Abstract
Transcription of the Escherichia coli biotin (bio) operon is regulated by BirA, a protein that is not only the repressor that regulates bio operon expression by DNA binding but also the enzyme that covalently attaches biotin to its cognate acceptor proteins. Binding of BirA to the bio operator requires dimerization of the protein that is triggered by BirA-catalyzed synthesis of biotinoyl-adenylate (bio-AMP), the obligatory intermediate of the attachment reaction. The current model postulates that the unmodified acceptor protein binds the monomeric BirA:bio-AMP complex and thereby blocks assembly (dimerization) of the form of BirA that binds DNA. We report that expression of fusion proteins that carry synthetic biotin accepting peptide sequences was as effective as the natural acceptor protein in derepression of bio operon transcription. These peptide sequences have sequences that are remarkably dissimilar to that of the natural acceptor protein and thus our data argue that the regulatory switch does not require the extensive protein-protein interactions postulated in the current model.
Keywords: biotin, BirA, repressor/enzyme, growth rate control, biotin protein ligase, biotinylation
Introduction
Biotin (vitamin H) is an enzyme cofactor required for central metabolism throughout the biological world. The cofactor is a component of essential carboxylases and decarboxylases whose function requires covalent attachment of biotin to the enzyme protein (the free form of biotin is not a physiologically relevant enzyme cofactor). Biotin protein ligases (BPLs) are the enzymes that attach biotin to its cognate enzymes (Fig. 1A) and a single ligase is responsible for biotinylation of the ε-amino group of a specific lysine residue of each of the biotin-dependent enzymes of an organism (Chapman-Smith and Cronan, 1999). In mammals biotin is an essential nutrient whereas plants, most bacteria and most fungi synthesize the vitamin. The biotin synthetic pathway is long and metabolically expensive and thus tight regulation of biotin synthesis is expected. In Escherichia coli expression of the biotin synthetic (bio) operon is controlled by a simple, yet remarkably sophisticated, regulatory system in which the rate of operon transcription responds not only to the supply of biotin, but also to the supply of the enzyme proteins (called biotin acceptor proteins) that become modified by covalent attachment of biotin (Beckett, 2007; Chapman-Smith and Cronan, 1999; Cronan, 1989). This regulatory system is understood in considerable detail due to a combination of genetic, physiological, biochemical and biophysical investigations and provides a striking example of a transcriptional regulatory protein (BirA) that is also an enzyme, in this case the BPL that catalyzes the covalent attachment of biotin to the essential fatty acid synthetic protein, AccB. Moreover, regulation of the E. coli biotin operon is probably the best understood example of transcriptional regulation by an enzyme unrelated to nucleic acid metabolism. The biotin regulatory system superficially resembles the classical TrpR regulation of the E. coli tryptophan operon where the Trp repressor protein binds to the trpEDCBA operator only when complexed with the corepressor, tryptophan. However, in bio operon regulation, the repressor (called BirA in E. coli) is also the sole cellular BPL and the corepressor is not biotin, but biotinoyl-5′-AMP (bio-AMP), the product of the first partial reaction of BirA catalysis (Fig. 1) (Beckett, 2007; Chapman-Smith and Cronan, 1999; Cronan, 1989; Prakash and Eisenberg, 1979). Thus, BirA catalyzes synthesis of its regulatory ligand. It is these novel features that give this regulatory system its sophisticated properties. It should be noted that a distantly related bacterium, Bacillus subtilis, is reported to similarly regulate its biotin synthetic pathway (Bower et al., 1995; Bower et al., 1996) and bioinformatic analyses argue that this regulatory system is widespread in bacteria (Rodionov et al., 2002).
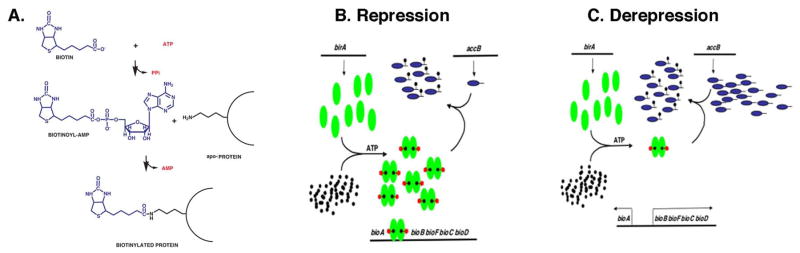
Figure 1.
(A) The biotin protein ligase (BPL) reaction and the general model of bio operon regulation. Biotin is attached to small (60–80 residues) well-conserved domains that are generally found at the C-termini of the acceptor proteins. (B & C) General model of repression and derepression of E. coli bio operon transcription by AccB supply. Green ovals denote BirA, tailed blue ovals are AccB, black dots are biotin and black dots with red pentagons are bio-AMP. (B) Under conditions of low AccB synthesis such as slow growing cells, the BirA:bio-AMP monomeric complex accumulates resulting in dimerization. The dimers bind the operator DNA and repress transcription. (C) Under conditions where the AccB synthetic rate is high, the monomeric BirA:bio-AMP complex does not accumulate due to biotinylation of AccB to give the active protein. Hence, dimerization does not occur and the bio promoters remain free to bind RNA polymerase. The bio genes are transcribed to produce the biotin biosynthetic proteins which act to replace the biotin consumed in biotinylation and thereby match biotin synthesis to its consumption in protein biotinylation (Beckett, 2007).
In the general model of BirA regulation (Fig. 1B) the maximal rates of E. coli bio operon transcription (derepression) occur when the biotin supply is severely limited (e.g., biotin starvation) or when high levels of a biotin acceptor protein are present (Beckett, 2007; Chapman-Smith and Cronan, 1999; Cronan, 1989). Under these conditions any bio-AMP synthesized is rapidly consumed in biotinylation of the acceptor proteins (AccB) and thus the BirA:bio-AMP complex cannot accumulate. Hence, BirA remains largely monomeric and transcription is maximal because the bio operator is seldom occupied (Fig. 1C). When all of the available AccB has been biotinylated, newly synthesized biotin-AMP remains bound in the BirA active site and the BirA:bio-AMP complex accumulates and dimerizes. These dimers then bind the operator sequence of the biotin operon resulting in repression of biotin biosynthetic gene transcription (Fig. 1B). The two conditions that result in derepression therefore act by a common mechanism in that both result in decreased levels of the BirA:bio-AMP complex required for binding the bio operator. However, it should be noted that of the two regulatory ligands, biotin and AccB, the latter plays the key regulatory role in vivo because it allows biotin synthesis to match AccB synthesis (James and Cronan, 2004; Li and Cronan, 1993)). Note that AccB and AccB-87, the last 87 residues of the protein which comprise the biotinylation domain, are used in their formal sense, as the primary translation products unmodified by biotin attachment.
The mechanism whereby accumulation of the unmodified biotin domain leads to increased transcription of the bio operon remains unclear. What is the mechanism of the regulatory switch and how is it triggered? The current model, that of Beckett, Matthews and coworkers (Fig. 2A), postulates that the unmodified acceptor protein binds a monomeric BirA:bio-AMP complex and thereby competes with formation of the dimeric complex, the species required for effective repression. The BirA surface used to form the heterodimer is proposed to be extensive and to be the same surface used in forming the BirA homodimer (Weaver et al., 2001). Hence, in this model competing protein-protein interactions are responsible for derepression triggered by accumulation of unmodified biotin acceptor domains. Implicit in this model is that the heterodimer must be sufficiently long-lived to compete with homodimerization of BirA:bio-AMP and thus as in the case of the homodimer, direct detection of the heterodimer might be expected. However, direct detection of the postulated AccB plus BirA:bio-AMP heterodimer has not been reported and only indirect evidence for its existence is available (Zhao and Beckett, 2008). In our view this is a major caveat to the model. An argument against the competing protein-protein interactions model is that the time scale of enzyme substrate interactions is generally short such that the lifetime of the postulated heterodimer would so brief that its formation could not effectively compete with BirA homodimer formation. However, enzyme-substrate interactions can be relatively long lived (Frieden, 1970, 2008).
Figure 2.
The competing protein interactions model of the biotin operon regulatory switch and alignments of biotin-accepting sequences. (A) Shown are the regulatory effects engendered by differing levels of AccB expression. The symbols are as in Fig. 1. In the competing protein-protein interaction model a BirA:bio-AMP monomer is proposed to interact with AccB and thereby inhibit dimerization of the BirA:bio-AMP complex, the species required for DNA binding (Beckett, 2007). When the rate of AccB synthesis and demand for biotin decreases BirA:bio-AMP dimers accumulate and repress bio operon transcription. (B) Alignment of the residues neighboring the biotin attachment site (asterisk) of E. coli AccB and the five biotin acceptor proteins from species whose BPLs are reported to biotinylate E. coli AccB, but not peptide 85 fusion proteins. For comparison the sequences of peptides 85 and ME are also given. Residues conserved in at least four of the sequences are boxed. The natural sequences are: Ec, E. coli AccB; Bs, Bacillus subtilis AccB: Mj, Methanocaldococcus jannaschii pyruvate carboxylase; Sc, Saccharomyces cerevesiae pyruvate carboxylase 1; At, Arabidopsis thaliana AccB and Hs, Homo sapiens, pyruvate carboxylase. Note that the in the libraries that yielded the two peptide sequences of interest, the codons of the conserved alanine residue located two resides upstream of the biotinylated lysine together with the upstream and downstream glutamate residues were 97% enriched.
We expected that a strong test of the competing protein-protein interaction model would be provided by the small peptide substrates that undergo BirA-catalyzed biotinylation isolated from peptide libraries by Schatz (Schatz, 1993) as a biotechnology tool. When fused to either end of a wide variety of proteins, these sequences of 14 to 30 residues are efficiently biotinylated by BirA (Cull and Schatz, 2000; Schatz, 1993). Surprisingly, these peptides have sequences remarkably unlike that of the natural substrate, AccB (Fig. 2B). Indeed, although certain peptide library residues were fixed to match those bracketing the AccB lysine residue that becomes biotinylated, many of these residues failed to survive the selection for biotinylation (Schatz, 1993). Despite the lack of sequence similarity to AccB, one of these peptides (peptide 85-14) was shown to be as active a biotin acceptor as AccB-87 (Beckett et al., 1999) which, in turn, is as good an acceptor as full-length AccB (Nenortas and Beckett, 1996). The lack of sequence conservation with AccB argues strongly that these peptide sequences bind to BirA by modes that differ markedly from that used by AccB. Compelling support for this argument comes from reports that ligases from other prokaryotic organisms (B. subtilis and the archaeon, Methanococcus jannaschii) and from eukaryotes (the yeast, plant, insect and human enzymes) are unable to biotinylate peptide 85 fusion proteins (Chen et al., 2007; de Boer et al., 2003; Duffy et al., 1998; Grosveld et al., 2005; Kaikkonen et al., 2008; Slavoff et al., 2008; Tissot et al., 1998) whereas each of these ligases modifies AccB (Chen et al., 2007; Leon-Del-Rio et al., 1995; Polyak et al., 1999; Polyak et al., 2001; Tissot et al., 1998). It seems clear that the Schatz peptide sequences cannot adopt structures similar to that of AccB, otherwise they would act as general BPL substrates. Moreover, due to the short lengths of the acceptor peptide sequences the extended interactions proposed for the AccB-BirA interface would be impossible. Therefore, if extensive protein-protein interactions are required for derepression of bio operon transcription, then expression of Schatz peptide fusion proteins should result in little or no increase in bio operon expression. On the other hand if the fusion proteins were effective in derepression, this would argue that competing protein-protein interactions are not the regulatory switch and that the switch mechanism must lay elsewhere. We report that in vivo production of fusion proteins containing the peptide 85 sequence or a second Schatz peptide sequence both resulted in powerful and efficient derepression of biotin operon expression.
Results
Our experimental system consisted of two maltose binding protein (MBP) fusion proteins in which either the 15-mer version of the peptide 85 sequence which we call Pep-85 (Schatz, 1993) or an unnamed peptide later described by Cull and Schatz (Cull and Schatz, 2000) (which we call Pep-ME) comprised the carboxyl termini of the fusion proteins (Fig. 2B). We also tested the peptide 8 sequence (Schatz, 1993), but these analyses were compromised by intracellular degradation of the fusion protein and will not be further discussed. A control plasmid in which the AccB-87 sequence was similarly fused to MBP was also constructed (the MBP constructs lack the periplasmic targeting sequence and thus are cytosolic proteins). A negative control plasmid in which the AccB-87 partner of the fusion protein had been inactivated by substituting methionine for the lysine residue that is the site of biotin attachment was also constructed.
Fusion to the carboxyl terminus of MBP was chosen because this was the system that Schatz originally used to analyze his peptide sequences (Cull and Schatz, 2000; Schatz, 1993) and biotinylation of a similar MBP-peptide 85 fusion protein has been studied in vitro (Beckett et al., 1999). These plasmids together with the MBP vector plasmid lacking a biotinylation sequence were introduced into E. coli CY1740, a strain in which biotin operon expression drives expression of LacZ β-galactosidase activity (Abdel-Hamid and Cronan, 2007; Barker and Campbell, 1980). The strain is also a biotin auxotroph (due to insertion of the lac genes into the bio operon) thus permitting manipulation of intracellular biotin concentrations and labeling with [3H]-biotin without dilution by endogenously synthesized biotin (Barker and Campbell, 1980; Cronan, 1988).
The first indication that expression of the two Schatz peptide fusion proteins gave efficient derepression of bio operon transcription was their behavior on MacConkey indicator plates containing 115 nM biotin (Fig. 4A). The red color of the colonies expressing the biotin accepting peptide sequences or AccB-87 fusion proteins is due to increased transcription from the bioBFCD promoter that drives expression of the lacZ and lacY genes. The color is due to increased transcription of the bioBFCD promoter which results in lactose utilization, the acidic products of which react with indicator dyes in the medium. In contrast the same host strain carrying either the empty vector plasmid or a version of the AccB-87 fusion protein (called K122M) in which the biotin accepting lysine residue (K122 of native AccB) had been changed to methionine gave colorless colonies indicating that lactose was not utilized (Fig. 3A). Derepression relative to the vector control was seen upon expression of the three biotin-accepting fusion proteins over a 100-fold range of biotin concentrations (Fig. 3B). At 100 nM biotin, a concentration that normally fully represses bio operon transcription (Fig. 4B) (Barker and Campbell, 1980; Cronan, 1988), expression of each of the three fusion proteins resulted in a ca. 200-fold derepression of bio operon expression relative to the host strain producing the K122M mutant accepting protein or the unfused maltose binding protein (Fig. 3C). Although in a given experiment one of the biotin-accepting fusion proteins often gave a 10–20% greater level of derepression than the others, when assessed over all of our experiments, no clear pecking order emerged. Since the expression of a biotin accepting protein will necessarily deplete intracellular bio-AMP levels, we also measured the levels of protein biotinylation in the presence of 100 nM [3H]-biotin (Fig. 3D). The peptide fusion proteins were found to be somewhat more efficient than the MBP-AccB-87 fusion when the level of bio operon derepression per protein biotinylation event was calculated. Western blotting with anti-MBP antibody indicated that the plasmids encoding the fusion proteins all produced comparable levels of MBP (Fig. 4A) and western blotting with streptavidin also demonstrated biotinylation of all three fusion proteins (Fig. 4B). The high expression level of the fusion proteins was designed to insure an excess of unbiotinylated fusion protein. To test if this was indeed the case, we performed experiments using unlabeled biotin at 100 nM and then washed the cells free of biotin. Cell extracts were prepared and incubated with ATP plus [3H]-biotin to allow biotinylation of any remaining acceptor protein by the BirA present in the extract. Since the [3H]-biotin had the same specific activity as that used in vivo, we could directly calculate the levels of unbiotinylated acceptor proteins remaining in the cell extracts by comparison of the in vitro data with those obtained by incorporation of [3H]-biotin in vivo in a parallel experiment. These experiments showed that about half of each of the biotin acceptor proteins remained unbiotinylated in cells grown with 100 nM biotin (data not shown).
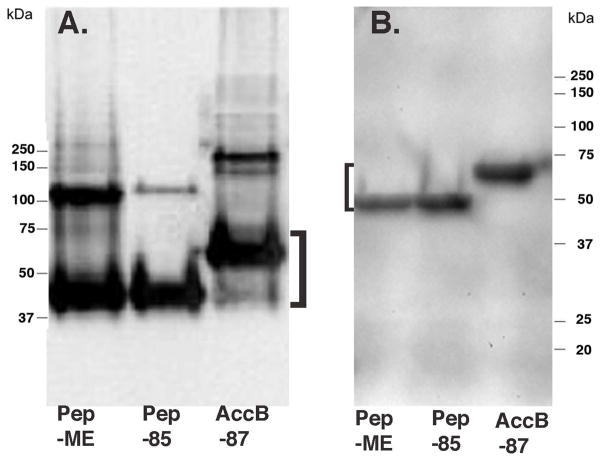
Fig. 4.
Levels of total MBP-fusion proteins and biotinylated MBP-fusion proteins. Polyacrylamide gels (12%) run in the presence of sodium dodecyl sulfate from two different experiments are shown. Both sets of cultures were grown with 100 nM biotin. (A) Detection with anti-MBP antibody. The upper bands appear to be the product of insufficient denaturation (dimers?) rather than cross-reacting contaminants because their apparent sizes vary with those of the major band in the lane and because in other experiments they were found to bind streptavidin thereby indicating biotinylation (B). Detection by streptavidin binding to the biotinylated proteins. The fusion proteins are denoted by brackets and the migration positions of the molecular weight markers (Kaleidoscope Protein standard from Bio-Rad) are given alongside the gels. The AccB band encoded from the chromosome was not seen at this exposure due to the abundance of the fusion proteins relative to AccB plus the competition for biotinylation that results in decreased biotinylation of AccB [37]. The electrophoresis time of the gel (A) was longer than that of gel (B) to increase resolution. The proteins were transferred to Immobilon-P membranes (Millipore) and detected as described in Experimental Procedures.
Figure 3.
Derepression of bio operon transcription by expression of biotin accepting proteins. In host strain CY1740 which carries the φ(bioFC-lacZ)501 fusion, the lacZ and lacY genes are fused to the rightward bio promoter (that of bioBFCD) resulting a lactose-positive phenotype when bio operon transcription is derepressed and a lactose-negative phenotype when the operon is repressed. This strain also carries a deletion of the chromosomal lactose operon and ia a biotin auxotroph due to the insertion of the lactose utilization genes into bioF. (A). The host strain was transformed with either the empty vector plasmid or plasmids encoding the Pep-ME, Pep-85 or AccB-87 fusion proteins. Also included was a strain CY1740 derivative that carried a plasmid encoding a mutant AccB-87 fusion protein (K122M) in which the lysine targeted for biotinylation had been replaced with methionine. Four independent transformants from each transformation were patched onto an LB-ampicillin plate that was incubated at 37°C for 8 h. The cells were then transferred by replica plating to a plate of MacConkey agar supplemented with 100 nM biotin (ca. 115 nM total) and ampicillin. After overnight incubation at 37°C derepression of bio operon transcription results in bright red colonies due to lactose utilization whereas repressed colonies remain colorless due to their inability to metabolize lactose. (B) Expression at various biotin concentrations of derivatives of strain CY1740 carrying plasmids encoding maltose binding protein fusions to biotin accepting peptides 85 (■) or ME (●). The maltose binding protein fusion to AccB-87 (▲) and the vector plasmid encoding a maltose binding protein that lacked a fusion partner (◆) are also shown. β-Galactosidase activities were assayed 4 h after induction of the maltose binding protein fusions with IPTG (1 mM). Note that the abscissa is a logarithmic scale. (C) Reproducibility of derepression. Four independent transformants from each of the above transformations were streaked on minimal medium plates supplemented with 100 nM biotin. Colonies from these plates were then used to inoculate overnight cultures on the same medium which in turn were diluted 1:100 into fresh medium containing 100 nM biotin and IPTG. These cultures were allowed to grow to late log phase prior to β-galactosidase assays. (D) Biotin attachment to the fusion proteins. Biotinylation of the acceptor proteins was assayed by incorporation of [3H]-biotin into trichloroacetic acid insoluble material (Abdel-Hamid and Cronan, 2007; Chapman-Smith et al., 1999; Cronan, 1988) 4 h after IPTG induction of expression of the maltose binding protein fusions. In the strain carrying the vector plasmid the attachment of biotin is to the essential chromosomally encoded AccB protein.
Discussion
We have shown that two fusion proteins containing synthetic biotin accepting sequences are as efficient in derepression of bio operon transcription as the natural acceptor domain AccB-87. Therefore, although the competing protein-protein interactions model of BirA transcriptional control has several attractive features, our data argue that it cannot play an important role in the regulatory switch. Our conclusions are based on the several lines of evidence indicating that the Schatz peptide sequences bind BirA in a markedly different manner than does AccB. The main lines of evidence are:
As noted in the Introduction six BPLs from diverse organisms biotinylate AccB, but cannot modify proteins carrying the peptide 85 sequence. Since BPLs and their natural acceptor proteins are generally interchangeable across biology (Cronan and Reed, 2000), if the peptide 85 sequence mimicked structural attributes of AccB, the sequence should be biotinylated by BPLs other than BirA. However, BirA is the only ligase known to biotinylate the peptide 85 sequence.
The Schatz peptides and AccB-87 differ markedly. Peptides 85 and ME lack the last two of the AccB residues (residues 122–128) postulated to interact with the main BirA dimerization surface (Weaver et al., 2001) and no residue other than the biotinylated lysine is conserved between these peptides and AccB residues 122–128. Moreover, the other AccB regions (residues 88–90 and residue 145) thought to be important in the competing protein-protein interactions model (Weaver et al., 2001) are absent because are located outside the sequences chosen by Schatz in his design of the peptide libraries. Moreover, a recent crystal structure of a complex of a biotinylated protein similar to AccB-87 and its cognate BPL from the archaeon, Pyrococcus horikoshii, shows that aside from the biotinylated lysine region, the residue 88–90 region is the major site of interaction between the two proteins (Bagautdinov et al., 2008). The importance of this region is supported by the finding that its deletion from natural biotin acceptor proteins greatly weakens BirA-catalyzed biotinylation (Cronan and Reed, 2000; Leon-Del-Rio and Gravel, 1994). Therefore, the Schatz peptides would be expected to be very weak biotin acceptors because they lack the AccB residue 88–90 sequence. However, as noted above, peptide 85 derivatives are biotinylated as efficiently as is AccB-87 arguing that AccB and the Schatz peptides must bind BirA by different modes. It should be noted that, although the peptides ME and 85 have related sequences, the former peptide lacks the N-terminal glycine of peptide 85, removal of which causes a severe (ca. 10-fold) decrease in the rate of biotinylation is vitro (Beckett et al., 1999). Since we find that the fusion proteins containing the two peptide sequences derepress bio operon transcription similarly and have similar biotinylation rates in vivo, it seems possible that even these two related peptides may bind BirA differently. It should be noted that circular dichroism spectra (Beckett et al., 1999) indicate that the 85-14 peptide is unstructured in solution, thus precluding modeling of its interactions with BirA,
Biotinylation of AccB-87 has been shown to be sensitive to substitutions of the residues neighboring the biotinylated lysine residue (Reche et al., 1998; Reche and Perham, 1999) whereas the neighboring residues of the Schatz biotin-accepting peptides are remarkably diverse (Schatz, 1993). For example, a glutamate residue positioned three residues before the target lysine residue is very highly conserved in naturally occurring biotinylated proteins (Bagautdinov et al., 2008). This residue makes a hydrogen bond with the main chain nitrogen of a lysine residue believed important in recognition of the biotin domain by BPL (Bagautdinov et al., 2008) and mutagenesis of this residue in AccB-87 abolishes biotinylation (Chapman-Smith et al., 1999). In contrast, Schatz isolated biotin-accepting peptides in which arginine, tyrosine, aspartate, asparagine, leucine, alanine or threonine were found at this position. Indeed, even in libraries constructed such that glutamate was strongly favored at this position, other residues emerged from the selection. In the most extreme case a library having 97% glutamate codons at this position gave biotinylated peptide sequences in which more than half of modified peptides contained a different residue at this position (Schatz, 1993). Since some of these residues have side chains that cannot participate in hydrogen bonding, it follows that the rules governing biotinylation are markedly different for AccB and the peptide sequences.
The above discussion aside, it might be argued that the Schatz peptide sequences are somehow able to form appropriately long lived complexes with BirA:bio-AMP that are as effective in derepression as the postulated complex with AccB. This possibility cannot be totally excluded, but seems extremely remote. For this to be the case the peptides would have to make interactions with BirA:bio-AMP that have the same binding strengths and kinetics as that of the natural acceptor protein despite their small size and markedly diverged sequences. This seems most unlikely, particularly since Schatz selected only for the ability to accept biotin and not for derepression (Schatz, 1993). Moreover, his selections were done in the presence of excess biotin, which precluded any indirect selection for derepression. Given our data we argue that the competing protein-protein interactions postulated in the Beckett model is not the regulatory switch of the bio operon. What then is the switch? An older model proposed that bio-AMP levels provide the switch (Cronan, 1989). In that model the switch was removal of bio-AMP from the BirA active site by protein biotinylation and thus AccB (or the Schatz peptides) would act only as classical enzyme substrates in which the interaction between AccB and BirA would be brief. The acceptor protein would bind, accept a biotin moiety from the BirA active site and then rapidly dissociate and leave the BirA protein as the monomeric form which would be unable to dimerize until it could synthesize another molecule of bio-AMP. Therefore, a high rate of acceptor protein production would result in a mainly monomeric pool of BirA and thus bio operon transcription would be derepressed (Zhao and Beckett, 2008). Our data are fully consistent with this model.
Note that BirA is not unique in utilizing small peptides as efficient biotin acceptors. Peptide sequences have been selected that are biotinylated by the yeast BPL, but not by BirA. Consistent with the E. coli results the sequences these peptides bear little resemblance to the natural yeast acceptor domains (Chen et al., 2007).
Experimental Procedures
The growth medium, E. coli strains and the assays of β-galactosidase activity and protein biotinylation were as described previously (Abdel-Hamid and Cronan, 2007). In vitro biotinylation was performed as previously described (Chapman-Smith et al., 1994). The two Schatz peptide MBP fusion proteins were constructed using cassettes made by annealing complementary oligonucleotides: 5′-AATTCGGAGGACTGAACGACATCTTTGAAGCGCAAAAAATCGAATGGCATTA plus 5′-AGCTTAATGCCATTCGATTTTTTGCGCTTCAAAGATGTCGTTCAGTCCTCCG for peptide 85 and 5′-AATTCGGACTGTGTGACATCTTCGAATCCCAGAAAATCGAATGGCATTCTGCGTA plus 5′-AGCTTACGCAGAATGCCATTCGATTTTCTGGGATTCGAAGATGTCACACAGTCCG for peptide ME. Annealing of the oligonucleotide pairs resulted in EcoRI and HindIII sticky ends that were ligated to pMAL-2c-E (New England Biolabs) cut with the same enzymes to give genes encoding the MBP-peptide fusion proteins. The MBP-AccB-87 fusion protein was constructed by cutting pCY142 (Reed and Cronan, 1991) with SalI plus XhoI and ligating the purified AccB-87 fragment to pMAL-2c-E cut with SalI. All constructs were verified by DNA sequencing. The plasmid encoding the K122M AccB-87 fusion protein was constructed by replacement of the NcoI-PstI fragment of the plasmid encoding the MBP-AccB-87 fusion protein with the Nco-NsiI fragment of the plasmid encoding the K122M mutant form of AccB constructed previously (Abdel-Hamid and Cronan, 2007). Western blotting with anti-MBP (New England Biolabs) were done according to the protocol provided with the reagent. The MBP-protein complexes were detected by the ECL system (Amersham) using a horseradish peroxidase-coupled sheep anti-mouse IgG (Amersham) as the secondary antibody whereas the streptavidin whereas blotting with a streptavidin-enzyme complex was done as described previously (Choi-Rhee et al., 2004). In both cases chemiluminescence detection was done on a Bio-Rad Chemidoc XRS system. The molecular mass markers were the Precision Plus.
Acknowledgments
This work was supported by National Institutes of Health grant AI15650.
Footnotes
Significance
Derepression of bio operon transcription is efficiently triggered by expression of synthetic short non-native biotin-accepting peptide sequences indicating that the regulatory switch does not depend on the extensive protein-protein interactions between AccB and the monomeric BirA-biotinoyl-AMP complex postulated in the current model.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Hamid AM, Cronan JE. Coordinate expression of the acetyl coenzyme A carboxylase genes, accB and accC, is necessary for normal regulation of biotin synthesis in Escherichia coli. J Bacteriol. 2007;189:369–376. doi: 10.1128/JB.01373-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagautdinov B, Matsuura Y, Bagautdinova S, Kunishima N. Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate. J Biol Chem. 2008;283:14739–14750. doi: 10.1074/jbc.M709116200. [DOI] [PubMed] [Google Scholar]
- Barker DF, Campbell AM. Use of bio-lac fusion strains to study regulation of biotin biosynthesis in Escherichia coli. J Bacteriol. 1980;143:789–800. doi: 10.1128/jb.143.2.789-800.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D. Biotin sensing: universal influence of biotin status on transcription. Annu Rev Genet. 2007;41:443–464. doi: 10.1146/annurev.genet.41.042007.170450. [DOI] [PubMed] [Google Scholar]
- Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower S, Perkins J, Yocum RR, Serror P, Sorokin A, Rahaim P, Howitt CL, Prasad N, Ehrlich SD, Pero J. Cloning and characterization of the Bacillus subtilis birA gene encoding a repressor of the biotin operon. J Bacteriol. 1995;177:2572–2575. doi: 10.1128/jb.177.9.2572-2575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower S, Perkins JB, Yocum RR, Howitt CL, Rahaim P, Pero J. Cloning, sequencing, and characterization of the Bacillus subtilis biotin biosynthetic operon. J Bacteriol. 1996;178:4122–4130. doi: 10.1128/jb.178.14.4122-4130.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman-Smith A, Cronan JE., Jr The enzymatic biotinylation of proteins: a post-translational modification of exceptional specificity. Trends Biochem Sci. 1999;24:359–363. doi: 10.1016/s0968-0004(99)01438-3. [DOI] [PubMed] [Google Scholar]
- Chapman-Smith A, Morris TW, Wallace JC, Cronan JE., Jr Molecular recognition in a post-translational modification of exceptional specificity. Mutants of the biotinylated domain of acetyl-CoA carboxylase defective in recognition by biotin protein ligase. J Biol Chem. 1999;274:1449–1457. doi: 10.1074/jbc.274.3.1449. [DOI] [PubMed] [Google Scholar]
- Chapman-Smith A, Turner DL, Cronan JE, Jr, Morris TW, Wallace JC. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem J. 1994;302:881–887. doi: 10.1042/bj3020881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I, Choi YA, Ting AY. Phage display evolution of a peptide substrate for yeast biotin ligase and application to two-color quantum dot labeling of cell surface proteins. J Am Chem Soc. 2007;129:6619–6625. doi: 10.1021/ja071013g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi-Rhee E, Schulman H, Cronan JE. Promiscuous protein biotinylation by Escherichia coli biotin protein ligase. Protein Sci. 2004;13:3043–3050. doi: 10.1110/ps.04911804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE., Jr Expression of the biotin biosynthetic operon of Escherichia coli is regulated by the rate of protein biotination. J Biol Chem. 1988;263:10332–10336. [PubMed] [Google Scholar]
- Cronan JE., Jr The E. coli bio operon: transcriptional repression by an essential protein modification enzyme. Cell. 1989;58:427–429. doi: 10.1016/0092-8674(89)90421-2. [DOI] [PubMed] [Google Scholar]
- Cronan JE, Jr, Reed KE. Biotinylation of proteins in vivo: a useful posttranslational modification for protein analysis. Methods Enzymol. 2000;326:440–458. doi: 10.1016/s0076-6879(00)26069-2. [DOI] [PubMed] [Google Scholar]
- Cull MG, Schatz PJ. Biotinylation of proteins in vivo and in vitro using small peptide tags. Methods Enzymol. 2000;326:430–440. doi: 10.1016/s0076-6879(00)26068-0. [DOI] [PubMed] [Google Scholar]
- de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc Natl Acad Sci U S A. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy S, Tsao KL, Waugh DS. Site-specific, enzymatic biotinylation of recombinant proteins in Spodoptera frugiperda cells using biotin acceptor peptides. Anal Biochem. 1998;262:122–128. doi: 10.1006/abio.1998.2770. [DOI] [PubMed] [Google Scholar]
- Frieden C. Kinetic aspects of regulation of metabolic processes. The hysteretic enzyme concept. J Biol Chem. 1970;245:5788–5799. [PubMed] [Google Scholar]
- Frieden C. A lifetime of kinetics. J Biol Chem. 2008;283:19873–19878. doi: 10.1074/jbc.X800003200. [DOI] [PubMed] [Google Scholar]
- Grosveld F, Rodriguez P, Meier N, Krpic S, Pourfarzad F, Papadopoulos P, Kolodziej K, Patrinos GP, Hostert A, Strouboulis J. Isolation and characterization of hematopoietic transcription factor complexes by in vivo biotinylation tagging and mass spectrometry. Ann N Y Acad Sci. 2005;1054:55–67. doi: 10.1196/annals.1345.008. [DOI] [PubMed] [Google Scholar]
- James ES, Cronan JE. Expression of two Escherichia coli acetyl-CoA carboxylase subunits is autoregulated. J Biol Chem. 2004;279:2520–2527. doi: 10.1074/jbc.M311584200. [DOI] [PubMed] [Google Scholar]
- Kaikkonen MU, Viholainen JI, Narvanen A, Yla-Herttuala S, Airenne KJ. Targeting and purification of metabolically biotinylated baculovirus. Hum Gene Ther. 2008;19:589–600. doi: 10.1089/hum.2007.177. [DOI] [PubMed] [Google Scholar]
- Leon-Del-Rio A, Gravel RA. Sequence requirements for the biotinylation of carboxyl-terminal fragments of human propionyl-CoA carboxylase alpha subunit expressed in Escherichia coli. J Biol Chem. 1994;269:22964–22968. [PubMed] [Google Scholar]
- Leon-Del-Rio A, Leclerc D, Akerman B, Wakamatsu N, Gravel RA. Isolation of a cDNA encoding human holocarboxylase synthetase by functional complementation of a biotin auxotroph of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:4626–4630. doi: 10.1073/pnas.92.10.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Cronan JE., Jr Growth rate regulation of Escherichia coli acetyl coenzyme A carboxylase, which catalyzes the first committed step of lipid biosynthesis. J Bacteriol. 1993;175:332–340. doi: 10.1128/jb.175.2.332-340.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenortas E, Beckett D. Purification and characterization of intact and truncated forms of the Escherichia coli biotin carboxyl carrier subunit of acetyl-CoA carboxylase. J Biol Chem. 1996;271:7559–7567. doi: 10.1074/jbc.271.13.7559. [DOI] [PubMed] [Google Scholar]
- Polyak SW, Chapman-Smith A, Brautigan PJ, Wallace JC. Biotin protein ligase from Saccharomyces cerevisiae. The N-terminal domain is required for complete activity. J Biol Chem. 1999;274:32847–32854. doi: 10.1074/jbc.274.46.32847. [DOI] [PubMed] [Google Scholar]
- Polyak SW, Chapman-Smith A, Mulhern TD, Cronan JE, Jr, Wallace JC. Mutational analysis of protein substrate presentation in the post-translational attachment of biotin to biotin domains. J Biol Chem. 2001;276:3037–3045. doi: 10.1074/jbc.M003968200. [DOI] [PubMed] [Google Scholar]
- Prakash O, Eisenberg MA. Biotinyl 5′-adenylate: corepressor role in the regulation of the biotin genes of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1979;76:5592–5595. doi: 10.1073/pnas.76.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche P, Li YL, Fuller C, Eichhorn K, Perham RN. Selectivity of post-translational modification in biotinylated proteins: the carboxy carrier protein of the acetyl-CoA carboxylase of Escherichia coli. Biochem J. 1998;329:589–596. doi: 10.1042/bj3290589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reche P, Perham RN. Structure and selectivity in post-translational modification: attaching the biotinyl-lysine and lipoyl-lysine swinging arms in multifunctional enzymes. Embo J. 1999;18:2673–2682. doi: 10.1093/emboj/18.10.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed KE, Cronan JE., Jr Escherichia coli exports previously folded and biotinated protein domains. J Biol Chem. 1991;266:11425–11428. [PubMed] [Google Scholar]
- Rodionov DA, Mironov AA, Gelfand MS. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 2002;12:1507–1516. doi: 10.1101/gr.314502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (N Y) 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Slavoff SA, Chen I, Choi YA, Ting AY. Expanding the substrate tolerance of biotin ligase through exploration of enzymes from diverse species. J Am Chem Soc. 2008;130:1160–1162. doi: 10.1021/ja076655i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissot G, Pepin R, Job D, Douce R, Alban C. Purification and properties of the chloroplastic form of biotin holocarboxylase synthetase from Arabidopsis thaliana overexpressed in Escherichia coli. Eur J Biochem. 1998;258:586–596. doi: 10.1046/j.1432-1327.1998.2580586.x. [DOI] [PubMed] [Google Scholar]
- Weaver LH, Kwon K, Beckett D, Matthews BW. Competing protein:protein interactions are proposed to control the biological switch of the E coli biotin repressor. Protein Sci. 2001;10:2618–2622. doi: 10.1110/ps.32701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Beckett D. Kinetic partitioning between alternative protein-protein interactions controls a transcriptional switch. J Mol Biol. 2008;380:223–236. doi: 10.1016/j.jmb.2008.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]