Abstract
Benzylsuccinate synthase is a member of the glycyl radical family of enzymes. It catalyzes the addition of toluene to fumarate to form benzylsuccinate as the first step in the anaerobic pathway of toluene fermentation. The enzyme comprises three subunits α, β and γ that in Thauera Aromatica T1 strain are encoded by the tutD, tutG and tutF genes respectively. The large α-subunit contains the essential glycine and cysteine residues that are conserved in all glycyl radical enzymes. However, the function of the small β- and γ-subunits has remained unclear. We have over-expressed all three subunits of benzylsuccinate synthase in E. coli, both individually and in combination. Co-expression of the γ-subunit (but not the β-subunit) is essential for efficient expression of the α-subunit. The benzylsuccinate synthase complex lacking the glycyl radical could be purified as an α2β2γ2 hexamer by nickel-affinity chromatography through a ‘His6’ affinity tag engineered onto the C-terminus of the α-subunit. Unexpectedly, BSS was found to contain two iron-sulfur clusters, one associated with the β-subunit and the other with the γ-subunit that appear to be necessary for the structural integrity of the complex. The spectroscopic properties of these clusters suggest that they are most likely [4Fe-4S] clusters. Removal of iron with chelating agents results in dissociation of the complex; similarly a mutant γ-subunit lacking the [4Fe-4S] cluster is unable to stabilize the α-subunit when the proteins are co-expressed.
Keywords: toluene metabolism, free radicals, Thauera. aromatica, glycyl radical enzyme, iron-sulfur clusters
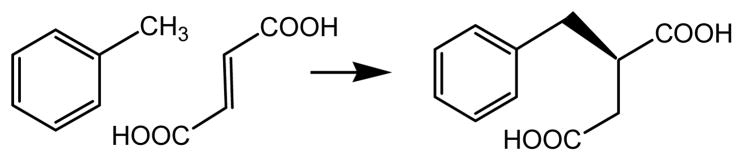
Benzylsuccinate synthase1 (BSS) catalyzes a most unusual chemical transformation in which toluene is added across the double bond of fumarate to produce (R)-benzylsuccinate (scheme 1). This is the first step in the anaerobic pathway of toluene metabolism that allows various denitrifying and sulfate-reducing bacteria such as Thauera aromatica and Desulfobacula toluolica to live on toluene as their sole source of carbon and energy under anaerobic conditions (1, 2). The metabolism of toluene and related aromatic compounds is of particular interest both because these compounds represent an important class of pollutants that are long-lived in the environment and have relatively high water solubility, and because of they involve novel approaches to the activation of unreactive C–H bonds (3, 4).
Scheme 1.
Reaction catalyzed by benzylsuccinate synthase
Sequence similarities initially identified BSS as a member of the glycyl radical-containing group of enzymes that includes pyruvate formate-lyase and anaerobic ribonucleotide reductase (5–10). Later EPR studies show that the resting enzyme harbors an organic radical similar to that observed in these enzymes (11–13). Like other glycyl radical enzymes, BSS is extremely oxygen sensitive, and exposure to air results in oxidative cleavage of the α subunit at the site of the presumed glycyl radical (5). The glycyl radicals in these enzymes are generated from the cognate glycine residue by specific activases that are members of the family of S- adenosylmethionine radical enzymes (9, 14, 15).
The proposed mechanism for the BSS-catalyzed reaction (3, 16) involves transfer of the glycyl radical to an active site cysteine residue, a feature common to both ribonucleotide reductase and pyruvate formate lyase, which then abstracts hydrogen from toluene to generate a benzylic radical. The benzylic radical subsequently undergoes addition to fumarate to generate the C-3 radical of (R)-benzylsuccinate. The abstracted hydrogen is transferred to the product to generate benzylsuccinate and the cysteinyl radical, and lastly the radical is transferred back to glycine.
Mechanistic studies in our laboratory have focused on providing evidence for putative radical intermediates in the reaction and probing the energetics of the reaction. Evidence for the formation of the C-3 radical of benzylsuccinate as an intermediate comes from experiments to investigate the stereochemistry of hydrogen transfer from toluene to fumarate and to the alternate co-substrate maleate (17). When fumarate was the co-substrate, syn addition of toluene to the double bond of fumarate was observed. However, when maleate was the co-substrate, the addition of toluene occured in an anti fashion. These observations are explained by the formation of the C-3 radical of benzylsuccinate in which rotation about the C-2, C-3 bond can occur to relieve the sterically unfavorable cis conformation of the carboxylate groups when maleate is the co-substrate.
Using deuterated toluene, we have undertaken steady-state deuterium kinetic isotope effect measurements to probe the free energy profile of the BSS reaction (18). By comparing the relative magnitudes of the DV and D(V/K) isotope effects, measured experimentally, with those predicted by a density functional theoretical study (19), we obtained evidence that the enzyme altered the free energy profile such that the starting glycyl radical and the intermediate cysteinyl and benzylic radicals appear to be much closer in energy than calculated. This suggests that the enzyme plays a role in stabilizing the cysteinyl and benzylic radicals relative to the glycyl radical, which may be an important and general mechanism for controlling the reactivity of radicals in this class of enzymes.
Further insight into the unusual energetics of the BSS reaction comes examining the exchange reaction of p-cresol with the benzyl portion of benzylsuccinate to form (4-hydroxybenzyl)-succinate (20). This allowed us to investigate the kinetics of the reverse reaction: disproportionation of benzylsuccinate to toluene and fumarate. Even though the equilibrium constant for the reverse reaction is extremely unfavorable, Keq~ 8 × 10−11 M at 4 °C, the enzyme catalyzes the reverse reaction at rate only 250-fold slower than the forward reaction. During the reverse reaction partial exchange of the migrating hydrogen with the solvent is observed. This provides the first direct evidence that the hydrogen is transferred to a labile site on the protein during catalysis, which is consistent with the participation of the proposed active site cysteine residue in the mechanism of BSS.
The rapid loss of activity encountered during attempts to purify the enzyme and its extreme oxygen sensitivity (5) has severely hindered efforts to study the enzyme. The above experiments could all be conducted with cell-free extracts of BSS because of the lack of alternate reaction pathways for toluene. However, for many studies it is necessary to have large amounts of pure enzyme and for some purposes the ability to generate BSS radical-free would be extremely useful. Here we report the over-expression of the BSS subunits in E. coli and characterization of the properties of the radical-free enzyme. Unexpectedly, the β- and γ-subunits are revealed to be iron-sulfur proteins and the iron sulfur clusters appear to play a key role in the assembly of the holo-enzyme.
MATERIALS AND METHODS
Materials
DNA-modifying enzymes and reagents were purchased from New England Biolabs (Beverly, MA). Oligonucleotide primers were obtained either from IDT Integrated DNA Technologies (Coralville, IA) or from Invitrogen (Carlsbad, CA). E. coli BL21(λDE3) and expression vectors pET-28b, pACYC-Duet, pET-Duet, pRSF-Duet were purchased from Novagen (Madison, WI); Pfu turbo DNA polymerase and E. coli XL1-Blue were from Stratagene (Cedar Creak, TX).. Bovine gamma globulin standard and Bradford reagent were purchased from Bio-Rad Laboratories, Inc (Hercules, CA) and used for routine protein concentration determination. Nickel nitrilotriacetic acid (Ni-NTA) resin and QIAquick PCR purification kit were purchased from Qiagen (Valencia, CA). Sephadex G-25 resin and Superose 6 pre-packed FPLC column were purchased from GE Biosciences (Piscataway, NJ). All other chemicals used were of the highest grade commercially available.
Subcloning of tut genes into expression vectors the BSS proteins were overexpressed in E. coli using the commercially available pET series of vectors to place the genes under control of the T7 promoter and, where desired, introduce a His6 tag at either the N- or C-terminus. Standard PCR and site-directed mutagenesis techniques were used to introduce appropriate restriction sites into the genes of interest to facilitate their subcloning into the desired expression vectors. A summary description of the plasmids generated in this study is given in Table 1; a detailed description of the construction of these plasmids is included as supporting material.
Table 1.
Summary of BSS expression constructs used in this study
| Plasmid name | Vector used | Gene inserted | Cloning sites | Description |
|---|---|---|---|---|
| pET28_F | pET28b | tutF | NdeI/EcoRI | Expression of BSS-γ subunit with a N-terminus His6 tag |
| pET28_G | pET28b | tutG | NdeI/HindIII | Expression of BSS-β subunit with a N-terminus His6 tag |
| pET28_D | pET28b | tutD | NdeI/HindIII | Expression of BSS-α subunit with a N-terminus His6 tag |
| pET-Duet_F | pET-Duet | tutF | NcoI/HindIII | Expression of BSS-γ subunit with no tag (negative control for monitoring BSS α subunit expression in cell-free extract) |
| pET-Duet_D_F | pET-Duet |
tutD tutF |
NdeI/KpnI NcoI/HindIII |
Expression of BSS-α and γ subunits with a C-terminus His6 tag on the α subunit |
| pRSF-RSF_G | pRSF-Duet | tutG | NdeI/XhoI | Expression of BSS-β subunit with no tag |
| pET-Duet_D_F_stop | pET-Duet | tutD | NdeI/KpnI | Expression of BSS-α subunit with a C-terminus His6 tag |
| pET-Duet_D_F_C9S | pET-Duet |
tutD tutF_C9S |
NdeI/KpnI NcoI/HindIII |
Expression of BSS-α and γ subunits with a subunit carrying a C-terminus His6 tag and γ subunit a C9S mutation |
| pRSF-Duet_G_C29S | pRSF-Duet | tutG_C29S | NdeI/XhoI | Expression of BSS-β subunit with a C29S mutation |
| pACYC-Duet_F_C9S | pACYC-Duet | tutF_C9S | BamHI/XhoI | Expression of BSS-γ subunit with a N-terminus His6 tag and a C9S mutation |
| pET28_G_C29S | pET28b | tutG_C29S | NdeI/HindIII | Expression of BSS-β subunit with a N-terminus His6 tag and a C29S mutation |
Expression of BSS proteins
The expression constructs containing the various tut genes were transformed into E. coli BL21(λDE3) cells. A single colony of transformed cells was used to inoculate 5 mL of LB medium containing the appropriate antibiotic to maintain selection for the plasmid. The cultures were grown to saturation at 37°C with vigorous shaking and then used to inoculate 1 L of antibiotic-containing LB medium. Upon reaching early log phase (OD600 = 0.8), expression of the genes was induced by addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. The cultures were grown for 4 h before harvesting by centrifugation (6000 rpm, 15 min at 4 °C). The media was decanted and the cell pellet stored at − 20 °C.
For the expression of the BSS holo-enzyme complex it was found advantageous to modify the expression protocol as follows. The medium was supplemented with 150 mg Fe(NH4)2(SO4)2 per liter of culture. Upon reaching OD600 = 0.8, the culture was allowed to cool to ambient temperature before induced by addition IPTG to a final concentration of 0.5 mM. The culture was grown for 3 more hours at room temperature with gentle stirring, to minimize oxygenation, before harvesting by centrifugation.
Purification of BSS proteins
To purify BSS-β and BSS-γ, the cell pellet from 1 L of culture was re-suspended in 30 mL of lysis buffer containing 50 mM Tris, 300 mM NaCl, 10 mM imidazole, 5 mM β-mercaptoethanol, 1 mM PMSF at pH 8.0. The cells were lysed by sonication on ice and centrifuged at 17,000 rpm for 20 min at 4°C. The supernatant was loaded onto a 1.5 mL Ni- NTA-agarose column and moved into a Coy anaerobic glove box. The rest of the protein purification steps were carried out under an inert atmosphere. The column was washed with 20 volumes of the buffer containing 50 mM Tris, 300 mM NaCl, 20 mM imidazole, at pH 8.0. The tagged BSS proteins were eluted with the same buffer but containing 300 mM imidazole. The eluted proteins were yellow-brown colored and were determined be essentially homogenous by SDS-PAGE on a 12% tricine gel stained with Coumassie blue. BSS was eluted with the same buffer but containing 100 mM imidazole. Proteins were dialyzed three times against the buffer containing 25 mM Tris and 300 mM NaCl at pH 8.0 and stored at −20 °C. A typical purification yielded 30 – 40 mg protein/L of cell culture. To purify the complex of BSS-α with BSS-β and BSS-γ the protocol was modified such that 100 mM imidazole was used to elute the proteins and the protein purity was assessed by SDS-PAGE using 10 % tricine gels.
Purification and refolding of BSS-α from inclusion bodies
Despite surveying a number of conditions, BSS-a protein, when expressed alone, was always produced as inclusion bodies and could only be purified by Ni-NTA chromatography in 8M urea. Briefly, inclusion bodies from 1L media were solublized in 100 mM NaH2PO4 buffer, pH 8.0, containing 10 mM Tris/Cl and 8.0 M urea. The resulting protein was loaded onto 1.5 mL Ni-NTA-agarose column and non- specifically bound proteins removed by washing with the same buffer adjusted to pH 6.3. The His-tagged a subunit was eluted by lowering the pH of the buffer further to pH 4.5. The protein obtained by this procedure was >95% homogeneous as judged by SDS-PAGE. Typically, the purification yielded 50 mg of unfolded protein per liter of LB media.
BSS-α protein was refolded by dialysis of the purified, urea-denatured protein against progressively lower concentrations of urea in a buffer containing 10% glycerol and 5 mM fumarate. Importantly, it was found that the presence of fumarate was essential to obtain good yields of refolded protein.
Determination of protein concentrations
Protein concentrations were calculated based on the absorption of aromatic residues at 280 nm in the presence of 6 M guanidine hydrochloride using the method of Gill and von Hippel (21).
Iron determination
Iron content was determined using o-bathophenanthroline (OBP) under reductive conditions after digestion of the protein in 0.8% KMnO4/1.2 N HCl as described by Fish (22). Iron standards were prepared from commercially available ferric chloride and ferrous ammonium sulfate.
Reconstitution of iron-sulfur clusters iron-sulfur clusters were reconstituted using a modified protocol described by Jarrett (23). The reactions were set up in an anaerobic glove box and contained DTT (5 mM), FeCl3 (500 μM), and Na2S (500 μM) in 1 mL of protein solution. The solutions were incubated for 20 hrs at 4 °C. Excess reagents were removed by dialysis three times against a buffer containing 25 mM Tris-Cl, 300 mM NaCl at pH 8.0. The extent of cluster formation was assessed by UV-visible spectroscopy (spectra taken using an anaerobic cuvette) and from the iron content determined as described above.
Size exclusion chromatography 250 μl of a 50 μM solution of BSS was loaded onto a Superose 6 FPLC column pre-equilibrated in 25 mM Tris/Cl, 300 mM NaCl, 10 mM β-mercaptoethanol, pH 8.0 at 4 °C. The column was eluted at 0.1 mL/min and the eluant monitored at 280 nm. The column was calibrated using standard proteins for molecular weight determination (BioRad).
Removal of iron-sulfur clusters from BSS iron was removed from the protein by treating it with the chelating agent OBP at a final concentration of 200 mM (24). The reaction was typically allowed to proceed at room temperature for 3 h in air or at 4 °C for 16 hrs in an anaerobic chamber. Occasionally some precipitation occurred during the reaction which as removed by centrifugation at 13000 rpm for 15 min at 4 °C.
EPR spectroscopy
Samples were prepared in an anaerobic glove box. Proteins were dialyzed twice against 50 mM Tris-HCl, 200 mM NaCl, 15% glycerol, 0.03% sarkosyl, pH 8.0. Dialyzed protein samples were concentrated to ~ 30 μM using an Amicon Microcon centrifugal ultrafiltration device (Bedford, MA) according to the manufacturer’s instructions. Samples were reduced by addition of sodium dithionite, 2 mM final concentration and immediately transferred to an EPR tube, fitted with an airtight septum and frozen and stored in liquid nitrogen. All EPR spectra were obtained on a Bruker EMX electron spin resonance spectrometer equipped with a Bruker 4102-ST general purpose cavity (rectangular TE102) and an Oxford liquid helium cryostat. Conditions for EPR spectra acquisition were as follow: temperature, 10K; microwave frequency, 9.41 GHz; microwave power, 2.05 milliwatt; modulation frequency, 100 kHz; modulation amplitude, 10.00 G; conversion time and time constant, 40.96 ms; 16 scan average. The data were analyzed using the Bruker Win-EPR data manipulation program.
RESULTS
Although BSS is expressed at quite high levels in T. aromatica, the protein has proven extremely hard to purify from this organism. In part, this is because the wild-type enzyme contains a reactive glycyl radical which renders it unstable to air, but significant losses in activity were encountered even during purification under rigorously anaerobic conditions in a glove box ((16); M.S. Huhta and E.N.G. Marsh unpublished results). To circumvent this problem, we sought to over-express and purify BSS from E. coli where the protein could be produced without the catalytic glycyl radical. This would render the protein air stable and thereby greatly facilitate purification and physical characterization of the BSS complex, as well as allowing the individual protein subunits to be purified and characterized. In particular, we wished to investigate the function of the small β and γ subunits that have been shown to be essential for enzyme activity (25, 26).
Expression of the tut genes
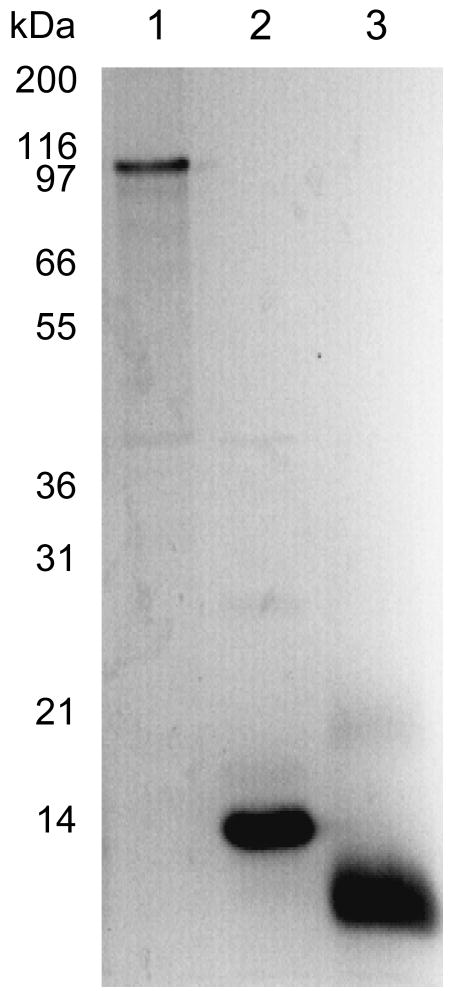
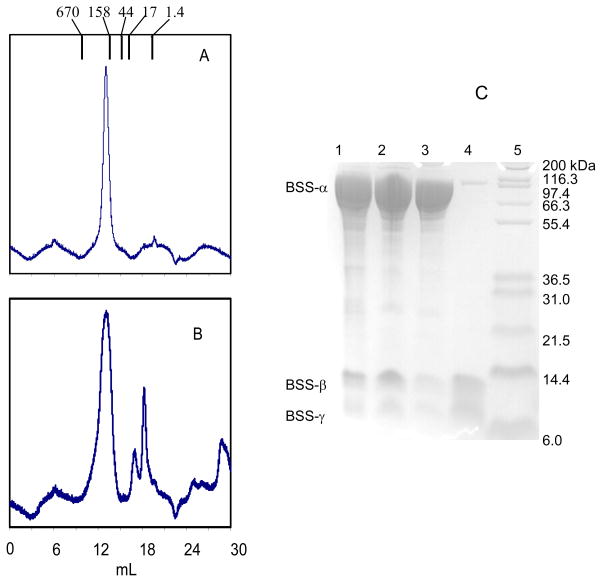
Initially, the tutD, tutF, and tutG genes (encoding the BSS-α, γ and β subunits respectively) were each subcloned into the expression vector pET28b (constructs pET28_D, pET28_F, pET28_G, Table 1) which placed them under control of the T7 promoter and also introduced a His6 affinity tagging sequence at the N-terminus of each protein. Each gene was strongly over-expressed from this vector in E. coli. The BSS-β and γ subunits were produced as soluble proteins, and could be purified straightforwardly by chromatography on a Ni-NTA column (Figure 1). However, the α subunit was always produced as insoluble inclusion bodies. Attempts to express BSS-α in soluble form by varying expression conditions, by, for example, lowering growth temperature, reducing the concentration of IPTG or inducing expression under anaerobic conditions, met with no success. Therefore, the protein was solublized with urea and purified from inclusion bodies by Ni-NTA chromatography under denaturing conditions; the protein was then allowed to refold by removing the urea by dialysis to give homogenous, soluble protein (Figure 1).
Figure 1.
Over-expression and purification of individual BSS subunits. Lane 1: BSS-α, purified from inclusion bodies and refolded; lane 2: BSS-β, purified as soluble protein by Ni-NTA chromatography; lane 3: BSS-γ, purified as soluble protein by Ni-NTA chromatography.
Interestingly, it was found that inclusion of 5 mM fumarate in the refolding buffer significantly improved the yield of soluble BSS-α, suggesting that this substrate may be stabilizing the refolded protein, or possibly nucleating the protein folding. However, surprisingly, the refolded protein did not bind to a Ni-NTA column, suggesting that the N- terminal His6 tag may be sequestered within the refolded protein. This raised the possibility that the His tag might interfere with the correct folding of the protein.
We therefore considered the possibility that the β and γ subunits might be required for the correct folding of the α subunit. To investigate whether this was the case we used the “Duet” expression system developed by Novagen to express each of the genes from its own T7 promoter. The tutD and tutF genes were subcloned into the two multiple cloning sites of pETDuet™ (plasmid pET-Duet_D_F), which is under the control of the ColE1 replicon and selected by ampicillin resistance. The tutG gene was subcloned into the second multiple cloning site of pRSFDuet™ (plasmid pRSF-Duet_G), which is under the control of the RSF1030 replicon and selected by kanamycin resistance. As part of this cloning strategy a sequence encoding His6 affinity tag was introduced at the 3′-terminus of the tutD gene.
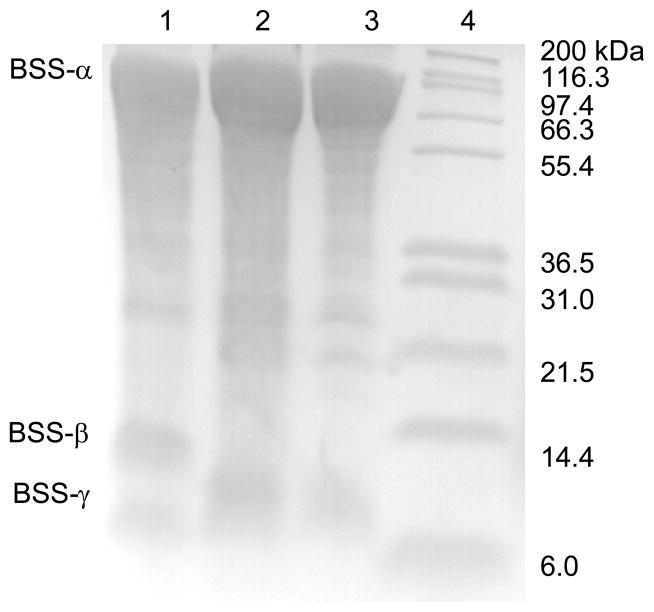
Induction of E. coli BL21 harboring the plasmids described above with IPTG at room temperature with limited aeration (gentle stirring) resulted in all three genes being expressed as soluble proteins. Furthermore, the intact BSS-αβγ complex could easily be purified through the His6 tag on the α subunit by chromatography on a Ni-NTA column, as shown in Figure 2. This result implies that the BSS-β and γ subunits are tightly bound to the α subunit since the small subunits were not themselves tagged and further suggested that one or both of them might be important for the correct folding of the α subunit.
Figure 2.
Over-expression and purification of BSS-αβγ and BSS-αγ complexes. Lane 1: BSS-α2β2γ2 complex, purified by Ni-NTA chromatography; lane 2: BSS-α2γ2, complex purified by Ni-NTA chromatography; lane 3: BSS-α2γ2, complex after gel filtration; lane 4: molecular weight standards. Note: the lanes have been heavily over-loaded so that the weakly staining β and γ subunits can be seen.
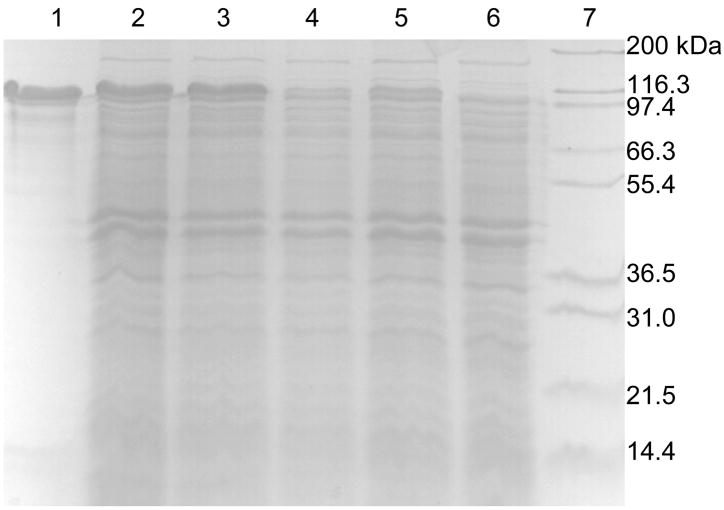
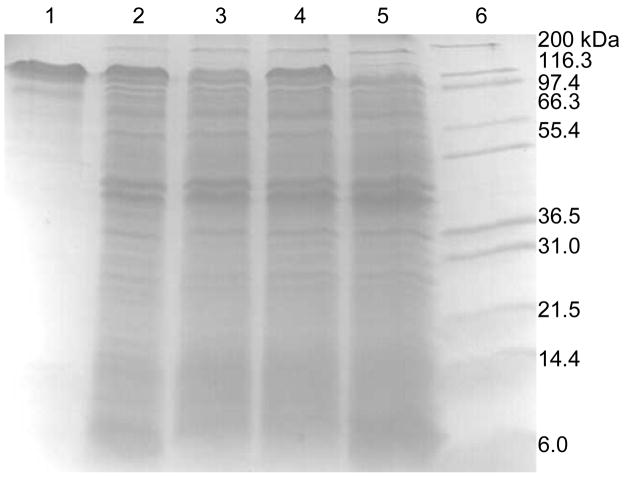
To investigate the effects of BSS-β and BSS-γ on the folding of BSS-α, E. coli strains were constructed in which either BSS-β or BSS-γ were absent. For BSS-γ this was achieved by introducing a stop codon after the Phe13 codon in the tutF gene (plasmid pET-Duet_D_F_stop, Table 1); for BSS-β this was simply achieved using cells only transformed with plasmid pET-Duet_D_F. The genes were induced by addition of 0.5 mM IPTG to cultures, which were gently stirred at room temperature for 3 hours. Cell free extracts were prepared and the amount of soluble BSS-α produced evaluated by SDS PAGE. As shown in Figure 3, the absence of the β subunit appears to have little effect on the level of soluble BSS-α. However, disruption of BSS-γ resulted in a significant reduction in the level at which BSS-α accumulated in the cells. Consistent with this, the complex of BSS-α with BSS-γ could be purified from the supernatant by Ni-NTA chromatography (Figure 2), whereas attempts to purify BSS-α by Ni-NTA chromatography from cells lacking BSS-γ were unsuccessful.
Figure 3.
Effect of the BSS-β and BSS-γ proteins on the expression of BSS-α in E. coli. Lane 1: BSS-α standard; lane 2: co-expression of tutD, tutF and tutG genes (BSS-α strongly expressed); lane 3: co-expression of tutD and tutF genes (BSS-α strongly expressed); lane 4: co-expression of tutD and tutG genes (BSS-α weakly expressed); lane 5: expression of only tutD gene (BSS-α weakly expressed); lane 6: expression of only tutF gene (negative control for BSS-α expression); lane 7: molecular weight standards.
Characterization of iron-sulfur clusters in BSS
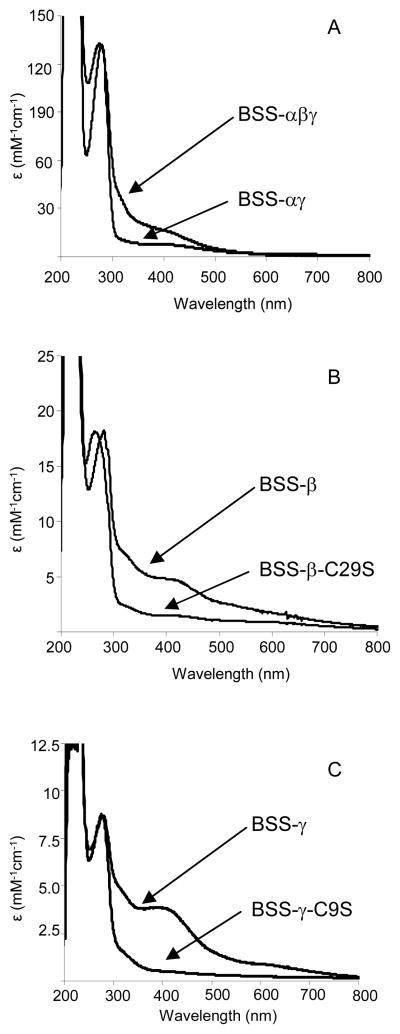
The purified BSS-αβγ protein complex was yellow-brown in color and the u.v.-visible spectrum of the enzyme exhibited a maximum at 420 nm, suggesting the presence of one or more iron sulfur clusters in the protein (Figure 4). Analysis of the iron content of the protein (Table 2) gave a value of 7.1 ± 0.3 iron atoms per αβγ trimer, which, together with the characteristic absorption at 420 nm, pointed to the complex most likely containing two [4Fe-4S]2+ clusters. Sequence analysis of the BSS proteins identified two cysteine-rich ferridoxin-like motifs, shown below, that could serve as binding sites for [4Fe-4S] clusters in the sequences of BSS-β and BSS-γ.
Figure 4.
U.V.-visible spectra of BSS proteins. A: Spectra of the BSS-αβγ complex and the BSS-αγ complex as isolated from E. coli; B: Spectra of BSS-β protein and the BSS-β-C29S mutant after reconstitution of iron-sulfur clusters; C: Spectra of BSS-γ protein and the BSS-γ-C9S mutant after reconstitution of iron-sulfur clusters.
Table 2.
Iron content and extinction coefficients of BSS subunits
| Protein or protein complex | Fe content | ε420 (mM−1cm−1) |
|---|---|---|
| BSS-αβγ | 7.1 ± 0.3 a | 18500 a |
| BSS-αγ | 2.9 ± 0.3 a | 8100 a |
| BSS-γ | 1.5 ± 0.2 b 0.5 ± 0.2 a |
3800 b |
| BSS-γ-C9S | 0.2 ± 0.12 b < 0.11 a |
700 b |
| BSS-β | 2.0 ± 0.2 b 0.7 ± 0.2 a |
6200 b |
| BSS-β-C29S | 0.4 ± 0.2 b ~ 0.1 a |
1800 b |
Iron content of protein as isolated
Iron content after reconstitution of iron-sulfur cluster under anaerobic conditions
| BSS-β | 26C-X-X29C-X14-44C-X22-67C |
| BSS-γ | 6C-X-X9C-X11-29C-X23-53C |
Consistent with this, the BSS-αγ complex exhibited a lower ε420 (Table 2) and contained only 2.9 ± 0.3 iron atoms per αγ heterodimer, indicating that the remaining iron-sulfur was associated with the β subunit.
The u.v.-visible spectra of the individually expressed β and γ subunits, purified by Ni-NTA chromatography from recombinant E. coli, each exhibited characteristic maxima at 420 nm suggesting that they contained iron-sulfur clusters (Figure 4). However, the extinction coefficients for the clusters were rather small and iron analysis indicated that less than one iron atom was bound per protein molecule (Table 2), suggesting that most of the iron may have been lost during purification. The iron-sulfur clusters could be partially reconstituted by incubating the proteins with Fe(II) and sulfide ions under reducing conditions, following established protocols described in the Materials and Methods section. After reconstitution and purification the iron content increased to 1.5 – 2.0 iron atoms per protein molecule, with a corresponding increase in ε420; this is still significantly lower than the iron content of the holo-enzyme and suggests that in the absence of BSS-α the iron-sulfur clusters are very labile. Possibly, when BSS-β, BSS-γ and BSS-α associate together the small proteins become more structured, and thus the binding site for the iron-sulfur cluster better pre-organized, which would stabilize the metal cluster.
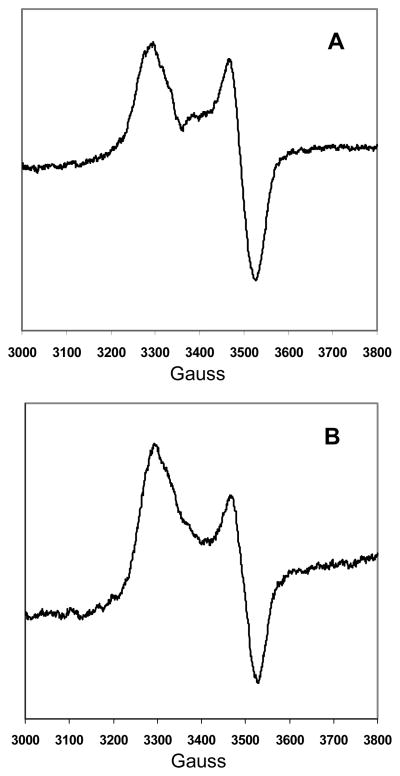
Further evidence for the identity of the iron-sulfur clusters comes from the EPR spectra of the sodium dithionite-reduced BSS-αβγ complex and the BSS-αγ complex, shown in figure 5. These spectra are characteristic of [4Fe-4S]+ iron sulfur clusters, with g|| = 2.04 and g⊥ = 1.94. However, the isolated BSS-β and BSS-γ proteins when reduced gave no EPR signals. The isolated BSS-β and BSS-γ were reduced immediately upon addition of sodium dithionite, whereas the BSS-αβγ complex and the BSS-αγ complex were reduced slowly over the course of ~ 30 min, as determined by bleaching of the sample color. This suggests that the clusters in the isolated BSS-β and BSS-γ proteins are much more exposed to solvent and that reduction results in the loss of the cluster from the protein, consistent with the lability of the unreduced clusters noted above.
Figure 5.
EPR spectra of sodium dithionite reduced BSS complexes as isolated from E. coli. A: Spectrum of the BSS-αβγ complex; B: Spectrum of the BSS-αγ complex
The finding that BSS-β and γ subunits contained iron-sulfur clusters was unexpected because there is no obvious requirement for a redox cofactor in the mechanism of the radical reaction catalyzed by BSS. We therefore considered whether the clusters may play a structural role. The molecular weight of the recombinant BSS was estimated by gel filtration chromatography using a Superose-6 FPLC column. The protein migrated in a single symmetrical peak (Figure 6) with apparent Mr = 230,000, which is consistent with an α2β2γ2 subunit structure for the enzyme, as was reported for the enzyme purified from T. aromatica (16). Analysis of the protein peak after gel filtration confirmed that all three subunits were present (Figure 5).
Figure 6.
Dissociation of BSS upon treatment with iron chelators. A: Chromatography of BSS on Superose-6 gel filtration column. The intact BSS complex elutes with apparent Mr = 230000 (elution volumes of molecular weight standards are indicated above chromatogram). B: elution profile of BSS after treatment with iron chelator. The first peak contains primarily BSS- α; the second BSS-β and BSS-γ; the third peak contains no proteins and is probably chelating agent. C: analysis of column fractions by SDS-PAGE; lane 1: BSS purified by chromatography on Ni-NTA column; lane 2: BSS after gel filtration chromatography - material from peak in chromatograph A; lane 3: BSS after treatment with iron chelator and gel filtration chromatography - material from first peak (BSS-α) in chromatograph B; lane 4: BSS after treatment with iron chelator and gel filtration chromatography - material from second peak (BSS-β and γ) in chromatograph B. lane 5: molecular weight standards.
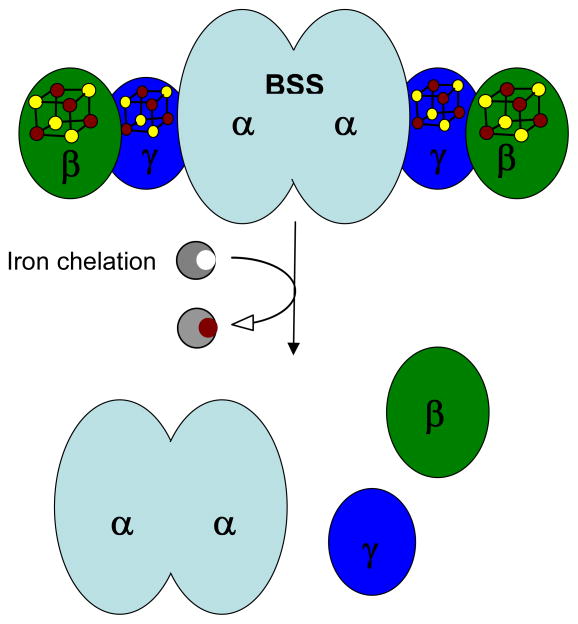
To investigate the effect of removing the iron-sulfur clusters on the quaternary structure of BSS, the protein was incubated with the iron chelating agent OBP overnight under anaerobic conditions, followed by analysis by gel filtration on a Superose-6 column. The OBP-treated protein eluted as two peaks (Figure 6) from the column. SDS-PAGE analysis showed that the first peak is mainly BSS-α, with traces of BSS-β and γ remaining; elution volume of this peak indicates that the isolated α subunit remains dimeric. The second peak contained BSS-β and γ subunits. Thus removal of the iron-sulfur clusters from the small subunits causes them to dissociate from the large α subunit.
To further investigate the role of the iron-sulfur clusters in the folding and stability of BSS, mutations were introduced to disrupt the binding sites for the clusters. The putative cysteine ligands, C29 in BSS-β and C9 in BSS-γ were mutated to serine (plasmids pACYC-Duet_F_C9S and pET28_G_C29S) and the mutant proteins expressed and purified from E. coli. Consistent with these cysteine residues providing the ligands to iron, the mutant proteins contained almost no iron ( ~ 0.1 Fe/protein; Table 2) and attempts to reconstitute the iron-sulfur clusters in these proteins resulted in less than 0.5 Fe/protein being incorporated (Table 2 and Figure 4). Such low levels of iron incorporation may well be due to non-specific binding of the metal to the protein.
We then examined the effect of co-expressing the mutant BSS-β and γ proteins on the stability of BSS-α expressed in E. coli. This was achieved by co-transforming E. coli BL12 with either plasmids pET-Duet_D_F and pRSF-Duet_G_C29S or with pET-Duet_D_F_C9S and pRSF-Duet_G. Similar to the experiments described above, the expression of the tut genes was induced with IPTG for 3 hours, cell-free extracts prepared and the amount of soluble BSS-α protein produced examined by SDS-PAGE. The results (Figure 7) mirrored those obtained from the gene deletion experiments. The C9S mutation in BSS-γ resulted in a noticeable reduction the amount of BSS-α produced; in contrast the C29S in BSS-β did not significantly alter BSS-α expression. The result indicates that the iron-sulfur cluster in BSS-γ is necessary for this subunit to bind to BSS-α, which is consistent with the gel filtration data that shows that removal of the clusters results in dissociation of the subunits.
Figure 7.
Effect of mutations that remove the iron-sulfur clusters in BSS-β and BSS-γ on expression of BSS-α in E. coli. Lane 1: BSS-α standard; lane 2: co-expression of tutD, tutF and tutG genes (BSS-α strongly expressed); lane 3: co-expression of tutD, tutG and tutF-C9S genes (BSS-α weakly expressed); lane 4: co-expression of tutD, tutF and tutG-C29S genes (BSS-α strongly expressed); lane 5: expression of only tutF gene (negative control for BSS-α expression); lane 6: molecular weight standards.
DISCUSSION
Here we report the expression and purification from E. coli and initial physical characterization of BSS from T. aromatica. The recombinant enzyme is produced without the catalytic glycyl radical, resulting in a stable protein that facilitates its handling and characterization. In this form the protein is, of course, inactive so that its catalytic properties cannot be examined, however that fact that recombinant BSS can be isolated as the iron-sulfur-containing hetero-hexameric complex strongly suggests that it is correctly folded. Generation of the radical-containing form of BSS will require the requisite activase enzyme be co-expressed in E. coli; experiments to accomplish this are currently on-going in our laboratory. Although we thought it might be possible that the endogenous E. coli activase enzymes for pyruvate formate lyase and anaerobic ribonucleotide reductase, which are expressed when E. coli is grown anaerobically, could recognize and activate BSS, it appears that this is not the case (L. L. and D. P. P., unpublished results).
BSS is has a more complex structure than other better-characterized glycyl radical enzymes: pyruvate formate lyase, anaerobic ribonucleotide reductase and the B12-independent glycerol dehydratase are all homodimers and contain no metal clusters (27–29). In contrast, BSS comprises three subunits, α, β and γ, and we have presented evidence that both the β and γ subunits contain [4Fe-4S] clusters. Genetic experiments in Coschigano’s laboratory have demonstrated that both small subunits are essential for T. aromatica to grow on toluene, indicating that they are required for the large subunit to be active (26). In this study we sought to gain insight into their biochemical function.
One function of BSS-γ appears to be to stabilize the structure of the active site-containing BSS-α subunit. Thus in the absence of the γ subunit BSS-α was expressed poorly, if at all, in E. coli and could not be purified from induced cells by Ni-NTA affinity chromatography. The iron- sulfur cluster appears to be essential for BSS-γ to bind BSS-α as chelation of the iron results in dissociation of the subunits. Furthermore, mutation of the putative iron ligand cysteine-9 to alanine abolished both formation of the cluster and expression of BSS-α in cells containing the mutant BSS-γ protein. This mutation has also been shown to abolish the ability of T. aromatica to grow on toluene (25).
The biochemical function of BSS-β is less clear. The iron-sulfur cluster appears to be important for BSS activity because T. aromatica carrying the BSS-β C29S mutation cannot grow on toluene (25). However, neither deletion of this subunit unit, nor mutation of the iron-sulfur cluster away from the protein appears to affect the stability of the α subunit as judged by the levels of expression of BSS-α in E. coli. The BSS-αγ complex can purified as a stable protein in the absence of BSS-β, whereas in the absence of BSS-γ the BSS-αβ complex cannot. Taken together, these results tentatively suggest that the γ-subunit may serve as a linker between the α and β-subunits, as illustrated in Figure 8.
Figure 8.
Structural model for BSS, illustrating the location of the [4Fe-4S] clusters and the proposed quaternary structure of the enzyme. Chelation of iron by OBP results in dissociation to the dimeric α subunit and monomeric β and γ subunits.
Structurally, the most closely related glycyl radical enzyme to BSS appears to be 4- hydroxyphenylacetate (HPA) decarboxylase which catalyzes the formation of p-cresol in various Clostridia (10). This enzyme comprises two subunits and is active as an α4β4 hetero-octamer. The large (~ 100 kDa) subunit contains the catalytic glycyl radical, whereas the small (~ 9.5 kDa) subunit was recently shown to contain a [4Fe-4S] cluster by Selmer and co-workers (30), who percipiently speculated the BSS-β and γ subunits might similarly be iron-sulfur proteins. It also was found that co-expression of the HPA decarboxylase β-subunit was essential to obtain soluble α-subunit in E. coli, (31) suggesting that HPA decarboxylase-β and BSS-γ subunits have similar functions.
Whereas the [4Fe-4S] clusters may simply play a structural role in BSS, as the γ-subunit appears to, this seems somewhat unlikely, especially for the cluster associated with the β-subunit. It has been suggested that in HPA decarboxylase the β-subunit may serve to regulate the enzyme by reducing the catalytic glycyl radical and thereby deactivating the enzyme, as the glycyl radical in this enzyme has a fairly short lifetime (30). It is plausible that the small subunits in BSS play a similar role and that perhaps BSS-β interacts with a further regulatory protein involved in sensing toluene concentrations.
Lastly, we note also that serine phosphorylation has been implicated in the regulation of HPA decarboxylase activity (30, 31) suggesting a complicated mechanism for the control of p-cresol production. This raises the intriguing question of whether phosphorylation may play a role in the activation of BSS. It appears that BSS and HPA decarboxylase form a subclass of glycyl radical enzymes that is distinct both structurally and mechanistically from the better understood paradigm represented by pyruvate formate lyase.
Supplementary Material
Acknowledgments
We thank Prof. Stephen Ragsdale and Dr. Gunes Bender for assistance with obtaining EPR spectra and Andrew Markham for valuable technical assistance.
Footnotes
The abbreviations used are: BSS, benzylsuccinate synthase; NTA, nitrilotriacetic acid; OBP, o-bathophenanthroline; HPA, 4-hydroxyphenylacetic acid
This research was supported by grants from the National Institutes of Health: GM 59227 to E.N.G.M. and K99 ES 017177 to L.L.; D.P.P. acknowledges the support of NIH funded Chemistry Biology Interface training grant T32 GM008597
Supporting material: A detailed description of the construction of the plasmids used in this study is available as supporting material with the web version of this manuscript: http://pubs.acs.org.
References
- 1.Biegert T, Fuchs G, Heider F. Evidence that anaerobic oxidation of toluene in the denitrifying bacterium Thauera aromatica is initiated by formation of benzylsuccinate from toluene and fumarate. Eur J Biochem. 1996;238:661–668. doi: 10.1111/j.1432-1033.1996.0661w.x. [DOI] [PubMed] [Google Scholar]
- 2.Rabus R, Heider J. Initial reactions of anaerobic metabolism of alkylbenzenes in denitrifying and sulfate reducing bacteria. Arch Microbiol. 1998;170:377–384. [Google Scholar]
- 3.Spormann AM, Widdel F. Metabolism of alkylbenzenes, alkanes, and other hydrocarbons in anaerobic bacteria. Biodegradation. 2000;11:85–105. doi: 10.1023/a:1011122631799. [DOI] [PubMed] [Google Scholar]
- 4.Heider J, Spormann AM, Beller HR, Widdel F. Anaerobic bacterial metabolism of hydrocarbons. FEMS Microbiol Rev. 1998;22:459–473. [Google Scholar]
- 5.Leuthner B, Heider J. A two-component system involved in regulation of anaerobic toluene metabolism in Thauera aromatica. FEMS Microbiol Lett. 1998;166:35–41. doi: 10.1111/j.1574-6968.1998.tb13180.x. [DOI] [PubMed] [Google Scholar]
- 6.Coschigano PW. Transcriptional analysis of the tutE tutFDGH gene cluster from Thauera aromatica strain T1. Appl Environ Microbiol. 2000;66:1147–1151. doi: 10.1128/aem.66.3.1147-1151.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Achong GR, Rodriguez AM, Spormann AM. Benzylsuccinate synthase of Azoarcus sp strain T: Cloning, sequencing, transcriptional organization, and its role in anaerobic toluene and m-xylene mineralization. J Bacteriol. 2001;183:6763–6770. doi: 10.1128/JB.183.23.6763-6770.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frey PA. Radical mechanisms of enzymatic catalysis. Ann Rev Biochem. 2001;70:121–148. doi: 10.1146/annurev.biochem.70.1.121. [DOI] [PubMed] [Google Scholar]
- 9.Marsh ENG, Huhta MS, Patwardhan A. S-adenosylmethionine radical enzymes. Bioorganic Chem. 2004;32:326–340. doi: 10.1016/j.bioorg.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Selmer T, Pierik AJ, Heider J. New glycyl radical enzymes catalysing key. metabolic steps in anaerobic bacteria. Biol Chem. 2005;386:981–988. doi: 10.1515/BC.2005.114. [DOI] [PubMed] [Google Scholar]
- 11.Duboc-Toia C, Hassan AK, Mulliez E, Ollagnier-de Choudens S, Fontecave M, Leutwein C, Heider J. Very high-field EPR study of glycyl radical enzymes. J Am Chem Soc. 2003;125:38–39. doi: 10.1021/ja026690j. [DOI] [PubMed] [Google Scholar]
- 12.Verfurth K, Pierik AJ, Leutwein C, Zorn S, Heider J. Substrate specificities and electron paramagnetic resonance properties of benzylsuccinate synthases in anaerobic toluene and m-xylene metabolism. Arch Microbiol. 2004;181:155–162. doi: 10.1007/s00203-003-0642-4. [DOI] [PubMed] [Google Scholar]
- 13.Krieger CJ, Roseboom W, Albracht SPJ, Spormann AM. A stable organic free radical in anaerobic benzylsuccinate synthase of Azoarcus sp strain T. J Biol Chem. 2001;276:12924–12927. doi: 10.1074/jbc.M009453200. [DOI] [PubMed] [Google Scholar]
- 14.Frey PA, Hegerman AD, Reed GH. Free radical mechanisms in enzymology. Chem Rev. 2006;106:3302–3316. doi: 10.1021/cr050292s. [DOI] [PubMed] [Google Scholar]
- 15.Walsby CJ, Ortillo D, Yang J, Nnyepi MR, Broderick WE, Hoffman BM, Broderick JB. Spectroscopic approaches to elucidating novel iron-sulfur chemistry in the “Radical-SAM” protein superfamily. Inorg Chem. 2005;44:727–741. doi: 10.1021/ic0484811. [DOI] [PubMed] [Google Scholar]
- 16.Leuthner B, Leutwein C, Schulz H, Horth P, Haehnel W, Schiltz E, Schagger H, Heider J. Biochemical and genetic characterization of benzylsuccinate synthase from Thauera aromatica: a new glycyl radical enzyme catalysing the first step in anaerobic toluene metabolism. Mol Microbiol. 1998;28:615–628. doi: 10.1046/j.1365-2958.1998.00826.x. [DOI] [PubMed] [Google Scholar]
- 17.Qiao CH, Marsh ENG. Mechanism of benzylsuccinate synthase: Stereochemistry of toluene addition to fumarate and maleate. J Am Chem Soc. 2005;127:8608–8609. doi: 10.1021/ja051972f. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Marsh ENG. Deuterium isotope effects in the unusual addition of toluene to fumarate catalyzed by benzylsuccinate synthase. Biochemistry. 2006;45:13932–13938. doi: 10.1021/bi061117o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Himo F. Catalytic mechanism of benzylsuccinate synthase, a theoretical study. J Phys Chem B. 2002;106:7688–7692. [Google Scholar]
- 20.Li L, Marsh ENG. Mechanism of Benzylsuccinate Synthase Probed by Substrate Exchange. J Am Chem Soc. 2006;128:16056–16057. doi: 10.1021/ja067329q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gill SC, Vonhippel PH. Calculation of Protein Extinction Coefficients from Amino-Acid Sequence Data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 22.Fish WW. Rapid colorimetric micromethod for the quantitation of complexed iron in biological samples. Method Enzymol. 1988;158:357–364. doi: 10.1016/0076-6879(88)58067-9. [DOI] [PubMed] [Google Scholar]
- 23.Ugulava NB, Gibney BR, Jarrett JT. Biotin synthase contains two distinct iron-sulfur cluster binding sites: chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry. 2001;40:8343–8351. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll KS, Gao H, Chen H, Leary JA, Bertozzi CR. Investigation of the iron-sulfur cluster in Mycobacterium tuberculosis APS reductase: implications for substrate binding and catalysis. Biochemistry. 2005;44:14647–14657. doi: 10.1021/bi051344a. [DOI] [PubMed] [Google Scholar]
- 25.Bhandare R, Calabro M, Coschigano PW. Site-directed mutagenesis of the Thauera aromatica strain T1 tutE tutFDGH gene cluster. Biochem Biophys Res Comm. 2006;346:992–998. doi: 10.1016/j.bbrc.2006.05.199. [DOI] [PubMed] [Google Scholar]
- 26.Coschigano PW. Construction and characterization of insertion/deletion mutations of the tutF, tutD, and tutG genes of Thauera aromatica strain T1. FEMS Microbiol Lett. 2002;217:37–42. doi: 10.1111/j.1574-6968.2002.tb11453.x. [DOI] [PubMed] [Google Scholar]
- 27.Becker A, Fritz-Wolf K, Kabsch W, Knappe J, Schultz S, Wagner AFV. Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase. Nat Struct Biol. 1999;6:969–975. doi: 10.1038/13341. [DOI] [PubMed] [Google Scholar]
- 28.Logan D, Andersson J, Sjoberg B-M, Nordlund P. A glycyl radical site in the crystal structure of a class III ribonucleotide reductase. Science. 1999;283:1499–1504. doi: 10.1126/science.283.5407.1499. [DOI] [PubMed] [Google Scholar]
- 29.O’Brien JR, Raynaud C, Croux C, Girbal L, Soucaille P, Lanzilotta WN. Insight into the mechanism of the B12-independent glycerol dehydratase from Clostridium butyricum: Preliminary biochemical and structural characterization. Biochemistry. 2004;43:4635–4645. doi: 10.1021/bi035930k. [DOI] [PubMed] [Google Scholar]
- 30.Yu L, Blaser M, Andrei PI, Pierik AJ, Selmer T. 4- hydroxyphenylacetate decarboxylases: Properties of a novel subclass of glycyl radical enzyme systems. Biochemistry. 2006;45:9584–9592. doi: 10.1021/bi060840b. [DOI] [PubMed] [Google Scholar]
- 31.Andrei PI, Pierik AJ, Zauner S, Andrei-Selmer LC, Selmer T. Subunit composition of the glycyl radical enzyme p-hydroxyphenylacetate decarboxylase - A small subunit, HpdC, is essential for catalytic activity. Eur J Biochem. 2004;271:2225–2230. doi: 10.1111/j.1432-1033.2004.04152.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.