Abstract
The aryl hydrocarbon receptor (AhR), a ligand-dependent transcription factor, mediates toxicity of several classes of xenobiotics and also has important physiological roles in differentiation, reproduction, and immunity, although the endogenous ligand(s) mediating these functions is/are as yet unidentified. One candidate endogenous ligand, 2-(1′H-indolo-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE), is a potent AhR agonist in vitro, activates the murine AhR in vivo, but does not induce toxicity. We hypothesized that ITE and the toxic ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), may modify transcription of different sets of genes to account for their different toxicity. To test this hypothesis, primary mouse lung fibroblasts were exposed to 0.5μM ITE, 0.2nM TCDD, or vehicle for 4 h, and total gene expression was evaluated using microarrays. After this short-term and low-dose treatment, several hundred genes were changed significantly, and the response to ITE and TCDD was remarkably similar, both qualitatively and quantitatively. Induced gene sets included the expected battery of AhR-dependent xenobiotic-metabolizing enzymes, as well as several sets that reflect the inflammatory role of lung fibroblasts. Real time quantitative RT-qPCR assay of several selected genes confirmed these microarray data and further suggested that there may be kinetic differences in expression between ligands. These data suggest that ITE and TCDD elicit an analogous change in AhR conformation such that the initial transcription response is the same. Furthermore, if the difference in toxicity between TCDD and ITE is mediated by differences in gene expression, then it is likely that secondary changes enabled by the persistent TCDD, but not by the shorter lived ITE, are responsible.
Keywords: aryl hydrocarbon receptor (AhR), endogenous ligand, TCDD, microarray, mouse lung fibroblasts, gene expression
Although the aryl hydrocarbon receptor (AhR) has been most extensively studied as a mediator of toxicity of several classes of environmental contaminants, it is also recognized as having important endogenous functions. For example, in its absence (in AhR−/− mice), vascular differentiation is severely disrupted, reproductive ability is impaired, and liver and immune abnormalities are observed (McMillan and Bradfield, 2007). It also has a role in regulation of inflammation (Baglole et al., 2008; Thatcher et al., 2007), in cell cycle (Elferink, 2003), and impacts numerous other pathways of cell signaling (Beischlag et al., 2008). The many physiological roles identified for the AhR also support the presumed existence of endogenous ligands for this transcription factor, and much effort has been expended on the search for such molecules. Although no definitive endogenous ligand has yet been identified, several classes of naturally occurring compounds as well as physiological compounds have been found that bind the AhR with varying affinities and have varying abilities to activate or antagonize its transcriptional function (Denison and Nagy, 2003; Nguyen and Bradfield, 2008).
Among the stronger candidates for putative endogenous AhR ligands are various photoproducts and/or metabolic products of tryptophan, including 6-formylindolo[3,2-b]carbazole (FICZ) and 2-(1′H-indolo-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE), as well as several arachidonic acid derivatives (Chiaro et al., 2008a,b; Schaldach et al., 1999). FICZ is a potent AhR activator and a good substrate for Cytochromes P450 (CYPs) 1A1, 1A2, and 1B1; sulfoconjugates of a hydroxylated metabolite of FICZ were recently identified in human urine samples (Wincent et al., 2009). ITE was originally isolated and identified from porcine lung tissue and found to be a potent AhR activator (Song et al., 2002). Compared with the prototypical toxic ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), ITE binds the AhR and elicits in vitro DNA (aryl hydrocarbon response element, AhRE) binding with similar potency (Henry et al., 2006). ITE injected iv into pregnant transgenic mice is able to cross the placenta to activate the AhR-dependent LacZ reporter in fetal tissues but does not induce the teratogenic effects typical of prenatal TCDD exposure such as cleft palate and hydronephrosis. Also unlike TCDD, ITE has no effect on the thymus after in vivo dosing of adult mice, whereas it induces thymic toxicity in fetal thymic organ culture. This difference in potency between TCDD and ITE in vivo is consistent with their relative chemical stabilities in tissues—unlike the sustained presence and activity of TCDD, available data suggest that ITE is rapidly degraded (Bemis and Gasiewicz, unpublished data). Although the breakdown products have not yet been identified, it is likely that ITE is a substrate for cytochrome P450s as reported for FICZ (Wincent et al., 2009). Indeed, it is expected that endogenous activators would be relatively short-lived, unlike the xenobiotic toxic ligands typified by TCDD.
Another possible explanation for the difference in toxicity is that the binding of different ligands to the receptor elicits subtly different protein conformations that may in turn differ qualitatively as well as quantitatively in their binding to DNA and interactions with other cofactors. This could result in some variation in the gene expression profile induced by the AhR and hence the ultimate toxicity or lack thereof. Such an hypothesis is supported by research on steroid-receptor complexes (McDonnell et al., 1995; Wijayaratne and McDonnell, 2001), as well as by studies in which individual AhR-dependent genes and/or toxic responses are reported to differ between ligands (Gouedard et al., 2004a,b; Matikainen et al., 2001; Quintana et al., 2008). The goal of the current study was to compare the changes in gene expression elicited by short-term exposure to ITE and TCDD to determine whether there was a disparity that could explain the difference in toxicity. We chose to use mouse lung fibroblasts for this study since they are a primary culture rather than a transformed cell line and because of previous data suggesting a major role for AhR in regulation of inflammation (Vogel and Matsumura, 2009), which in the lung is partially mediated by lung fibroblasts (Baglole et al., 2008). Our results indicate that the short-term response to ITE and TCDD is essentially identical. However, validation of several of the affected genes shows some differences in kinetics consistent with the more rapid degradation of ITE compared to the persistence of TCDD.
MATERIALS AND METHODS
Cell culture.
Primary mouse lung fibroblasts from C57BL/6 mice were obtained and cultured as described previously (Baglole et al., 2005, 2008). Cells were grown in six-well plates in Modified Eagle's Medium (MEM) + 10% fetal bovine serum (FBS) and supplemented with pyruvate, glutamine, sodium bicarbonate, and gentamicin. Serum was removed for 24 h before treatment with ITE or TCDD when cells were nearing confluence. Ligands were added in dimethylsulfoxide (DMSO) vehicle (1 μl/ml medium).
Western blot analysis.
At the end of the exposure time, culture media were removed, cells washed twice with PBS, and lysed (Reporter Lysis Buffer, Promega, Madison, WI). Lysates were stored at −80°C. Lysate proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (8.4% acrylamide resolving gel) and transferred to polyvinylidene fluoride (PVDF) membrane which was then blocked using 5% nonfat milk in wash buffer (50mM Tris base, 150mM NaCl, and 0.2% Tween 20, pH 7.5). Antibodies used were Cox-2 (rabbit polyclonal, Cayman Chemical), CYP1B1 (Santa Cruz Biotechnology), AhR (mouse monoclonal Rpt-1 and ascites prepared in our laboratory), and β-actin (rabbit polyclonal, Sigma, St Louis, MO). The appropriate horseradish peroxidase-conjugated secondary antibodies were purchased from Jackson Immunoresearch (West Grove, PA). Proteins were visualized by chemiluminescence using LumiGlo reagents (KPL, Gaithersburg, MD).
RNA extraction.
RNA was prepared from individual wells using Qiashredder and RNeasy kits (Qiagen, Valencia, CA) with DNase treatment (RNase-free DNase [Qiagen]). Concentrations were measured using a ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA was stored at −20°C until used.
Microarrays.
RNA was prepared from five replicate wells for each treatment (DMSO, ITE, TCDD; 4 h exposure). Gene expression analysis was performed by the Functional Genomics Center at University of Rochester using the Affymetrix Mouse M430 2.0 array. RNA from each well was analyzed separately (i.e., five microarrays per treatment group). Instrumentation used was GeneChip Fluidics Station 450 and GeneChip Scanner 3000 7G. Background subtraction and quantile normalization of arrays was done with the GeneChip Robust Multiarray Average method (Wu and Irizarry, 2004). Probe sets showing no expression according to the Affymetrix MAS5 presence/absence algorithm were eliminated from further calculations. Fold induction by ITE or TCDD compared to DMSO was calculated for each gene probe. The Significance Analysis of Microarrays plug-in (www-stat.stanford.edu/∼tibs/SAM/) was used to determine the false discovery probability (q values). Nominal p values were calculated by the Microsoft Excel program. Further examination of the overall patterns of gene expression in these data was done using Gene Set Enrichment Analysis (GSEA, Broad Institute, MIT) (Subramanian et al., 2005). For this analysis, 1000 iterations, permuting by gene set, were used for comparing each pair of treatments (ITE vs. DMSO, TCDD vs. DMSO, and ITE vs. TCDD). Gene sets included in the analysis were from the curated collection in the Molecular Signature Database, MsigDB, maintained by Broad Institute.
Quantitative real-time PCR.
RNA (250 or 500 ng of each sample) was reverse transcribed using Superscript III and oligo(dT) primers (Invitrogen, Carlsbad, CA). Applied Biosystems Gene Expression Assays (Taqman chemistry) and the iCycler (Bio-Rad, Hercules, CA) were used to quantify expression of selected genes. Relative expression of the several target genes was determined from the measured PCR efficiencies for each gene and the ΔCt values (DMSO-treated) and normalized to the equivalent expression of Gapdh in each sample, as described by Pfaffl (2001).
RESULTS
Optimization of Doses and Exposure Time
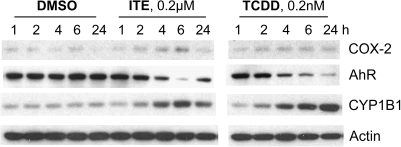
Initial experiments were performed to characterize the dose and time dependence of expression of selected AhR-responsive gene products in order to determine an appropriate dose and exposure time for subsequent microarray analyses. Cell lysates from mouse lung fibroblasts exposed to a range of exposure times and doses of ITE or TCDD were compared by Western blot (Fig. 1). Levels of AhR (which is rapidly degraded in response to agonist) were partially restored within 24 h after ITE, but not after TCDD (Fig. 1). Based on several experiments evaluating levels of AhR and products of two AhR-responsive genes, CYP1B1 and Cox-2, we determined that TCDD at 0.2nM and ITE at 0.5μM elicited comparable effects on these proteins. Short-term exposure (4 h) was chosen in order to focus on the primary transcriptional response to these two AhR ligands.
FIG. 1.
Western blot of several proteins in mouse lung fibroblasts exposed to ITE (0.2μM) or TCDD (0.2nM). Cells were grown to near confluence, cultured for 24 h without FBS, and then treated with ligand or DMSO vehicle as indicated. Whole-cell extracts were separated by SDS-PAGE; proteins were identified by Western blot. Representative of results from three experiments.
Short-Term Exposure to TCDD or ITE Induces the Same Changes in Gene Expression
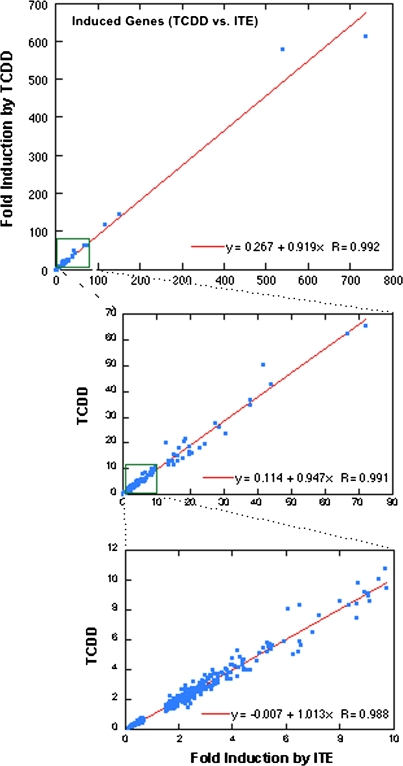
RNA extracted from mouse lung fibroblasts exposed to ITE, TCDD, or DMSO vehicle for 4 h was analyzed for changes in overall gene expression using the mouse M430 2.0 array. Depending on the criteria used to determine significance (as indicated in Table 1), several hundred genes (probe sets) were identified as induced or repressed by ITE or TCDD compared with DMSO (Table 1, A and B). However, none of these showed a difference between ITE and TCDD at q < 5% (Table 1, C). This pattern is clearly illustrated by Figure 2 in which fold-change values for TCDD-treated samples are plotted against fold change by ITE for all the significantly changed genes. The correlation coefficient in each range of this plot was 0.98–0.99. This indicates that in these cells and under these conditions, there was essentially no difference between the genes affected at 4 h after treatment with ITE compared with TCDD. In Table 1, the numbers in each category differ slightly between ITE and TCDD; this reflects the fact that there were some probe sets in each case for which (1) a similar mean fold change relative to DMSO (n = 5) was observed for both ligands, but for one or the other treatment, the p value or q value did not quite meet the criterion value or (2) the fold change by one ligand exceeded the indicated cutoff value but for the other ligand was slightly less than this cutoff.
TABLE 1.
Summary of Microarray Results
| (A) Filtered by ITE/DMSO | Total: 4521 |
| Fold change | |
| > 2.5 or < 0.4 | 255 |
| > 2.0 or < 0.5 | 482 |
| > 1.5 or < 0.67 | 1271 |
| (B) Filtered by TCDD/DMSO | Total: 3257 |
| Fold change | |
| > 2.5 or < 0.4 | 269 |
| > 2.0 or < 0.5 | 458 |
| > 1.5 or < 0.67 | 1201 |
| (C) Filtered by ITE/TCDD | 21a |
| Fold change | |
| > 2.0 or < 0.5 | 0 |
| > 1.5 or < 0.67 | 0 |
Note. Microarray data were analyzed by Significance Analysis of Microarrays as described in Materials and Methods. In parts A and B, the total number of genes (probe sets) meeting the < 5% false discovery probability is shown in the first row of each table. These were further filtered by different magnitudes of fold change as shown. Calculated p-values were all < 0.015 at this estimated false discovery rate. In part C, criteria were relaxed as indicated.
For all but one of these, the q value was 14–42%; p values were all ≤ 0.0016.
FIG. 2.
Gene induction by ITE and TCDD is highly correlated. Calculated mean fold induction/decrease by TCDD compared with DMSO is plotted against mean fold induction/decrease by ITE for all genes that were significantly changed (> 1.5-fold or < 0.67-fold, q value < 5%). Linear regression and correlation coefficients are shown for each range of the plot.
The complete list of significantly altered genes (probability of false discovery [q value] of < 5%) and their fold induction by ITE and TCDD is included in Supplementary table 1. The cutoff for inclusion in this table was 1.5-fold (up- or downregulated). In this table are included those genes that may meet the above criteria for one compound but not for the other (e.g., 1.55-fold induced by ITE but only 1.45-fold by TCDD or vice versa). Such genes are generally found in the borderline range of 1.5- to 2-fold change in expression. Sorting the list of genes according to magnitude of either ITE- or TCDD-induced expression yielded very similar order, as expected from the very high correlation shown in Figure 2. Among the most highly induced genes (Table 2) are the expected AhR-dependent drug-metabolizing enzymes, including Aldh3a1 and Cyp1a1 (> 500-fold), Nqo1 (ninefold), Cyp1b1 and Adh (fourfold to fivefold; not included in Table 2), as well as AhRR (116-fold).
TABLE 2.
The Most Highly Induced/Repressed Genes Identified by Microarray
| Relative expression | ||||
| Symbol | ITE/DMSO | TCDD/DMSO | ITE/TCDD | Description |
| Aldh3a1 | 735.916 | 611.531 | 1.203 | Aldehyde dehydrogenase family 3, subfamily A1 |
| Cyp1a1 | 538.552 | 577.663 | 0.932 | Cytochrome P450, family 1, subfamily a, polypeptide 1 |
| Megf10 | 150.877 | 146.849 | 1.027 | Multiple EGF-like domains 10 |
| Ahrr | 116.436 | 119.109 | 0.978 | AhR repressor |
| Tac1 | 72.179 | 65.510 | 1.102 | Tachykinin 1 |
| Nsg2 | 66.602 | 62.499 | 1.066 | Neuron-specific gene family, member 2 |
| Tnfrsf19 | 43.960 | 42.908 | 1.025 | Tumor necrosis factor receptor superfamily, member 19 |
| St8sia1 | 41.767 | 50.470 | 0.828 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| Slc1a2 | 37.978 | 37.087 | 1.024 | Solute carrier family 1 (glial high-affinity glutamate transporter), member 2 |
| Slc2a6 | 37.937 | 34.988 | 1.084 | Solute carrier family 2 (facilitated glucose transporter), member 6 |
| Nsg2 | 29.508 | 39.405 | 0.749 | Neuron-specific gene family, member 2 |
| Retn | 28.617 | 26.067 | 1.098 | Resistin |
| Ccnu | 27.563 | 27.670 | 0.996 | Cyclin U |
| Tgfbi | 24.449 | 19.460 | 1.256 | Transforming growth factor, beta induced |
| Nmnat2 | 22.617 | 17.992 | 1.257 | Nicotinamide nucleotide adenylyltransferase 2 |
| Mpp4 | 19.698 | 18.798 | 1.048 | Membrane protein, palmitoylated 4 (MAGUK p55 subfamily, member 4) |
| Tgfbi | 19.584 | 16.833 | 1.163 | Transforming growth factor, beta induced |
| Tiparp | 18.753 | 21.690 | 0.865 | TCDD-inducible poly(ADP-ribose) polymerase |
| St8sia1 | 18.274 | 20.822 | 0.878 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| Nr4a3 | 17.642 | 14.125 | 1.249 | Nuclear receptor subfamily 4, group A, member 3 |
| Rgnef | 16.680 | 17.953 | 0.929 | Rho-guanine nucleotide exchange factor |
| Olfm1 | 16.109 | 15.298 | 1.053 | Olfactomedin 1 |
| Slc1a2 | 16.099 | 13.291 | 1.211 | Solute carrier family 1 (glial high-affinity glutamate transporter), member 2 |
| Slc14a1 | 15.117 | 11.633 | 1.300 | Solute carrier family 14 (urea transporter), member 1 |
| Lrp4 | 14.549 | 13.108 | 1.110 | Low-density lipoprotein receptor–related protein 4 |
| Prss35 | 13.718 | 11.407 | 1.203 | Protease, serine, 35 |
| Tspan33 | 13.631 | 12.905 | 1.056 | Tetraspanin 33 |
| St8sia1 | 12.878 | 20.101 | 0.641 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
| Afp | 12.311 | 13.211 | 0.932 | Alpha fetoprotein |
| Nos3 | 9.832 | 12.234 | 0.804 | Nitric oxide synthase 3, endothelial cell |
| Cpox | 9.690 | 10.815 | 0.896 | Coproporphyrinogen oxidase |
| Tiparp | 9.428 | 10.120 | 0.932 | TCDD-inducible poly(ADP-ribose) polymerase |
| Nqo1 | 9.113 | 8.614 | 1.058 | NAD(P)H dehydrogenase, quinone 1 |
| Bmpr1b | 0.268 | 0.210 | 1.279 | Bone morphogenetic protein receptor, type 1B |
| Crct1 | 0.267 | 0.307 | 0.869 | Cysteine-rich C-terminal 1 |
| Kcnh1 | 0.247 | 0.243 | 1.019 | Potassium voltage-gated channel, subfamily H (eag related), member 1 |
| Chst1 | 0.242 | 0.357 | 0.680 | Carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 |
| Fgfr2 | 0.240 | 0.254 | 0.947 | Fibroblast growth factor receptor 2 |
| Edn1 | 0.240 | 0.240 | 1.000 | Endothelin 1 |
| Ivl | 0.227 | 0.207 | 1.097 | Involucrin |
| Ccdc85a | 0.219 | 0.271 | 0.810 | Coiled-coil domain containing 85A |
| Sprr1a | 0.184 | 0.185 | 0.997 | Small proline-rich protein 1A |
| Klhl29 | 0.180 | 0.203 | 0.888 | Kelch-like 29 (Drosophila) |
For all genes shown, q < 5%. Omitted from this list were several probe sets identified only by riken no. or expressed sequence no. and two additional Tgfbi (they are included in the Supplementary table 1).
Evaluation of Altered Gene Pathways
The major objective of this study was to determine differences (or lack thereof) between the spectrum of genes responding to ITE compared with TCDD; clearly, the initial transcriptional activation by these two ligands was very similar in these cells. Since this is the first time to our knowledge that gene array studies have been reported for mouse lung fibroblasts, we evaluated the patterns of alterations induced by these AhR ligands in more detail. Simply focusing on the most highly up- or downregulated genes (such as listed in Table 2 or in Supplementary table 1) does not necessarily provide much information regarding the whole spectrum of biological pathways that may be affected (e.g., pathways in which many individual components may not be highly changed). A number of tools have been developed to analyze genome-wide expression data to extract more physiological information. We chose to use GSEA (Subramanian et al., 2005). This technique is designed to determine whether observed patterns of gene expression show evidence of alterations in groups of genes that share biological functions or pathways.
A summary of the GSEA results is presented in Table 3. In this analysis, 1442 gene sets were included (containing 14–500 genes per set). Comparing ITE or TCDD data with the DMSO control data showed nominal enrichment of a large number of gene sets. However, of these, only 13 and 9 showed enrichment in ITE and TCDD samples, respectively, at a false discovery rate (FDR) of < 25% (Table 3) (an FDR of 25% indicates a three in four chance that the result is valid; this level is considered appropriate for the stated purpose of hypothesis generation from GSEA [Subramanian et al., 2005]). The 13 gene sets that met the 25% FDR criterion for the ITE versus DMSO comparison are identified and described in Table 4. Among these enriched sets are the expected “metabolism of xenobiotics by cytochrome P450,” as well as several that reflect the inflammatory role of lung fibroblasts (“prostaglandin and leukotriene metabolism,” “arachidonic acid metabolism,” and “eicosanoid synthesis”). Consistent with the high correlation between ITE and TCDD results noted above for individual genes, the top-ranked gene sets for TCDD samples were largely the same ones as for ITE. Note that the rank order as well as the differences in rank order between ligands are not considered to be biologically meaningful. Indeed, since the analysis involves random permutation of phenotype or gene set labels in order to determine significance of enrichment results, each analysis produces slightly different ordering. However, the fact that out of > 1400 gene sets ITE and TCDD samples were found to contain a similar group of affected sets based on these calculations is considered to strongly support the conclusion that these ligands act very similarly. Additionally, GSEA identified no gene sets that were significantly enriched in the ITE compared with TCDD datasets (Table 3).
TABLE 3.
Summary of GSEA
| ITE versus DMSO (DMSO vs. ITE) | TCDD versus DMSO (DMSO vs. TCDD) | ITE versus TCDD (TCDD vs. ITE) | |
| # Gene sets upregulated (downregulated) | 676 (766) | 714 (728) | 524 (918) |
| # Gene sets significantly enriched at FDR < 25% | 13 (0) | 9 (0) | 0 (0) |
| # Gene sets enriched at nominal p < 1% | 21 (22) | 21 (29) | 2 (10) |
Microarray data were subjected to GSEA as described in “Materials and Methods” section using the three indicated comparisons (ITE vs. DMSO, TCDD vs. DMSO, and ITE vs. TCDD); 21,891 genes (after collapsing the 45,101 array features into gene symbols since many genes are represented by two or more probe sets on the arrays); 1442 gene sets were included in analysis (14–500 genes per set); analyses performed with 1000 iterations.
TABLE 4.
Gene Sets Enriched in ITE Samples (vs. DMSO)
| Rank ITEa | Rank TCDDb | Gene set name | Descriptionc | # Genes in setd | FDR q valuee | Nominal p value |
| 1 | 6 | Dendritic cell pathway | ANPEP, several interleukins, TLRs, interferons, and CD molecules | 21 (2) | 0.047 | 0.000 |
| 2 | 3 | Arachidonic acid metabolism | PTGS1,2; carbonyl reductases; GGT; PTGES, PTGDS; phospholipases, Cyp2U1 and 2E1; and GPXs, … | 40 (10) | 0.138 | 0.002 |
| 3 | 4 | IDX TSA DN cluster1 | Genes downregulated 48–96 h during 3T3-L1 fibroblast differentiation into adipocytes with IDX (insulin, dex, and isobutylxanthine) | 40 (12) | 0.094 | 0.000 |
| 4 | 25 | γHexachlorocyclohexane degradation | Acid and alkaline phosphatases, Cyp3a's, and paraoxonases | 14 (4) | 0.118 | 0.000 |
| 5 | 2 | Fatty acid metabolism | AcylCoA dehydrogenases, oxidases, synthetases; ADHs, AldHs, many P450s, … | 60 (10) | 0.101 | 0.000 |
| 6 | 5 | Metabolism of xenobiotics by cytochrome P450, … | Many AldHs, Cyps, GSTs, epoxide hydrolase, and UDPGTs | 34 (7) | 0.116 | 0.002 |
| 7 | 1 | Prostaglandin and leukotriene metabolism | Lipoxygenases, Cyp4F's, carbonyl reductases, PTGES, and phospholipases, … | 28 (7) | 0.193 | 0.000 |
| 8 | 15 | Greenbaum E2A DN | E2A target genes in lymphocyte development | 25 (8) | 0.170 | 0.004 |
| 9 | 9 | Eicosanoid synthesis | PG synthases, arachidonate lipoxygenases, PTGS1,2 | 16 (7) | 0.153 | 0.004 |
| 10 | 10 | Butanoate metabolism | 27 (8) | 0.231 | 0.006 | |
| 11 | 43 | Pomeroy desmoplasmic versus classic MD_DN | Genes expressed in classic medulloblastomas | 37 (2) | 0.236 | 0.002 |
| 12 | 13 | Nicotinate and nicotinamide metabolism | 22 (8) | 0.241 | 0.006 | |
| 13 | 14 | Valine, leucine, and isoleucine degradation | Transferases, dehydrogenases, … | 35 (10) | 0.225 | 0.007 |
| (6)f | (11) | TGFβ signaling pathway | TGFβs, TGFRs, LTBP1; bone morphogenic proteins, Id1–4; MAPK1,3; protein phosphatase 2’s; CDK inhibitor 2B | 82 (27) | 0.510 | 0.002 |
Note. Listed are the gene sets which were identified by GSEA to be enriched in ITE-treated cells compared with DMSO treated, with FDR < 25%. ADHs, alcohol dehydrogenases; AldHs, aldehyde dehydrogenases; ANPEP, alanyl aminopeptidase; GGT, g-glutamyl transpeptidase (or gamma-glutamyl transpeptidase); GPXs, glutathione peroxidases; GSTs, glutathione S-transferases; MAPK, mitogen-activated protein kinase; PG, prostaglandin; PTGS, prostaglandin endoperoxide synthase; PTGES, prostaglandin E synthase; PTGDS, prostaglandin D synthase; TLR, toll-like receptor; UDPGTs, UDP-glucuronyltransferases.
Rank within the enriched gene sets, based on normalized enrichment score (ITE vs. DMSO). Rank differs among repeated runs of GSEA due to random permutation process of analysis.
Rank within enriched gene sets (TCDD vs. DMSO).
Some of the genes/classes of genes included in indicated gene set.
Total number of genes in set (number individually induced in ITE samples).
FDR, estimated probability that set represents a false positive.
One of the sets downregulated by ITE and TCDD relative to DMSO; FDR > 25% (as it is for all downregulated sets).
Generally, within each enriched gene set, not all the member genes are actually significantly induced/repressed. The analysis looks at the distribution of all the genes in the set within the list of all genes on the array (ranked by magnitude of change relative to control) to determine whether they are distributed randomly throughout or concentrated within the upper or lower extremes of the ranked list. Not all that are listed as showing “core enrichment” in a particular enriched gene set are actually identified as significantly affected based on simple fold change by ITE (or TCDD) compared to DMSO as calculated based on individual mean values and shown in Supplementary table 1.
There is some overlap of gene members among sets, not surprisingly, since few biomolecules are involved in only one unique pathway, and therefore some genes that are induced are members of multiple gene sets. In this experiment, relatively few genes determined the calculations that yielded the conclusion that certain gene sets are significantly induced. For example, Aldh3a1, Adh7, Ptgs1 and 2, Cyp1a1, and other Cyps are in several of the enriched gene sets. Many more genes that were individually up- or downregulated are members of one or more gene sets in the database, but those gene sets were not determined to be “enriched” in this experiment or were not included in this analysis (14–500 genes per set). Notably, one of the most highly induced genes (Ahrr) happens not to be in any of the induced gene sets, including metabolism of xenobiotics by CytP450, perhaps because the function of AhRR is not yet fully understood.
Quantitative-PCR Confirmation of Expression Changes of Selected Genes
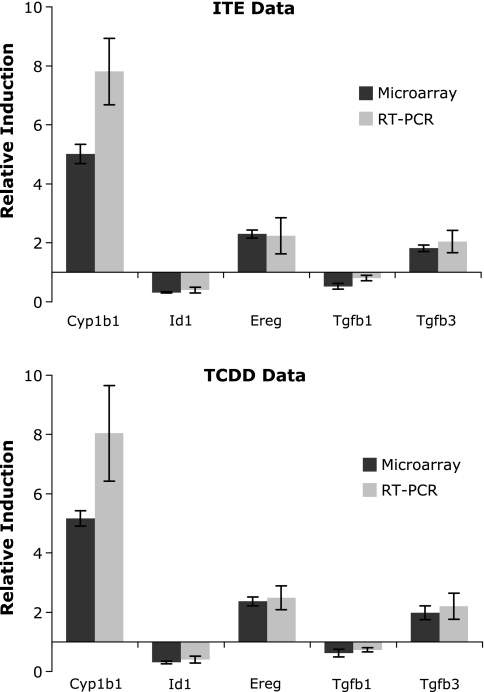
Several genes identified by microarray to be affected by ligand treatment were chosen for validation of results using real-time PCR quantitation. Genes chosen represented highly induced (Cyp1a1 and Cyp1b1), modestly induced (epiregulin [Ereg] and Tgfb3), and decreased (Id1 and Tgfb1) according to microarray analysis. Ereg is a recently identified AhR target gene, is a member of the epidermal growth factor family of growth regulators, and its promotor includes an AhRE sequence (Patel et al., 2006). The cytokine transforming growth factor β (TGFβ) impacts many cellular processes, and its cross talk with AhR signaling has been widely reported, though not yet well understood (reviewed by Gomez-Duran et al., 2009). In the present experiment, Tgfb1 and 2 were modestly downregulated, whereas Tgfb3 was upregulated; promotors of all three forms are reported to contain AhRE sites (Sun et al., 2004). The Id (inhibitor of DNA binding) genes were among the more prominently downregulated genes in our cells, as well as in human dermal fibroblasts (Akintobi et al., 2007). Using the same RNA as analyzed on the microarrays, the correlation between microarray and RT-qPCR was very strong for these genes (Fig. 3). Induction of Cyp1a1 also was highly correlated between RT-qPCR and microarrays and between ITE and TCDD but is not shown in Figure 3 because its magnitude of induction is far off the scale used for the other genes (inductions of > 500-fold in all cases).
FIG. 3.
RT-qPCR confirms gene expression data from microarrays. Fold induction by ITE and TCDD relative to DMSO as calculated from microarrays and RT-qPCR using the same RNA samples is shown for selected genes. Cyp1a1 also showed equivalent induction by RT-qPCR and microarray, but actual values are off this scale and are not shown. All values are normalized to expression of Gapdh in the same samples; mean ± SD (n = 5).
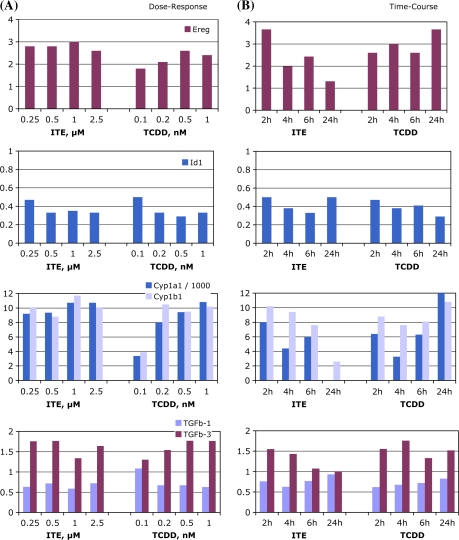
Since responses to ITE and TCDD were so similar at 4 h, and yet clearly these ligands have different toxicity (at least in mice), it is possible that divergent patterns of gene expression would be detectable after longer exposure periods. Therefore, these selected genes were also quantified by RT-qPCR analysis of RNA extracted from separate cultures of mouse lung fibroblasts exposed over a range of time intervals or to a range of doses to determine time-course and dose-response of these changes (Fig. 4). These data were consistent with the microarray results, although over the dose range used, a clear dose-response pattern was not observed for all the tested genes. Between 2 and 24 h after ITE treatment, Ereg, Cyp1a1, and Cyp1b1 and Tgfb3 induction diminished over time, whereas these levels were maintained by TCDD up to 24 h. The downregulation of Id1 was maintained over the 24 h by both ligand treatments; the modest decrease in Tgfb1 appeared to be more transient.
FIG. 4.
Time-course and dose responsiveness of transcription of selected genes. Mouse lung fibroblasts were grown and treated as described in “Materials and Methods” section, with a range of concentrations of ITE or TCDD or with DMSO vehicle for 4 h (A), or were treated with DMSO, 0.5μM ITE, or 0.5nM TCDD for 2–24 h (B). Total RNA extraction and RT-qPCR were performed as described. Expression levels shown are relative to DMSO and are normalized to Gapdh assayed concurrently in the same samples. Representative of at least two experiments.
DISCUSSION
These results show a very close similarity in short-term gene expression changes in response to ITE and TCDD. At least in these lung fibroblast cells in culture, we find no evidence that the two ligands elicit divergent patterns of gene expression that could account for their very different in vivo toxicity. Since our original hypothesis was that when AhR is activated by different ligands a different spectrum of genes may be induced/repressed, we used a short exposure period (4 h) for microarray analysis so that observed gene expression changes can be considered to reflect a primary ligand-induced transcriptional response. This primary response was remarkably similar between ligands, both qualitatively and quantitatively, strongly suggesting that ITE and TCDD elicit the same, or at least a sufficiently similar, conformational change in the AhR to initiate the similar transcriptional responses. The specific genes that respond to AhR ligand treatment are expected to differ among cell type and developmental stage at time of exposure, particularly in vivo, since the role of the AhR differs among cells/tissues and with their state of differentiation. However, we predict that the overall similarity in the total spectrum of initial gene expression changes in response to equivalent AhR-activating levels of ITE and TCDD would be observed consistently.
These data further suggest that the disparity in toxicity between these two high-potency AhR ligands is more likely related to their different chemical stability (in vivo) and resultant difference in kinetics. This is consistent with earlier findings that whereas ITE and TCDD have similar AhR-binding affinity and similar AhR·DRE–binding potency in vitro, ITE is several orders of magnitude less potent than TCDD in inducing reporter gene activity in treated cells or mice (Henry et al., 2006). The preliminary RT-qPCR time-course results in Figure 4 suggest an analogous kinetic difference in ligand-induced transcription of several genes. Furthermore, the fact that certain TCDD-sensitive genes remain up- or downregulated while ITE effects are less persistent suggests that the spectrum of later secondary gene expression likely differs substantially between ligands. The combination of more prolonged primary gene changes elicited by TCDD and subsequent secondary gene changes (due to prolonged occupancy and activation of the AhR that would not be produced by ITE) likely lead to its toxic actions. Additional studies would be needed to verify such an hypothesis, but much evidence has accumulated to indicate markedly different sequelae from sustained versus transient AhR activation (e.g., Mitchell and Elferink, 2009). Relevant to this concept, the present data also imply that in order to identify those gene expression changes that may be associated with or mediate TCDD toxicity, longer term exposure must be included in the experimental design.
Since the goal of this study was to compare the transcriptional effects of ITE and TCDD, the identity of individual affected genes is considered less important than the remarkable similarity of the whole spectrum of changes elicited by the two ligands. Indeed, the subset of genes whose expression is detectably altered by any particular treatment would depend on the cell type studied and the specific conditions (dose and duration) of exposure. Nonetheless, it is interesting to consider our findings in perspective with published microarray data from other cell types and in vivo data on AhR activity. Another putative endogenous ligand, indirubin, has been compared with TCDD with respect to gene expression and metabolism in HepG2 cells (Adachi et al., 2004). In that study, both ligands were at 10nM for 8 h, and the microarray used was limited to 1176 genes relevant to “toxicology.” Few genes were affected by either TCDD or indirubin; of the 28 listed as changed and that were also present on our microarrays, only Cyp1a1 induction was in common with our results. Similar to ITE, Cyp1a1 induction by indirubin was transient, and indirubin was degraded by CYP1A1 (Adachi et al., 2004). In a recent study comparing human and mouse AhR, it was reported that the human AhR (hAhR) expressed in transgenic mouse liver binds indirubin (and quercetin) with higher affinity than the C57(wild-type) mouse AhR (mAhR) and that indirubin activates the hAhR more potently than the mAhR (Flaveny et al., 2009). Although we found that ITE showed a similar dose-response and time-course of reporter gene induction in cultured human (HepG2) and mouse (Hepa1c1c7) cells (Henry and Gasiewicz, 2006, unpublished data), the data of Flaveny et al. (2009) suggest an interesting species-specific ligand selectivity that must be kept in mind when evaluating AhR ligands.
The expected induction of xenobiotic-metabolizing enzymes (including Cyps, Aldh, and Nqo1), as' well as Ahrr, was prominent in these mouse lung fibroblasts (Table 2), and the GSEA indicated enrichment of such a gene set (Table 4). Several other defined sets that were enriched reflect the inflammatory role of lung fibroblasts (Table 4). These sets share a number of genes that are markers of inflammation, including prostaglandin synthases, phospholipases, and carbonyl reductases. These sets are of particular interest since arachadonic acid derivatives such as eicosanoids may serve as endogenous AhR ligands (Chiaro et al., 2008a,b). Also included in Table 4, although it did not meet the 25% FDR criterion, is the gene set “TGFβ signaling pathway,” which was downregulated compared with DMSO. Of the 82 member genes, 27 were listed with core enrichment and were downregulated, including Tgfb1, Tgfb2, Tgfbr1, Id1–4, Mapk3, Bmp 4 and 6, among many others. Tgfb3 is also in this set but was induced. In contrast to the downregulation of a TGFβ gene set, one of the most highly induced genes in our study (but not included in this TGFβ gene set) was Tgfbi, a ubiquitously expressed protein induced by TGFβ (type unspecified) that is a component of extracellular matrix, and loss or downregulation of which is associated with tumor cell lines and primary tumor specimens. The arrays also included several probe sets for Ltbp (latent TGF–binding proteins), but none were affected by ITE or TCDD. The fact that many TGFβ pathway genes were enriched is interesting given the known interaction between AhR and TGFβ both in cell culture and in vivo and both in the presence and absence of xenobiotic ligand (Gomez-Duran et al., 2009; Haarmann-Stemmann et al., 2009; Puga et al., 2005). TGFβ3 is also the only member of the AHRE-II gene battery listed by Boutros et al. (2004) that was changed by ITE/TCDD in the current study. The Id proteins (inhibitor of DNA binding, Id1–4) were all repressed by ITE and TCDD and contributed to the enrichment of this TGFβ gene set. These helix-loop-helix (HLH) proteins dimerize with other HLH proteins, thereby inhibiting their transcriptional activities. TCDD was also reported to downregulate Id1 and Id3 in human dermal fibroblasts (Akintobi et al., 2007).
Based on observations that IL1R receptor (IL1R) and tumor necrosis factor receptor 1 and 2 (TNFR1,2) knockout mice are resistant to some types of TCDD hepatotoxicity and liver inflammation, though still show Cyp induction, hydropic degeneration, and hepatomegaly, Pande et al. (2005) concluded that IL1R-like cytokines mediate some (though not all) responses to TCDD. Since the cells we used are involved in inflammation, it is notable that several IL1R- and TNF receptor–related genes on the array were significantly affected by ITE and TCDD (Table 5). (The curated database of gene sets currently used in the GSEA, however, did not include a set containing this grouping of genes, although some of the individual genes were present in a few sets that were not enriched in this experiment.)
TABLE 5.
Some IL1R- and TNFR-Related Genes Are Altered by ITE/TCDD
| Gene | Gene name | Fold induction by ITEa |
| Il1rap | IL1R accessory protein | 0.65 |
| Irak2 | IL1R receptor–associated kinase 2 | 1.83 |
| IL33 (IL-1F11) | Interleukin 33 | 0.345 |
| IL1rl2 | IL1R-like 2 | (1.2) |
| Tnfrsf19 | TNF receptor superfamily, member 19 | 44 |
| Tnfrsf18 GITR | TNF receptor superfamily, member 18 | 2.18 |
| Tnfrsf12a | TNF receptor superfamily, member 12a | 0.5 |
| Traf5 | TNF receptor–associated factor 5 | 2.86 |
| Clqtnf3 | Clq- and TNF-related protein 3 | 8.9 |
Numbers shown are fold induction by ITE relative to DMSO from microarray analysis.
From our study, in which a large number of genes are induced or repressed, it is not possible to say how many of these changes are biologically significant or would be observed consistently or in different cell types. Only 8 out of 22 genes with defined, functional AhREs (as listed in a recent review, Gasiewicz et al., 2008) and on the arrays were affected by ITE and TCDD. Most of these eight are in the xenobiotic-metabolizing family of genes. Many more of the genes identified as up- or downregulated (Supplementary table 1) are obviously involved in a wide variety of basic cellular pathways, which is consistent with the very extensive spectrum of genes that was recently reported to comprise AhR-binding targets as determined by a combination of genetic and in silico investigations (Sartor et al., 2009). The promoter regions of many of these targets (including Tgfb1, 2, and 3, Tgfbi, and IL1rap) have AhRE sequences determined computationally but not as yet verified to be functional (Sun et al., 2004).
Microarrays, as utilized for this study, assess the genomic route of response to AhR ligands. There is also evidence for AhR-dependent but nongenomic effects (Matsumura, 2009). Some of these effects occur more rapidly than genomic responses, for example, increased intracellular Ca++ is reported to occur within 15–30 min of TCDD addition in a variety of cell types. Calcium influx is proposed to be the initiator of inflammatory responses such as cytosolic phospholipase A2 and Cox-2 induction. These putative nongenomic responses can occur faster because they are mediated by processes such as protein phosphorylation and dephosphorylation rather than the more complex transcription/translation/posttranslational modification. It seems plausible that endogenous AhR ligands such as ITE would also mediate such nongenomic responses, and there may be differences among ligands in ability to initiate such effects. To our knowledge, these signals have not been investigated following treatments with ligands other than TCDD or related chemicals.
In summary, we conclude that the putative endogenous AhR ligand, ITE, and the prototypical xenobiotic ligand, TCDD, despite their disparate chemical structures, likely elicit an analogous change in AhR conformation such that the initial transcriptional response is essentially the same (as analyzed in mouse lung fibroblasts). If differences in gene expression do mediate the difference in toxicity of these ligands, then it must be later, secondary changes in gene transcription that are enabled by the persistence of TCDD compared with the more short-lived ITE.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
FUNDING
National Institute of Environmental Health Sciences (ES015775, Training Grant ES07026, Center Grant ES01247).
Supplementary Material
Acknowledgments
We thank Dr Carolyn Baglole for supplying the mouse lung fibroblasts and for her advice on their culture.
References
- Adachi J, Mori Y, Matsui S, Matsuda T. Comparison of gene expression patterns between 2,3,7,8-tetrachlorodibenzo-p-dioxin and a natural arylhydrocarbon receptor ligand, indirubin. Toxicol. Sci. 2004;80:161–169. doi: 10.1093/toxsci/kfh129. [DOI] [PubMed] [Google Scholar]
- Akintobi AM, Villano CM, White LA. 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD) exposure of normal human dermal fibroblasts results in AhR-dependent and -independent changes in gene expression. Toxicol. Appl. Pharmacol. 2007;220:9–17. doi: 10.1016/j.taap.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Baglole CJ, Maggirwar SB, Gasiewicz TA, Thatcher TH, Phipps RP, Sime PJ. The aryl hydrocarbon receptor attenuates tobacco smoke-induced cyclooxygenase-2 and prostaglandin production in lung fibroblasts through regulation of the NF-kB family member RelB. J. Biol. Chem. 2008;283:28944–28957. doi: 10.1074/jbc.M800685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglole CJ, Reddy JY, Pollock SJ, Feldon SE, Sime PJ, Smith TJ, Phipps RP. Isolation and phenotypic characterization of lung fibroblasts. Methods Mol. Med. 2005;117:115–127. doi: 10.1385/1-59259-940-0:115. [DOI] [PubMed] [Google Scholar]
- Beischlag TV, Morales JL, Hollingshead BD, Perdew GH. The aryl hydrocarbon receptor complex and the control of gene expression. Crit. Rev. Eukaryot. Gene Expr. 2008;18:207–250. doi: 10.1615/critreveukargeneexpr.v18.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutros PC, Moffat ID, Franc MA, Tijet N, Tuomisto J, Pohjanvirta R, Okey AB. Dioxin-responsive AHRE-II gene battery: identification by phylogenetic footprinting. Biochem. Biophys. Res. Commun. 2004;321:707–715. doi: 10.1016/j.bbrc.2004.06.177. [DOI] [PubMed] [Google Scholar]
- Chiaro CR, Morales JL, Prabhu KS, Perdew GH. Leukotriene A4 metabolites are endogenous ligands for the Ah receptor. Biochemistry. 2008a;47:8445–8455. doi: 10.1021/bi800712f. [DOI] [PubMed] [Google Scholar]
- Chiaro CR, Patel RD, Perdew GH. 12(R)-hydroxy-5(Z),8(Z),10(E),14(Z)-eicosatetraenoic acid [12(R)-HETE], an arachidonic acid derivative, is an activator of the aryl hydrocarbon receptor. Mol. Pharmacol. 2008b;74:1649–1656. doi: 10.1124/mol.108.049379. [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Elferink CJ. Aryl hydrocarbon receptor-mediated cell cycle control. Prog. Cell Cycle Res. 2003;5:261–267. [PubMed] [Google Scholar]
- Flaveny CA, Murray IA, Chiaro CR, Perdew GH. Ligand selectivity and gene regulation by the human aryl hydrocarbon receptor in transgenic mice. Mol. Pharmacol. 2009;75:1412–1420. doi: 10.1124/mol.109.054825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasiewicz TA, Henry EC, Collins LL. Expression and activity of aryl hydrocarbon receptors in development and cancer. Crit. Rev. Eukaryot. Gene Expr. 2008;18:279–321. doi: 10.1615/critreveukargeneexpr.v18.i4.10. [DOI] [PubMed] [Google Scholar]
- Gomez-Duran A, Carvajal-Gonzalez JM, Mulero-Navarro S, Santiago-Josefat B, Puga A, Fernandez-Salguero PM. Fitting a xenobiotic receptor into cell homeostasis: how the dioxin receptor interacts with TGFβ signaling. Biochem. Pharmacol. 2009;77:700–712. doi: 10.1016/j.bcp.2008.08.032. [DOI] [PubMed] [Google Scholar]
- Gouedard C, Barouki R, Morel Y. Dietary polyphenols increase paraoxonase 1 gene expression by an aryl hydrocarbon receptor-dependent mechanism. Mol. Cell. Biol. 2004a;24:5209–5222. doi: 10.1128/MCB.24.12.5209-5222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouedard C, Barouki R, Morel Y. Induction of the paraoxonase-1 gene expression by resveratrol. Arteroscler. Thromb. Vasc. Biol. 2004b;24:2378–2383. doi: 10.1161/01.ATV.0000146530.24736.ce. [DOI] [PubMed] [Google Scholar]
- Haarmann-Stemmann T, Bothe H, Abel J. Growth factors, cytokines and their receptors as downstream targets of arylhydrocarbon receptor (AhR) signaling pathways. Biochem. Pharmacol. 2009;77:508–1250. doi: 10.1016/j.bcp.2008.09.013. [DOI] [PubMed] [Google Scholar]
- Henry EC, Bemis JC, Henry O, Kende AS, Gasiewicz TA. A potential endogenous ligand for the aryl hydrocarbon receptor has potent agonist activity in vitro and in vivo. Arch. Biochem. Biophys. 2006;450:67–77. doi: 10.1016/j.abb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu H-Y, Laine J, Sakai T, Korsmeyer SJ, Casper RF, et al. Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nat. Genet. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- Matsumura F. The significance of the nongenomic pathway in mediating inflammatory signaling of the dioxin-activated Ah receptor to cause toxic effects. Biochem. Pharmacol. 2009;77:608–626. doi: 10.1016/j.bcp.2008.10.013. [DOI] [PubMed] [Google Scholar]
- McDonnell DP, Clemm DL, Herman T, Goldman ME, Pike JW. Analysis of estrogen receptor function in vitro reveals three distinct classes of antiestrogens. Mol. Endocrinol. 1995;9:659–669. doi: 10.1210/mend.9.6.8592512. [DOI] [PubMed] [Google Scholar]
- McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol. Pharmacol. 2007;72:487–498. doi: 10.1124/mol.107.037259. [DOI] [PubMed] [Google Scholar]
- Mitchell KA, Elferink CJ. Timing is everything: consequences of transient and sustained AhR activity. Biochem. Pharmacol. 2009;77:947–956. doi: 10.1016/j.bcp.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem. Res. Toxicol. 2008;21:102–116. doi: 10.1021/tx7001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pande K, Moran SM, Bradfield CA. Aspects of dioxin toxicity are mediated by interleukin 1-like cytokines. Mol. Pharmacol. 2005;67:1393–1398. doi: 10.1124/mol.105.010983. [DOI] [PubMed] [Google Scholar]
- Patel RD, Kim DJ, Peters JM, Perdew GH. The aryl hydrocarbon receptor directly regulates expression of the potent mitogen epiregulin. Toxicol. Sci. 2006;89:75–82. doi: 10.1093/toxsci/kfi344. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-qPCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga A, Tomlinson CR, Xia Y. Ah receptor signals cross-talk with multiple developmental pathways. Biochem. Pharmacol. 2005;69:199–207. doi: 10.1016/j.bcp.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E, Caccamo M, Oukka M, Weiner HL. Control of Tregand TH17 cell differentiation by the aryl hydrocarbon receptor. Nature. 2008;453:65–71. doi: 10.1038/nature06880. [DOI] [PubMed] [Google Scholar]
- Sartor MA, Schnekenburger M, Marlowe JL, Reichard JF, Wang Y, Fan Y, Ma C, Karyala S, Halbleib D, Liu X, et al. Genomewide analysis of aryl hydrocarbon receptor binding targets reveals an extensive array of gene clusters that control morphogenetic and developmental programs. Environ. Health Perspect. 2009;117:1139–1146. doi: 10.1289/ehp.0800485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaldach CM, Riby J, Bjeldanes LF. Lipoxin A4: a new class of ligand for the Ah receptor. Biochemistry. 1999;38:7594–7600. doi: 10.1021/bi982861e. [DOI] [PubMed] [Google Scholar]
- Song J, Clagett-Dame M, Peterson RE, Hahn ME, Westler WM, Sicinski RR, DeLuca HF. A ligand for the aryl hydrocarbon receptor isolated from lung. Proc. Natl Acad. Sci. USA. 2002;99:14694–14699. doi: 10.1073/pnas.232562899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski T. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–4523. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher TH, Maggirwar SB, Baglole CJ, Lakatos HF, Gasiewicz TA, Phipps RP, Sime PJ. Aryl hydrocarbon receptor-deficient mice develop heightened inflammatory responses to cigarette smoke and endotoxin associated with rapid loss of the nuclear factor-kB component RelB. Am. J. Physiol. 2007;170:855–864. doi: 10.2353/ajpath.2007.060391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel CFA, Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kB family. Biochem. Pharmacol. 2009;77:734–745. doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijayaratne AL, McDonnell DP. The human estrogen receptor-a is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J. Biol. Chem. 2001;276:35684–35692. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- Wincent E, Amini N, Luecke S, Glatt H, Bergman J, Crescenzi C, Rannug A, Rannug U. The suggested physiologic aryl hydrocarbon receptor activator and cytochrome P4501 substrate 6-formylindolo[3,2-b]carbazole is present in humans. J. Biol. Chem. 2009;284:2690–2696. doi: 10.1074/jbc.M808321200. [DOI] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA. Preprocessing of oligonucleotide array data. Nat. Biotechnol. 2004;22:656–658. doi: 10.1038/nbt0604-656b. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.