recently, a genome wide screening scheme was developed based on this phenomenon to look for new processing factors in Drosophila (Wagner et al., 2007). In this system, a fragment of the Drosophila histone H3 gene containing all necessary processing sequences but lacking the stop codon is followed by the in-frame coding sequence of green fluorescent protein (GFP) and a polyadenylation signal. Drosophila cells containing this reporter gene normally do not express GFP since the U7-dependent cleavage immediately after the stem-loop removes the GFP coding region from the hybrid pre-mRNA. However, any defect in U7-dependent processing results in transcription into the GFP ORF and expression of GFP, thus turning Drosophila cells green.
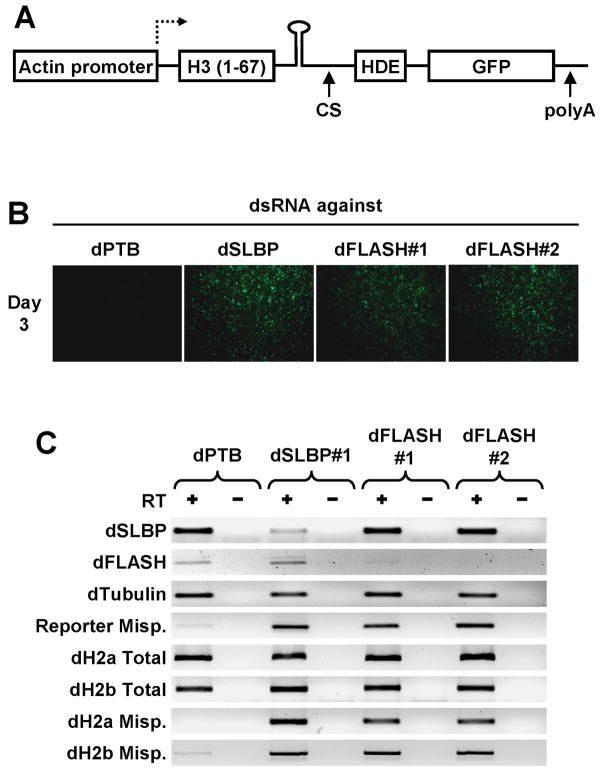
To determine whether dFLASH is involved in 3′ end processing of histone pre-mRNAs, we generated two double stranded (ds) RNAs, each approximately 500 base pairs long targeting different regions within the dFLASH open reading frame. Since it was possible that dFLASH is also required for transcription of histone genes, we used Drosophila cells stably expressing a reporter gene driven by a constitutive promoter from the actin 5C gene rather than by the histone gene promoter (Fig. 4A). Drosophila D.Mel-2 cells expressing the reporter gene were treated separately with each of the two dFLASH dsRNAs and analyzed for expression of GFP. Double stranded RNA against polypyrimidine tract binding protein (PTB), a known splicing factor, was used as a negative control, whereas a dsRNA against SLBP, an essential 3′end processing factor, was used as a positive control. Drosophila cells treated for 3 days with dsRNA targeting SLBP were clearly GFP positive, whereas cells treated with anti-PTB dsRNA did not express detectable levels of GFP (Fig. 4B). Most importantly, D.Mel-2 cells treated with either of the two dFLASH-specific dsRNAs exhibited robust fluorescence, suggesting that dFLASH is required for 3′ end processing of histone pre-RNAs in vivo.
Figure 4. Downregulation of dFLASH in Drosophila cells by RNAi activates expression of a GFP reporter gene.
A. A diagram of a reporter gene containing the actin 5C promoter, a region encoding the first 67 amino acids of Drosophila histone H3, a portion of the histone H3 3′ UTR including the stem-loop and the HDE, and an in-frame GFP ORF. The U7-dependent cleavage site (CS) and a polyA signal from the OpIE2 gene are indicated with arrows. B. Fluorescence images of D.Mel-2 cells expressing the reporter gene after three days of treatment with double stranded RNAs against indicated mRNAs. C. RT-PCR analysis of various mRNAs isolated from D.Mel-2 cells treated with double stranded RNAs, as indicated at the top. Total RNA was isolated from the treated cells and amplified by PCR with (+) or without (−) prior reverse transcription (RT).
To monitor the efficiency of RNAi we used an RT-PCR assay of total RNA isolated from cells after three days of treatment with dsRNAs. We also analyzed misprocessing of reporter mRNA and endogenous histone mRNAs using forward PCR primers within the H3 ORF and reverse primers targeting either the GFP ORF or a region located downstream of the U7-dependent cleavage site, respectively. The RT-PCR assay confirmed that dSLBP and dFLASH mRNAs were downregulated by their respective dsRNAs, whereas an unrelated mRNA encoding tubulin was unaffected (Fig. 4C). Both the reporter pre-mRNA and endogenous H2a and H2b pre-mRNAs were misprocessed in cells treated with the anti-dSLBP and anti-dFLASH dsRNAs as judged by accumulation of RNA species extended beyond the stem-loop structure (Fig. 4C). Misprocessing was also evident for the other three histone mRNAs (not shown). The total levels of the H2a and H2b mRNAs were not significantly changed in cells containing misprocessed mRNA, demonstrating that the reduction of normal U7-dependent processing was efficiently compensated by switching to cleavage/polyadenylation.
Direct detection of misprocessed endogenous histone mRNAs
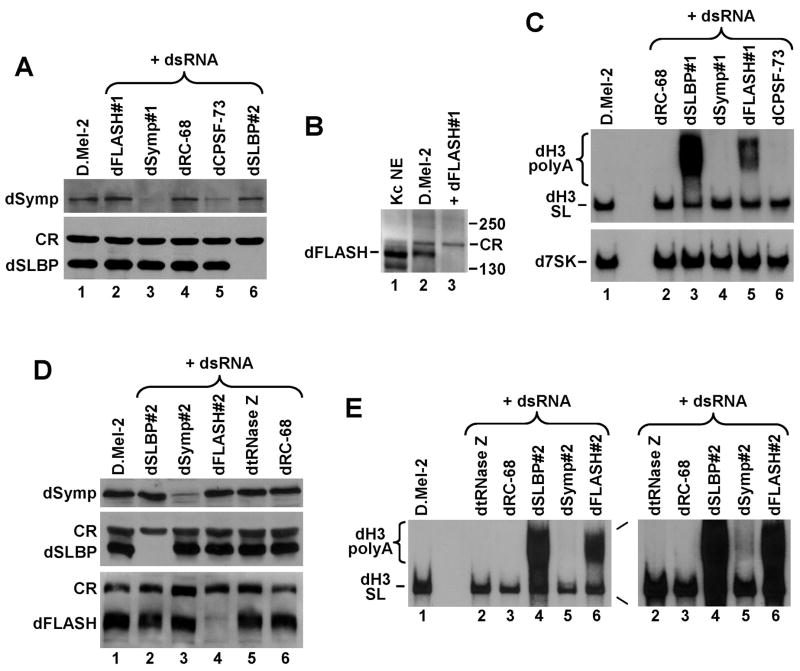
Misprocessed endogenous histone mRNAs containing a polyA tail can be readily resolved from properly cleaved, stem-loop terminated histone mRNAs by gel electrophoresis, with misprocessed mRNAs being longer and heterogeneous in size. We treated D.Mel-2 cells with dsRNAs against dFLASH and other factors involved in 3′ end processing of histone pre-mRNAs and analyzed total RNA from the treated cells by Northern blotting. As negative controls we used dsRNA targeting proteins involved in other RNA processing events. When possible, the level of downregulated proteins was monitored by specific antibodies. After 5 days of treatment with an anti-dSLBP dsRNA, D.Mel-2 cells contained no detectable dSLBP (Fig. 5A, lane 6) and the majority of the Drosophila histone H3 mRNA was polyadenylated and detected as a smear migrating slower than the stem-loop terminated species (Fig. 5C, lane 3).
Figure 5. Generation of polydenylated histone mRNAs in Drosophila cells in the absence of dFLASH.
A. Western blot analysis of lysates from untreated D.Mel-2 cells (lane 1), or cells treated with the indicated dsRNAs, including dFLASH#1 (lane 2) that targets an upstream region in the dFLASH mRNA. The blot was probed with an anti-dsymplekin antibody (top panel) or anti-SLBP antibody (bottom panel). CR band in the bottom panel corresponds to a cross-reacting protein that is detected by the anti-SLBP antibody and serves as an internal loading control. B. Western blot analysis of lysates from untreated D.Mel-2 cells (lane 1) or cells treated with the dFLASH#1 dsRNA (lane 2). An aliquot of a nuclear extract (NE) from Drosophila Kc cells was analyzed in lane 1. The blot was probed with an anti-dFLASH antibody. CR band corresponds to a cross-reacting protein that is detected by the anti-dFLASH antibody in the D.Mel-2 whole cell lysates but not in the Kc NE. C. Northern blot analysis of total RNA isolated from D.Mel-2 cells tested in panel A and B. The blot was hybridized with 32P-labeled probes directed against the Drosophila H3 (dH3) histone mRNA (top panel) and Drosophila 7SK RNA to control for equal loading (bottom panel). D. Western blot analysis of lysates from untreated D.Mel-2 cells (lane 1) or cells treated with the indicated dsRNAs, including the dFLASH#2 (lane 4) that targets a downstream region in the dFLASH mRNA. The blot was probed with an anti-dsymplekin antibody (top panel), anti-dSLBP antibody (middle panel), and anti-dFLASH antibody (bottom panel). E. Northern blot analysis of total RNA isolated from D.Mel-2 cells tested in panel D. The blot was hybridized with a 32P-labeled probe directed against Drosophila H3 histone mRNA. An overexposure of the part of the film containing lanes 2–6 is shown to the right.
Consistent with the results of the GFP assay, D.Mel-2 cells exposed for 5 days to anti-dFLASH dsRNA#1 accumulated a readily detectable amount of polyadenylated histone H3 mRNA (Fig. 5C, lane 5). We generated an antibody against the first 178 amino acids of the dFLASH protein and observed that a protein migrating at about 150 kDa and most likely corresponding to dFLASH was missing in the treated extract (Fig. 5B, lane 3). This mobility was significantly slower than expected for dFLASH (95 kDa). Synthesis of dFLASH using an in vitro translation system confirmed that this protein has an unusual electrophoretic mobility and indeed migrates at ~150 kDa (Fig. S3).
A different effect was seen for two other factors involved in 3′ end processing of histone pre-mRNAs: symplekin and CPSF-73. These two proteins are also required for cleavage/polyadenylation and therefore their depletion by specific dsRNAs while inhibiting correct U7-dependent processing, at the same time prevents efficient polyadenylation of histone mRNAs (Fig. 5C, lanes 4 and 6). The appearance of polyadenylated histone mRNAs in cells depleted of CPSF-73 and symplekin can be detected only after prolonged exposure times (Fig. 5E, right panel, lane 5). No detectable changes in the pattern of histone pre-mRNA processing were observed in D.Mel-2 cells treated with a dsRNA against RC-68, which is a homolog of CPSF-73 involved in 3′ end processing of pre-snRNAs, or in untreated cells after 5 days of cultivation (Fig. 5C, lanes 1 and 2). Consistent with the results of RT-PCR assay (Fig. 4C), the overall level of the H3 mRNA was not reduced in cells depleted of dFLASH (Fig. 5C, lane 5), suggesting that dFLASH does not play a role in transcription of histone genes. To control for equal loading, the same Northern blot was probed against Drosophila 7SK RNA, an approximately 550-nucleotide RNA synthesized by RNA polymerase III (Fig. 5C, bottom panel).
We repeated the RNAi experiment using a dsRNA targeting a different region in the dFLASH mRNA (dFLASH#2). We also used a dsRNA targeting a different region of dsymplekin mRNA (Symp#2) and a dsRNA that targets dtRNase Z, an endonuclease involved in 3′ end processing of pre-tRNAs, as a negative control. The results of this RNAi experiment were identical to those observed in the first experiment, with the anti-SLBP and anti-dFLASH#2 dsRNAs resulting in a robust polyadenylation of the H3 mRNA (Fig. 5E, lanes 4 and 6), and the dsRNA targeting dtRNase Z having no effect (Fig. 5E, lane 2). The level of dFLASH protein was significantly reduced only in D.Mel-2 cells treated with anti-dFLASH#2 dsRNA (Fig. 5D, lane 4). The same selective effect on protein levels was observed for dsRNAs directed against SLBP and symplekin (Fig. 5D, lanes 2 and 3).
Overall, these results strongly support the notion that Drosophila FLASH is a factor involved in 3′ end processing of histone pre-mRNAs in vivo. Moreover, the observation that depletion of Drosophila FLASH results in generation of readily detectable levels of polyadenylated histone mRNAs suggests that FLASH, in contrast to CPSF-73 and symplekin, participates only in 3′ end processing of histone pre-mRNAs and has no role in generation of polyadenylated mRNAs. Surprisingly, in spite of the robust polyadenylation of histone mRNAs consistently observed in dFLASH-deficient cells, dFLASH escaped detection as a 3′ end processing factor in the original genome wide reporter screen (Wagner et al., 2007). It is possible that some factors were overlooked in the screen due to lower quality of dsRNAs and/or dsRNAs targeting mRNA regions more resistant to RNAi.
dFLASH interacts with Lsm11 in vivo and localizes to histone locus bodies
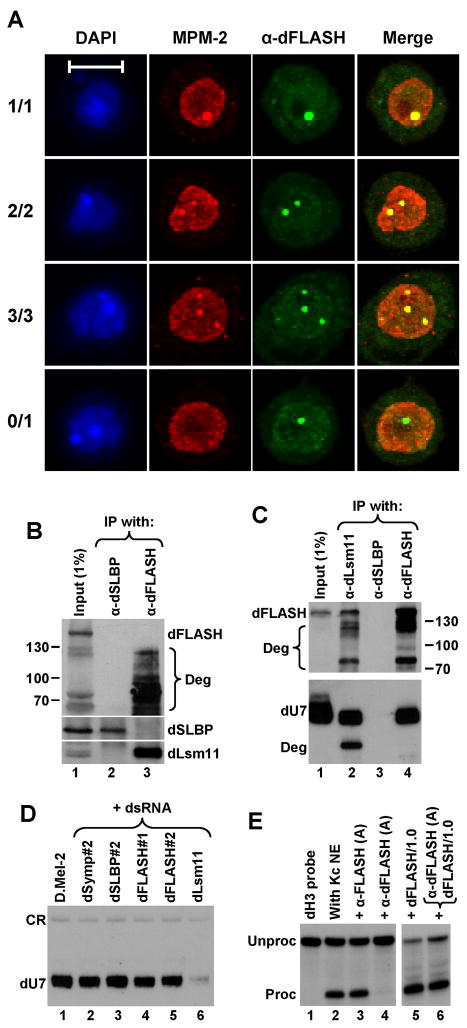
We analyzed the intracellular localization of dFLASH in D.Mel-2 cells. Nuclei of Drosophila embryos contain sites of concentrated U7 snRNP that assemble near histone gene loci (Liu et al., 2006). These sites are distinct from Cajal bodies and have been designated histone locus bodies (HLBs). HLBs in S-phase Drosophila cells are recognized by the MPM-2 monoclonal antibody, which interacts with unidentified Drosophila phosphoproteins (White et al., 2007). We examined whether dFLASH colocalizes with the MPM-2 epitope by co-staining D.Mel-2 cells with the rabbit anti-dFLASH antibody and the MPM-2 mouse monoclonal antibody. In the majority of cells the anti-dFLASH antibody detected a single pronounced structure per nucleus with the rest of the nucleus displaying more diffuse staining (Fig. 6A). We also frequently observed nuclei with two and rarely three structures containing concentrated dFLASH. More importantly, the structures containing dFLASH were 100% coincident with structures containing the MPM-2 epitope, demonstrating that dFLASH is localized to HLBs (Fig. 6A). In addition to nuclei with both proteins colocalized in HLBs, we observed nuclei that contained dFLASH structures but lacked any detectable MPM-2-stained structures (Fig. 6A), and those that did not display any focal staining with either antibody (not shown). It is likely that the first class represents cells in early G1 phase before the MPM-2 epitope is phosphorylated. Cells of the other class might be in late G2 phase when HLBs are becoming disassembled. The varying number of structures detected with the anti-dFLASH and MPM-2 antibodies likely reflects the pairing status of homologous chromosomes (White et al., 2007) and/or the presence of additional histone gene loci in some cells.
Figure 6. dFLASH interacts with dLsm11 in vivo and localizes to histone locus bodies.
A. Immunofluorescence images of Drosophila D.Mel-2 cells indirectly stained with both the MPM-2 monoclonal antibody (red) and anti-dFLASH antibody (green). Nuclei were visualized with DAPI. Four different D.Mel-2 cells represent four different co-staining patterns observed with the MPM-2 and anti-dFLASH antibodies. The bar represents 10 μm. B. Western blot analysis of proteins precipitated from Drosophila Kc cell lysates using anti-dSLBP (lane 2) or anti-dFLASH antibodies (lane 3). Proteins were detected using anti-dFLASH (top panel), anti-dSLBP (middle panel), or anti-dLsm11 antibodies (bottom panel). The input lysate (1%) is shown in lane 1. Degradation products of dFLASH (Deg) are indicated by a bracket. C. Top panel: Western blot analysis of proteins precipitated from Drosophila Kc cell lysates using anti-dLsm11 (lane 2), anti-dSLBP (lane 3), or anti-dFLASH antibodies (lane 4). Proteins were detected using anti-dFLASH antibody. The input lysate (1%) is shown in lane 1. Bottom panel: A fraction of precipitates was analyzed by Northern blotting for the presence of Drosophila U7 snRNA (dU7). “Deg” is a degradation product of U7 snRNA precipitated by the anti-dLsm11 antibody. D. Northern blot analysis of dU7 snRNA isolated from untreated D.Mel-2 cells (lane 1) or D.Mel-2 cells treated with indicated dsRNAs (lanes 2–6). A cross-reacting RNA species indicated at the top (CR) served as a loading control. E. In vitro 3′ end processing of Drosophila H3 (dH3) histone pre-mRNA in a nuclear extract from Kc cells. Processing was carried out under control conditions (lane 2) or in the presence of the indicated protein A-purified antibodies (lanes 3 and 4) and/or 1μg of the N-terminal dFLASH protein fused to GST (lanes 5 and 6). Lane 1 contains the input dH3 substrate.
We tested whether the anti-dFLASH antibody can co-precipitate Drosophila Lsm11 (dLsm11), which also resides in HLBs (Liu et al., 2006), or dSLBP, a processing factor that displays a more diffuse staining pattern throughout the nucleoplasm (Lanzotti et al., 2004). Drosophila Kc cells were lysed in the presence of 0.5% NP-40 buffer and incubated with the anti-dFLASH antibody, or an antibody against dSLBP. As determined by Western blot analysis using the anti-dFLASH antibody, in some experiments the majority of the dFLASH protein underwent substantial proteolysis during immunoprecipitation (Fig. 6B, top panel, lane 3). However, regardless of this degradation, the precipitate was invariably enriched in dLsm11 (Fig. 6B, bottom panel, lane 3), indicating that these two proteins form a stable complex in Drosophila Kc cells. Note, that the anti-dLsm11 antibody detects two closely migrating forms of dLsm11 in Drosophila extracts (Fig. 6B, bottom panel, lane 1) and the two forms were present in the anti-dFLASH precipitate (lane 3).
In contrast to dLsm11, no detectable amount of dSLBP was precipitated by the anti-dFLASH antibody suggesting that only a limited number of processing factors stably associate with dFLASH (Fig. 6B, middle panel, lane 3). The anti-dSLBP antibody specifically precipitated dSLBP (Fig. 6B, middle panel, lane 2). However, the precipitate did not contain any detectable amount of dFLASH or dLsm11, further confirming that dSLBP is not part of a stable complex with the other two processing factors. We used the reciprocal approach and found that the anti-dLsm11 precipitated dFLASH (Fig. 6C, top panel, lane 2), although as expected the amount of the precipitated protein was lower compared to the amount precipitated with the anti-dFLASH antibody (lane 4). Again, the anti-dSLBP antibody did not precipitate any dFLASH (Fig. 6C, top panel, lane 3).
We used Northern blotting to test a fraction of each precipitate shown in the top panel of Fig. 6C for the presence of Drosophila U7 snRNA (dU7 snRNA). As expected, the anti-dLsm11 precipitate was enriched with the dU7 snRNA (Fig. 6C, bottom panel, lane 2). In contrast, the anti-dSLBP antibody failed to precipitate dU7 snRNA, consistent with its inability to precipitate dLsm11 (Fig. 6C, bottom panel, lane 3). Significantly, the anti-dFLASH antibody precipitated at least as much dU7 snRNA as did the anti-Lsm11 antibody (Fig. 6C, bottom panel, lane 4). Thus, dFLASH forms a tight complex with the entire U7 snRNP rather than with dLsm11 alone.
To test whether dFLASH is an integral part of the U7 snRNP, we analyzed the level of dU7 snRNA in D.Mel-2 cells depleted of Drosophila symplekin, SLBP, FLASH (Fig. 5) and Lsm11 (Fig. S4, lane 2). RNAi-mediated depletion of Lsm11 resulted in readily detectable polyadenylation of histone pre-mRNAs (Fig. S4, lane 2). Moreover, depletion of dLsm11, as expected for a stable component of the U7-specific Sm ring, resulted in nearly complete disappearance of the dU7 snRNA (Fig. 6D, lane 6). In contrast, depletion of dsymplekin and dFLASH only moderately reduced the level of the dU7 snRNA (Fig. 6D, lanes 2 and 4–5) and depletion of dSLBP had no effect (Fig. 6D, lane 3). We conclude that dFLASH is not required for the integrity of the dU7 snRNP.
We examined whether the anti-dFLASH antibody inhibits 3′ end processing in a nuclear extract from Drosophila Kc cells, using as a negative control an antibody against human FLASH (see below). Importantly, the protein A-purified anti-dFLASH fully blocked processing of the Drosophila histone H3 substrate (Fig. 6E, lane 4), whereas the antibody against human N-terminal FLASH had no effect (Fig. 6E, lane 3). Addition of the competing N-terminal dFLASH protein fused to GST (the protein that was used to generate the antibody) completely reversed the negative effect of the anti-dFLASH antibody, proving that the effect was specific (Fig. 6E, lane 6). Thus, dFLASH is required for 3′ end processing of histone pre-mRNAs in Drosophila nuclear extracts.
FLASH is required for in vitro 3′ end processing of mammalian histone pre-mRNAs
In human cells, FLASH is present in nuclear foci that are 100% coincident with structures containing NPAT, a 220 kDa co-activator of histone genes (Barcaroli et al., 2006; Bongiorno-Borbone et al., 2008). Since NPAT localizes near histone gene clusters (Ma et al., 2000; Zhao et al., 2000), these structures are considered as histone locus bodies (HLBs). In addition to NPAT, mammalian HLBs also contain the U7 snRNP with its two unique components, Lsm10 and Lsm11 (Ghule et al., 2008). HLBs are often associated, although not identical, with the coilin-containing Cajal bodies (CBs) and the extent of co-localization depends on both the cell type (primary versus transformed) and cell cycle phase (Bongiorno-Borbone et al., 2008).
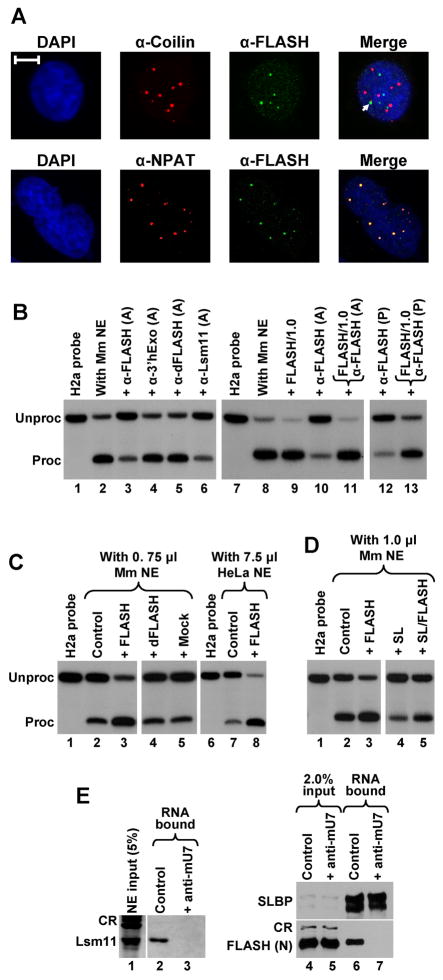
We generated a rabbit antibody against the N-terminal region of human FLASH (amino acids 1–139) fused to GST and tested its activity in immunofluorescence experiments to analyze the intracellular localization of FLASH in HeLa cells. The antibody stained a number of nuclear structures in HeLa cells that only occasionally overlapped with Cajal bodies detected with an anti-coilin antibody (Fig. 7A, top). Importantly, nuclear structures stained with the anti-FLASH antibody colocalized with NPAT-positive structures (Fig. 7A, bottom). Thus, as previously shown with other anti-FLASH antibodies (Barcaroli et al., 2006; Bongiorno-Borbone et al., 2008), human FLASH localizes to HLBs.
Figure 7. FLASH localizes to histone locus bodies and is required for 3′ end processing in mammalian nuclear extracts.
A. Immunofluorescence images of HeLa cells indirectly stained with anti-FLASH polyclonal antibody (green) and either the anti-coilin monoclonal antibody (red) or anti-NPAT monoclonal antibody (red). Nuclei were visualized with DAPI. The arrow indicates colocalization of Cajal and FLASH bodies. The bar represents 10 μm. B. The anti-FLASH antibody inhibits in vitro 3′ end processing of the mouse H2a histone pre-mRNA in a nuclear extract from mouse myeloma cells. Processing was carried out under control conditions (lanes 2 and 8) or in the presence of protein A-purified (A) or affinity-purified (P) antibodies, as indicated at the top of each lane. Processing reactions shown in lanes 11 and 13 contain both the antibody and 1 μg of the competing N-terminal FLASH (amino acids 1–139) fused to GST, whereas the processing reaction shown in lane 9 contains only 1 μg of the protein. Lanes 1 and 7 contain the input H2a substrate. C. The N-terminal fragment of human FLASH stimulates 3′ end processing of the H2a histone pre-mRNA in 0.75 μl of a mouse myeloma nuclear extract (lanes 2–5) and in 7.5 μl of a HeLa nuclear extract (lanes 7–8). Processing was carried out under control conditions (lanes 2 and 7), in the presence of 40 ng of the human N-terminal FLASH (lanes 3 and 8), or in the presence of 40 ng of control GST fusion proteins (lanes 4 and 5). D. Stimulation of 3′ end processing in 1 μl of a mouse nuclear extract by the N-terminal fragment of human FLASH (40 ng) in the absence (lane 3) or in the presence of a molar excess of the stem-loop RNA (lane 5). E. Processing complexes were assembled on a 5′ biotinylated histone pre-mRNA (0.25 μg) under normal conditions (lanes 2 and 6) or in the presence of the anti-mU7 oligonucleotide (lanes 3 and 7) and purified on streptavidin beads. The presence of individual proteins bound to histone pre-mRNA was detected by specific antibodies. Processing complexes analyzed in lanes 6 and 7 were assembled in a mouse nuclear extract supplemented with the N-terminal fragment of FLASH. CR indicate cross-reacting proteins recognized by an anti-Lsm11 antibody (lane 1) or anti-FLASH antibody (lane 4 and 5).
We next tested the ability of the anti-FLASH antibody to directly inhibit the in vitro processing reaction. Protein A-purified anti-FLASH antibody reduced the efficiency of processing of H2a pre-mRNA in a mouse nuclear extract from 80–95% (Fig. 7B, lanes 2 and 8) to less than 20% (Fig. 7B, lanes 3 and 10), while protein A-purified antibodies against 3′hExo and dFLASH had no effect (Fig. 7B, lanes 4 and 5). Protein A-purified antibody against the N-terminal region of Lsm11 was as effective in inhibiting processing as the anti-FLASH antibody (Fig. 7B, lane 6). The bacterially expressed N-terminal FLASH fused to GST fully neutralized the inhibitory effect of the protein A-purified anti-FLASH antibody (Fig. 7B, lane 11), demonstrating that the inhibition resulted from the antibody blocking FLASH present in the extract. An affinity purified anti-FLASH antibody also strongly inhibited in vitro processing in a mouse nuclear extract and its inhibitory effect was fully neutralized by the N-terminal fragment of FLASH fused to GST (Fig. 7B, lanes 12 and 13, respectively). Overall, these data demonstrate that FLASH localizes to HLBs and is required for 3′ end processing of histone pre-mRNAs in mammalian nuclear extracts.
The N-terminal region of FLASH stimulates in vitro processing
The bacterially expressed N-terminal fragment of human FLASH (amino acids 1–139) fused to GST moderately stimulated cleavage of the mouse H2a substrate (Fig. 7B, compare lanes 8 and 9). We studied this effect in more detail using suboptimal amounts of mouse nuclear extracts in the processing reaction. Lowering the amount of a very active mouse nuclear extract from 7.5 μl to 0.75 μl per reaction resulted in a dramatic reduction of processing efficiency from ~95% (Fig. 7B, lane 8) to ~10% (Fig. 7C, lane 2). Remarkably, addition of 40 ng of the recombinant N-terminal FLASH fused to GST restored the efficiency of cleavage to more than 75% (Fig. 7C, lane 3) and the stimulatory effect was observed with as little as 4 ng of the recombinant protein (Fig. S5). Addition of GST itself (not shown) or control GST fusion proteins, including dFLASH, had no effect on processing (Fig. 7C, lanes 4 and 5).
We also used 7.5 μl of a weakly active HeLa nuclear extract and observed strong stimulation of processing with 40 ng of the N-terminal FLASH fragment (Fig. 7C, lanes 7 and 8). Thus, both in human and mouse nuclear extracts, FLASH is a limiting factor. More importantly, these results demonstrate that this short fragment (amino acids 1–139) supports all necessary functions of human FLASH in 3′ end processing of histone pre-mRNAs in vitro. Since SLBP is an auxiliary factor that improves binding of the U7 snRNP to histone pre-mRNA in mammalian nuclear extracts (Dominski and Marzluff, 2007), it was possible that this protein mediates the stimulatory effect of FLASH. We tested whether the N-terminal FLASH functions in a mouse nuclear extract containing a large excess of the stem-loop RNA. This RNA tightly binds SLBP present in the extract and reduces the efficiency of processing of the H2a pre-mRNA (Fig. 7D, compare lanes 2 and 4). Importantly, FLASH stimulated 3′ end processing in the presence of the stem-loop RNA (Fig. 7D, compare lanes 4 and 5). We conclude that the stimulatory effect of FLASH is independent of SLBP and is most likely achieved by the recruitment of a different, yet uncharacterized 3′ end processing factor.
We next asked whether FLASH can interact with Lsm11 in the histone pre-mRNA processing complex. We incubated a 5′ biotinylated histone pre-mRNA (0.25 μg) in a mouse nuclear extract (750 μl) to assemble stable processing complexes containing the U7 snRNP (Yang et al., 2009). The assembly reaction was carried out in the presence of 0.1% NP-40 to inhibit the cleavage reaction and processing complexes were purified by binding to streptavidin sepharose beads. As a negative control, one reaction contained a 2′ O-methyl oligonucleotide (anti-mU7) that blocks the 5′ end of the U7 snRNA and inhibits the recruitment of the U7 snRNP to histone pre-mRNA (Fig. 7E, lane 3). In agreement with our previous results (Yang et al., 2009), Lsm11 was detected by Western blotting only in the assembly reaction lacking the anti-mU7 oligonucleotide (Fig. 7E, lane 2). Surprisingly, no FLASH could be detected in the same purified sample, presumably because of the low concentrations of the endogenous FLASH (not shown). We therefore repeated the experiment using a mouse nuclear extract (750 μl) supplemented with the N-terminal FLASH (7.5 μg) (Fig. 7E, lanes 4 and 5). The recombinant protein was readily detected in the processing complexes assembled under normal conditions (Fig. 7E, bottom panel, lane 6) and was missing when binding of the U7 snRNP to histone pre-mRNA was prevented by the anti-mU7 oligonuceotide (lane 7). Importantly, the 5′ biotinylated histone pre-mRNA was purified from both reactions with a similar efficiency, as demonstrated by the presence of the same amount of SLBP, which binds the stem-loop in the substrate independently of the U7 snRNP (Fig. 7E, top panel, lanes 6 and 7). We conclude that the N-terminal FLASH (1–139) interacts with Lsm11 in the context of the pre-processing complex containing the complete U7 snRNP and that FLASH is limiting in mammalian nuclear extracts.
DISCUSSION
Conversion of replication-dependent histone pre-mRNAs to histone mRNAs requires a single endonucleolytic cleavage to form the mature 3′ end. This 3′ end processing reaction is guided by the U7 snRNP, which binds downstream of the cleavage site and recruits the 3′ endonuclease CPSF-73 (Dominski and Marzluff, 2007). The most distinctive feature of the U7 snRNP is the composition of its Sm ring, which contains Lsm11 and Lsm10 rather than SmD1 and SmD2 found in the spliceosomal snRNPs (Schumperli and Pillai, 2004). Compared to other proteins of the Sm family, Lsm11 has an unusually long N-terminal domain that is essential for 3′ end processing of histone pre-mRNAs. Here, we show that the N-terminal domain of Lsm11 interacts with FLASH and that FLASH is required for 3′ end processing of histone pre-mRNAs in vertebrates and invertebrates.
FLASH (for “FLICE-associated huge” protein), also known as CASP8AP2 (for “caspase-8 associated protein 2”), was originally identified as a protein that binds and activates caspase-8 in response to activation of Fas/CD95 receptors (Imai et al., 1999). Subsequently, FLASH was shown to localize to nuclear structures also containing NPAT (Barcaroli et al., 2006) that assemble in the vicinity of histone gene loci on chromosomes 1 and 6 (Ma et al., 2000; Zhao et al., 2000) and are now referred to as histone locus bodies (HLBs). In cells stimulated for apoptosis by activation of Fas/CDC95 receptor, FLASH translocates from nuclear bodies to the cytoplasm (Milovic-Holm et al., 2007). In the cytoplasm, FLASH is predominantly targeted to mitochondria, where it associates with and likely activates caspase-8, thus triggering apoptosis. Consistent with this, depletion of FLASH by RNA interference inhibits CD95-induced apoptosis. At the mitochondria, FLASH and caspase-8 can form a ternary complex with the anti-apoptotic adenoviral protein E1B19K and formation of this complex renders cells resistant against Fas/CD95-mediated apoptosis (Milovic-Holm et al., 2007).
The localization of FLASH in HLBs for the first time pointed to a possible role of FLASH in histone gene expression, an interpretation substantiated by the observation that downregulation of FLASH by RNAi results in delayed S-phase progression and reduced expression of histone genes (Barcaroli et al., 2006). Our results demonstrate that FLASH participates in histone gene expression by playing an essential role in 3′ end processing of histone pre-mRNAs. FLASH and Lsm11 tightly interact with each other through their N-terminal regions in both the yeast two-hybrid system and in vitro. Through a very limited sequence similarity we identified the ortholog of human FLASH in Drosophila (dFLASH). dFLASH interacts with Drosophila Lsm11 and this interaction can be readily detected in Drosophila cells and in vitro. The N-terminal fragment of human FLASH is efficiently recruited to histone pre-mRNA by the U7 snRNP and antibodies to dFLASH precipitate U7 snRNA from Drosophila extracts. These results indicate that FLASH and dFLASH can interact with Lsm11 in the context of the fully assembled Sm ring. FLASH and dFLASH each localize to HLBs and depletion of dFLASH in Drosophila cells results in robust polyadenylation of histone mRNAs, indicating that dFLASH is required for 3′ end processing of histone pre-mRNAs in vivo. In addition, antibody inhibition experiments indicate that both orthologs are required for 3′ end processing of histone pre-mRNAs in vitro.
All the biochemical requirements for human FLASH to function in histone pre-mRNA processing in vitro are provided by the first 139 amino acids of the overall 1982 amino acids. Addition of this fragment to mammalian nuclear extracts stimulates 3′ end processing, indicating that FLASH is a limiting processing factor. This interpretation is supported by our previous inability to detect FLASH in processing complexes assembled on histone pre-mRNA using large amounts of a highly active mouse nuclear extract (Yang et al., 2009).
What are the biochemical function of FLASH in histone pre-mRNA 3′ end processing and a rationale for embedding a pro-apoptotic protein into this critical step of histone gene expression? The stimulation of 3′ end processing does not depend on SLBP and hence does not occur through a more efficient recruitment of the U7 snRNP to histone pre-mRNA. A plausible hypothesis is therefore that one region in the N-terminal fragment of FLASH interacts with Lsm11, whereas another region makes contacts with an unidentified component of the processing machinery that in turn communicates with the 3′ endonuclease CPSF-73. This interaction may serve to recruit CPSF-73 to histone pre-mRNA and/or to activate the endonuclease mode of CPSF-73 that appears latent in vitro (Mandel et al., 2006).
The involvement of a 3′ end processing factor in apoptosis is surprising although it most likely reflects the importance of histone gene expression in cell cycle progression and severity of consequences if cells do not properly coordinate DNA replication with the accumulation of histone mRNAs. Future studies on the biochemical role of FLASH in both vertebrates and invertebrates should shed more light on the unusual mechanism of 3′ end processing of histone pre-mRNAs and its links to apoptosis and both DNA replication and chromatin assembly.
EXPERIMENTAL PROCEDURES
Yeast two-hybrid screen
The open reading frame for human Lsm11 was cloned into the pGBKT7 vector (Clontech) and used to screen a pre-transformed HeLa cDNA library (Clontech), as suggested by the manufacturer. Diploid cells expressing proteins potentially interacting with Lsm11 were initially selected on plates containing 2.5 mM 3-aminotriazole (3-AT) and subsequently tested on plates containing up to 100 mM 3-AT.
RNA substrates
RNA substrates were chemically synthesized by Dharmacon (Lafayette, Co) or generated by transcription using appropriate DNA templates and T7 RNA polymerase. Chemically synthesized RNAs were labeled at the 5′ end with 32P using T4 polynucleotide kinase (NEB). RNAs generated by T7 transcription were treated with calf intestinal phosphatase (NEB) to remove the 5′ triphosphate prior to 5′ labeling.
In vitro processing of histone pre-mRNAs
A standard processing reaction in a total volume of 10 μl contained ~0.5 ng of a 5′ labeled RNA substrate (0.05 pmol), 0.5–7.5 μl of a nuclear extract and 20 mM EDTA (pH 8). Processing reactions containing mammalian (HeLa or mouse myeloma) or Drosophila Kc nuclear extracts were incubated for 60 min at 32°C or room temperature, respectively, treated for 60 min with proteinase K and analyzed on 8% polyacrylamide/7M urea denaturing gels. Antibodies (1.5 μl) were either directly added to processing reactions or after premixing with 1 μg of competing protein fragments.
Other Methods and Materials
See Supplemental Data for detailed description on the following methods and materials: Cell cultures and nuclear extracts, Construction of Act5Cp/H3/GFP reporter gene and generation of a stable D.Mel-2 cell line, GST pull-down assay, Immunoprecipitations, RNA-mediated purification of processing complexes, Northern blot analysis, Antibodies, Immunofluorescence, Generation of dsRNAs and RNA interference, RT-PCR Analysis.
Supplementary Material
Acknowledgments
We thank the following researchers for providing reagents: J. Gall (Carnegie Institution, Baltimore) for anti-Drosophila Lsm11 and Lsm10; M. Steiniger (UNC Chapel Hill) for anti-Drosophila symplekin; R. Lerner (UNC Chapel Hill) for GST and GST fusion proteins; G. Matera (UNC Chapel Hill) for mouse anti-coilin and anti-NPAT; S. Crews (UNC Chapel Hill) for Drosophila cDNA clones, and O. Gabrielsen (University of Oslo) for FLASH cDNA clones. We also thank T.K. Rajendra (UNC Chapel Hill) for help with IF experiments and E. Palazzolo (UNC Chapel Hill) for help with yeast directed two-hybrid experiments. This work was supported by NIH grants GM29832 and GM58921.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alm-Kristiansen AH, Saether T, Matre V, Gilfillan S, Dahle O, Gabrielsen OS. FLASH acts as a co-activator of the transcription factor c-Myb and localizes to active RNA polymerase II foci. Oncogene. 2008;27:4644–4656. doi: 10.1038/onc.2008.105. [DOI] [PubMed] [Google Scholar]
- Azzouz TN, Gruber A, Schumperli D. U7 snRNP-specific Lsm11 protein: dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids Res. 2005;33:2106–2117. doi: 10.1093/nar/gki516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De LV. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A. 2006;103:14808–14812. doi: 10.1073/pnas.0604227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De LV. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle. 2008;7:2357–2367. doi: 10.4161/cc.6344. [DOI] [PubMed] [Google Scholar]
- Dominski Z. Nucleases of the metallo-beta-lactamase family and their role in DNA and RNA metabolism. Crit Rev Biochem Mol Biol. 2007;42:67–93. doi: 10.1080/10409230701279118. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The Polyadenylation Factor CPSF-73 Is Involved in Histone-Pre-mRNA Processing. Cell. 2005a;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Wagner EJ, Marzluff WF. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol Cell Biol. 2005b;25:1489–1500. doi: 10.1128/MCB.25.4.1489-1500.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friend K, Lovejoy AF, Steitz JA. U2 snRNP binds intronless histone pre-mRNAs to facilitate U7-snRNP-dependent 3′ end formation. Mol Cell. 2007;28:240–252. doi: 10.1016/j.molcel.2007.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, Van Wijnen AJ, Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:16964–16969. doi: 10.1073/pnas.0809273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM. Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev. 2005;19:2517–2521. doi: 10.1101/gad.1378105. [DOI] [PubMed] [Google Scholar]
- Imai Y, Kimura T, Murakami A, Yajima N, Sakamaki K, Yonehara S. The CED-4-homologous protein FLASH is involved in Fas-mediated activation of caspase-8 during apoptosis. Nature. 1999;398:777–785. doi: 10.1038/19709. [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieghoff E, Milovic-Holm K, Hofmann TG. FLASH meets nuclear bodies: CD95 receptor signals via a nuclear pathway. Cell Cycle. 2007;6:771–775. doi: 10.4161/cc.6.7.4046. [DOI] [PubMed] [Google Scholar]
- Lanzotti DJ, Kupsco JM, Yang XC, Dominski Z, Marzluff WF, Duronio RJ. Drosophila stem-loop binding protein intracellular localization is mediated by phosphorylation and is required for cell cycle-regulated histone mRNA expression. Mol Biol Cell. 2004;15:1112–1123. doi: 10.1091/mbc.E03-09-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma TL, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220NPAT by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes and Development. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature. 2006;444:953–956. doi: 10.1038/nature05363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milovic-Holm K, Krieghoff E, Jensen K, Will H, Hofmann TG. FLASH links the CD95 signaling pathway to the cell nucleus and nuclear bodies. EMBO J. 2007;26:391–401. doi: 10.1038/sj.emboj.7601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell. 2007;26:349–365. doi: 10.1016/j.molcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- Schumperli D, Pillai RS. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell. 2007;28:692–699. doi: 10.1016/j.molcel.2007.10.009. [DOI] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Torres MP, Marzluff WF, Dominski Z. Three proteins of the U7-specific Sm ring function as the molecular ruler to determine the site of 3′-end processing in mammalian histone pre-mRNA. Mol Cell Biol. 2009;29:4045–4056. doi: 10.1128/MCB.00296-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao JY, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes and Development. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.