Abstract
The latest development of protein engineering allows the production of proteins having desired properties and large potential markets, but the clinical advances of therapeutical proteins are still limited by their fragility. Nanotechnology could provide optimal vectors able to protect from degradation therapeutical biomolecules such as proteins, enzymes or specific polypeptides. On the other hand, some proteins can be also used as active ligands to help nanoparticles loaded with chemotherapeutic or other drugs to reach particular sites in the body. The aim of this review is to provide an overall picture of the general aspects of the most successful approaches used to combine proteins with nanosystems. This combination is mainly achieved by absorption, bioconjugation and encapsulation. Interactions of nanoparticles with biomolecules and caveats related to protein denaturation are also pointed out. A clear understanding of nanoparticle-protein interactions could make possible the design of precise and versatile hybrid nanosystems. This could further allow control of their pharmacokinetics as well as activity, and safety.
Keywords: nanoparticles, drug delivery, proteins, polypeptides, absorption, bioconjugation, encapsulation
Introduction and background
Nanotechnology has the potential to create new materials and devices with wide-ranging applications in medicine,1–3 agriculture,4 and energy or electronic production.5,6
The size-dependent optical, electrical, and magnetic properties of nanoparticles make nanotechnology a promising candidate for bioapplications such as in vivo imaging, sensing, catalysis, therapeutics, and cell targeting.7–9
Based on different approaches, physicians, physicists, chemists, biologists as well as bioengineers share a common interest to treat severe diseases through nanotechnology. Theoretically, nanoparticles can be tailored to reach the right target at the right time. Pathogenic agents such as viruses or bacteria, and cancer cells could be precisely targeted and affected without disturbing healthy tissues. This crucial task has been one of the highest priorities for the past 10 years.
Among several medical applications, nanoparticles could be largely employed as carriers of therapeutical biomolecules.10 The combination of nanoparticles with biomolecules such as proteins or specific polypeptides offers opportunities for the design of very precise and versatile hybrid systems mostly useful in helping to fight cancer and immunological diseases.11–13
There are more than 50,000 different proteins in the human body.14 Proteins are present in complex biological processes such as muscle contraction, immune protection and transmission of nerve impulses. All enzymes and most hormones are proteins; hence, proteins are vital sources for the body’s metabolism and their lack can result in several diseases (eg, lack of insulin in type 1 diabetes).
The latest development of protein engineering allows the production of proteins having desired properties and great potential market;15 however, protein fragility is one of the major drawbacks for their utilization. Consequently, the discovery and development of new therapeutic proteins have also created new opportunities for drug-delivery systems involving the design of appropriate nanocarriers such as liposomes, micro-, and nanoparticles.16–19
The oral route is a comfortable way for drug administration especially when repeated or routine dosing is necessary.20 Nevertheless, the development of oral carriers for many proteins remains a challenge due to the fact that bioavailability of these molecules is limited.21 Indeed, most polypeptides and proteins are quickly degraded in the gastrointestinal (GI) tract by proteolytic enzymes.22,23 Moreover, the intestinal epithelium is a major barrier to the absorption of hydrophilic drugs that cannot easily diffuse across the cells through the lipid-bilayer cell membranes.
Numerous investigations have shown that nanocarriers can improve the stability of therapeutic agents against enzymatic degradation and achieve desired therapeutic levels in target tissues for the required duration. Nanoparticle drug-delivery systems (nano-DDS) could permit an optimal pharmacokinetic profile and meet specific needs. For example, nanoparticles as oral protein carriers could protect the active ingredient in the GI tract and/or prolong the residence time of its contents on the mucous membrane. After administration, nano-DDS can be taken up and transported across the intestinal mucosa by enterocytes or M cells in the Peyer’s patches because of their small size.24
Several articles and reviews on the use of nanoparticles or microparticles for oral drug delivery are dedicated to insulin.25–28 In 1980, Couvreur and colleagues performed the first study on hypoglycemic effects after oral and parenteral administration of insulin-loaded nanoparticles to diabetic rats.26
Proteins are also difficult to be delivered via topical or transdermal routes and therefore their parenteral administration is still largely applied.
Besides the general complications of the parenteral route (such as local infections, thrombophlebitis, rarely tissue necrosis), small proteins (<30 kD) are quickly filtered out by the kidneys. Without an appropriate drug carrier, proteins can also cause unwanted allergic reactions, can be targeted by the immune system and be rapidly degraded.
For example, rapid clearance from the circulation can be an explanation of the modest in vivo antitumor effects of the antiangiogenic RGD (Arg–Gly–Asp) peptides.29
Bone morphogenetic proteins (BMPs) induce bone formation after implantation; their orthopedic application in repair of bone fractures and defects is focused in local device and spinal fusion procedures. The problem of BMP is its rapid diffusion from the administration site when applied without a carrier. Currently, one of the most effective and biocompatible carriers for BMP delivery is the type I bovine absorbable collagen sponge (ACS). However the BMP release rate is difficult to control and to maintain constant for long term because of a high initial burst release of this device. The use of new nanotechnologies could maintain BMPs at the treatment site preventing extraneous bond formation and optimizing the drug release.30
Synthetic antigenic peptides are specific sections or a variant sequence of viral/bacterial proteins able to induce an immune response in the host. These small peptides are very useful in the vaccine development compared to the use of the whole protein/antigen. To date, several antigenic peptides have been identified but delivery problems still limit their application. Even in this case, the design of an effective delivery system is an important challenge in nanotechnology field.
An efficient protein carrier should solve different problems allowing the access to the target sites, at the right time and for the proper duration. In order to choose the best nanosystem, five factors must be considered: nature of the protein, route of administration, pattern of drug release, method of delivery and formulation.31,32
Proteins such as albumin, antibody, growth factors, transferrin, cytokines and low-density lipoprotein can be also used as active ligands to help nanoparticles loaded with chemotherapeutic or other drugs to reach particular sites in the body.33–35 Abraxane® (Abraxis Bioscience, Los Angeles, CA; AstraZeneca, Wilmington, DE), albumin–bound nanoparticle of paclitaxel, is an example of US Food and Drug Administration (FDA)-approved protein-based active ligand for the treatment of metastatic breast cancer.35
Monoclonal antibodies (mAb) has been widely used as bioprobes in diagnostics as well as delivery drug to specific tumors.36,37 OX26 mAb can help nanoparticles to cross the blood–brain barrier and diffuse in the brain tissue in order to transport drugs (eg, the anticaptase peptide, Z-DEVD-FMK) for the treatment of neurological and psychiatric disorders.38 Nanoparticles can be also coated with mAb for cell surface antigen and used as a bait for detection or isolation of various kind of cells including lymphocyte and tumor cells.39,40
Despite many potential applications, the interaction of nanoparticles with biomolecules and living systems is still not fully understood.41–50 Continuous study on this subject contributes to the current knowledge and stimulates the development of novel therapies such as nonviral vectors for gene therapies or as precise anticancer molecules.37,51–53 Furthermore, by clarifying these aspects, specific protein-based nanovectors with optimized functions could be developed. This review aims to provide an overall picture on current progress and general aspect of the most successful approaches used to combine proteins with nanosystems. This combination is mainly achieved by absorption, bioconjugation or strong binding via avidin–biotin technology and encapsulation. These methods and the correlated problems of protein denaturation are discussed in turn in this review.
Nanoparticle–protein absorption, bioconjugation, and encapsulation
Absorption of proteins on nanoparticles surface
The interaction between biological and synthetic materials impacts on a vast range of medical issues from implants to pharmacokinetic aspects. The study of the materials bio-compatibility starts, therefore, with the analysis of protein absorption on surfaces.54 Synthetic materials for biomedical applications are immediately covered by proteins when put in contact with a biological environment.55,56 After protein binding, nanoparticles are quickly cleared by the mononuclear phagocytic system (MPS), also known as the reticuloendothelial system (RES).57,58 These macrophages, which are typically Kupffer cells of the liver, cannot directly identify the nanoparticles themselves, but rather recognize specific opsonin proteins bound to the surface of the particles.59
The interaction between proteins and nanoparticles surface leads to the formation of proteins “corona” around nanoparticles that largely defines their biological identity as well their potential toxicity.49,50,58,60–64 Recently, Lynch and Dawson postulated the importance of the “protein corona” as the vehicle and the biological identity of a nanoparticle for its transport through cell membranes.60
The nanoparticle surface is immediately occupied by proteins with high concentrations and high association rate constants and successively by proteins having lower concentrations but a higher affinity.47 Competitive absorption of proteins is influenced by several factors such as electrostatic interactions, protein stability, and kinetic parameters.65
As the protein corona could affect the nanoparticle behavior, including its biological effect, the nanoparticle could also have an effect on the protein behavior. Some nanoparticles seem able to promote the protein assembly into amyloid fibrils in vitro by assisting the nucleation process.66 Bellezza and colleagues found that nanoparticles affect the morphology of the myoglobin absorbed onto phosphate-grafted zirconia nanoparticles, inducing prefibrillar-like aggregates.67 This phenomenon could have important implications for medical application of nanoparticles because the self-assembly of a variety of proteins and peptides is known to be the cause of human amyloid diseases where fibrous protein aggregates are formed, resulting in amyloid plaque deposition in the extra-cellular tissues.68–74 Moreover, fibrillar structure seems to be related to heavy human disorders such as Alzheimer’s disease, Parkinson’s disease, and spongiform encephalopathies.
However this action seems strictly related to the type of nanosystems chosen. For instance, there are nanosystems such as C60 hydrated fullerenes that can relax fibrillar structures.60,68 Certainly, the control of the protein absorption on nanoparticle surfaces is an important issue to control their fate in biological systems.75
In order to prevent or control the opsonization, several methods of disguising nanoparticles have been developed. In these methods, generally, nanoparticles are coated with biocompatible polymers that have the double function of preventing their aggregation and retarding the protein absorption.57,76,77
A common strategy to improve blood compatibility and to increase the blood circulation half-life of the nanoparticles is the construction of a protein-coated surface resistant to the absorption of the other opsonines.78 A thin layer of protein appears to minimize adhesion and aggregation of nanoparticles, avoiding subsequent macrophage recognition or, in the worst case, a thrombus formation. Moreover, it is possible to properly tune the cells uptake of the nanoparticles using specific proteins.35
Proteins are mainly amphiphatic molecules that typically adhere to the surface of a biomaterial in a nonspecific way. In various cases, this nonspecific adhesion is sufficient to artificially immobilize proteins on the nanoparticles surface, and no surface modification is necessary.
Despite of the large number of studies, the absorption of a protein on whatever the solid surface is still a complex and not well understood process.60,79–85 In the case of nanoparticles, size and radius of curvature become significant when compared to the protein size resulting in new interactions not shown with the bulk materials.47
The high hydrophobicity of many proteins seems to play an important role in their absorption on the nanoparticles surface.86 Several models of protein absorption on surfaces identify two main steps in the process. The first step could involve the arrival of the protein at the interface, through a diffusion process following the Brownian law of motion, and its further collision with the solid surface. Depending on the balance of the energetic interaction, proteins can remain on the solid surface or return to solution. If the protein has been absorbed, the second step could lead to conformational changes (because of van der Waals interactions), surface charge, protein dipole moment, and protein size or solution ionic strength.84,85,87–90 This second step often involves irreversible changes in the protein structure up to denaturation.91–93
Proteins can be divided in two groups: hard and soft proteins. The first group includes proteins with high internal stability, while proteins in the second group have a low internal stability. Soft proteins seem to be able to change their conformation better than the hard ones. This characteristic results in a gain in conformational entropy when absorbed on solid surfaces, improving the efficacy of the absorption process when compared to the hard proteins. On the other hand, it seems that some degree of denaturation upon absorption is more probable for soft proteins than for the hard ones, especially on hydrophobic surfaces.84,94–97
During the artificial absorption of protein to nanoparticles surface, the use of a large excess of the target material could allow the retention of sufficient biological activity and native epitopes, even if some proteins are denatured. However, problems associated with denaturation of the protein over time, or its exchange with other proteins in solution, could make this strategy satisfactory only for short-term uses.
The success of an absorption strategy to deliver drug or therapeutical proteins using protein-based nanoparticles as a carrier can be influenced by several factors such as the type of nanoparticles, delivery route and the nature of proteins to be absorbed. For this reason, nanoparticle–protein affinity needs to be intensely examined case-by-case.
The knowledge of how the protein-based nanoparticles interact with other proteins present in the blood is fundamental for the understanding of their biological and toxicological properties.77 Many methods based on established techniques could be applied such as size-exclusion chromatography, isothermal titration calorimetry, surface plasmon resonance, atomic force microscopy, differential scanning calorimeter, and circular dichroisim (CD) spectroscopy.47,67
Even if several existing characterization methods for measuring the nature and the amount of absorbed protein on solid surfaces could be applied to nanoparticle systems,96 the development of new physical and biophysical methods may be necessary to fully understand the relationship between proteins and nanomaterials.
Bioconjugation of proteins on nanoparticle surfaces
Conjugation of biomolecules on nanoparticle surfaces has attracted widespread interest in biotechnology and medicine.7,98–100 The conjugation of specific proteins with nanoparticles has introduced a new advancement in molecular and cellular biology which has further led to a vast improvement of in vivo gene delivery, clinical diagnosis, medical/cancer imaging, receptor-targeted delivery.40,101–105
A preferred method used in many areas of biochemistry to couple specific protein to solid surface is the bioconjugation by covalent binding. While protein absorption on solid surfaces such as nanoparticles can be reversible depending on pH, salt concentration, temperature or other environment physicochemical characteristics, protein covalent bounds are highly stable. To fulfill the purpose of stable covalent binding, a large number of reactions have been proposed and many protein modifications using new techniques have been developed.7,106–111
The choice of the bioconjugation procedure depends strictly on physicochemical and biochemical properties of nanomaterials and proteins. Protein made by various side chains and residues can interact by multiple coating ligands with the same nanoparticles or even with more nanoparticles. Moreover, nanoparticles can be more or less polydispersed and have different physicochemical surface properties such as area, porosity, and charge. These aspects are very important since the hydrophobicity, charge and site affinity could affect the interaction and thus jeopardize the stability of final covalent-coupled products.
The most popular approach for coupling covalently nanoparticle to protein is based on the existence on proteins of specific and reactive functional groups such as amino–NH2 (lysine), carboxylic acid–COOH (aspartic, glutamic), hydroxyl–OH (serine, tyrosine) and –SH (cysteine).112
Proteins can be chemically coupled to different kinds of nanoparticles using established reagents such bifunctional cross-linker molecules. In this case, nanoparticles need to be functionalized with functional groups such as carboxylic acid, hydroxyl, sulfhydryl and amino groups.
Proteins, including antibodies, generally have several primary amines in the side chain of lysine residues and the N-terminus of each polypeptide that are available as targets for N-hydroxysuccinimide-ester and carbodiimide reagents. Cysteine residues on proteins can react with maleimides and iodoacetamides reagents to give thioether-coupled products.113 These reagents react rapidly at physiological pH and can be usually coupled with thiol groups selectively in the presence of amine groups. Maleimides and iodoacetamides have the same application but the first reagent seems to have better selectivity than the second one, not apparently reacting with histidine or methionine.
Cross-linking reagents contain reactive ends to specific functional groups (such as primary amines, sulfhydryls) on proteins or other molecules. They can be divided into homobifunctional (same reactive groups) and heterobifunctional (different reactive groups) which chemical cross-links may or may not be reversed.114 Homobifunctional cross-linkers have a disadvantage of potentially connecting two neighboring groups, either on the nanoparticle surface or on the protein inducing undesired cross-linking. Heterobifunctional crosslinkers allow sequential conjugations, minimizing polymerization. For example, sulfosuccinimidyl-4-(N-maleimidomethyl)-cyclohexane-1-carboxylate (sulfo-SMCC) can be used to couple thiol-containing biomolecules with amine–coated nanoparticles, or vice versa. Whereas the hetero-bifunctional cross-linker 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) is commonly used to link –NH2 and –COOH groups (Table 1).114–118
Table 1.
The most popular cross-linker reagents for coupling protein to nanoparticle based on their respective functions
| Reactive groups | Eg of functional cross-linker | Functional groups on nanoparticles/proteins | Functional groups on proteins/nanoparticles |
|---|---|---|---|
| –NHS ester Maleimide or Iodoacetamides | SIAB, SMCC, SPDP, SPMB, MBS | –NH2 | –SH |
| Carbodiimide | EDC or EDAC + sulfo-NHS stabilizer | –COOH | –NH2 |
| –NHS ester | EGS, DSP, DSS, BS3 | –NH2 | –NH2 |
| Maleimide | BMME | –SH | –SH |
Abbreviations: SIAB, N-succinimidyl(4-iodoacetyl)aminobenzoate; SMCC, succinimidyl-4-(N-maleimidomethyl)cyclohexana-1-carboxylate; MBS, m-maleimidobenzoyl-N-hydroxysuccimide ester; SPDP, succinimidyl 3-(2-pyridyldithio)propionate; SPMB, succinimidyl (4-p-maleimidophenyl)butyrate; EDC, 1-Ethyl-3-[3-dimethylaminopropyl] carbodiimide hydrochloride; EGS, ethylene glycolbis(succinimidylsuccinate); BS3, bis-(sulfosuccinimidyl) suberate; BMME, bis(maleimido methyl) ether; DSS, disuccinimidyl suberate; DSP, dithiobis (succinimidyl propionate).
Many cross-linkers are available in the market and they can be chosen for specific needs (such as chemical specificity, spacer arm length, cleavability). Among several cross-linkers, the zero-length ones such as carbodiimides are widely used allowing covalent bonds between nanoparticles and proteins without insertion of an exogenous spacer. Nevertheless, the direct attachment of a protein to a surface without a spacer can cause steric constraint modifying the protein reactivity compared to the protein in solution. In addition, without a spacer, multiple contacts between protein and nanoparticle surface are more probable favoring total or partial protein denaturation and thus decreasing protein activity.119
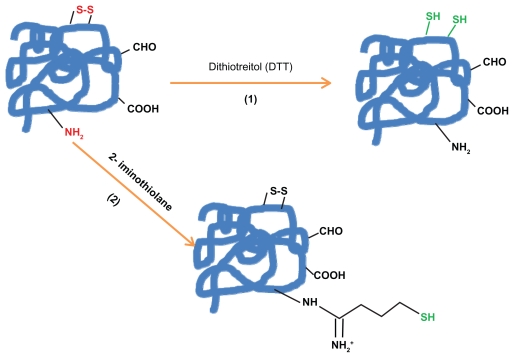
When protein does not have the suitable residue necessary for the specific conjugation, the most common way to get it is the chemical introduction of sulfhydryl groups. This process (Figures 1a and 1b) can be mainly made by the following four methods: 1) reduction of protein disulfide bonds using reductive agents such as dithiotreitol (DTT = Clelands reagent). 2) Coupling of protein primary amino groups with 2-iminothiolane (Trauts reagent). 3) Quenching of reactive protein aldehyde residues with cystaminiumdichloride reagents or 4) coupling of cystaminiumdichloride to carboxyl groups via 1-ethyl-3-(3-dimethyl-aminopropyl)carbodiimide (EDC); both cases followed by the disulfide bonds reduction with DTT as outlined above.112,120–123
Figure 1a.
The introduction of sulfhydryl groups by: 1) the reduction of protein disulfide bonds using reductive agents such as dithiotreitol (DTT = Cleland’s reagent). 2) Coupling protein primary amino groups with 2-iminothiolane (Traut; s reagent).
Figure 1b.
The introduction of sulfhydryl groups by: 3) Quenching of reactive protein aldehyde residues with cystaminiumdichloride reagents or 4) coupling of cystaminiumdichloride to carboxyl groups via 1-ethyl-3-(3-dimethyl-aminopropyl)carbodiimide (EDC); both cases followed by the disulfide bonds reduction with DTT.
The avidin/streptavidin–biotin bound is the strongest noncovalent biological interaction known; for this reason this technology is commonly used in biological labs.124,125
Biotinylated proteins/antibodies/enzymes can be efficiently coupled on amino nanoparticle surfaces by streptavidin-biotin technology accomplished by streptavidin activation through carbodiimide (EDC) chemistry. Biotin binds strongly to this biochemically modified surface in the most specific and sensitive way. Furthermore, streptavidin through carbodiimide (EDC) chemistry can be covalently coupled with different ligands such as mAb and enzymes which make the biotin–streptavidin system widely used in a variety of biotinylated nanoparticles.38,126–128
Proteins having cysteine residues can be directly attached to some metal nanoparticle surfaces such as gold and silver by stable metal–sulfur bonds.129,130 In the other cases, the covalent coupling of proteins on nanoparticle surfaces is always a long experimental procedure.
Covalent bioconjugation procedure can be summarized in: 1) Coating of nanoparticles with the selected active functional groups. 2) Chemical activation of thiol groups on the protein side with specific reductive agents, if necessary. 3) Total removal of the reduction agent in excess; this step can create unplanned reactions and spoil the whole coupling process. 4) Post conjugation procedures such as removal of unbound protein/remnant excess.
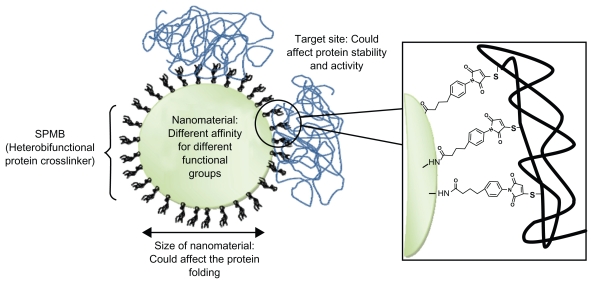
In addition to the disadvantage of the long experimental procedure, covalent bioconjugation can affect the protein structure and function resulting in its partial denaturation (Figure 2). Moreover, modification of enzymes under strong denaturing conditions can result in their complete loss of activity.
Figure 2.
Protein–nanoparticle interactions: main factors that can affect proteins resulting in their denaturation. In this example, proteins are conjugated on amino functionalized nanoparticles using the cross-linker SPMB.
Proteins can be denaturated during manipulations or formulations mainly by two mechanisms: conformational denaturation (eg, reversible unfolding and irreversible aggregation via noncovalent interactions) and chemical denaturation (covalent bonds such as deamidation, hydrolysis, oxidation, β-elimination, incorrect disulfide formation, Maillard reaction, and transamidation).
Even if the first denaturation mechanism can happen during the nanoparticles bioconjugation process, the second one is often necessary to obtain high efficacy of the coupling. For example, the –SH or –S-groups in cysteine –SH or disulfide –S–S– bridges are important in maintaining the conformation of the proteins. As a result, the engineering introduction of sulfhydryl groups in the protein changes its natural disulfide bonds resulting in partial conformational and chemical denaturation.
The DTT reagent, widely used to reduce disulfide bonds in biochemical systems, can alter protein function not only by thiol-disulfide exchanging but also by interacting with protein domains in the absence of cysteine residues.131
While carboxyl groups seem to play an important role in enzymes catalytic activity,132 their modification likely results in a change of protein secondary and tertiary structure.
The ɛ-amino groups of lysine are often specifically targeted because of their high reactivity and their modification seems to have fewer effects on protein properties. Unfortunately, the high abundance of these groups in many proteins can lead to increased heterogeneity and restricted conformational flexibility owing to multipoint attachment on a nanoparticles surface.
It is also possible that other reagents used during the coupling chemical process can contribute to the protein denaturation and to its activity loss. Therefore biological function checking as well as close monitoring of the quality and quantity of conjugated protein are extremely important to be assessed before being used.111
Gold or silver nanoparticles too, due to the similar strength bond between Au/Ag–S and S–S, can potentially break up protein disulfide –S–S– bridges leading to denaturation.
Specific ELISA kits can be used to explore the activity of the proteins coupled to nanosystems. However there are nanoparticles such as quantum dots (QD) that can have an overlap in the absorption spectra and the ELISA essay end product. In this case the proper specific activity of the protein needs to be assessed directly by in vitro testing.133
Protein encapsulation
Therapeutic biomolecules based on peptides, proteins or enzymes can be extremely fragile and easily aggressed by external agent such as proteases. Encapsulation of these fragile drugs in nanocarriers is a possible strategy for preventing their aggression and denaturation. This process can also improve the drug pharmacokinetic pathway and reduce immunological reactions.19
An optimal drug delivery system should be biocompatible, biodegradable and should not cause any immunological adverse reaction in the human body. Among several candidates, liposomes are considered as the most promising vectors for proteins delivery due to their biocompatibility and their capacity to improve the drug pharmacokinetic. Liposomes, also known as lipid-based vesicles, are generally composed of concentric amphiphilic lipids, such as phospholipids, containing a water compartment. These carriers are versatile and their physico–chemical characteristics can be properly tuned.19 Liposomes synthesized from dehydrated–rehydrated vesicles are widely used due to the ease of this preparation process and the low amount of stress applied to the proteins.134
Liposome formulations are most frequently considered for parental administration of the drug, but may also be a potential formulation principle for alternative routes such as topical and nasal administration.
Several liposomes have immunoadjuvant properties and their application in vaccines based on recombinant protein subunits and synthetic–peptide antigens is attractive. The first liposome based vaccine (against hepatitis A) that has been licensed for human use is commercially known as Epaxal Berna® vaccine.135
The main drawback of liposomes is their instability in biological media as well as their sensitivity to many external parameters such as temperature or osmotic pressure. Theoretically, it could be possible to increase their stability following several strategies such as the polymerization of a two–dimensional network in the hydrophobic core of the membrane, coating the liposome with a polyelectrolyte shell or adding surface active polymers to form mixed vesicular structures.136–138 However, poor loading and partial protein/enzyme denaturation during the entrapment process can occur.
Another well established technique to encapsulate biological species such as enzymes, antibodies and other proteins in a functional state is based on the sol–gel chemistry method.139 Silica is indeed considered a very appealing material for drug delivery systems because it is relatively inexpensive, chemically inert, thermally stable, and biocompatible. Amorphous silica, used for decades as a food additive and for specific applications, is generally regarded as safe. Up until now, the FDA has not established if existing silica safety data can be applied to nanoscale forms of the material. In this approach, polypeptides, especially enzymes, could be entrapped inside silica matrix allowing the retention of enzymatic activity.139–141
On the other hand, process difficulties such as uncontrolled release, denaturation and the hardness control of the protein orientation can be found.142,143 The control of the drug release of such silica nanoparticles is the most important and difficult parameter that needs to be properly tuned. The encapsulation efficacy of insoluble protein is greatly different compared to the soluble one and the existence of soluble and insoluble part of polypeptides in the same therapeutic protein subunit complicates the synthesis process. Additionally, in the crowded environment of a silica matrix, the physical and chemical properties of the silica can directly influence protein structure and activity. Furthermore, functional activity of proteins entrapped into the sol-gel matrix needs to be accurately analyzed case-by-case using several techniques such as CD spectropolarimetry.144,145
The drug release and the capability of the carrier to be metabolized can be important factors to be considered when chronic or repeated treatments are necessary. The disadvantage associated with inorganic and synthetic carriers are the poor or slow biodegradability and possible inflammatory responses.146
Biodegradable polymers nanosystems are an attractive alternative to liposomes since they have the advantages of longer circulation in the blood stream and generally higher drug carrying capacity.147 Polymers such as poly(lactic acid) (PLA), poly(lactic-co-glycolic acid) (PLGA) have been extensively investigated for their biocompatibility and potential capability of releasing therapeutically proteins in a controlled way even over a prolonged period of time.148–154 These polymers are degradable by bulk erosion through hydrolysis of the ester bonds. The hydrolysis rate depends on several nanoparticles physicochemical parameters and can be tailored according to the desired release pattern of the protein to be incorporated.
PLA and PLGA are FDA-approved as excipients to achieve sustained release of the active ingredient. However, their application in protein delivery systems is often characterized by low entrapment efficiency, burst release, instability of encapsulated hydrophilic protein and partial protein release.155–158 To improve the performance of these polymer nanoparticles, polysaccharides such as alginate (ALG) and chitosan (CS) could be applied.151,159 CS and its derivatives have been intensively studied as carriers for proteins and drugs. More specifically these nanoparticles can be totally made by CS or used in several copolymer combinations.25,160
Copolymers made by the combination of CS/ALG are able to generate a more “friendly” environment which protects peptides and proteins from stressing conditions and allows their stabilization during encapsulation, storage and release.161–166
Glycol chitosan nanoparticles modified with hydrophobic bile acid analogs self-assemble into polymeric nanoparticles with hydrophilic shells of glycol chitosan and hydrophobic cores of bile acid derivatives have been reported as possible vehicle for RGD (Arg–Gly–Asp) peptide.29,30
Regardless of the nanomaterial chosen for protein encapsulation, an important issue that needs to be considered is the understanding of protein–protein interactions. There are large numbers of transient protein–protein interactions that occur in the cell, which in turn control a large number of cellular processes. These transient interactions of protein complexes can cause several effects such as activation/inactivation of certain proteins, resulting in the formation of a new binding site.167,168 Kinetics properties of enzymes can be also altered by denaturation during the entrapment process allowing potential change of the protein specificity to its substrate.169–171
Gaining a clear picture of these basics knowledge will definitely lead to a change of object design to increase the protein load, to control the protein release and to retain the protein integrity and efficacy.
Conclusion
Although new proteins are available for medical purposes, their administration as therapeutics still remains difficult. Nanosystems seem to be the optimal solution to improve protein bioavailability, biodistribution and safety. Moreover, the combination of nanoparticles with proteins could also be a valid system to achieve the design of efficient nanovectors for drug delivery. Indeed, nanoparticles can be properly tuned for specific applications and could be precisely designed to meet biological needs. However, to completely fulfill this purpose, it is necessary to better clarify the nature of interaction between nanoparticles and biomolecules. The control of the protein denaturation is another important parameter that needs a deeper understanding. Further investigations should help to manage these hybrid nanosystems, opening new therapeutic and diagnostic perspectives as well as new challenges in the near future.
Figure 3.
Entrapment of enzymes using sol-gel chemistry: A schematic overview of the sol-gel process. Several silicate precursors can be used to modify the surface chemistry of the sol-gels such as TMSO, APTES, MTMOS, and ETMOS.
Abbreviations: TMSO, tetramethyl orthosilicate; APTES, 3-aminopropyltriethoxysilane; MTMOS, methyltrimethoxysilane; ETMOS, ethyltrimethoxysilane.
Footnotes
Disclosures
The authors report no conflicts of interest in this work.
References
- 1.Navalakhe RM, Nandedkar TD. Application of nanotechnology in biomedicine. Indian J Exp Biol. 2007;45:160–165. [PubMed] [Google Scholar]
- 2.Engel E, Michiardi A, Navarro M, et al. Nanotechnology in regenerative medicine: the materials side. Trends Biotechnol. 2008;26:39–47. doi: 10.1016/j.tibtech.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Heath JR, Davis ME. Nanotechnology and Cancer. Annu Rev Med. 2008;59:251–265. doi: 10.1146/annurev.med.59.061506.185523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melendi PG, Ferna R, Pacheco RF, et al. Nanoparticles as smart treatment–delivery systems in plants: Assessment of different techniques of microscopy for their visualization in plant tissues. Ann Bot. 2008;101:187–195. doi: 10.1093/aob/mcm283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gidaspow D, Jiradilok V. Nanoparticle gasifier fuel cell for sustainable energy future. J Power Source. 2007;166:400–410. [Google Scholar]
- 6.Chen D, Qiao X, Qiu X, et al. Synthesis and electrical properties of uniform silver nanoparticles for electronic applications. J Mat Sci. 2009;44:1076–1081. [Google Scholar]
- 7.Medintz IL, Uyeda HT, Goldman ER, et al. Quantum dot bioconjugates for imaging, labeling and sensing. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 8.Corot C, Robert P, Idée JM, et al. Recent advances in iron oxide nanocrystal technology for medical imaging. Adv Drug Deliv Rev. 2006;58:1471–1504. doi: 10.1016/j.addr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 9.De la Fuente JM, Berry CC, Riehle MO, et al. Nanoparticle targeting at cells. Langmuir. 2006;22:3286–3293. doi: 10.1021/la053029v. [DOI] [PubMed] [Google Scholar]
- 10.Alonso MJ. Nanoparticulate drug carrier technology. In: Cohen S, Bernstein H, editors. Microparticulate Systems for the Delivery of Proteins and Vaccines. New York, NY: Marcel Dekker; 1996. pp. 203–242. [Google Scholar]
- 11.Drexler KE. Molecular engineering: an approach to the development of general capabilities for molecular manipulation. Proc Natl Acad Sci U S A. 1981;78:5275–5278. doi: 10.1073/pnas.78.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rajagopal K, Schneider JP. Self-assembling peptides and proteins for nanotechnological applications. Curr Opin Struct Biol. 2004;14:480–486. doi: 10.1016/j.sbi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Sinha R, Kim GJ, Nie S, Shin DM. Nanotechnology in cancer therapeutics: bioconjugated nanoparticles for drug delivery. Mol Cancer Ther. 2006;5:1909–1917. doi: 10.1158/1535-7163.MCT-06-0141. [DOI] [PubMed] [Google Scholar]
- 14.Nelson DL, Cox MM. Lehninger Principles of Biochemistry. 4th Edition. New York, NY: W H Freeman; 2004. [Google Scholar]
- 15.Liang FH, Hong MH, Ho RM, et al. Novel method using a temperature– sensitive polymer (methylcellulose) to thermally gel aqueous alginate as a pH-sensitive hydrogel. Biomacromolecules. 2004;5:1917–1925. doi: 10.1021/bm049813w. [DOI] [PubMed] [Google Scholar]
- Tan ML, Choong PFM, Dass CR. Recent developments in liposomes, microparticles and nanoparticles for protein and peptide drug delivery. Peptides. Oct 9; doi: 10.1016/j.peptides.2009.10.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Ferrari M, Downing G. Medical nanotechnology: shortening clinical trials and regulatory pathways. Biodrugs. 2005;19:203–210. doi: 10.2165/00063030-200519040-00001. [DOI] [PubMed] [Google Scholar]
- 18.Almeida AJ, Souto E. Solid lipid nanoparticles as a drug delivery system for peptides and proteins. Adv Drug Delivery Rev. 2007;59:478–490. doi: 10.1016/j.addr.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Martins Sarmento B, Ferreira DC, Souto EB. Lipid-based colloidal carriers for peptide and protein delivery: liposomes versus lipid nanoparticles. Int J Nanomed. 2007;2:595–607. [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, Langer R. Oral particulate delivery: status and future trends. Adv Drug Del. 1998;34:339–350. doi: 10.1016/s0169-409x(98)00047-7. [DOI] [PubMed] [Google Scholar]
- 21.Delie F, Blanco–Príeto MJ. Polymeric particulates to improve oral bioavailability of peptide drugs. Molecules. 2005;10:65–80. doi: 10.3390/10010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borchard G, Luegen HL, de Boer AG, et al. The potential of mucoadhesive polymers in enhancing intestinal peptide drug absorption. III: Effects of chitosan–glutamate and carbomer on epithelial tight junctions in vitro. J Control Release. 1996;39:131–138. [Google Scholar]
- 23.Malik DK, Baboota S, Ahuja A, et al. Recent advances in protein and peptide drug delivery systems. Curr Drug Deliv. 2007;4:141–151. doi: 10.2174/156720107780362339. [DOI] [PubMed] [Google Scholar]
- 24.des Rieux A, Fievez V, Garinot M, et al. Nanoparticles as potential oral delivery systems of proteins and vaccines: A mechanistic approach. J Control Release. 2006;116:1–27. doi: 10.1016/j.jconrel.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Rekha MR, Sharma CP. Synthesis and evaluation of lauryl succinyl chitosan particles towards oral insulin delivery and absorption. J Control Release. 2009;135:144–151. doi: 10.1016/j.jconrel.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Couvreur P, Lenaerts V, Kante B, et al. Oral and parenteral administration of insulin associated to hydrolysable nanoparticles. Acta Pharm Technol. 1980;26:220–222. [Google Scholar]
- 27.Aboubakar M, Couvreur P, Pinto-Alphandary H, et al. Insulin-loaded nanocapsules for oral administration: in vitro and in vivo investigation. Drug Develop Res. 2000;49:109–117. [Google Scholar]
- 28.Carino GP, Jacob JS, Mathiowitz E. Nanospheres based oral insulin delivery. J Control Release. 2000;65:261–269. doi: 10.1016/s0168-3659(99)00247-3. [DOI] [PubMed] [Google Scholar]
- 29.Kim JH, Kim YS, Park K, et al. Self–assembled glycol chitosan nanoparticles for the sustained and prolonged delivery of antiangiogenic small peptide drugs in cancer therapy. Biomaterials. 2008;29:1920–1930. doi: 10.1016/j.biomaterials.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, Doschak MR, Uluda H. Pharmacokinetics and bone formation by BMP-2 entrapped in polyethylenimine–coated albumin nanoparticles. Biomaterials. 2009;30:5143–5155. doi: 10.1016/j.biomaterials.2009.05.060. [DOI] [PubMed] [Google Scholar]
- 31.Sinha VR, Trehan A. Biodegradable microspheres for protein delivery. J Control Release. 2003;90:261–280. doi: 10.1016/s0168-3659(03)00194-9. [DOI] [PubMed] [Google Scholar]
- 32.Frokjaer S, Hovgaard L. Pharmaceutical Formulation Development of Peptides and Proteins. Andover, UK: Taylor and Francis; 2000. [Google Scholar]
- 33.Mo Y, Lim LY. Paclitaxel-loaded PLGA nanoparticles: potentiation of anticancer activity by surface conjugation with wheat germ agglutinin. J Control Release. 2005;108:244–262. doi: 10.1016/j.jconrel.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Acharya S, Dilnawaz F, Sahoo SK. Targeted epidermal growth factor receptor nanoparticle bioconjugates for breast cancer therapy. Biomaterials. 2009;30:5737–5750. doi: 10.1016/j.biomaterials.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 35.Miele E, Spinelli GP, Miele E, et al. Albumin-bound formulation of paclitaxel (Abraxane® ABI–007) in the treatment of breast cancer. Int J Nanomed. 2009;4:99–105. doi: 10.2147/ijn.s3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huh YM. In vivo magnetic resonance detection of cancer by using multifunctional magnetic nanocrystals. J Am Chem Soc. 2005;127:12387–12391. doi: 10.1021/ja052337c. [DOI] [PubMed] [Google Scholar]
- 37.DeNardo SJ, De Nardo GL, Miers LA, et al. Development of tumor targeting bioprobes (In–chimeric L6 mAb nanoparticles) for alternating magnetic field cancer therapy. Clin Cancer Res. 2005;11:7087–7092. doi: 10.1158/1078-0432.CCR-1004-0022. [DOI] [PubMed] [Google Scholar]
- 38.Aktaş Y, Yemisci M, Andrieux K, et al. Development and brain delivery of chitosan–PEG nanoparticles functionalized with the monoclonal antibody OX26. Bioconjug Chem. 2005;16:1503–1511. doi: 10.1021/bc050217o. [DOI] [PubMed] [Google Scholar]
- 39.Zhao X, Hilliard LR, Merchery SJ, et al. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticle. Proc Natl Acad Sci U S A. 2004;101:5027–5032. doi: 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Natarajan A, Xiong CY, Gruattner C, et al. Development of multivalent radioimmunonanoparticles for cancer imaging and therapy. Cancer Biol Ther. 2008;23:82–90. doi: 10.1089/cbr.2007.0410. [DOI] [PubMed] [Google Scholar]
- 41.Bhatia SN, Balis UJ, Yarmush ML, et al. Effect of cell–cell interactions in preservation of cellullar phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–1900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 42.Anderson JM. Biological responses to materials. Annu Rev Mater Res. 2001;31:81–110. [Google Scholar]
- 43.Hench LL, Polak JM. Third-generation biomedical materials. Sci. 2002;295:1014–1017. doi: 10.1126/science.1067404. [DOI] [PubMed] [Google Scholar]
- 44.Allen LT, Fox EJP, Blute I, et al. Interaction of soft condensed Interaction of soft condensed materials with living cells: phenotype/transcriptome correlations for the hydrophobic effect. Proc Natl Acad Sci U S A. 2003;100:6331–6336. doi: 10.1073/pnas.1031426100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heungsoo S, Seongbong J, Mikos AG. Biomimetic materials for tissue engineering. Biomaterials. 2003;24:4353–4364. doi: 10.1016/s0142-9612(03)00339-9. [DOI] [PubMed] [Google Scholar]
- 46.Tsuda Y, Kikuchi A, Yamato M, et al. Control of cell adhesion and detachment using temperature and thermoresponsive copolymer grafted culture surfaces. J Biomed Mater Res. 2004;69A:70–78. doi: 10.1002/jbm.a.20114. [DOI] [PubMed] [Google Scholar]
- 47.Cedervall T, Lynch I, Lindman S, et al. Understanding the nanoparticle–protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci U S A. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311:622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 49.Lynch I, Dawson KA, Linse S. Detecting cryptic epitopes created by nanoparticles. Sci STKE. 2006;327:pe14. doi: 10.1126/stke.3272006pe14. [DOI] [PubMed] [Google Scholar]
- 50.Lynch I, Cedervall T, Lundqvist M, et al. The nanoparticle-protein complex as a biological entity; a complex fluids and surface science challenge for the 21st century. Adv Colloid Interface Sci. 2007:134–135. 167–174. doi: 10.1016/j.cis.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 51.Ahmad A, Evans HM, Ewert K, et al. New multivalent cationic lipids reveal bell curve for transfection efficiency versus membrane charge density: lipid-DNA complexes for gene delivery. J Gene Med. 2005;7:739–748. doi: 10.1002/jgm.717. [DOI] [PubMed] [Google Scholar]
- 52.Hapca AC, Nobs LB, Buchegger F, et al. Differential tumor cell targeting of anti–HER2 (Herceptin®) and anti-CD20 (Mabthera®) coupled nanoparticles. Inter J Pharm. 2007;331:190–196. doi: 10.1016/j.ijpharm.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Kocbek P, Obermajer N, Cegnar M, et al. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J Control Release. 2007;120:18–26. doi: 10.1016/j.jconrel.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 54.Tirrel M, Kokkoli E, Biesalski M. The role of surface science in bioengineered materials. Surf Sci. 2002;500:61–83. [Google Scholar]
- 55.Vroman L, Adams AL. Identification of rapid changes at plasma–solid interfaces. J Biomed Mater Res. 1969;3:43–67. doi: 10.1002/jbm.820030106. [DOI] [PubMed] [Google Scholar]
- 56.Young BR, Lambrecht LK, Mosher DF, et al. Plasma proteins: Their role in initiating platelet and fibrin deposition on biomaterials. In: Cooper SL, Peppas NA, editors. Biomaterials: Interfacial phenomena and applications. Vol. 199. Washington, DC: American Chemical Society; 1982. pp. 317–349. [Google Scholar]
- 57.Gref R, Minamitake Y, Peracchia MT, et al. Biodegradable long– circulating polymeric nanospheres. Science. 1994;263:1600–1603. doi: 10.1126/science.8128245. [DOI] [PubMed] [Google Scholar]
- 58.Owens DE, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int J Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 59.Frank M, Fries L. The role of complement in inflammation and phagocytosis. Immunol Today. 1991;12:322–326. doi: 10.1016/0167-5699(91)90009-I. [DOI] [PubMed] [Google Scholar]
- 60.Lynch I, Dawson KA. Protein–nanoparticle interactions. Nano Today. 2008;3:40–47. [Google Scholar]
- 61.Dutta D, Sundaram SK, Teeguarden JG, et al. Adsorbed proteins influence the biological activity and molecular targeting of nanomaterials. Toxicol Sci. 2007;100:303–315. doi: 10.1093/toxsci/kfm217. [DOI] [PubMed] [Google Scholar]
- 62.Illum L, Hunneybal IM, Davis SS. The effect of hydrophilic coatings on the uptake of colloidal particles by the liver and by peritonealmac-rophages. Int J Pharm. 1986;29:53–65. [Google Scholar]
- 63.Kaul G, Amiji M. Long-circulating poly(ethylene glycol)-modified gelatin nanoparticles for intracellular delivery. Pharm Res. 2002;19:1061–1067. doi: 10.1023/a:1016486910719. [DOI] [PubMed] [Google Scholar]
- 64.Aggarwal P, Hall JB, McLeland CB, et al. Nanoparticle interaction with plasma proteins as it relates to particle biodistribution, biocompatibility and therapeutic efficacy. Adv Drug Del Rev. 2009;61:428–437. doi: 10.1016/j.addr.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arai T, Norde W. The behavior of some model proteins at solid–liquid interfaces. II. Sequential and competitive absorption. Colloid Surf. 1990;51:17–28. [Google Scholar]
- 66.Linse S, Cabaleiro–Lago C, Xue WF, et al. Nucleation of protein fibrillation by nanoparticles. Proc Natl Acad Sci U S A. 2007;104:8691–8696. doi: 10.1073/pnas.0701250104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bellezza F, Cipiciani A, Quotadamo MA, et al. Structure, stability, and activity of myoglobin adsorbed onto phosphate–grafted zirconia nanoparticles. Langmuir. 2007;23:13007–13012. doi: 10.1021/la7015269. [DOI] [PubMed] [Google Scholar]
- 68.Podolski IY, Podlubnaya ZA, Kosenko EA, et al. Effects of hydrated forms of C60 fullerene on amyloid b–peptide fibrillization in vitro and performance of the cognitive task. J Nanosci Nanotechnol. 2007;7:1479–1485. doi: 10.1166/jnn.2007.330. [DOI] [PubMed] [Google Scholar]
- 69.Chien P, Weissman JS, DePace AH. Emerging principles of conformation–based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 70.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Ann Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 71.Dobson CM. The structural basis of protein folding and its link with human disease. Philos Trans R Soc Lond B Biol Sci. 2001;356:133–145. doi: 10.1098/rstb.2000.0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koo EH, Lansbury PT, Jr, Kelly JW. Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc Natl Acad Sci U S A. 1999;96:9989–9990. doi: 10.1073/pnas.96.18.9989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Floege J, Ehlerding G. Beta–2–microglobulin associated amyloidosis. Nephron. 1996;72:9–26. doi: 10.1159/000188801. [DOI] [PubMed] [Google Scholar]
- 74.SCENIHR (Scientific Committee on Emerging and Newly Identified Health Risks) Risk Assessment of Products of Nanotechnologies. Brussels, Belgium: European Commission; 2009. pp. 1–71. [Google Scholar]
- 75.Chun AL. Nanoparticles: Coat of proteins. Nat Nanotechnology. 2008 [Google Scholar]
- 76.Klein J. Probing the interactions of proteins and nanoparticles. Proc Natl Acad Sci U S A. 2007;104:2029–2030. doi: 10.1073/pnas.0611610104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lundqvist M, Stigler J, Elia G, et al. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci U S A. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martins MCL, Ratner BD, Barbosa MA. Protein absorption on mixtures of hydroxyl-and methyl-terminated alkanethiols self–assembleted monolayers. J Biomed Mater Res Pt A. 2003;67A:158–171. doi: 10.1002/jbm.a.10096. [DOI] [PubMed] [Google Scholar]
- 79.Phillips MC, Evans MTA, Graham DE, et al. Structure and properties of protein films adsorbed at the air–water interface. Colloid Polym Sci. 1975;253:424–427. [Google Scholar]
- 80.Brash JL, Horbett TA, editors. Proteins at Interfaces: Physicochemical and biochemical studies. Washington, DC: American Chemical Society; 1987. Advanced Symposium Series, 343. http://catalogue.nla.gov.au/Record/36034. [Google Scholar]
- 81.Horbett TA, Brash JL, editors. Proteins at Interfaces II: Fundamentals and application. Washington, DC: American Chemical Society; 1995. Advanced Symposium Series, 602. http://searchworks.stanford.edu/view/3082531. [Google Scholar]
- 82.Hlady V, Buijs J. Protein absorption on solid surfaces. Curr Opin Biotechnol. 1996;7:72–77. doi: 10.1016/s0958-1669(96)80098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sigal GB, Mrksich M, Whitesides GM. Effect of surface wetability on the absorption of proteins and detergents. J Am Chem Soc. 1998;120:3464–3473. [Google Scholar]
- 84.Waner MJ, Gilchrist M, Schindler M, et al. Imaging the molecular dimensions and oligomerization of proteins at liquid/solid interfaces. J Phys Chem B. 1998;102:1649–1657. [Google Scholar]
- 85.Gray JJ. The interaction of protein with solid surfaces. Curr Opin Struct Biol. 2004;14:110–115. doi: 10.1016/j.sbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 86.Wang D, Douma M, Swift B, et al. The absorption of globular protein onto a fluorinated PDMS surface. J Colloid Interface Sci. 2009;331:90–97. doi: 10.1016/j.jcis.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 87.Andrade JD, Hlady V, Wei AP. Absorption of complex proteins at interfaces. Pure Appl Chem. 1992;64:1777–1781. [Google Scholar]
- 88.Andrade JD, Hlady V. Protein absorption and materials biocompatibility: A tutorial review and suggested hypothesis. Adv Polym Sci. 1986;79:1–63. [Google Scholar]
- 89.Collier TO, Jenney CR, Defife KM, et al. Protein absorption on chemically modified surfaces. Biomed Sci Instrum. 1997;33:178–183. [PubMed] [Google Scholar]
- 90.Moulin AM, O’Shea SJ, Badley RA, et al. Measuring surface–induced conformational changes in proteins. Langmuir. 1999;15:8776–8779. [Google Scholar]
- 91.Holt SA, McGillivray DJ, Poon S, et al. Protein deformation and surfactancy at an interface. J Phys Chem B. 2000;104:7431–7438. [Google Scholar]
- 92.Henderson MJ, Perriman AW, Robson–Marsden H, et al. Protein–poly(silicic)acid interactions at the air/solution interface. J Phys Chem B. 2005;109:20878–20886. doi: 10.1021/jp051908k. [DOI] [PubMed] [Google Scholar]
- 93.Perriman AW, Henderson MJ, Holt SA, et al. Effect of the air–water interface on the stability of β–Lactoglobulin. J Phys Chem B. 2007;111:13527–13537. doi: 10.1021/jp074777r. [DOI] [PubMed] [Google Scholar]
- 94.Norde W, MacRitchie F, Nowicka G, et al. Protein absorption at solid liquid interfaces: Reversibility and conformation aspects. J Colloid Interface Sci. 1986;112:447–456. [Google Scholar]
- 95.Hayens CA, Norde W. Structures and stabilities of adsorbed proteins. J Colloid Interface Sci. 1995;169:313–328. [Google Scholar]
- 96.Nakanishi K, Sakiyama T, Imaura K. On the absorption of proteins on solid surfaces, a common but very complicated phenomenon. J Biosc Bioeng. 2001;91:233–244. doi: 10.1263/jbb.91.233. [DOI] [PubMed] [Google Scholar]
- 97.Roach P, Farrar D, Perry CC. Interpretation of protein absorption: surface–induced conformational changes. J Am Chem Soc. 2005;127:8168–8173. doi: 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- 98.Niemeyer CM. Semi-synthetic DNA–protein conjugates: Novel tools in analytics and nanobiotechnology. Nucleic acids chemistry and biology. Biochem Soc Trans. 2004;32:51–53. doi: 10.1042/bst0320051. [DOI] [PubMed] [Google Scholar]
- 99.Shinkai M. Functional magnetic particles for medical application. J Biosci Bioeng. 2002;94:606–613. [PubMed] [Google Scholar]
- 100.Wang L, Zhao W, Tan W. Bioconjugated silica nanoparticles: Development and applications. Nano Res. 2008;1:99–115. [Google Scholar]
- 101.Bharali DJ, Ilona K, Ewa KS, et al. Organically modified silica nanoparticles: A nonviral vector for in vivo gene delivery and expression in the brain. Proc Natl Acad Sci U S A. 2005;102:11539–11544. doi: 10.1073/pnas.0504926102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arruebo M, Fernandez-Pacheco R, Velasco B, et al. Antibody-functionalized hybrid superparamagnetic nanoparticles. Adv Funct Mater. 2007;17:1473–1479. [Google Scholar]
- 103.Aytur T, Foley J, Anwar M, et al. A novel magnetic bead bioassay platform using a microchip–based sensor for infectious disease diagnosis. J Immunol Methods. 2006;314:21–29. doi: 10.1016/j.jim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 104.Zhao X, Tapec-Dytioco R, Wang K, et al. Collection of trace amounts of DNA/mRNA molecules using genomagnetic nanocapturers. Anal Chem. 2003;75:3476–3483. doi: 10.1021/ac034330o. [DOI] [PubMed] [Google Scholar]
- 105.Thirumamagal BTS, Zhao XB, Bandyopadhyaya AK, et al. Receptor-targeted liposomal delivery of boron-containing cholesterol mimics for boron neutron capture therapy (BNCT) Bioconjug Chem. 2006;17:1141–1150. doi: 10.1021/bc060075d. [DOI] [PubMed] [Google Scholar]
- 106.Hermanson GT. Bioconjugate Techniques. San Diego, CA: Academic Press; 1996. [Google Scholar]
- 107.Graham DE, Harich KC, White RH. Reductive dehalogenation of monobromobimane by tris(2–carboxyethyl)phosphine. Anal Biochem. 2003;318:325–328. doi: 10.1016/s0003-2697(03)00239-2. [DOI] [PubMed] [Google Scholar]
- 108.Simonian AL, Good TA, Wang SS, et al. Nanoparticle-based optical biosensors for the direct detection of organophosphate chemical warfare agents and pesticides. Anal Chim Acta. 2005;534:69–77. [Google Scholar]
- 109.Sun EY, Josephson L, Kelly KA, et al. Development of nanoparticle libraries for biosensing. Bioconjug Chem. 2006;17:109–113. doi: 10.1021/bc050290e. [DOI] [PubMed] [Google Scholar]
- 110.Grüttner C, Müller K, Teller J, et al. Synthesis and antibody conjugation of magnetic nanoparticles with improved specific power absorption rates for alternating magnetic field cancer therapy. J Magn Magn Mater. 2007;311:181–186. [Google Scholar]
- 111.Aubin-Tam ME, Schifferli KA. Structure and function of nanoparticle-protein conjugates. Biomed Mater. 2008;3(3):034001. doi: 10.1088/1748-6041/3/3/034001. [DOI] [PubMed] [Google Scholar]
- 112.Weber C, Reiss S, Langer K. Preparation of surface modified protein nanoparticles by introduction of sulfhydryl groups. Int J Pharm. 2000;211:67–78. doi: 10.1016/s0378-5173(00)00590-1. [DOI] [PubMed] [Google Scholar]
- 113.Derek B, Smyth DG. Reactions of N-ethylmaleimide with peptides and amino acids. Biochem J. 1964;91:589–595. doi: 10.1042/bj0910589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jameson DM, Wong S. Chemistry of Protein Conjugation and Cross-linking. 2nd Ed. Baton Rouge, FL: CRC Press; 2009. [Google Scholar]
- 115.Madison LD, Rosenzweig SA, Jamieson JD. Use of the heterobifunctional cross–linker rn–maleimidobenzoyl N–hydroxysuccinimide ester to affinity label cholecystokinin binding proteins on rat pancreatic plasma membranes. J Biol Chem. 1984;259:1481–1482. [PubMed] [Google Scholar]
- 116.Mattson G, Conklin E, Desai S, et al. A practical approach to crosslinking. Mol Biol Reports. 1992;17:167–183. doi: 10.1007/BF00986726. [DOI] [PubMed] [Google Scholar]
- 117.Golemis E. Protein–Protein Interactions. A molecular cloning manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 118.Xing Y, Rao Y. Quantum dot bioconjugates for in vitro diagnostics and in vivo imaging. Cancer Biomark. 2008;4:307–319. doi: 10.3233/cbm-2008-4603. [DOI] [PubMed] [Google Scholar]
- 119.Jonkheijm P, Weinrich D, Schröder H, et al. Chemical strategies for generating protein biochips. Angew Chem Int Ed. 2008;47:9618–9647. doi: 10.1002/anie.200801711. [DOI] [PubMed] [Google Scholar]
- 120.Cleland WW. Dithiothreitol, a new protective reagent for SH groups. Biochemistry. 1964;3:480–482. doi: 10.1021/bi00892a002. [DOI] [PubMed] [Google Scholar]
- 121.Traut RR, Bollen A, Sun TT, et al. Methyl 4-mercaptobutyrimidate as a cleavable cross–linking reagent and its application to the Escherichia coli 30S ribosome. Biochemistry. 1973;12:3266–3273. doi: 10.1021/bi00741a019. [DOI] [PubMed] [Google Scholar]
- 122.Langer K, Coester C, Weber C, et al. Preparation of avidin–labeled protein nanoparticles as carriers for biotinylated peptide nucleic acid. Eur J Pharm Biopharm. 2000;49:303–307. doi: 10.1016/s0939-6411(00)00068-0. [DOI] [PubMed] [Google Scholar]
- 123.Wang S, Mamedova N, Kotov NA, et al. Antigen/antibody immunocomplex from CdTe nanoparticle bioconjugates. Nano Lett. 2002;2:817–822. [Google Scholar]
- 124.Gonzalez M, Bagatolli LA, Echabe I, et al. Interaction of biotin with streptavidin. Thermostability and conformational changes upon binding. J Biol Chem. 1997;272:11288–11294. doi: 10.1074/jbc.272.17.11288. [DOI] [PubMed] [Google Scholar]
- 125.Green NM. Avidin and streptavidin. Method Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 126.Parka JA, Lee JJ, Kim IS, et al. Magnetic and MR relaxation properties of avidin–biotin conjugated superparamagnetic nanoparticles. Colloid Surf A. 2008:313–314. 288–291. [Google Scholar]
- 127.Langer K, Coester C, Weber C, et al. Preparation of avidin–labeled protein nanoparticles as carriers for biotinylated peptide nucleic acid. Eu J Pharm Biopharm. 2000;49:303–307. doi: 10.1016/s0939-6411(00)00068-0. [DOI] [PubMed] [Google Scholar]
- 128.Coester C, Kreuter J, von Briesen H, et al. Preparation of avidin– labelled gelatin nanoparticles as carriers for biotinylated peptide nucleic acid (PNA) Int J Pharm. 2000;196:147–149. doi: 10.1016/s0378-5173(99)00409-3. [DOI] [PubMed] [Google Scholar]
- 129.Gole A, Dash C, Soman C, et al. On the preparation, characterization, and enzymatic activity of fungal protease–gold colloid bioconjugates. Bioconjug Chem. 2001;12:684–690. doi: 10.1021/bc0001241. [DOI] [PubMed] [Google Scholar]
- 130.Kudelski A. Influence of electrostatically bound proteins on the structure of linkage monolayers: absorption of bovine serum albumin on silver and gold substrates coated with monolayers of 2-mercaptoethanesulphonate. Vib Spectrosc. 2003;33:197–204. [Google Scholar]
- 131.Alliegro MC. Effects of dithiothreitol on protein activity unrelated to thiol-disulfide exchange: for consideration in the analysis of protein function with Cleland’s reagent. Anal Biochem. 2000;282:102–106. doi: 10.1006/abio.2000.4557. [DOI] [PubMed] [Google Scholar]
- 132.Shenoy BC, Appu Rao AG, Raghavendra Rao MR. Effect of chemical modification on structure and activity of glucoamylase from Aspergillus candidus and Rhizopus species. J Biosci. 1987;11:339–350. [Google Scholar]
- 133.Diagaradjane P, Orenstein-Cardona JM, Colon-Casasnovas NE, et al. Imaging epidermal growth factor receptor expression in vivo: pharmacokinetic and biodistribution characterization of a bioconjugated quantum dot nanoprobe. Clin Cancer Res. 2008;14:731–741. doi: 10.1158/1078-0432.CCR-07-1958. [DOI] [PubMed] [Google Scholar]
- 134.Kisel MA, Kulik LN, Tsybovsky IS, et al. Liposomes with phosphatidylethanol as a carrier for oral delivery of insulin: studies in the rat. Int J Pharm. 2001;216:105–115. doi: 10.1016/s0378-5173(01)00579-8. [DOI] [PubMed] [Google Scholar]
- 135.Gluck R. Liposomal hepatitis A vaccine and liposomal multiantigen combination vaccines. J Lipo Res. 1995;5:467–469. [Google Scholar]
- 136.Gregoriadis G. Engineering liposomes for drug delivery: progress and problems. Trends Biotechnol. 1995;13:527–537. doi: 10.1016/S0167-7799(00)89017-4. [DOI] [PubMed] [Google Scholar]
- 137.Colletier JP, Chaize B, Winterhalter M, et al. Protein encapsulation in liposomes: efficiency depends on interactions between protein and phospholipid bilayer. BMC Biotechnol. 2002;2:1–8. doi: 10.1186/1472-6750-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ruysschaert T, Germain M, da Silva Gomes JFP. Liposome-based nanocapsules. IEEE Trans Nanobioscience. 2004;3:49–55. doi: 10.1109/tnb.2004.824273. [DOI] [PubMed] [Google Scholar]
- 139.Besanger TR, Brennan JD. Entrapment of membrane proteins in Sol Gel derived silica. J Sol Gel Sci Techn. 2006;40:209–225. [Google Scholar]
- 140.Tan W, Wang K, He X, et al. Bionanotechnology based on silica nanoparticles. Med Res Rev. 2004;24:621–638. doi: 10.1002/med.20003. [DOI] [PubMed] [Google Scholar]
- 141.Nandiyanto ABD, Kim SG, Iskandar F, et al. Synthesis of spherical mesoporous silica nanoparticles with nanometer–size controllable pores and outer diameters. Micropor Mesopor Mat. 2009;120:447–453. [Google Scholar]
- 142.Lu B, Smyth MR, O’Keneddy JR. Immunological activities of IgG antibody on pre-coated Fc receptor surfaces. Anal Chim Acta. 1996;331:97–102. [Google Scholar]
- 143.Vanderberg ET, Brown RS, Krull UJ. Immobilized enzymes and cells in biochemical reactions. In: Veliky IE, McLean RJC, editors. Immobilized Biosystems: Theory and Practical Applications. Amsterdam, Holland: Elsevier; 1983. [Google Scholar]
- 144.Narayanan SS, Sarkar R, Pal SK. Structural and functional characterization of enzyme quantum dots conjugates: covalent attachment of CdS nanocryrtal to α-chemo trypsin. J Phys Chem C. 2007;111:11539–11543. [Google Scholar]
- 145.Menaa B, Herrero M, Rives V, Lavrenko M, Eggers DK. Favourable influence of hydrophobic surfaces on protein structure in porous organically-modified silica glasses. Biomaterials. 2008;29:2710–2718. doi: 10.1016/j.biomaterials.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Seeherman H, Wozney J, Li R. Bone morphogenetic protein delivery systems. Spine. 2002;27:16–23. doi: 10.1097/00007632-200208151-00005. [DOI] [PubMed] [Google Scholar]
- 147.Janes KA, Calvo P, Alonso MJ. Polysaccharide colloidal particles as delivery systems for macromolecules. Adv Drug Deliv Rev. 2001;47:83–97. doi: 10.1016/s0169-409x(00)00123-x. [DOI] [PubMed] [Google Scholar]
- 148.Gombotz VR, Pettit DK. Biodegradable polymers for protein and peptide drug delivery. Bioconjug Chem. 1995;6:332–351. doi: 10.1021/bc00034a002. [DOI] [PubMed] [Google Scholar]
- 149.Anderson JM, Shive MS. Biodegradation and biocompatibility of PLA and PLGA microspheres. Adv Drug Deliv Rev. 1997;28:5–24. doi: 10.1016/s0169-409x(97)00048-3. [DOI] [PubMed] [Google Scholar]
- 150.Crotts G, Park TG. Protein delivery from poly(lactic-coglycolic acid) biodegradable microspheres: release kinetics and stability issues. J Microencapsul. 1998;15:699–713. doi: 10.3109/02652049809008253. [DOI] [PubMed] [Google Scholar]
- 151.Zheng CH, Goa JQ, Zhang YP, Liang WQ. A protein delivery system: biodegradable alginate-chitosan-poly(lactic-co-glycolic acid) composite microspheres. Biochem Biophys Res Commun. 2004;323:1321–1327. doi: 10.1016/j.bbrc.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 152.Sun-Woong K, Oju J, Byung-Soo K.Tissue Eng 200511438–447.15869422 [Google Scholar]
- 153.Lee ES, Park KH, Park IS, et al. Glycol chitosan as a stabilizer for protein encapsulated into poly(lactide–co–glycolide) microparticle. Int J Pharm. 2007;338:310–316. doi: 10.1016/j.ijpharm.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 154.Xu P, Gullotti E, Tong L, et al. Intracellular drug delivery by poly(lactic–co–glycolic acid) nanoparticles, revisited. Mol Pharm. 2009;6:190–201. doi: 10.1021/mp800137z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Fu K, Griebenow K, Hsieh L, et al. FTIR characterization of the secondary structure of proteins encapsulated with PLGA microspheres. J Control Release. 1999;58:357–366. doi: 10.1016/s0168-3659(98)00192-8. [DOI] [PubMed] [Google Scholar]
- 156.Diwan M, Park TG. Pegylation enhances protein stability during encapsulation in PLGA microspheres. J Control Release. 2001;73:233–244. doi: 10.1016/s0168-3659(01)00292-9. [DOI] [PubMed] [Google Scholar]
- 157.Kang J, Schwendeman SP. Comparison of the effects of Mg(OH)2 and sucrose on the stability of bovine serum albumin encapsulated in injectable poly(d,l-lactide-co-glycolide) implants. Biomaterials. 2002;23:239–245. doi: 10.1016/s0142-9612(01)00101-6. [DOI] [PubMed] [Google Scholar]
- 158.Van de Weert M, Hennink WE, Jiskoot W. Protein instability in poly(lactic–co–glycolic acid) microparticles. Pharm Res. 2004;17:1159–1167. doi: 10.1023/a:1026498209874. [DOI] [PubMed] [Google Scholar]
- 159.Vila A, Sanchez A, Tobio M, et al. Design of biodegradable particles for protein delivery. J Control Release. 2002;78:15–24. doi: 10.1016/s0168-3659(01)00486-2. [DOI] [PubMed] [Google Scholar]
- 160.Calvo P, Remunan–López C, Vila–Jato JL, et al. Chitosan and chitosan/ethylene oxide-propylene oxide block copolymer nanoparticles as novel carriers for proteins and vaccines. Pharm Res. 1997;14:1431–1436. doi: 10.1023/a:1012128907225. [DOI] [PubMed] [Google Scholar]
- 161.Calvo P, Remunan-Lopez C, Vila-Jato JL, et al. Novel hydrophilic chitosan-polyethylene oxide nanoparticles as protein carriers. J Appl Polymer Sci. 1997;63:125–132. [Google Scholar]
- 162.Calvo P, Remunan-Lopez C, Vila-Jato JL, et al. Chitosan and chitosan/ethylene oxide–propylene oxide block copolymer nanoparticles as novel carriers for proteins and vaccines. Pharm Res. 1997;14:1431–1436. doi: 10.1023/a:1012128907225. [DOI] [PubMed] [Google Scholar]
- 163.Amidi M, Romeijn SG, Borchard G, et al. Preparation and characterization of protein–loaded N-trimethyl chitosan nanoparticles as nasal delivery system. J Control Release. 2006;111:107–116. doi: 10.1016/j.jconrel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 164.Sarmento B, Ferreira DC, Jorgensen L, et al. Probing insulin’s secondary structure after entrapment into alginate/chitosan nanoparticles. Eur J Pharm Biopharm. 2007;65:10–17. doi: 10.1016/j.ejpb.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 165.Li T, Shi XW, Du YM, et al. Quaternized chitosan/alginate nanoparticles for protein delivery. J Biomed Mater Res A. 2007;83:383–390. doi: 10.1002/jbm.a.31322. [DOI] [PubMed] [Google Scholar]
- 166.Schoubben A, Blasi P, Giovagnoli S, et al. Novel composite microparticles for protein stabilization and delivery. Eur J Pharm Sci. 2009;36:226–234. doi: 10.1016/j.ejps.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 167.Nakai H, Richardson CC. The gene 1.2 protein of bacteriophage T7 interacts with the E. coli dGTP triphosphohydrolase to form a GTP-binding protein. J Biol Chem. 1990;265:4411–4419. [PubMed] [Google Scholar]
- 168.Evans PR, Farrants GW, Hudson PJ. Phosphofructokinase: Structure and control. Philos Trans R Soc Lond B Biol Sci. 1981;293:53–62. doi: 10.1098/rstb.1981.0059. [DOI] [PubMed] [Google Scholar]
- 169.Hill RL, Brew K. Lactose synthetase. Adv Enzymol. 1975;43:411–490. doi: 10.1002/9780470122884.ch5. [DOI] [PubMed] [Google Scholar]
- 170.Srere PA. Complexes of sequential metabolic enzyme. Ann Rev Biochem. 1987;56:89–124. doi: 10.1146/annurev.bi.56.070187.000513. [DOI] [PubMed] [Google Scholar]
- 171.Prelich G, Tan CK, Kostura M, et al. Functional identity of proliferating cell nuclear antigen and a DNA Polymerase-δ auxiliary protein. Nature. 1989;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]