Abstract
Background
Immune responses to parasites, which start with pathogen recognition, play a decisive role in the control of the infection in mosquitoes. Peptidoglycan recognition proteins (PGRPs) are an important family of pattern recognition receptors that are involved in the activation of these immune reactions. Pathogen pressure can exert adaptive changes in host genes that are crucial components of the vector's defence. The aim of this study was to determine the molecular evolution of the three short PGRPs (PGRP-S1, PGRP-S2 and PGRP-S3) in the two main African malaria vectors - Anopheles gambiae and Anopheles arabiensis.
Results
Genetic diversity of An. gambiae and An. arabiensis PGRP-S1, PGRP-S2 and PGRP-S3 was investigated in samples collected from Mozambique and Tanzania. PGRP-S1 diversity was lower than for PGRP-S2 and PGRP-S3. PGRP-S1 was the only gene differentiated between the two species. All the comparisons made for PGRP-S1 showed significant P-values for Fst estimates and AMOVA confirming a clear separation between species. For PGRP-S2 and PGRP-S3 genes it was not possible to group populations either by species or by geographic region. Phylogenetic networks reinforced the results obtained by the AMOVA and Fst values. The ratio of nonsynonymous substitutions (Ka)/synonymous substitutions (Ks) for the duplicate pair PGRP-S2 and PGRP-S3 was very similar and lower than 1. The 3D model of the different proteins coded by these genes showed that amino acid substitutions were concentrated at the periphery of the protein rather than at the peptidoglycan recognition site.
Conclusions
PGRP-S1 is less diverse and showed higher divergence between An. gambiae and An. arabiensis regardless of geographic location. This probably relates to its location in the chromosome-X, while PGRP-S2 and PGRP-S3, located in chromosome-2L, showed signs of autosomal introgression. The two short PGRP genes located in the chromosome-2L were under purifying selection, which suggests functional constraints. Different types of selection acting on PGRP-S1 and PGRP-S2 and S3 might be related to their different function and catalytic activity.
Background
Mosquito immune responses to Plasmodium play an important role in the natural control of the infection and are initiated when pathogen-associated molecular patterns (PAMPs) are recognized by pattern recognition receptor (PRR) molecules [1]. Peptidoglycan recognition proteins (PGRPs) are one family of PRR, which contain a domain very similar to bacterial amidase. The Anopheles PGRPs can be divided into two different classes: short and long. In Drosophila short PGRPs are small extracellular proteins about 200 amino acids long and 18-20 kDa that are present in the hemolymph and cuticle. They are constitutively synthesized or induced mainly in the fat-body and, to a lesser extent, also in the epidermal cells, in the gut and in the hemocytes. Long PGRPs, have long transcripts and are either intracellular or membrane-spanning proteins, expressed mainly in the hemocytes [1]. Seven PGRP genes are known in the Anopheles gambiae genome [2]. Of these, three genes code for the short PGRPs namely PGRP-S1, PGRP-S2 and PGRP-S3. Short-PGRP genes from An. gambiae differ in their structure, as PGRP-S2 and S3 have predicted amidase activity while PGRP-S1 does not. The presence of catalytic activity in Drosophila short-PGRPs determines its function. Drosophila has two noncatalytic short-PGRPs, PGRP-SA and PGRP-SD, which are involved in recognition of bacteria and activation of the Toll pathway [3,4], while PGRP-SC1/2 have catalytic activity and can specifically control the level of activation of the IMD signalling pathway [5]. Transcription alteration of short-PGRPs in response to Plasmodium infection [6] have been reported. However, the way these molecules are involved in the response to the parasite remains unknown. The most plausible explanation in view of recent findings [7,8] is that gut microbiota modulate mosquito response to Plasmodium.
PGRP genes form clusters in the genome suggesting that they may have been originated by gene duplication [9]. This phenomenon is also observed in other PRR, like TEP-1, in which gene conversion plays a determinant role on their evolution [10]. The reasons why duplications occur are diverse. Once a gene is duplicated, the new gene might degenerate into a pseudogene due to recurrent deleterious mutations. However, if the duplication is advantageous for the organism, the gene might evolve new functions ("neofuntionalization") since the first copy maintains the original function. In addition duplicated genes can also have the same function but which is partially or fully subdivided between them ("subfunctionalization").
Molecular evolution of immune related genes is determined by their interaction with pathogens. In the present study we analysed patterns of evolution in three short PGRPs of the malaria vectors An. gambiae and Anopheles arabiensis, the main malaria vectors in sub-Sahara Africa, and considered the type of selective pressure acting on them.
Results
Polymorphism and diversity
A total of 237 sequences were analysed, 146 from An. gambiae (all identified as S-form) and 91 from An. arabiensis. Eighty eight sequences were obtained for PGRP-S1 (genbank accession nos FJ821900 - FJ821987), 47 for PGRP-S2 (genbank accession nos FJ821988 - FJ822034) and 102 for PGRP-S3 (genbank accession nos FJ821798 - FJ821899). There were 15 An. gambiae and 18 An. arabiensis for which all three genes were successfully sequenced.
In the PGRP-S1 gene, a fragment of 1182 bp that includes the coding region (552 bp) was amplified. For this gene the nucleotide diversity (π) was higher for An. arabiensis from Mozambique than in the other groups especially in the 5' upstream and 3' downstream non-coding regions (0.013 and 0.023 respectively) (Table 1). Overall nucleotide diversity was lower for PGRP-S1 (0.000-0.008) when compared to PGRP-S2 (0.009-0.022) and PGRP-S3 (0.002-0.027).
Table 1.
Intraspecific polymorphism and neutrality tests for the three Anopheles short PGRP genes.
| 5' upstream | Exon 1 | 3' downstream | ||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PGRP | Population | sequence | UTR | Translated region | Intron | Exon 2 | sequence | Total | Neutrality | |||||||||||||||||||
| L | S | π | L | S | π | L | S | π | L | S | π | L | S | π | L | S | π | L | S | π | πa/πs | Tajima's | Fu & Li | |||||
| s | n | s | n | D | D' | F | ||||||||||||||||||||||
| Ag_Tz | 5 | 0.002 | - | - | 0 | 1 | 0.000 | 1 | 0.003 | 1 | 0 | 0.001 | 0 | 0.000 | 8 | 0.001 | 0.338 | -1.03* | -2.32* | -2.51* | ||||||||
| Ag_Mz | 1 | 0.000 | - | - | 0 | 0 | 0.000 | 0 | 0.000 | 0 | 0 | 0.000 | 0 | 0.000 | 1 | 0.000 | - | -1.11 n/s | -1.81 n/s | -1.86 n/s | ||||||||
| S1 | Aa Tz | 534 | 1 | 0.000 | - | - | - | 255 | 0 | 0 | 0.000 | 74 | 0 | 0.000 | 297 | 0 | 0 | 0.000 | 32 | 0 | 0.000 | 1192 | 1 | 0.000 | - | -1.16 n/s | -1.45 n/s | -1.57 n/s |
| Aa Mz | 31 | 0.013 | - | - | 3 | 0 | 0.003 | 1 | 0.004 | 4 | 1 | 0.004 | 3 | 0.023 | 43 | 0.008 | - | -1.04 n/s | 1.19 n/s | 0.65 n/s | ||||||||
| Ag_Tz | 8 | 0.039 | 19 | 0.021 | 14 | 3 | 0.014 | - | - | - | - | - | 9 | 0.027 | 54 | 0.020 | 0.073 | 2.50* | 1.19 n/s | 1.82*** | ||||||||
| Ag_Mz | 10 | 0.041 | 27 | 0.022 | 18 | 3 | 0.014 | - | - | - | - | - | 17 | 0.038 | 75 | 0.022 | 0.049 | 0.34 n/s | 0.62 n/s | 0.62 n/s | ||||||||
| S2 | Aa Tz | 107 | 1 | 0.002 | 409 | 5 | 0.005 | 567 | 11 | 1 | 0.007 | - | - | - | - | - | - | - | 172 | 14 | 0.024 | 1255 | 32 | 0.009 | 0.030 | -0.34 n/s | -0.39 n/s | -0.43 n/s |
| Aa Mz | 5 | 0.023 | 7 | 0.007 | 9 | 2 | 0.007 | - | - | - | - | - | 6 | 0.015 | 29 | 0.010 | 0.061 | 0.86 n/s | 0.89 n/s | 0.99/s | ||||||||
| Ag_Tz | 1 | 0.001 | - | - | 10 | 1 | 0.003 | - | - | - | - | - | 0 | 0.000 | 12 | 0.002 | 0.015 | -1.53 n/s | -1.78 n/s | -1.99 n/s | ||||||||
| Ag_Mz | 10 | 0.043 | - | - | 28 | 10 | 0.025 | - | - | - | - | - | 6 | 0.019 | 54 | 0.027 | 0.141 | 1.08 n/s | 1.21 n/s | 1.38 n/s | ||||||||
| S3 | Aa Tz | 84 | 8 | 0.038 | - | - | - | 567 | 16 | 1 | 0.007 | - | - | - | - | - | - | - | 61 | 6 | 0.028 | 712 | 31 | 0.012 | 0.038 | -0.08 n/s | 0.31 n/s | 0.22 n/s |
| Aa Mz | 10 | 0.014 | - | - | 23 | 9 | 0.010 | - | - | - | - | - | 5 | 0.019 | 47 | 0.011 | 0.042 | -1.66 n/s | -1.97 n/s | -2.19 n/s | ||||||||
*, **, ***represent P < 0.05, P < 0.01, and P < 0.001, n/s not significant. Ag_Tz, An. gambiae from Tanzania; Ag_Mz, An. gambiae from Mozambique; Aa_Tz, An. arabiensis from Tanzania; Aa_Mz, An. arabiensis from Mozambique; L, length; S, segregating site n - total number of nonsynonymous changes; s - total number of synonymous changes; π, nucleotide diversity.
A total of 1255 bp was amplified in the PGRP-S2 gene, which included the coding region (567 bp). In this gene the π values varied between 0.007 and 0.014 in the coding region and between 0.002 and 0.041 in the non-coding regions (Table 1). For PGRP-S3 gene a fragment of 712 bp, that included the coding region (567 bp), was amplified. The π values varied between 0.003 and 0.025 in the coding-region and between 0.000 and 0.043 in the non-coding regions.
For the three genes the total number of segregating sites was estimated for each geographic sample (Table 1). These were very similar in PGRP-S2 and PGRP-S3 genes, and much higher than in PGRP-S1. The number of synonymous and nonsynonymous substitutions was always higher in PGRP-S2 and PGRP-S3 genes than in PGRP-S1 and for those two genes synonymous changes were always higher than nonsynonymous changes. This is reflected in the πa/πs ratios that were always below one (Table 1).
Species divergence and population differentiation
Species divergence and population differentiation were calculated for each gene of An. gambiae and An. arabiensis from two different East-Africa locations, Tanzania and Mozambique.
For the PGRP-S1 gene all pairwise Fst estimates were significant (p < 0.05). The lowest Fst estimates were obtained in intraspecific comparisons, indicating that differentiation was higher between species than between geographic populations (Table 2). For the other two genes within and between species Fst estimates were lower but still significant in almost all comparisons made (Table 2). Exceptions were for PGRP-S2 gene between An. gambiae_Tanzania and An. gambiae_Mozambique; An. arabiensis_Tanzania and An. arabiensis_Mozambique and for PGRP-S3 gene between An. arabiensis_Tanzania and An. gambiae_Mozambique.
Table 2.
Matrix of pairwise comparisons of F st for the four Anopheles populations studied.
| Genes | Gamb_Mz | Gamb_Tz | Arab_Mz | Arab_Tz | |
|---|---|---|---|---|---|
| PGRP-S1 | Gamb_Mz | - | |||
| Gamb_Tz | 0.196* | - | |||
| Arab_Mz | 0.950* | 0.882* | - | ||
| Arab_Tz | 0.989* | 0.997* | 0.034* | - | |
| PGRP-S2 | Gamb_Mz | - | |||
| Gamb_Tz | 0.100NS | - | |||
| Arab_Mz | 0.153* | 0.317* | - | ||
| Arab_Tz | 0.153* | 0.317* | -0.111NS | - | |
| PGRP-S3 | Gamb_Mz | - | |||
| Gamb_Tz | 0.109* | - | |||
| Arab_Mz | 0.161* | 0.347* | - | ||
| Arab_Tz | 0.041NS | 0.284* | 0.116* | - |
NS - not significant; * P < 0.05; Arab, Anopheles arabiensis; Gamb, Anopheles gambiae; Mz, Mozambique; Tz, Tanzania.
To better understand the relationship between species and geographic regions, a hierarchical analysis of molecular variance (AMOVA) was performed (Table 3). For PGRP-S2 and PGRP-S3 genes most of the variation was distributed within populations, but for PGRP-S1 the remaining variation was distributed between species. These results corroborate the previous analyses indicating that for this gene the major variation occurred between species and not between different geographic regions.
Table 3.
Hierarchical analysis of molecular variance (AMOVA) among the An. gambiae and An. arabiensis groups.
| Source of variation | Hierarchical AMOVA for An. gambiae and An. arabiensis | ||
|---|---|---|---|
| PGRP-S1 | PGRP-S2 | PGRP-S3 | |
| Among groups | 95.23 | 23.53 | 8.88 |
| Among populations within groups | 0.45 | 2.68 | 10.28 |
| Within populations | 4.31 | 73.8 | 80.84 |
| Fcs (population/group) | 0.095*** | 0.035*** | 0.113*** |
| Fst (population/total) | 0.957** | 0.262NS | 0.192** |
| Fct (group/total) | 0.952* | 0.235* | 0.089* |
NS- not significant; * P < 0.05; ** P < 0.01 and *** P < 0.001
Phylogeny
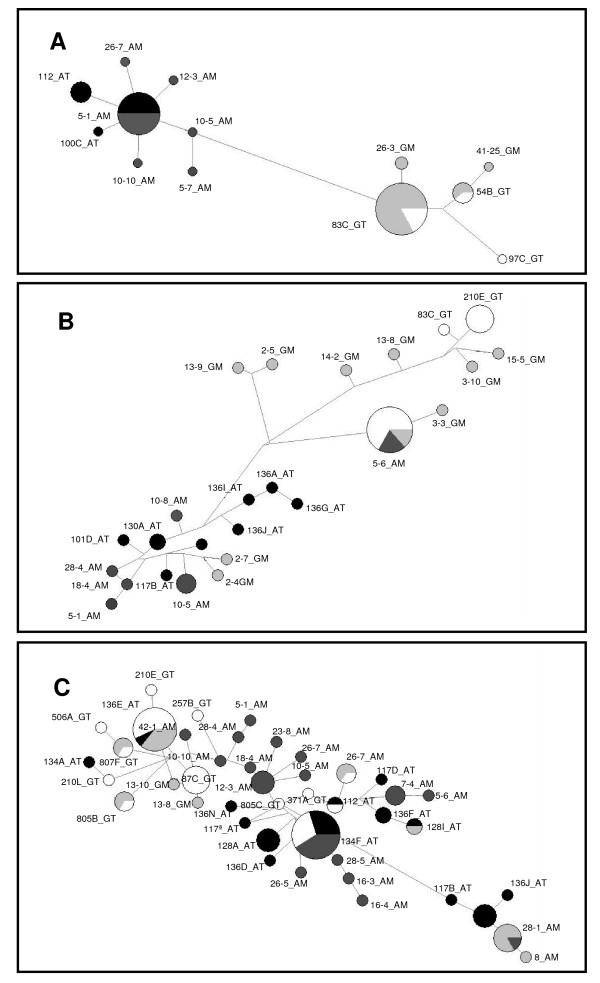
The median-joining network based on the PGRP-S1 haplotypes showed a clear interspecific separation (Figure 1A). Each species presented one haplotype at a higher frequency, 5-1_AM for An. arabiensis and 83C_GT for An. gambiae. Both were present in the 2 geographic regions and probably correspond to the ancestral haplotype. For An. arabiensis the other 7 haplotypes were unique for each geographic sample. In An. gambiae a lower number of haplotypes was found (5 versus 8) and a second haplotype was shared by the An. gambiae - Mozambique and An. gambiae - Tanzania populations.
Figure 1.
Median-joining network for PGRP-S1, PGRP-S2 and PGRP-S3 genes. Network was based on 13 haplotypes for PGRP-S1 gene, 25 haplotypes for PGRP-S2 gene and 43 haplotypes for PGRP-S3. The area of circles is proportional to the frequency of the haplotypes. Black - An. arabiensis, Tanzania; Dark-grey - An. arabiensis, Mozambique; Light-grey - An. gambiae, Mozambique and White - An. gambiae, Tanzania.
The network based on the PGRP-S2 haplotypes (Figure 1B) showed a higher number of haplotypes when compared to the PGRP-S1 network (26 versus 13), and the separation between species was not as clear as in the network of the PGRP-S1. There was a single high frequency haplotype, 5-6_AM, that was shared by more than one population and by both species. All the other haplotypes were unique for each population and species, and showed low frequency. All 13 low-frequency haplotypes specific of An. arabiensis were more closely related in the network. For An. gambiae, 9 haplotypes grouped together in the network but two (2-7_GM, 2-4_GM) were closer to An. arabiensis haplotypes.
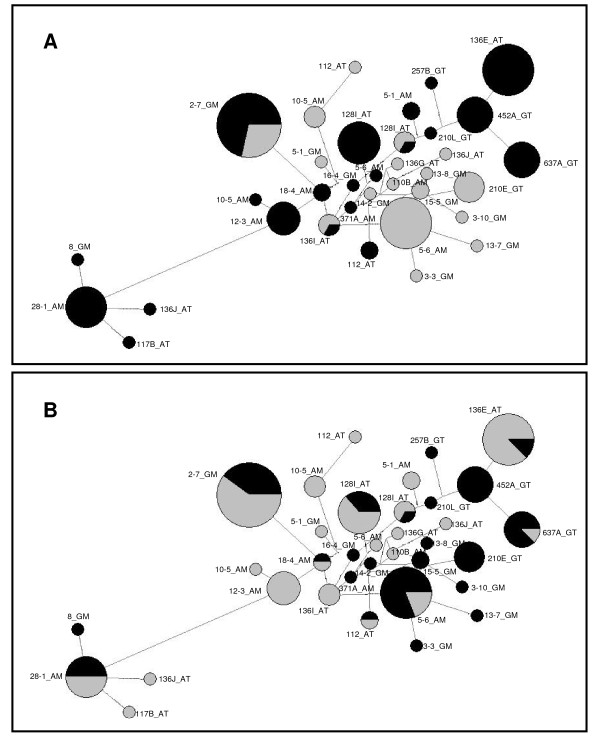
The network based on PGRP-S3 haplotypes was the most complex. As illustrated in Figure 1C, this network presented a high number of haplotypes (N = 43). Most of the haplotypes were unique, although 5 of them were shared between species and three between geographic populations within An. gambiae. No clear separation was observed either between species or between geographic regions. A network based on pooled PGRP-S2 and PGRP-S3 haplotypes showed interspecific common haplotypes between these two genes (Figure 2A), namely the most frequent haplotype 2-7_GM and another 2 less frequent (136I_AT and 128I_AT). All remaining haplotypes were exclusive.
Figure 2.
Median-joining network for PGRP-S2 and PGRP-S3 coding regions of both An. arabiensis and An. gambiae. Network was based on thirty four haplotypes. The area of circles is proportional to the frequency of the haplotypes. A: Black - PGRP-S3, Grey - PGRP-S2; B: Black - An. gambiae, Grey - An. arabiensis.
Another network based on pooled An. gambiae and An. arabiensis haplotypes (with sequences from both genes) (Figure 2B) was made to evaluate if clustering was predominately due to species or to homologous loci, in order to infer for concerted evolution. In case of concerted evolution we would expect that PGRP-S2 and PGRP-S3 would be more similar within the same species than to the homologous gene in the other species. This was not observed suggesting that gene conversion between PGRP-S2 and PGRP-S3 is not a major determinant of diversity of these genes.
Tests for selection
For the three short PGRPs genes, neutrality tests were performed for each of the four populations separately. The Tajima's D test detected one gene, PGRP-S1, with significant departure from neutrality in the An. gambiae-Tanzania population. For the other genes, the results were consistent with neutral evolution (Table 1). Similarly, in Fu & Li's D and F tests only the PGRP-S1 gene showed a significant departure from neutrality for An. gambiae-Tanzania population. Tests for departure from neutrality can be affected by various factors such as population expansion, which would result in negative Tajima D values for all loci. However, since only one gene displayed this pattern, population expansion is thus unlikely
Synonymous (πs) and nonsynonymous (πa) nucleotide diversity was calculated for each gene within each group (Table 1) revealing values below one for PGRP-S2 and PGRP-S3, suggesting purifying selection (Table 1). A Fisher's exact test of neutrality based on the number of synonymous and nonsynonymous substitutions between sequence pairs of An. gambiae and An. arabiensis was conducted in MEGA 4.1 [11] for each collection site. P-values were equal to one for the vast majority of pairwise comparisons confirming purifying selection (data not shown). Evidence of purifying selection was also confirmed by synonymous (Ks) and nonsynonymous (Ka) divergence rates. Interspecific comparisons had Ka/Ks ratios lower than one for the three genes, particularly for PGRP-S2 and PGRP-S3, which is compatible with purifying selection (Table 4).
Table 4.
The ratio of nonsynonymous substitutions per site (Ka) and the number of synonymous substitutions per site (Ks) in short PGRP genes between the two Anopheles species.
| Genes/species | Ka/Ks* | |
|---|---|---|
| PGRP-S1 | An. gambiae - An. arabiensis | 0.763 |
| PGRP-S2 | An. gambiae - An. arabiensis | 0.044 |
| PGRP-S3 | An. gambiae - An. arabiensis | 0.058 |
* Ka and Ks were estimated by DnaSP 4.0 [38] for the total coding regions
Protein diversity
There were 14 types of proteins present in our sample [see Additional file 1: Figure S1] representing 168 individuals, six different protein sequences for PGRP-S2 and PGRP-S3 and two that were shared by both genes. An. arabiensis had more protein sequences at lower frequencies while the opposite was observed in An. gambiae. The most frequent proteins were those shared by both species (protein type 5 and 7 for PGRP-S2 and 2, 4, 9 and 10 for PGRP-S3) [see Additional file 2: Table S1].
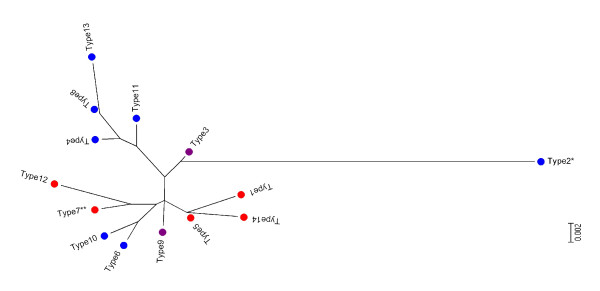
The phylogenetic analysis using different methods (Minimum Evolution, Maximum, Neighbor-Joining and UPGMA, with or without an out group - PGRP-S1) show the same structure (data not shown). An isolated branch corresponding to PGRP-S3 type 2 was always present and was the only one with a bootstrap confidence above 70% (Figure 3).
Figure 3.
Evolutionary relationships of the fourteen PGRP-S2/3 protein types. The evolutionary history was inferred using the Neighbor-Joining method. The bootstrap consensus tree was inferred from 1000 replicates. Tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. Red dots represent PGRP-S2 proteins, blue dots represent PGRP-S3 proteins and purple represent protein types shared by PGRP-S2 and PGRP-S3.
Sequences were allocated to each protein type [see Additional file 3: Table S2] and protein types associated to haplotypes in the network (Figure 2A) revealing that the most isolated cluster of the network correspond to protein type 2 while haplotypes clustered at the opposite corner are associated with protein type 4.
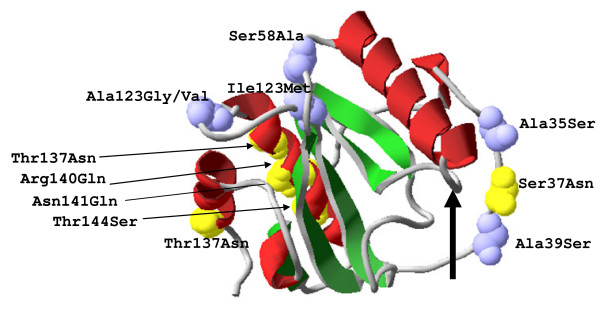
In order to understand if different protein sequences for PGRP-S2 and PGRP-S3 display different 3D configurations, proteins were modelled and visualized with the Swiss-PdB viewer v. 4.0.1. [12-14]. The best fitting 3D model for PGRP-S2 and PGRP-S3 was based on the crystal structure of the human PGRP-Iα (2aphB). The homology model shows the presence of three α helices, five β strands and coils (Figure 4).
Figure 4.
Structural model of PGRP-S2 and PGRP-S3 proteins. Three-dimensional (3D) structural localization of mutated amino acids represented as yellow and blue (Van de Walls spheres). The PGRP domain has three α helices (red), five β strands (green) and coils (grey); Arrow indicates the specificity-determining residues responsible for the muramyl pentapeptide - MPP-Dap recognition.
Amino acid substitutions were mainly present at the periphery in coils. The exception was PGRP-S3 type 2 that presented 4 substitutions (Thr137Asn, Arg140Gln, Asn141 Gln, Thr144Ser) in α2 helices and 1 in α3 helices (Thr185Asn). No mutations were present in the β strands or at the recognition sites.
For PGRP-S1 there were 2 protein types, one corresponding to An. arabiensis and another to An. gambiae [see Additional file 4: Figure S2]. The 3D model showed that the amino acid substitutions between the species were also observed at the α2 helices (data not shown).
Discussion
Nucleotide diversity estimated for the three short PGRP genes was comparable with other studies made on immune related genes of Drosophila and Anopheles [15-19]. Cohuet et al. [20] analysed 72 immune related genes of An. gambiae among them PGRP-S1. The nucleotide diversity (π) found for this gene for An. gambiae S form was 0.008; twice as high as the maximum value found in our study. This may relate to differences in geographic location. Although differences were not observed when we compared Mozambique and Tanzania a greater geographical separation could account for differences in genetic diversity between East and West African samples [20].
Some differences were found in the three genes studied: PGRP-S2 and PGRP-S3 revealed higher nucleotide diversities when compared to PGRP-S1. With respect to species divergence and population differentiation, once again PGRP-S1 differed from the others. Fst values and AMOVA indicated a clear separation between An. gambiae and An. arabiensis for PGRP-S1 gene, which was not observed for the other genes. The phylogenetic networks reinforced the results obtained by the AMOVA and Fst values. The different patterns observed between the three genes may be due to their location on the genome. PGRP-S1 gene is located in the chromosome X (within the Xag inversion) and PGRP-S2 and PGRP-S3 are located in the chromosome 2L (within the 2La inversion). An. gambiae and An. arabiensis display species-specific paracentric inversion arrangements Xag and Xbcd, respectively at the chromosome X. This should contribute to a higher differentiation between the two species at this chromosome due to reduced recombination in the case of rare hybridization between the two species. A recent study has shown that introgressed alleles between An. arabiensis and An. gambiae in the chromosome X were lost in two generations but introgressed alleles were not lost at loci located in the autosomic chromosome 2L [21]. This may explain the higher genetic divergence between species found in PGRP-S1 compared to the other two short PGRPs analysed. On the other hand, PGRP-S2 and PGRP-S3 showed no clear separation between species suggesting either retention of ancestral polymorphism or introgression between An. gambiae and An. arabiensis. Introgression of genes in autosomal chromosomes through rare hybridisation between these two sibling species has been demonstrated previously, particularly for chromosome 2L [21,22].
PRGP-S2 and PGRP-S3 are duplicated genes that are physically close (ca 3 kb) at chromosome 2L http://www.ensembl.org/Anopheles_gambiae and their coding region have 95% homology. These two genes are both considered functional, since the ratio Ka/Ks presents values much lower than 0.5 [23] and seem to be under purifying selection. This suggests subfunctionalization, i.e., these genes share the same or very similar functions. Neofunctionalization has been normally accepted has the terminal fate of duplicated genes. However more recently Gibson & Goldberg [24] suggest that this might not be a dominant mechanism of protein evolution.
This agrees with the 3D models of the protein, where substitutions were concentrated at the periphery (Figure 3), even for protein type 2 which was the most phylogenetically separated (Figure 2), while retaining the amidase/PGRP activity on both duplicate genes. Mammal PGRPs are known to form dimmers [9] and in Drosophila PGRP-SD interact with GNBP in order to optimally activate the Toll pathway [25]. Amino acid substitutions at non-catalytic sites that might be responsible for PRR-PRR interactions might favour the maintenance of slightly different proteins that interact optimally with different peptidoglycans. An. gambiae PGRP-S2 and PGRP-S3 function is not known but transcription profiles of Plasmodium falciparum infected mosquitoes were down regulated in midgut and carcasses when compared with mosquitoes that had a blood meal with a P. falciparum strain unable to produce gametocytes [6]. However, no differences were observed in both expression profiles [6]. Once again these results suggest similar function/regulation of these genes.
We cannot determine with certainty when the duplication occurred, but since it is present in both species it is likely that it happened before An. gambiae and An. arabiensis split from a common ancestor. The higher complexity of the haplotype network for PGRP-S3, showing more diversity and a higher proportion of shared haplotypes between species suggests that this may be the ancestral gene from which PGRP-S2 may have originated.
The results of selection tests showed that PGRP-S1 was the only gene presenting significant departures from neutrality with negative values for Tajima's D and Fu & Li's D and F. Negative D values indicate an excess of low frequency mutations consistent with positive selection, however this can be due to effective population size expansion. Ka/Ks ration for PGRP-S1 was also high (Table 4) but this could be a result of a very small number of diverged sites and per se do not provide a strong evidence of positive selection. For PGRP-S2 and PGRP-S3, both tests presented a non-significant deviation from neutrality. Ka/Ks ratio showed values much lower than one, reflecting functional constrains on the encoded proteins, i.e., this kind of selection contributes to the elimination of amino acid variation. Therefore, these results suggest that the PGRP-S2 and PGRP-S3 genes are under purifying selection. This does not come as a surprise as accumulating evidence suggests that the majority of Anopheles immune related genes, studied so far, are also under purifying selection (e.g. [26,27]). Since PGRP-S2 and PGRP-S3 could be recently duplicated genes tests like Tajima's D and Fu & Li's D and F provide limited information [28] and analysis of synonymous and nonsynonymous substitutions should be more informative. These analyses also point out to purifying selection as the driven force of short PGRPs evolution, in the same way as described for other PGRP genes of An. gambiae [29] or Drosophila [29].
The different types of selection observed between PGRP-S1 and PGRP-S2 and S3 might be a consequence of their function as PGRP-S1 does not have predicted catalityc activity and is probably involved in recognition and subsequent activation of an effector pathway, as PGRP-SA and PGRP-SD in Drosophila [3,4]. While PGRP-S2 and S3 would exert antimicrobial activity like PGRP-SB1 in Drosophila [30] and/or modulate the response as does Drosophila PGRP-SC1 and PGRP-SC2 [5]. Further, specificity-determining residues (Sdr) (Figure 4) will determine that distinct classes of peptidoglycans are recognized by different PGRPs: PGRP-S2 and S3 are predicted to bind MPP-Dap type peptidoglycans [31]. This does not explain why silencing of PGRP-S1 and PGRP-S2/3 protects the mosquito from infection by Staphylococcus aureus, that displays Lys-type peptidoglycan, but not from infection by Escherichia coli (Dap-type peptidoglycan) [32]. PGRP-S3 is expressed in response to gut microbiota [33]. and microbiota can modulate the response to Plasmodium by inducing mosquito basal immunity, which is essential to control the infection. Therefore pathogen specificity will indirectly determine the faith of mosquito malaria infection and exert different selection pressure on short PGRP coding genes.
Conclusions
The three Anopheles short PGRP genes studied are involved in the recognition of pathogens. However, they show different evolutionary pathways. PGRP-S1 gene is located in the chromosome X while PGRP-S2 and PGRP-S3 are located in chromosome 2L. This explains why PGRP-S1 is less genetically diverse and shows higher divergence between An. gambiae and An. arabiensis regardless of geographic location. On the contrary, PGRP-S2 and PGRP-S3 are more diverse and less divergent due to autosomal introgression between An. arabiensis and An. gambiae. Data indicated that PGRP-S2 and PGRP-S3 are likely subject to purifying selection consistent with their role in recognising conserved PAMPs. The 3D model of the proteins showed that no mutations were present at the cleft that forms the peptidoglycan binding groove, once again implying strong evolutionary constrains probably because these proteins need to maintain their PAMP recognition site unaltered, while the periphery that interact with other molecules is more prone to accumulate variation. The lower diversity and apparently higher divergence between species for the PGRP-S2 gene suggests that this is duplication from PGRP-S3. Different types of selection acting on PGRP-S1 and PGRP-S2 and S3 might be a consequence of their different function and catalytic activity.
Methods
Mosquito sampling and collection methods
Samples of An. gambiae (82) and An. arabiensis (46) genomic DNA were analysed from two areas in East Africa: Mozambique and Tanzania. Mozambique samples were collected in Furvela (Inhambane province) in February/April of 2004 using light traps and samples from Tanzania were collected in Ifakara in 2000 using also light traps [34]. Mosquitoes were kept dry in single tubes with silica gel.
Dna extraction
Genomic DNA was extracted from individual specimens according to the protocol described by Ballinger-Crabtree et al. [35]. Species identification and determination of An. gambiae molecular forms was carried out by PCR as described in Fanello et al. [36].
Polymerase chain reaction and sequencing
The primers used to amplify PGRP-S1, PGRP-S2 and PGRP-S3 genes were designed based on the sequences annotated in the complete An. gambiae s.s. genome at Ensembl (AGAP000536; AGAP006343; AGAP006342 respectively). Name, sequence and product length of each pair of primers are represented in [see Additional file 5: Table S3].
Nested PCRs were performed in a MyCycler™ Thermal cycler (Biorad) with final reagent concentrations of 1× reaction buffer, 1.5 mM of MgCl2, 200 μl dNTPs, 0.5 μl of each primer and 1.25 U/μl of Taq DNA Polymerase (Fermentas) for all reactions. PGRP-S1 1st nested PCR cycle conditions were: initial denaturation at 95°C for 2 minutes, followed by 35 cycles of 95°C for 45 seconds, 58°C for 60 seconds and 72°C for 2 minutes, with a final extension at 72°C for 5 minutes. PGRP-S1 2nd nested PCR cycle conditions were: initial denaturation at 95°C for 2 minutes, followed by 35 cycles of 95°C for 45 seconds, 58°C for 30 seconds and 72°C for 60 seconds and a final extension of 72°C for 5 minutes.
PGRP-S2 and PGRP-S3 1st nested PCR cycle conditions were similar to those used for PGRP-S1 amplification except for the annealing that was performed at 60°C. PGRP-S2 2nd nested PCR cycle conditions were: an initial denaturation step at 94°C for 2 minutes, followed by 35 cycles of 94°C for 45 seconds, 60°C for 30 seconds and 72°C for 60 seconds, with a final step of 72°C for 5 minutes. PGRP-S3 2nd nested PCR cycle conditions were similar to 1st nested but the annealing temperature was 55°C.
PCR products were examined on a 2% agarose gel and products of the expected length were sequenced in both directions after purification with the SureClean Kit (Bioline) according to manufacturer's recommendations. Products were commercially sequenced by Macrogen, Korea.
Genetic diversity and selection tests
Sequence alignments were performed using the BioEdit Sequence Alignment Editor version 7.0.5.2 [37]. Basic population genetic analyses and haplotype statistics were performed in DnaSP version 4.50.1 [38]. For each population sequence diversity was quantified by nucleotide diversity (π). To test deviation from neutrality Tajima's D test was performed. D is expected to be zero under neutrality with constant population size; Fu & Li's D and F tests were also performed; these are similar to Tajima's D test. The main differences between these tests concern the different estimators of the genetic diversity used. The Tajima's D test estimates the difference between η (total number of mutations that occurred in the entire genealogy) and πn (average number of nucleotide differences between two sequences) whereas in the Fu & Li's D and F tests the difference used is between the ηi (numbers of mutations in internal branches) and ηe (numbers of mutations in external branches) or between ηe and πn.
The tests referred above give little information in the case of young genes [39]. Therefore the Ka/Ks ratio was determined which compares the number of nonsynonymous (Ka) substitutions and the number of synonymous (Ks) substitutions per site between DNA sequences. Ka and Ks ratios were estimated by DnaSP version 4.50.1 [38]. Ka/Ks ratios equal to one are expected in genes under neutrality, Ka/Ks ratios less than one indicates purifying selection and Ka/Ks ratios higher than one indicate positive selection.
Protein diversity
Amino acid sequences were obtained using the BioEdit Sequence Alignment Editor version 7.0.5.2 [37] and aligned using the Clustal W program. Each protein sequence was modelled using swissmodel at http://swissmodel.expasy.org/workspace/index.php. The best fitting 3D model for PGRP-S2 and PGRP-S3 was based on the crystal structure of the human PGRP-Iα (2aphB) and for PGRP-S1 was based on Drosophila PGRP-SA (1sxrB). Nonsynonymous mutations were visualized on the models using the Swiss-PdB viewr v. 4.0.1. [14,13,12] in order to identify possible structural alterations and if location was within protein activity sites. Phylogenetic trees were constructed using MEGA 4.1 software [11].
Genetic structure and population differentiation
The genetic structure within and among An. gambiae and An. arabiensis populations was examined with analysis of molecular variance (AMOVA) [40]. The test was performed considering each species as a distinct group. Genetic differentiation between populations was estimated by sequence-based F statistics (Fst) according to Hudson et al. [41]. Significance of Fst estimates was assessed by pairwise genetic distances. These tests were performed in Arlequin software version 3.11 [42].
Phylogenetic analysis
To better understand phylogenetic relationships between intraspecific data, which normally consist in very similar sequences, we connected haplotypes on a median-joining network [43] using NETWORK 4.5.0.0 program based on default parameters.
Authors' contributions
CM, RF, MAS and JL performed the experiments; CM, RF, JP and HS analyzed and interpreted the data; JP, JDC and VER contributed with samples. CM, RF, JP and HS wrote the paper. HS conceived and designed the experiments. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Multiple alignment of deduced amino acid sequences of An. gambiae and An. arabiensis PGRP-S2 and PGRP-S3.
Table S1. Frequencies of PGRP-S2 and PGRP-S3 protein types of An. arabiensis and An. gambiae from Mozambique and Tanzania.
Table S2. DNA sequences grouped by protein type.
Figure S2. Multiple alignment of deduced amino acid sequence of An. gambiae and An. arabiensis PGRP-S1.
Table S3. Sequences of primers used to amplify the three Anopheles short PGRP genes.
Contributor Information
Cristina Mendes, Email: cristina.mendes@ihmt.unl.pt.
Rute Felix, Email: rfelix@ihmt.unl.pt.
Ana-Margarida Sousa, Email: am_sousa@sapo.pt.
Joana Lamego, Email: joana_lamego@hotmail.com.
Derek Charlwood, Email: jdcharlwood@gmail.com.
Virgílio E do Rosário, Email: virgilio.rosario@ihmt.unl.pt.
João Pinto, Email: jpinto@ihmt.unl.pt.
Henrique Silveira, Email: hsilveira@ihmt.unl.pt.
Acknowledgements
We are grateful to C. Drakeley and P. Alonso that were responsible for mosquito sampling in Tanzania. This work was supported by Fundação para a Ciência e a Tecnologia (FCT), through research funds from project POCTI/IMI/59489/2004. RF is a recipient of a FCT grant (SFRH/BD/28024/2006).
References
- Dziarski R. Peptidoglycan recognition proteins (PGRPs) Mol Immunol. 2004;40:877–886. doi: 10.1016/j.molimm.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Christophides GK, Zdobnov E, Barillas-Mury C, Birney E, Blandin S, Blass C, Brey PT, Collins FH, Danielli A, Dimopoulos G, Hetru C, Hoa NT, Hoffmann JA, Kanzok SM, Letunic I, Levashina EA, Loukeris TG, Lycett G, Meister S, Michel K, Moita LF, Müller HM, Osta MA, Paskewitz SM, Reichhart JM, Rzhetsky A, Troxler L, Vernick KD, Vlachou D, Volz J, von Mering C, Xu J, Zheng L, Bork P, Kafatos FC. Immunity-related genes and gene families in Anopheles gambiae. Science. 2002;298:159–165. doi: 10.1126/science.1077136. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5:1175–1180. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Aguilar R, Xi Z, Warr E, Mongin E, Dimopoulos G. Anopheles gambiae immune responses to human and rodent Plasmodium parasite species. PLoS Pathog. 2006;2:e52. doi: 10.1371/journal.ppat.0020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister S, Agianian B, Turlure F, Relogio A, Morlais I, Kafatos FC, Christophides GK. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5:e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziarski R, Gupta D. The peptidoglycan recognition proteins (PGRPs) Genome Biol. 2006;7:232. doi: 10.1186/gb-2006-7-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obbard DJ, Callister DM, Jiggins FM, Soares DC, Yan G, Little TJ. The evolution of TEP1, an exceptionally polymorphic immunity gene in Anopheles gambiae. BMC Evol Biol. 2008;8:274. doi: 10.1186/1471-2148-8-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Labate JA, Biermann CH, Eanes WF. Nucleotide variation at the runt locus in Drosophila melanogaster and Drosophila simulans. Mol Biol Evol. 1999;16:724–731. doi: 10.1093/oxfordjournals.molbev.a026157. [DOI] [PubMed] [Google Scholar]
- Jiggins FM, Hurst GD. The evolution of parasite recognition genes in the innate immune system: purifying selection on Drosophila melanogaster peptidoglycan recognition proteins. J Mol Evol. 2003;57:598–605. doi: 10.1007/s00239-003-2506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlais I, Poncon N, Simard F, Cohuet A, Fontenille D. Intraspecific nucleotide variation in Anopheles gambiae: new insights into the biology of malaria vectors. Am J Trop Med Hyg. 2004;71:795–802. [PubMed] [Google Scholar]
- Braverman JM, Lazzaro BP, Aguade M, Langley CH. DNA sequence polymorphism and divergence at the erect wing and suppressor of sable loci of Drosophila melanogaster and D. simulans. Genetics. 2005;170:1153–1165. doi: 10.1534/genetics.104.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard F, Licht M, Besansky NJ, Lehmann T. Polymorphism at the defensin gene in the Anopheles gambiae complex: testing different selection hypotheses. Infect Genet Evol. 2007;7:285–292. doi: 10.1016/j.meegid.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohuet A, Krishnakumar S, Simard F, Morlais I, Koutsos A, Fontenille D, Mindrinos M, Kafatos FC. SNP discovery and molecular evolution in Anopheles gambiae, with special emphasis on innate immune system. BMC Genomics. 2008;9:227. doi: 10.1186/1471-2164-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman MA, della TA, Calzetta M, Powell JR. Differential introgression of chromsomal regions between Anopheles gambiae and An. arabiensis. Am J Trop Med Hyg. 2005;73:326–335. [PubMed] [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, Toure Y, Sagnon N. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci USA. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton K, Long M. Rapid divergence of gene duplicates on the Drosophila melanogaster X chromosome. Mol Biol Evol. 2002;19:918–925. doi: 10.1093/oxfordjournals.molbev.a004149. [DOI] [PubMed] [Google Scholar]
- Gibson TA, Goldberg DS. Questioning the ubiquity of neofunctionalization. PLoS Comput Biol. 2009;5:e1000252. doi: 10.1371/journal.pcbi.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Gilbert RJ, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. Peptidoglycan recognition protein-SD provides versatility of receptor formation in Drosophila immunity. Proc Natl Acad Sci USA. 2008;105:11881–11886. doi: 10.1073/pnas.0710092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann T, Hume JC, Licht M, Burns CS, Wollenberg K, Simard F, Ribeiro JM. Molecular evolution of immune genes in the malaria mosquito Anopheles gambiae. PLoS ONE. 2009;4:e4549. doi: 10.1371/journal.pone.0004549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmakelis A, Slotman MA, Marshall JC, Awono-Ambene PH, Antonio-Nkondjio C, Simard F, Caccone A, Powell JR. The molecular evolution of four anti-malarial immune genes in the Anopheles gambiae species complex. BMC Evol Biol. 2008;8:79. doi: 10.1186/1471-2148-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Selection and gene duplication: a view from the genome. Genome Biol. 2002;3:reviews1012. doi: 10.1186/gb-2002-3-5-reviews1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little TJ, Cobbe N. The evolution of immune-related genes from disease carrying mosquitoes: diversity in a peptidoglycan- and a thioester-recognizing protein. Insect Mol Biol. 2005;14:599–605. doi: 10.1111/j.1365-2583.2005.00588.x. [DOI] [PubMed] [Google Scholar]
- Mellroth P, Steiner H. PGRP-SB1: an N-acetylmuramoyl L-alanine amidase with antibacterial activity. Biochem Biophys Res Commun. 2006;350:994–999. doi: 10.1016/j.bbrc.2006.09.139. [DOI] [PubMed] [Google Scholar]
- Swaminathan CP, Brown PH, Roychowdhury A, Wang Q, Guan R, Silverman N, Goldman WE, Boons GJ, Mariuzza RA. Dual strategies for peptidoglycan discrimination by peptidoglycan recognition proteins (PGRPs) Proc Natl Acad Sci USA. 2006;103:684–689. doi: 10.1073/pnas.0507656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister S, Agianian B, Turlure F, Relogio A, Morlais I, Kafatos FC, Christophides GK. Anopheles gambiae PGRPLC-mediated defense against bacteria modulates infections with malaria parasites. PLoS Pathog. 2009;5:e1000542. doi: 10.1371/journal.ppat.1000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Manfredini F, Dimopoulos G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009;5:e1000423. doi: 10.1371/journal.ppat.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakeley C, Schellenberg D, Kihonda J, Sousa CA, Arez AP, Lopes D, Lines J, Mshinda H, Lengeler C, Armstrong Schellenberg J, Tanner M, Alonso P. An estimation of the entomological inoculation rate for Ifakara: a semi-urban area in a region of intense malaria transmission in Tanzania. Trop Med Int Health. 2003;8:767–774. doi: 10.1046/j.1365-3156.2003.01100.x. [DOI] [PubMed] [Google Scholar]
- Ballinger-Crabtree ME, Black WC, Miller BR. Use of genetic polymorphisms detected by the random-amplified polymorphic DNA polymerase chain reaction (RAPD-PCR) for differentiation and identification of Aedes aegypti subspecies and populations. Am J Trop Med Hyg. 1992;47:893–901. doi: 10.4269/ajtmh.1992.47.893. [DOI] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, della TA. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16:461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser (Oxf) 1999;41:95–98. [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Thornton KR. The neutral coalescent process for recent gene duplications and copy-number variants. Genetics. 2007;177:987–1000. doi: 10.1534/genetics.107.074948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schwede S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005. pp. 47–50. [PMC free article] [PubMed]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Multiple alignment of deduced amino acid sequences of An. gambiae and An. arabiensis PGRP-S2 and PGRP-S3.
Table S1. Frequencies of PGRP-S2 and PGRP-S3 protein types of An. arabiensis and An. gambiae from Mozambique and Tanzania.
Table S2. DNA sequences grouped by protein type.
Figure S2. Multiple alignment of deduced amino acid sequence of An. gambiae and An. arabiensis PGRP-S1.
Table S3. Sequences of primers used to amplify the three Anopheles short PGRP genes.