Summary
Histone deacetylase (HDAC) inhibitors are in clinical development for several diseases, including cancers and neurodegenerative disorders. HDACs1 and 2 are among the targets of these inhibitors and are part of multisubunit protein complexes. HDAC inhibitors (HDACi) block the activity of HDACs by chelating a zinc molecule in their catalytic sites. It is not known if the inhibitors have any additional functional effects on the multisubunit HDAC complexes. Here, we find that suberoylanilide hydroxamic acid (SAHA), the recently FDA approved HDACi, causes the dissociation of the PHD-finger containing ING2 subunit from the Sin3 deacetylase complex. Loss of ING2 disrupts the in vivo binding of the Sin3 complex to the p21 promoter, an important target gene for cell growth inhibition by SAHA. Our findings reveal a new molecular mechanism by which HDAC inhibitors disrupt deacetylase function.
Introduction
Histone deacetylases (HDACs) remove acetyl groups from histones as well as non-histone proteins. Histone hyperacetylation is generally correlated with gene expression and HDACs often work to repress gene expression. Inhibitors to HDACs (HDACi) show promise as anticancer agents as well as in therapies for neurodegenerative diseases (Khan and La Thangue, 2008; Wiech et al., 2009). The hydroxamic acid, SAHA, is currently used as a treatment for advanced and refractory cutaneous T cell lymphoma (Khan and La Thangue, 2008; Mann et al., 2007). HDAC inhibitors can inhibit cancer progression through a number of mechanisms, including inducing apoptosis, arresting cells in G1/S or G2/M and causing cells to differentiate (Frew et al., 2009; Marks and Xu, 2009; Smith and Workman, 2009). One of the mechanisms by which HDACi work is through modulation of gene expression by acetylation of histones, to produce a transcriptional program that is favorable for cell cycle arrest or apoptosis (Frew et al., 2009; Marks and Xu, 2009; Smith and Workman, 2009). Overall, HDAC inhibitors cause a small percentage of genes to be misregulated transcriptionally and in this subset of genes some are upregulated while some are downregulated (Smith, 2008; Van Lint et al., 1996). In addition, HDAC inhibitors mediate acetylation of many non-histone proteins, though this also appears to be a rather small subset of all possibly acetylated proteins (Choudhary et al., 2009; Spange et al., 2009).
There are four classes of histone deacetylases. Classes I, II and IV are zinc-dependent hydrolases while Class III HDACs are NAD-dependent enzymes called sirtuins (Yang and Seto, 2008). There are eleven known zinc-dependent HDACs (Class I: HDACs 1–3 and 8; Class II: HDACs 4–7, 9 and 10; Class IV: HDAC11) (Yang and Seto, 2008). Many inhibitors being tested as anticancer agents affect several of these enzymes. Crystal structures have been solved for a bacterial Class I homolog and for human HDACs 7 and 8 in complex with hydroxamic acid inhibitors trichostatin A (TSA) and SAHA (Finnin et al., 1999; Schuetz et al., 2008; Vannini et al., 2004). These inhibitors work by chelating a zinc molecule in the active site of the HDACs through their hydroxamic acid moieties (Finnin et al., 1999; Schuetz et al., 2008; Vannini et al., 2004). Since these molecules also contain aliphatic chains that extend out through the normal acetyl lysine binding pockets in the HDACs, they also may inhibit binding of the HDAC to their normal acetyl lysine substrates (Finnin et al., 1999; Schuetz et al., 2008; Vannini et al., 2004). Many inhibitors in clinical development affect several HDACs, therefore work has recently focused on understanding which HDACs are needed to mediate anti-cancer effects of the inhibitors (Balasubramanian et al., 2009; Witt et al., 2009). These efforts are to obtain cancer cell growth inhibiting properties while maximizing selectivity of the inhibitors. Studies suggest that in vivo, HDACs 1 and 2 play a role in mediating cell growth arrest by these molecules (Glaser et al., 2003; Haberland et al., 2009).
However, HDACs1 and 2 do not work alone, rather they reside in multisubunit chromatin modifying complexes, of which three have been characterized: Mi-2/NuRD which contains histone deacetylase, histone demethylase and chromatin remodeling activities, CoREST, which can repress neuronal specific genes in non-neuronal cells and Sin3 which has been implicated in cell cycle control (Wang et al., 2009; Yang and Seto, 2008). Residency in these complexes is important for full activity and specificity of these HDACs in the cell (Alland et al., 2002; Denslow and Wade, 2007). However, it is not known if HDACi act directly on these multisubunit complexes. The Sin3 complex is a 1.2 MDa complex implicated in cell cycle control through its interactions with the tumor suppressor protein Rb and can repress E2F-mediated transcription to prevent progression to S phase (Lai et al., 2001). The Sin3 complex is also implicated in controlling progression through the G2 phase of the cell cycle (David et al., 2003; Pile et al., 2002). Therefore, this complex is among the potential targets of HDACi that could mediate the growth arrest by these molecules. We set out to determine if HDAC inhibitors had any effects on the multisubunit Sin3 complex and if the complex was still intact after the HDACs were bound to the inhibitors.
Results
ING2 purified complexes are altered by HDACi
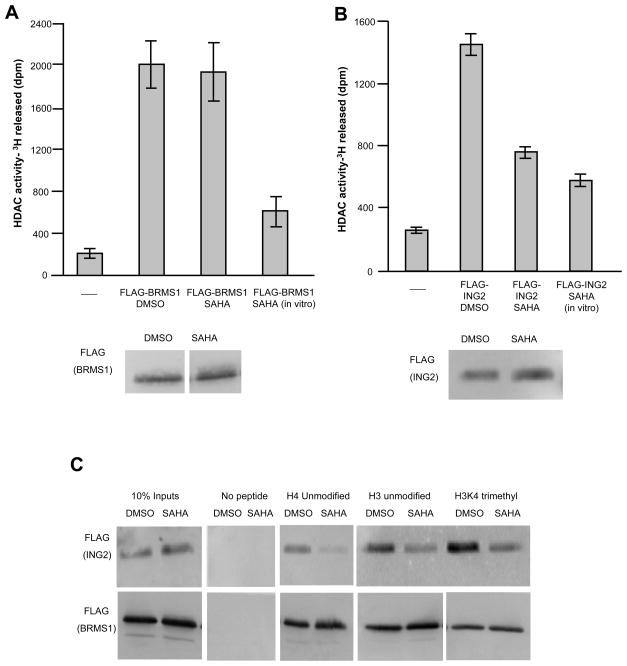
To determine if HDAC inhibitors alter the properties of the Sin3 complex, we purified the complex from 293T cells that stably expressed tagged subunits. We used two different known subunits of the Sin3 complex as baits for these purifications, inhibitor of growth 2, (ING2), which binds to H3K4 that is di and tri-methylated through its PHD finger and breast cancer metastasis suppressor 1 (BRMS1) which has an unknown function in the complex (Doyon et al., 2006; Meehan et al., 2004; Shi et al., 2006). The ING2 gene is deleted in some head and neck carcinomas, while BRMS1 is important for suppressing cancer metastasis, suggesting that their roles in the Sin3 complex could be related to cell growth and cancer progression (Seraj et al., 2000; Sironi et al., 2004). We performed purifications from cells treated with the HDAC inhibitor SAHA (7.5 μM), or DMSO and tested if there were differences in the HDAC activities of the complexes. Sin3 complexes purified through the BRMS1 subunit from SAHA treated cells still had HDAC activity on acetylated core histones, suggesting that the inhibitor was lost during the purification (Figure 1A). This result is consistent with kinetic analyses of these competitive inhibitors (Sekhavat et al., 2007). These complexes were still sensitive to SAHA however, since adding inhibitor in vitro to the HDAC assay reduced catalytic activity (Figure 1A). By contrast, complexes purified through ING2 from SAHA-treated cells had reduced HDAC activity, similar to that observed when SAHA was added directly to the HDAC assay (Figure 1B). We wondered if this was due solely to a change in catalytic activity or if it also affected the ability of the complex to bind directly to histones.
Figure 1. SAHA alters the biochemical properties of the Sin3 complex.
HDAC assays were performed on 3H acetylated core histones with a. FL-BRMS1 purified complexes (top panel) or b. FL-ING2 purified complexes (top panel). DMSO and SAHA labels indicate that complexes were purified from 293T cells treated for nine hours with these compounds. The SAHA (in vitro) label indicates that complexes were purified from untreated cells and SAHA was added directly to the deacetylation reaction. Amounts of complex used in the assays were normalized to levels of FL-BRMS1 (a) (lower panel), or to levels of FL-ING2 (b) (lower panel). c. Western blot analysis of FLAG tagged proteins bound to histone peptides.
To ask if histone binding was disrupted, we tested the binding of Sin3 complexes purified from SAHA/DMSO treated cells to histone peptides. The Sin3 complex preferentially binds to hypoacetylated histones through the RbAp46/48 subunits (Vermeulen et al., 2004; Yoon et al., 2005). It can also be recruited to chromatin through the H3K4-di/tri-methyl mark by ING1/2 (Pena et al., 2008; Shi et al., 2006). Therefore we tested the ability of the complexes to bind unmodified and H3K4 trimethylated peptides. We found that complexes purified through the ING2 subunit in the presence of SAHA were compromised for their ability to bind to histone peptides (Figure 1C). By contrast, complexes purified through BRMS1 retained the ability to bind to histone peptides after SAHA treatment (Figure 1C). Thus, Sin3 complexes purified through ING2 from cells treated with SAHA lost HDAC activity and histone binding ability while the BRMS1 purified complexes did not.
HDACi cause ING2 to dissociate from the Sin3 complex
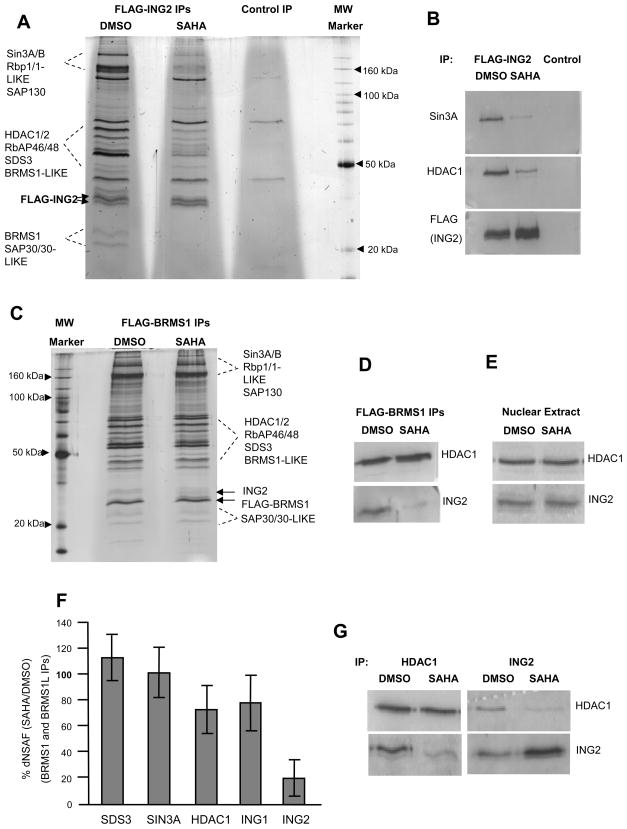
To elucidate the molecular basis for the altered properties of ING2-purified Sin3 complexes from cells treated with SAHA we examined the composition of the complexes. Silver stained gels of the purified complexes revealed that ING2 purified from SAHA-treated cells had reduced levels of other Sin3 complex subunits associated with it (Figure 2A). Multidimensional protein identification technology (MudPIT) (Paoletti et al., 2006) and Western blot analysis confirmed that this loss included reductions in HDAC1, Sin3a, SDS3 and other known subunits (Figure 2B, Table S1). This effect was not limited to SAHA since other HDAC inhibitors including trichostatin A (TSA) and a cyclic tetrapeptide, apicidin, also dissociated FLAG-ING2 from Sin3 complex subunits, however sodium butyrate and valproic acid did not (Figure S1A, B). Therefore, specific deacetylase inhibitors can mediate the dissociation of ING2 from the Sin3 deacetylase complex.
Figure 2. HDAC inhibitors dissociate ING2 from the Sin3 complex.
a. Silver stain analysis of FL-ING2 complexes purified from DMSO or SAHA-treated 293T cells (7.5 μM SAHA or an equal volume of DMSO was added to the cells for nine hours), b. Western blot analysis of FL-ING2 IPs from (a) in the presence of SAHA. c, silver stained gels of FL-BRMS1 complexes purified from DMSO or SAHA-treated 293T cells (7.5 μM SAHA or an equal volume of DMSO was added to the cells for nine hours). The arrows indicate the locations of endogenous ING2 and FLAG-BRMS1. d. Western blots of FLAG-BRMS1 purifications from (c). e. Western blots of nuclear extracts from DMSO or SAHA treated 293T cells. f. Relative percent dNSAF (distributed normalized spectral abundance factor) of proteins in SAHA versus DMSO purifications as determined by MudPIT analysis. Error bars represent +/− 1 standard deviation. g. Western blots of immunoprecipitated proteins from MDA-MB-231 breast carcinoma cells treated with 7.5 μM SAHA or DMSO for nine hours.
The tagged bait protein ING2 was overexpressed in these initial experiments, therefore we tested if the association of endogenous ING2 with the Sin3 complex was changed after SAHA treatment of 293T cells. We analyzed Sin3 complexes purified through BRMS1 and an additional subunit, BRMS1-LIKE (Nikolaev et al., 2004) by SDS-PAGE. BRMS1 purifications from cells treated with SAHA for nine hours showed reduced silver staining of a band at ~ 33kDa, which is the predicted size of ING2 (Figure 2C). Western blots and MudPIT analysis confirmed a reduction of endogenous ING2, but not ING1 in the BRMS1 and BRMS1-LIKE purifications (Figure 2D, F, Table S2) and this occurred as early as 3 hours post-drug treatment (Figure S1C). Total nuclear levels of ING2 and HDAC1 were not changed by SAHA treatment (Figure 2E). Thus, the ING2 protein was still intact in SAHA treated cells and SAHA was causing its dissociation from the Sin3 complex.
SAHA and other HDAC inhibitors have been shown to effectively halt growth of many cell types, including breast cancer cells (Huang and Pardee, 2000). Therefore, we wanted to know if the dissociation of ING2 also occurred in cancer cells. We treated MDA-MB-231 breast carcinoma cells with DMSO or SAHA and performed immunoprecipitations with antibodies to endogenous ING2 or HDAC1. Following treatment of these cells with SAHA, the amount of ING2 that co-precipitated with HDAC1 was reduced (Figure 2G). Conversely the amount of HDAC1 co-precipitated with ING2 was also reduced (Figure 2G). Thus, ING2 and HDAC1 were also dissociated after SAHA treatment of breast carcinoma cells suggesting this is a consequence of HDAC inhibitor treatment in diverse cell types.
Dissociation of ING2 occurs through a direct mechanism
HDAC inhibitors cause an accumulation of acetylated proteins in the cell. This raised the possibility that a subunit of the Sin3 complex became acetylated during SAHA treatment which could be responsible for ING2 dissociation. We tested if acetylated proteins were detected in the SAHA treated purifications, however we did not detect acetylated lysines in any Sin3 complex component after SAHA treatment (Figure S2A, B) This finding is consistent with a recently published study on the acetylome (Choudhary et al., 2009) which did not find large increases in acetylation of Sin3 subunits after HDAC inhibitor treatment. Next we asked if the dissociation between ING2 and the Sin3 complex could occur in vitro. We treated 293T whole cell extract with a panel of HDAC inhibitors and again observed the dissociation between ING2 and the Sin3 complex with SAHA, TSA and apicidin (Figure 3A, B, Figure S1D). Addition of Acetyl CoA in the presence of SAHA to the whole cell extract did not enhance the dissociation of ING2 (Figure S2C). Finally, we tested if HAT inhibitors could prevent ING2 dissociation from the Sin3 complex. However, we found that SAHA and TSA still caused dissociation of ING2 from the complex in a whole cell extract in the presence of a HAT inhibitor (Figure S2D). Together the results suggest that acetylation is not needed for ING2 dissociation to occur.
Figure 3. ING2 dissociates through a direct mechanism.
a. HDAC inhibitors or control compounds were added to whole cell extracts from FLAG-BRMS1 expressing 293T cells, purified with FLAG affinity beads and then probed by Western blot for the indicated proteins. b. quantification of (a), c. FL-BRMS1 purified complex was immobilized on FLAG beads and treated with TSA or control (ethanol) and then probed by Western blot for indicated proteins. d. quantification of (c), Bars in b and d represent the average of three experiments expressed as percent band intensity of ING2 in SAHA or TSA treated extract or complex compared to the intensity in the control treatment, normalized to intensity of Sin3a. Error bars represent +/− 1 standard deviation.
Since ING2 can tether the Sin3 complex to chromatin, we wondered if chromatin binding by ING2’s PHD finger was necessary for SAHA to mediate the dissociation. We expressed an ING2 lacking its PHD finger in 293T cells. This deletion mutant was previously shown to maintain association with the Sin3 complex (Shi et al., 2006). We found that ING2 lacking its PHD finger still dissociated from the Sin3 complex, after cells were treated with SAHA (Figure S2E). Consistent with this result, we also found that TSA did not prevent recombinant ING2 from binding to an H3K4 trimethylated peptide (Figure S2F). Together the data suggested that neither acetylation nor chromatin binding by ING2 were necessary for HDACi-mediated dissociation from the Sin3 complex.
To determine if ING2 dissociation from the Sin3 complex might be a direct consequence of the HDAC binding to the inhibitor, we asked if the dissociation could occur with purified Sin3 complex. To test this, we immobilized the Sin3 complex purified through the BRMS1 subunit to FLAG beads and incubated the complex with increasing amounts of TSA. We found that the addition of TSA to the purified complex resulted in dissociation of endogenous ING2 (Figure 3C, D). This suggests that the dissociation of ING2 could be mediated by a direct/physical disruption which does not require additional factors in cell extracts or living cells. Thus it appears that binding of the small molecule inhibitors to the catalytic sites of the HDAC enzymes leads to physical dissociation of ING2 from the Sin3 complex.
SAHA disrupts Sin3 complex binding at p21 through dissociation of ING2
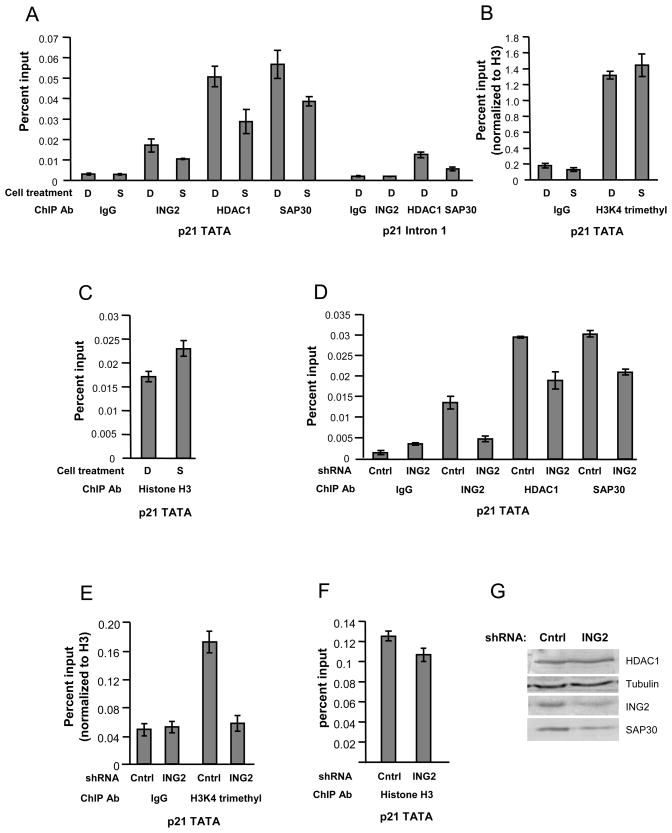
The Sin3 complex contains several proteins that can recruit or retain it at chromatin. These include RbAp46/48, SAP30 and SAP30-LIKE, Sin3 and ING1 and ING2 (Shi et al., 2006; Vermeulen et al., 2004; Viiri et al., 2009; Yoon et al., 2005). Therefore, ING2 is one of the many subunits through which the Sin3 complex can be tethered to chromatin. Since HDACi were effective at dissociating ING2 from the Sin3 complex, we hypothesized that SAHA could cause changes in occupancy of the Sin3 complex at promoters where binding was dependent on ING2. The cyclin dependent kinase inhibitor p21 is a tumor suppressor that is transcriptionally induced in response to HDACi (Richon et al., 2000; Smith and Workman, 2009). Previous studies have shown that the occupancies of HDAC1 and HDAC2 were reduced at the p21 TATA after HDAC inhibitor treatment (Gui et al., 2004; Lin et al., 2008). To test if this might be due to ING2 dissociation, we performed ChIP in 293T cells. We found that binding of ING2 and the Sin3 complex were enriched at the p21 TATA region compared to intron 1 in control treated cells (Figure 4A). SAHA treatment for ten hours caused a reduction in ING2, HDAC1 and SAP30 occupancies (Figure 4A) but did not affect the levels of H3K4 trimethylation (Figure 4B) or H3 occupancy (Figure 4C) at the p21 TATA. These results are consistent with our biochemical studies indicating that ING2 is dissociated from the Sin3 complex in the presence of SAHA.
Figure 4. ING2 dissociation causes the loss of the Sin3 complex from the p21 promoter.
a, b and c. ChIP was performed in 293T cells treated for 10 hours with DMSO (D) or SAHA (S) d, e and f. ChIP was performed in ING2 shRNA knock-down or non-effective control (cntrl) shRNA knock-down 293T cells. Bars (a through f) show the average from a representative experiment using primers adjacent to the p21 TATA or in intron 1 of p21. As a control in b and e, percent inputs were normalized to the amount of H3 immunoprecipitated for each treatment condition or cell line. Error bars represent +/− 1 standard deviation of triplicate or quadruplicate real-time PCR reactions. g. Total nuclear extracts from the ING2 knock-down cells or GFP shRNA control cells were probed for the indicated proteins.
Since the Sin3 complex can be recruited and tethered to chromatin through multiple subunits, we tested if occupancy of the Sin3 complex was dependent on ING2 at p21. To do this, we assessed the occupancy of the Sin3 complex in ING2 knock-down cells and compared this to control cells expressing non-effective shRNA. As expected, the occupancy of ING2 at the p21 TATA was reduced in ING2 knock-down cells (Figure 4D). Interestingly, the level of H3K4 trimethylation at this region was also reduced in the ING2 knock-down line (Figure 4E), but occupancy of H3 was not altered (Figure 4F) suggesting that ING2 binding could protect this region from demethylation. The occupancies of HDAC1 and SAP30 were also reduced at p21 in the ING2 knock-down cells compared to the control cell line showing that they were dependent on ING2 (Figure 4D). Total levels of HDAC1 were not changed in the ING2 knock-down cells however SAP30 protein levels were reduced in the ING2 knock-down line, but not in cells treated with SAHA (Figure 4G and data not shown). Thus, SAHA treatment promotes the loss of the Sin3 complex from the p21 promoter in vivo through the dissociation of the ING2 subunit (Figure 5A, B).
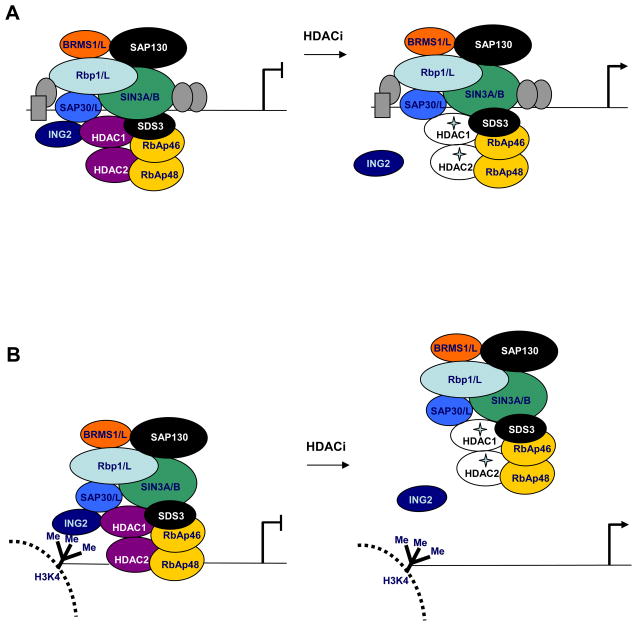
Figure 5. Models for gene activation after HDAC inhibitor treatment.
a. The Sin3 complex can be tethered to chromatin through interactions of Rbp1 or Sin3 subunits with transcription factors (gray ovals and rectangle) and through SAP30/SAP30-LIKE and RbAp46/48 subunits. At these promoters, HDAC inhibitors cause inactivation of the HDACs and dissociation of ING2 from the complex. b. ING2 is required for tethering the Sin3 complex at some promoters where H3K4 is di/tri-methylated. At these regions, HDAC inhibitor treatment causes inactivation of the HDACs as well as dissociation of the Sin3 complex from chromatin.
Discussion
Previous studies showed that the catalytic activities of HDACs are important for their association with other proteins (Hassig et al., 1998; Matsuoka et al., 2007). Catalytically inactive mutants of HDAC1 do not associate with RbAp48 or Sin3 (Hassig et al., 1998). However, here, we find that the FDA approved HDACi, SAHA does not alter HDAC1 or HDAC2 interactions with the Sin3 complex and this is consistent with a previous study (Sekhavat et al., 2007). Instead, we find that HDAC inhibitors affect the association of a non-enzymatic subunit important for chromatin targeting, ING2.
The inhibitor of growth family of proteins contains five known members in humans. ING1 and ING2 reside in Sin3 deacetylase complexes, while ING3, 4 and 5 are in histone acetyltransferase (HAT) complexes (Doyon et al., 2006). It is interesting that all of these proteins are thought to bind the same methylated histone mark, H3K4 that is di or tri-methylated. It is unclear what controls which ING protein will be bound to this epigenetic mark at a given time, and therefore, whether a HAT or HDAC complex will be recruited.
ING family members are of potential therapeutic interest (Unoki et al., 2009). ING2 is of interest due to its role in modulating p53 activity (Menendez et al., 2009; Nagashima et al., 2001). In addition, ING2 can recruit the repressive Sin3 complex to the cyclin D1 promoter after DNA damage (Shi et al., 2006). Here, we find that ING2 is also functionally targeted by HDAC inhibitors and that HDAC inhibitors cause ING2’s dissociation from the Sin3 HDAC1/2 complex. ING2 is likely required for Sin3 complex occupancy at a specific subset of Sin3 target genes. Therefore, the dissociation of ING2 is one possible mechanism that contributes to gene expression alterations after HDAC inhibitor treatment (Figure 5).
We have tested several possibilities to explain the mechanism by which ING2 is dissociating from the Sin3 complex. We found that TSA can cause ING2 to dissociate from purified Sin3 complex in vitro and does not require additional factors in the cell. In addition, we also showed that the dissociation does not require acetylation of specific proteins, nor does this dissociation involve the PHD finger of ING2 directly. We also showed that not all inhibitors are effective at dissociating ING2, suggesting dissociation is not simply due to inhibition of catalytic activity of the HDACs. Interestingly, the inhibitors that do mediate the dissociation (TSA, SAHA and apicidin) are larger and bulkier than those that do not (Na butyrate, valproic acid) suggesting that the ability to dissociate ING2 may involve the structure of the inhibitors themselves. All of these results together suggest that ING2 dissociates through a direct effect of the HDACs binding to the inhibitors in the context of the complex.
Our studies leave open the possibility that a conformational change occurs in the complex after inhibitor binding. Showing this will require further structural studies of the HDACs in the contexts of their complexes. To date, structural studies have only been carried out on isolated deacetylases in complex with inhibitors (Finnin et al., 1999; Schuetz et al., 2008; Vannini et al., 2004). Future studies will have to address several questions. First, it is unclear how the multisubunit complexes are assembled. Second, it is not known if the HDACs undergo structural changes when they are assembled in the complexes. Third, it is also unclear if inhibitor binding alters HDAC conformation in the context of their complexes. Our results suggest that the HDACs bind to the inhibitors and that this causes a conformational change in at least part of the Sin3 complex, which is sufficient for dissociation of ING2. Since the Sin3 complex largely stays intact and is active without ING2, this suggests that ING2 is not needed for the integrity of the complex as a whole or for catalytic activity. The data also suggest that ING2 resides on an outer surface of the complex and support a main role for ING2 in targeting the Sin3 complex to chromatin.
ING2 is considered a stable subunit of the Sin3 complex however, we show here that dynamic interactions can occur between ING2 and the Sin3 complex and this can be perturbed by HDAC inhibitors. ING2 has the ability to dynamically respond to several cellular signals, including phosphoinositides and DNA damage (Gozani et al., 2003; Shi et al., 2006). The model proposed by Shi et al. (Shi et al., 2006) suggests that ING2 can recruit the Sin3 complex for immediate gene repression when needed. Our results suggest that HDAC inhibitors cause the physical release of the Sin3 complex from promoters through dissociation of ING2. This may be an important step which allows recruitment of other factors and gene activation. Overall, our results implicate ING2 and the Sin3 complex in mediating the cellular response to HDAC inhibitors.
Disrupting just the ING2/Sin3 complex interaction should lead to a smaller subset of gene expression changes than treatment with pan-HDAC inhibitors do. Our results raise the question if modulating the ING2/Sin3 interaction may be sufficient to have any anti-cancer growth effects. It will be interesting to test if reduction of ING2 protein levels is sufficient for induction of p21 and other genes associated with SAHA-mediated growth inhibition. If so, disrupting this interaction may be of therapeutic value, either alone or in combination with other chemotherapeutic agents. Together, our findings reveal that the Sin3 complex is among the in vivo targets for HDAC inhibitors and describe a new mechanism by which HDAC inhibitors can alter HDAC function.
Significance
Histone deacetylase inhibitors, such as SAHA are being tested as treatments for cancers and neurodegenerative diseases. Despite the therapeutic successes of these molecules, we are just beginning to understand how they work. The recognized mode of action for these drugs is by binding to their target enzymes, HDACs and catalytically inhibiting their activity. This in turn, leads to changes in the expression of specific genes and produces a gene expression program that is overall favorable for cell cycle arrest, apoptosis or differentiation. Unexpectedly, our findings here show that HDAC inhibitors can also disrupt critical protein-protein interactions between HDACs 1 and 2 and ING2, a protein responsible for recruitment of the Sin3/HDAC1/2 complex to chromatin. Abrogation of this interaction disrupts the targeting of the Sin3/HDAC complex to chromatin. Therefore, HDAC inhibitors disrupt HDAC function not only by inhibiting HDAC catalytic activity, but also by disrupting HDAC complex subunit composition and chromatin targeting. These findings reveal that these small molecule inhibitors exert their effects through multiple molecular modes of action.
Experimental Procedures
Cell lines and culture
293T cells stably expressing FLAG tagged subunits of the Sin3 complex were made using the Flp-In system (Invitrogen). 293T FRT cells were a gift from Drs. Joan and Ron Conaway. MDA-MB-231 cells were obtained from ATCC. All cell lines were maintained in DMEM (Gibco) supplemented with 10% FBS, Pen/Strep and Glutamax (Gibco) in a humidified atmosphere at 37 degrees. For experiments involving drug treatment, HDAC inhibitors or appropriate vehicles (Ethanol or DMSO) were added directly to the culture media and cells were collected at the indicated times. TSA and SAHA were purchased from Cayman Chemical or BioVision, Inc., Apicidin, Valproic Acid and Sodium Butyrate were purchased from Sigma.
Purifications
293T whole cell extracts were made using a high salt extraction method (Mahrour et al., 2008). Anti-FLAG M2 agarose resin was added overnight to the soluble protein fraction with rotation according to manufacturer’s instructions (Sigma). Complexes were eluted with 3x FLAG peptide and then analyzed or used for MudPIT analysis or in biochemical assays. In purifications with drug treatment, HDAC inhibitors were included at all steps of the purifications except the elution step.
HDAC assays
HDAC assays were performed with purified HDAC complexes, essentially as described (Meehan et al., 2004), except HeLa core histones acetylated by yeast SAGA were used as a substrate and reactions were incubated for 1 hr at 37 degrees. DMSO or SAHA (7.5 μM) were included in the HDAC assays where indicated.
Peptide binding assays
Biotinylated histone peptides were either purchased from Upstate biotechnology or were a gift from Dr. Matthias Mann (Max-Planck Institute). Binding reactions were performed at 150mM NaCl in the presence of 0.1% Triton X-100 as described (Shi et al., 2006). For each assay, the entire bound sample was run on SDS-PAGE.
In vitro dissociation experiments
HDAC inhibitors and Garcinol (where indicated) were added to whole cell extracts from 293T cells stably expressing FLAG-BRMS1. FLAG resin beads were added concurrently with the HDAC/HAT inhibitors. In experiments with Acetyl-CoA, HDAC inhibitors and Acetyl-CoA were added at the same time with FLAG resin to whole cell extracts from 293T cells stably expressing FLAG-ING2. In both experiments, the IPs were incubated at 4 degrees overnight with rotation. The next day, unbound protein was removed and beads were washed four times with Buffer A (10 mM HEPES pH 7.5, 0.2% Triton X-100, 300 mM NaCl, 10 mM KCl and 1.5 mM MgCl2). For dissociation with purified complex, FL-BRMS1 complex was purified from untreated whole cell extracts, immobilized on FLAG beads and washed four times with Buffer A. Beads were then incubated in Buffer B (10 mM HEPES pH 7.5, 0.05% Triton X-100, 150 mM NaCl, 1.5 mM MgCl2) with ethanol or TSA for two hours at thirty degrees with rotation. Unbound protein was removed, and beads were washed four times with Buffer A.
Plasmids
Full length cDNAs encoding human BRMS1, BRMS1L or ING2, or ING2 lacking amino acids 208–280 (ING2ΔPHD) were cloned into pcDNA5/FRT (a gift from Drs. Joan and Ron Conaway) with a single N-terminal (BRMS1, ING2, ING2ΔPHD) or C-terminal (BRMS1L) FLAG tag. HuSH 29mer shRNA constructs against ING2 (cat# TG312145) or non-effective shRNA (cat# TR30008) were purchased from Origene Technologies and stably expressed in 293T cells.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described by Upstate Biotechnology, Inc., except antibodies were first bound to pre-blocked Protein A Sepharose (Sigma) or Protein G Sepharose (Amersham) in binding buffer (5 mM Tris pH 7.5, 250 mM NaCl, 1 mM EDTA, 0.05% NP-40). ChIPs were performed at least three independent times.
Antibodies
Antibodies used for ChIP, Western blotting and IPs are as follows: Anti-acetyl H4 (Upstate, 06-866), HDAC1 (Abcam-Ab7028), ING2 antibody was a gift from Dr. Or Gozani (Stanford) or was purchased from ProteinTech Group, Inc (#11560-1-AP), H3 antibody (Abcam-Ab1791), H3K4 trimethyl (Abcam-Ab8580), Sin3a (Abcam-Ab3479), Sap30 (Upstate-06-875), acetylated-lysine antibodies (Cell signaling technology, #9681, # 9441), FLAG-HRP (Sigma).
Real-time PCR
PCR was performed with a BioRad iCycler machine using SYBR green. Cycling conditions are as follows: 3 minutes at 95°C, then 41 cycles of: 10 seconds at 95°, 30 seconds at 55 or 60°, and 30 seconds at 72°, then followed by a melt curve. A standard curve of input DNA was used to determine relative ChIP sample abundance. Each sample was run in triplicate or quadruplicate. Primers adjacent to the p21 TATA or in intron 1 (negative control) were used for real-time PCR of ChIP samples. Sequences of primers are as follows: +85p21TATA-F: 5′-GATTCGCCGAGGCACCGAGGCA-3′, +218p21TATA-R: 5′-GAACACGCATCCTCGCGGACAC-3′, p21-IN1-F: 5′-GTGCCTGCCTAGATCCTAGTCCT-3′, p21-IN1-R: 5′-GGAGACACACTGGTATGTTTGAA-3′.
Quantification of signal intensity on Western blots
Western blots were developed with ECL-Plus (GE Healthcare) and then scanned on a Typhoon 9400 imaging system. Image Quant V 5.2 was used to quantify bands by the volume integration method. Background was normalized using a local average and a volume report was generated. ING2 band intensity was normalized to Sin3a band intensity per lane.
Supplementary Material
Acknowledgments
We thank Dr. Or Gozani (Stanford) for ING2 antibody, Dr. Matthias Mann (Max-Planck) for histone peptides, Drs. Joan and Ron Conaway (Stowers Institute) for 293T-FRT cell lines and vectors and for sharing of equipment and Dr. Kenneth Lee for providing acetylated histones. This work was supported by NIGMS grant R37 GM047867 and NCI fellowship 5F32CA130468. KTS is the American Cancer Society’s 2008 Cattle Baron’s Ball of Lubbock Postdoctoral Fellow (#PF-09-109-01-GMC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alland L, David G, Shen-Li H, Potes J, Muhle R, Lee HC, Hou H, Jr, Chen K, DePinho RA. Identification of mammalian Sds3 as an integral component of the Sin3/histone deacetylase corepressor complex. Molecular and cellular biology. 2002;22:2743–2750. doi: 10.1128/MCB.22.8.2743-2750.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Verner E, Buggy JJ. Isoform-specific histone deacetylase inhibitors: the next step? Cancer letters. 2009;280:211–221. doi: 10.1016/j.canlet.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science (New York, NY) 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- David G, Turner GM, Yao Y, Protopopov A, DePinho RA. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes & development. 2003;17:2396–2405. doi: 10.1101/gad.1109403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Molecular cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer letters. 2009;280:125–133. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- Glaser KB, Li J, Staver MJ, Wei RQ, Albert DH, Davidsen SK. Role of class I and class II histone deacetylases in carcinoma cells using siRNA. Biochemical and biophysical research communications. 2003;310:529–536. doi: 10.1016/j.bbrc.2003.09.043. [DOI] [PubMed] [Google Scholar]
- Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, et al. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/s0092-8674(03)00480-x. [DOI] [PubMed] [Google Scholar]
- Gui CY, Ngo L, Xu WS, Richon VM, Marks PA. Histone deacetylase (HDAC) inhibitor activation of p21WAF1 involves changes in promoter-associated proteins, including HDAC1. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1241–1246. doi: 10.1073/pnas.0307708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberland M, Johnson A, Mokalled MH, Montgomery RL, Olson EN. Genetic dissection of histone deacetylase requirement in tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7751–7755. doi: 10.1073/pnas.0903139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassig CA, Tong JK, Fleischer TC, Owa T, Grable PG, Ayer DE, Schreiber SL. A role for histone deacetylase activity in HDAC1-mediated transcriptional repression. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:3519–3524. doi: 10.1073/pnas.95.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Pardee AB. Suberoylanilide hydroxamic acid as a potential therapeutic agent for human breast cancer treatment. Molecular medicine (Cambridge, Mass) 2000;6:849–866. [PMC free article] [PubMed] [Google Scholar]
- Khan O, La Thangue NB. Drug Insight: histone deacetylase inhibitor-based therapies for cutaneous T-cell lymphomas. Nature clinical practice. 2008;5:714–726. doi: 10.1038/ncponc1238. [DOI] [PubMed] [Google Scholar]
- Lai A, Kennedy BK, Barbie DA, Bertos NR, Yang XJ, Theberge MC, Tsai SC, Seto E, Zhang Y, Kuzmichev A, et al. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Molecular and cellular biology. 2001;21:2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Lin JH, Chou CW, Chang YF, Yeh SH, Chen CC. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer research. 2008;68:2375–2383. doi: 10.1158/0008-5472.CAN-07-5807. [DOI] [PubMed] [Google Scholar]
- Mahrour N, Redwine WB, Florens L, Swanson SK, Martin-Brown S, Bradford WD, Staehling-Hampton K, Washburn MP, Conaway RC, Conaway JW. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. The Journal of biological chemistry. 2008;283:8005–8013. doi: 10.1074/jbc.M706987200. [DOI] [PubMed] [Google Scholar]
- Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. The oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- Marks PA, Xu WS. Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem. 2009;107:600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka H, Fujimura T, Hayashi M, Matsuda K, Ishii Y, Aramori I, Mutoh S. Disruption of HDAC4/N-CoR complex by histone deacetylase inhibitors leads to inhibition of IL-2 gene expression. Biochemical pharmacology. 2007;74:465–476. doi: 10.1016/j.bcp.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Meehan WJ, Samant RS, Hopper JE, Carrozza MJ, Shevde LA, Workman JL, Eckert KA, Verderame MF, Welch DR. Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription. The Journal of biological chemistry. 2004;279:1562–1569. doi: 10.1074/jbc.M307969200. [DOI] [PubMed] [Google Scholar]
- Menendez C, Abad M, Gomez-Cabello D, Moreno A, Palmero I. ING proteins in cellular senescence. Current drug targets. 2009;10:406–417. doi: 10.2174/138945009788185077. [DOI] [PubMed] [Google Scholar]
- Nagashima M, Shiseki M, Miura K, Hagiwara K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K, Harris CC. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:9671–9676. doi: 10.1073/pnas.161151798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev AY, Papanikolaou NA, Li M, Qin J, Gu W. Identification of a novel BRMS1-homologue protein p40 as a component of the mSin3A/p33ING1b/HDAC1 deacetylase complex. Biochemical and biophysical research communications. 2004;323:1216–1222. doi: 10.1016/j.bbrc.2004.08.227. [DOI] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena PV, Hom RA, Hung T, Lin H, Kuo AJ, Wong RP, Subach OM, Champagne KS, Zhao R, Verkhusha VV, et al. Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor. Journal of molecular biology. 2008;380:303–312. doi: 10.1016/j.jmb.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pile LA, Schlag EM, Wassarman DA. The SIN3/RPD3 deacetylase complex is essential for G(2) phase cell cycle progression and regulation of SMRTER corepressor levels. Molecular and cellular biology. 2002;22:4965–4976. doi: 10.1128/MCB.22.14.4965-4976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz A, Min J, Allali-Hassani A, Schapira M, Shuen M, Loppnau P, Mazitschek R, Kwiatkowski NP, Lewis TA, Maglathin RL, et al. Human HDAC7 Harbors a Class IIa Histone Deacetylase-specific Zinc Binding Motif and Cryptic Deacetylase Activity. The Journal of biological chemistry. 2008;283:11355–11363. doi: 10.1074/jbc.M707362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhavat A, Sun JM, Davie JR. Competitive inhibition of histone deacetylase activity by trichostatin A and butyrate. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2007;85:751–758. doi: 10.1139/o07-145. [DOI] [PubMed] [Google Scholar]
- Seraj MJ, Samant RS, Verderame MF, Welch DR. Functional evidence for a novel human breast carcinoma metastasis suppressor, BRMS1, encoded at chromosome 11q13. Cancer research. 2000;60:2764–2769. [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi E, Cerri A, Tomasini D, Sirchia SM, Porta G, Rossella F, Grati FR, Simoni G. Loss of heterozygosity on chromosome 4q32-35 in sporadic basal cell carcinomas: evidence for the involvement of p33ING2/ING1L and SAP30 genes. Journal of cutaneous pathology. 2004;31:318–322. doi: 10.1111/j.0303-6987.2004.0187.x. [DOI] [PubMed] [Google Scholar]
- Smith CL. A shifting paradigm: histone deacetylases and transcriptional activation. Bioessays. 2008;30:15–24. doi: 10.1002/bies.20687. [DOI] [PubMed] [Google Scholar]
- Smith KT, Workman JL. Histone deacetylase inhibitors: anticancer compounds. The international journal of biochemistry & cell biology. 2009;41:21–25. doi: 10.1016/j.biocel.2008.09.008. [DOI] [PubMed] [Google Scholar]
- Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. The international journal of biochemistry & cell biology. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Unoki M, Kumamoto K, Harris CC. ING proteins as potential anticancer drug targets. Current drug targets. 2009;10:442–454. doi: 10.2174/138945009788185059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene expression. 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- Vannini A, Volpari C, Filocamo G, Casavola EC, Brunetti M, Renzoni D, Chakravarty P, Paolini C, De Francesco R, Gallinari P, et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15064–15069. doi: 10.1073/pnas.0404603101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Carrozza MJ, Lasonder E, Workman JL, Logie C, Stunnenberg HG. In vitro targeting reveals intrinsic histone tail specificity of the Sin3/histone deacetylase and N-CoR/SMRT corepressor complexes. Molecular and cellular biology. 2004;24:2364–2372. doi: 10.1128/MCB.24.6.2364-2372.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viiri KM, Janis J, Siggers T, Heinonen TY, Valjakka J, Bulyk ML, Maki M, Lohi O. DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides. Molecular and cellular biology. 2009;29:342–356. doi: 10.1128/MCB.01213-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Chen Y, Sun Y, Yang F, Yu W, Liang J, Sun L, Yang X, Shi L, et al. LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell. 2009;138:660–672. doi: 10.1016/j.cell.2009.05.050. [DOI] [PubMed] [Google Scholar]
- Wiech NL, Fisher JF, Helquist P, Wiest O. Inhibition of histone deacetylases: a pharmacological approach to the treatment of non-cancer disorders. Current topics in medicinal chemistry. 2009;9:257–271. doi: 10.2174/156802609788085241. [DOI] [PubMed] [Google Scholar]
- Witt O, Deubzer HE, Milde T, Oehme I. HDAC family: What are the cancer relevant targets? Cancer letters. 2009;277:8–21. doi: 10.1016/j.canlet.2008.08.016. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HG, Choi Y, Cole PA, Wong J. Reading and function of a histone code involved in targeting corepressor complexes for repression. Molecular and cellular biology. 2005;25:324–335. doi: 10.1128/MCB.25.1.324-335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.