Abstract
To identify the vectors of bluetongue virus (BTV) in Germany, we monitored Culicoides spp. biting midges during April 2007–May 2008. Molecular characterization of batches of midges that tested positive for BTV suggests C. obsoletus sensu stricto as a relevant vector of bluetongue disease in central Europe.
Keywords: Culicoides, bluetongue disease, bluetongue virus, monitoring, epidemiology, RT-PCR, viruses, Germany, dispatch
Bluetongue disease (BT), discovered north of the Alps in Europe in August 2006 (1–5), causes massive losses of farm ruminants, particularly sheep. Epidemic BT has been caused by serotype 8 of bluetongue virus (BTV-8). The virus overwintered and spread over a large area in 2007 (5,6). Culicoides spp. biting midges can transmit BT. In the Mediterranean region, BT is mainly transmitted by C. imicola midges, a species that has so far not been detected north of the Alps (7,8). We aimed to determine the abundance of hematophagous Culicoides spp. biting midges and to identify putative vectors of BTV in Germany.
The Study
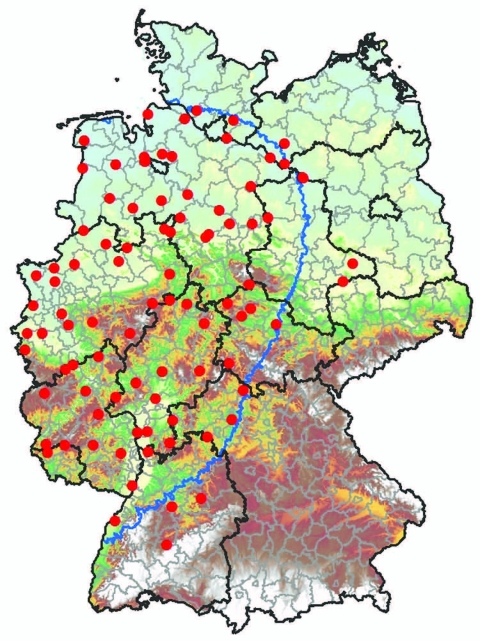
Biting midges were caught from April 2007 through May 2008 by using 89 black light traps (BG-Sentinel Trap; Biogents, Regensburg, Germany) distributed mostly in the German BT restriction zone of January 2007 (Figure 1). Most (n = 85) traps were placed in the vicinity of cow sheds, either adjacent to barns or in their entrance area; 4 additional traps were placed in pastures if the cattle were kept there day and night.
Figure 1.
Culicoides spp. monitoring area, 150-km zone restricted because of the occurrence of bluetongue disease in Germany as of January 2007 (blue border), and geographic positions of 89 black light traps (red dots).
Biting midges were caught during 7 consecutive nights in the first week of each month. We preserved them in 70% ethanol and sorted them under the dissection microscope according to wing patterns (9,10) into batches of <50 parous, nulliparous, or blood-fed female or male insects of the C. obsoletus group (including C. dewulfi), the C. pulicaris group, or other Culicoides spp. biting midges.
Batches of female biting midges were tested for BTV by real-time reverse transcription–PCR (rRT-PCR). Each batch was homogenized in 400 μL lysis buffer (NucleoSpin 96 Virus kit; Macherey-Nagel, Düren, Germany) with a 5-mm steel bead by using a TissueLyser instrument (QIAGEN, Hilden, Germany) for 2 min at 30 Hz. After short centrifugation at 12,000 × g, nucleic acids were extracted from 200 µL homogenate (NucleoSpin 96 Virus kit) on a Tecan Freedom EVO automatic platform (Tecan Deutschland GmbH, Crailsheim, Germany). RNA was analyzed by using the iScript One-Step RT-PCR Kit for Probes (Bio-Rad, Munich, Germany) in a duplex rRT-PCR (pan-BTV-duplex rRT-PCR) combining a BTV rRT-PCR that detects all known serotypes (11) with a PCR that detects all members of the genus Culicoides (pan-Culicoides assay) as an internal control for RNA extraction and amplification. For the pan-Culicoides assay, primers and a probe were selected from aligned sequences of the rDNA internal transcribed spacer 1 and 2 (Pan-Culi-ITS1+2-597F [5′-CAG GAC ACA CGA TCA TTG ACA-3′], Pan-Culi-ITS1+2-976R [5′-CAC ATG AGY TGA GGT CGT CAT-3′], Pan-Culi-ITS1+2-623HEX [5′-HEX-AAC GCA TAT TGC ACC CCA TGC GA-BHQ1-3′]). To confirm positive pan-BTV-duplex rRT-PCR results, we extracted RNA from the remaining homogenate of the batch and subjected it to BTV-8- rRT-PCR (5). Batches were considered BTV positive if results of both assays were positive. Batches of the C. obsoletus and C. pulicaris groups with high viral loads were further analyzed for Culicoides spp. by amplification of the mitochondrial cytochrome oxidase subunit I (12).
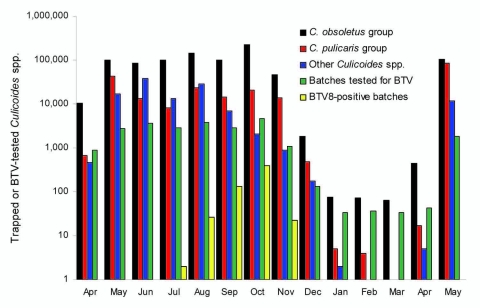
The overall number of biting midges caught started at a moderate level (11,577) in April 2007, peaked in October (246,882), decreased to low levels during December 2007–March 2008, and started to rise again (462) in April 2008 (Figure 2). Small numbers (66–81) of Culicoides spp. midges also were detected in some traps during January–March 2008. Members of the C. obsoletus group (including C. dewulfi) were most frequently trapped, followed by the C. pulicaris group. Biting midges of the C. pulicaris group were more often collected during spring and summer 2007 in discrete locations.
Figure 2.
Monthly catches of midges of the Culicoides obsoletus group (black), C. pulicaris group (red), and other Culicoides spp. (blue) captured with 89 black light traps in Germany during 7 consecutive nights in the first week of each month during the study period (April 2007–May 2008). Batches consisting of <50 female biting midges were tested for bluetongue virus (BTV) by real-time reverse transcription–PCR. The total number of batches (green) and the number of batches positive for BTV (gold) are shown.
Of 24,513 batches analyzed by rRT-PCR, 16,206 (66.1%) batches belonged to the C. obsoletus group, 5,796 (23.6%), to the C. pulicaris group, and 2,511 (10.2%) to other Culicoides spp. A total of 585 (2.4%) batches were positive for BTV by rRT-PCR, 562 (96.1%) of which belonged to the C. obsoletus group, 16 (2.7%), to the C. pulicaris group, and 6 (1.0%) to other Culicoides spp.; 1 was identified as C. achrayi. The number of positive pools varied considerably by month (Figure 2). All batches that were positive in the pan-BTV-rRT-PCR analysis with a cycle threshold (Ct) value <37 (n = 464) were confirmed as BTV positive. BTV-infected biting midges (C. pulicaris group) were first detected in June 2007, a few weeks after the first new infection of the year with BTV-8 had been discovered (5). The number of BTV-positive Culicoides batches peaked (n = 401) in October 2007, which coincided with the peak of midge abundance (Figure 2). During December 2007–May 2008, no BTV-positive batches were detected.
A total of 540 batches of biting midges carried a low or medium (Ct values >30–40), 38 a high (Ct values 25–30), and 7 a very high (Ct values <25) BTV genome load. Batches with a high virus genome load showed Ct values similar to that of highly positive, undiluted blood samples from cattle or sheep. Although ≈70–100 µL of cattle or sheep blood are used for the BTV genome detection, <1 µL blood remains in a biting midge after a blood meal. The uptake of highly BTV-positive blood can therefore only lead to a Ct value increased at least by 6 or 7 when the biting midge is tested by rRT PCR for BTV. Our findings provide strong evidence for virus replication in the biting midges in the highly positive pools.
All batches with very high and 36 batches with a high virus genome load consisted of midges of the C. obsoletus group. Only 2 batches with a high viral load belonged to the C. pulicaris group. These data clearly support the previously suggested role of species of the C. obsoletus group as competent vectors for BTV (13,14).
The species composition of batches of the C. obsoletus and C. pulicaris groups with a high viral genome load was further determined by PCRs of the mitochondrial cytochrome oxidase subunit I (10). Although our analyses showed that most batches consisted of several species (Table 1), C. obsoletus sensu stricto was identified in all investigated batches of the C. obsoletus group (Table 1). Furthermore, 18 BTV-positive batches morphologically classified as C. obsoletus group consisted exclusively of C. obsoletus sensu stricto. These findings indicate that C. obsoletus sensu stricto is involved in the transmission of BTV in Germany, but a role for other members of the C. obsoletus group in the transmission of BTV cannot be ruled out. In contrast, the characterization of the pools of the C. pulicaris group yielded inconclusive results (Table 2). More than 1 species-specific fragment was amplified in all tested pools, but C. punctatus could be identified in all investigated pools.
Table 1. Genetic characterization of batches of midges of the Culicoides obsoletus group, Germany, April 2007–May 2008*.
| Batch no. | rRT-PCR, Ct | C. obsoletus sensu stricto | C. scoticus | C. chiopterus | C. dewulfi |
|---|---|---|---|---|---|
| 270/38F | 20.10 | +++ | – | +++ | +++ |
| 276/2T | 20.74 | +++ | – | +++ | +++ |
| 306/12FN | 21.33 | +++ | – | +++ | +++ |
| 304/34B | 21.60 | +++ | – | +++ | – |
| 276/2O | 23.40 | +++ | – | +++ | +++ |
| 296/52Q | 23.87 | +++ | – | +++ | +++ |
| 296/44G | 24.00 | +++ | – | +++ | +++ |
| 296/22R | 25.90 | +++ | – | +++ | +++ |
| 296/52AL | 27.26 | +++ | – | +++ | +++ |
| 306/1X | 27.30 | +++ | – | – | – |
| 306/1A | 27.60 | +++ | – | – | – |
| 306/1E | 27.70 | +++ | – | – | – |
| 306/1Z | 27.70 | +++ | – | – | – |
| 270/45B | 27.76 | +++ | – | – | +++ |
| 306/1L | 27.90 | +++ | – | – | – |
| 306/1T | 27.90 | +++ | – | – | – |
| 263/46B | 28.00 | +++ | – | ++ | +++ |
| 306/1H | 28.00 | +++ | – | – | – |
| 306/1AA | 28.00 | +++ | – | – | – |
| 306/1S | 28.10 | +++ | – | – | – |
| 306/1AC | 28.10 | +++ | – | – | – |
| 18/156B | 28.18 | +++ | – | + | +++ |
| 346/29C | 28.19 | +++ | – | – | + |
| 306/1R | 28.30 | +++ | – | – | – |
| 306/1V | 28.30 | +++ | – | – | +++ |
| 306/1N | 28.40 | +++ | ++ | – | – |
| 276/10S | 28.41 | +++ | – | +++ | +++ |
| 306/1I | 28.50 | +++ | – | – | – |
| 306/1M | 28.70 | +++ | + | +++ | – |
| 263/46A | 28.80 | +++ | – | +++ | – |
| 296/51AH | 28.82 | +++ | ++ | +++ | +++ |
| 306/1D | 28.90 | +++ | – | – | – |
| 171/13E | 29.00 | +++ | – | – | +++ |
| 306/1C | 29.00 | +++ | +++ | – | – |
| 304/1H | 29.10 | +++ | +++ | +++ | |
| 306/1B | 29.20 | +++ | – | – | – |
| 306/1F | 29.20 | +++ | – | – | – |
| 306/1U | 29.20 | +++ | – | – | – |
| 336/24B | 29.21 | +++ | – | +++ | – |
| 306/12LR | 29.26 | +++ | – | +++ | +++ |
| 306/1K | 29.30 | +++ | – | – | – |
| 306/1J | 29.40 | +++ | – | – | – |
| 270/55I | 29.47 | +++ | – | +++ | +++ |
*rRT-PCR, real-time reverse transcription–PCR; Ct, cycle threshold; –, no visible band; +, faint; ++, distinct; +++, strong band after agarose gel electrophoresis of PCR products and staining with ethidium bromide.
Table 2. Genetic characterization of batches of midges of the Culicoides pulicaris group, Germany, April 2007–May 2008*.
| Batch no. | rRT-PCR, Ct | C. pulicaris s.s. | C. punctatus | C. impunctatus | C. grisescens | C. newsteadi |
|---|---|---|---|---|---|---|
| 263/49 A | 28.57 | – | ++ | + | + | – |
| 263/49 B | 29.46 | +++ | +++ | ++ | + | + |
| 276/46B | 31.37 | – | +++ | – | + | +++ |
| 276/62A | 34.43 | +++ | +++ | – | – | – |
| 292/4A | 33.97 | +++ | +++ | – | – | – |
| 304/63A | 32.90 | +++ | +++ | + | – | ++ |
*rRT-PCR, real-time reverse transcription–PCR; Ct, cycle threshold; –, no visible band; +, faint; ++, distinct; +++, strong band after agarose gel electrophoresis of PCR products and staining with ethidium bromide.
Conclusions
Our study yielded no evidence that C. imicola midges occurred in the study area in Germany. Members of the C. obsoletus group were detected in the entire monitoring area in high abundances and frequently contained BTV genome. Because of the detection of BTV in a high number of batches, which consisted of C. obsoletus sensu stricto, this species must be assumed to play a major role as a vector of BTV in Germany.
Acknowledgments
We thank Stefanie Bartsch, René Focke, Thomas Hörbrand, Sven Klimpel, Miriam Lang, Kathrin Lehmann, Christian Karl Meiser, Anja Stephan, Esther Timmermann, Bettina Vorsprach, Volker Walldorf, and Denis Wolf for their contributions. We also thank Petra Kranz for technical assistance in managing the midge database; Christoph Staubach for scientific advice; and Karin Lissek, Christian Korthase, and Katja Wittig for excellent technical assistance in the German National Reference Laboratory for Bluetongue Disease.
This study was supported by the German Federal Ministry of Food, Agriculture and Consumer Protection.
Biography
Dr Hoffmann is a veterinarian and senior scientist at the Institute of Diagnostic Virology, Friedrich-Loeffler-Institute in Greifswald-Insel Riems and head of the German National Reference Laboratory for BTV. His research interests focus on exotic animal diseases, diagnostics, pathogenesis studies, and development of new methods for molecular diagnostics.
Footnotes
Suggested citation for this article: Hoffmann B, Bauer B, Bauer C, Bätza H-J, Beer M, Clausen P-H, et al. Monitoring of putative vectors of bluetongue virus serotype 8, Germany. Emerg Infect Dis [serial on the Internet]. 2009 Sep [date cited]. Available from http://www.cdc.gov/EID/content/15/9/1481.htm
References
- 1.Toussaint JF, Vandenbussche F, Mast J, De Meester L, Goris N, Van Dessel W, et al. Bluetongue in northern Europe. Vet Rec. 2006;159:327. [DOI] [PubMed] [Google Scholar]
- 2.Elbers A, Backx A, van der Spek A, Ekker M, Leijs P, Steijn K, et al. Epidemiology of bluetongue virus serotype 8 outbreaks in the Netherlands in 2006. Tijdschr Diergeneeskd. 2008;133:222–9. [PubMed] [Google Scholar]
- 3.Toussaint JF, Sailleau C, Mast J, Houdart P, Czaplicki G, Demeestere L, et al. Bluetongue in Belgium, 2006. Emerg Infect Dis. 2007;13:614–6. 10.3201/eid1304.061136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saegerman C, Berkvens D, Mellor PS. Bluetongue epidemiology in the European Union. Emerg Infect Dis. 2008;14:539–44. 10.3201/eid1404.071441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conraths FJ, Gethmann JM, Staubach C, Mettenleiter TC, Beer M, Hoffmann B. Epidemiology of bluetongue virus serotype 8, Germany. Emerg Infect Dis. 2009;15:433–5. 10.3201/eid1503.081210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffmann B, Sasserath M, Thalheim S, Bunzenthal C, Strebelow G, Beer M. Bluetongue virus serotype 8 reemergence in Germany, 2007 and 2008. Emerg Infect Dis. 2008;14:1421–3. 10.3201/eid1409.080417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Purse BV, Mellor PS, Rogers DJ, Samuel AR, Mertens PPC, Baylis M. Climate change and the recent emergence of bluetongue in Europe. Nat Rev Microbiol. 2005;3:171–81. 10.1038/nrmicro1090 [DOI] [PubMed] [Google Scholar]
- 8.Meiswinkel R, Baldet T, de Deken R, Takken W, Delécolle JC, Mellor PS. The 2006 outbreak of bluetongue in northern Europe—the entomological perspective. Prev Vet Med. 2008;87:55–63. 10.1016/j.prevetmed.2008.06.005 [DOI] [PubMed] [Google Scholar]
- 9.Delecolle J-C. Nouvelle contribution à l’étude systématique et iconographique des espèces du genre Culicoides (Diptera:Ceratopogonidae) du Nord-Est de la France. Strasbourg (France): Université Louis Pasteur de Strasbourg; 1985. [Google Scholar]
- 10.Glukhova VM. Blood-sucking midges of the genera Culicoides and Forcipomyia (Ceratopogonidae) [in Russian]. Leningrad (Russia): Nauka; 1989. [Google Scholar]
- 11.Toussaint JF, Sailleau C, Breard E, Zientara S, De Clercq K. Bluetongue virus detection by two real-time RT-qPCRs targeting two different genomic segments. J Virol Methods. 2007;140:115–23. 10.1016/j.jviromet.2006.11.007 [DOI] [PubMed] [Google Scholar]
- 12.Nolan DV, Carpenter S, Barber J, Mellor PS, Dallas JF, Mordue Luntz AJ, et al. Rapid diagnostic PCR assays for members of the Culicoides obsoletus and Culicoides pulicaris species complexes, implicated vectors of bluetongue virus in Europe. Vet Microbiol. 2007;124:82–94. 10.1016/j.vetmic.2007.03.019 [DOI] [PubMed] [Google Scholar]
- 13.Mehlhorn H, Walldorf V, Klimpel S, Jahn B, Jaeger F, Eschweiler J, et al. First occurrence of Culicoides obsoletus–transmitted bluetongue virus epidemic in central Europe. [erratum in Parasitol Res 2007,101:833–4]. Parasitol Res. 2007;101:219–28. 10.1007/s00436-007-0519-6 [DOI] [PubMed] [Google Scholar]
- 14.Meiswinkel R, van Rijn P, Leijs P, Goffredo M. Potential new Culicoides vector in northern Europe. Vet Rec. 2007;161:564–5. [DOI] [PubMed] [Google Scholar]