Summary
MutLα, the heterodimeric eukaryotic MutL homolog, is required for DNA mismatch repair (MMR) in vivo. It has been suggested that conformational changes, modulated by adenine nucleotides, mediate the interactions of MutLα with other proteins in the MMR pathway, coordinating the recognition of DNA mismatches by MutSα and the activation of MutLα with the downstream events that lead to repair. Thus far, the only evidence for these conformational changes has come from x-ray crystallography of isolated domains, indirect biochemical analyses, and comparison to other members of the GHL ATPase family to which MutLα belongs. Using atomic force microscopy (AFM), coupled with biochemical techniques, we demonstrate that adenine nucleotides induce large asymmetric conformational changes in full-length yeast and human MutLα, and that these changes are associated with significant increases in secondary structure. These data reveal an ATPase cycle where sequential nucleotide binding, hydrolysis, and release modulate the conformational states of MutLα.
Introduction
DNA mismatch repair (MMR) is the mechanism by which base-base mismatches and insertion-deletion loops generated during DNA replication are repaired. This post-replicative repair increases the fidelity of DNA synthesis 100 – 1000 fold (Bellacosa, 2001; Iyer et al., 2006; Kunkel and Erie, 2005; Modrich, 2006; Modrich and Lahue, 1996; Schofield and Hsieh, 2003). The proteins involved in MMR also participate in meiotic and mitotic recombination, in apoptotic signaling, and in somatic hypermutation of immunoglobulin genes (Bellacosa, 2001; Iyer et al., 2006; Kunkel and Erie, 2005; Modrich and Lahue, 1996; O’Brien and Brown, 2006; Schofield and Hsieh, 2003; Stojic et al., 2004).
In both prokaryotes and eukaryotes, the mismatch repair process begins when MutS or a MutS homolog recognizes and binds to a mismatch. In E. coli, MutL is then recruited, and the resulting MutS-MutL complex signals to MutH and the other downstream proteins to remove the mismatched base and complete the repair process (Iyer et al., 2006; Kunkel and Erie, 2005). MutH recognizes and binds to hemi-methylated GATC sites, which serve as strand discrimination signals. The endonuclease activity of MutH must be activated by the MutS-MutL complex for it to incise the DNA, providing an entry point for the exonucleases and helicase involved in repair.
Eukaryotes, as well as some prokaryotes, have no known MutH homolog, and the sequence of events from mismatch recognition to repair for these organisms is not completely understood. As such, the role of the eukaryotic MutL homolog, MutLα, is not fully known. MutLα is a heterodimer comprised of Mlh1 and Pms2 (Pms1 in yeast) and contains a latent endonuclease activity that is activated in the presence of a mismatch, MutSα, PCNA, RFC and ATP (Kadyrov et al., 2006). In addition to this endonucleolytic activity, MutLα is able to suppress the activity of ExoI on homoduplex DNA (Genschel and Modrich, 2003; Jager et al., 2001; Nielsen et al., 2004), and it interacts with several other proteins in the MMR pathway (Jager et al., 2001; Kadyrov et al., 2006; Lee and Alani, 2006; Mendillo et al., 2005; Umar et al., 1996). It has been proposed that conformational changes in MutLα caused by ATP binding and/or hydrolysis modulate these varying protein interactions during the repair process (Guarne et al., 2001; Raschle et al., 2002; Tran and Liskay, 2000).
Like the homodimeric E. coli MutL (Guarne et al., 2004; Kosinski et al., 2005), each subunit in the heterodimeric MutLα contains a C-terminal dimerization domain (Pang et al., 1997) connected to an N-terminal ATPase domain (Pang et al., 1997; Tran and Liskay, 2000) via a linker region that is predicted to be disordered (Guarne et al., 2004). The ATPase activity of MutL and MutLα is required for MMR (Hall et al., 2002; Pang et al., 1997; Raschle et al., 2002; Spampinato and Modrich, 2000; Tran and Liskay, 2000) and is also required for the endonuclease activity of hMutLα (Kadyrov et al., 2006).
MutL and MutL homologs are members of the GHL ATPase family (Ban et al., 1999; Ban and Yang, 1998; Dutta and Inouye, 2000; Guarne et al., 2001; Hu et al., 2003), which is characterized by a non-traditional ATP binding fold (Bergerat et al., 1997). Other members of this family include the namesakes, DNA Gyrase and Hsp90, as well as Grp94 and the type II topoisomerases. MutL and other GHL family members have been shown to have slow rates (0.4 min−1 – 0.9 min−1) of ATP hydrolysis in the absence of other cofactors (Ban et al., 1999; Dutta and Inouye, 2000; Spampinato and Modrich, 2000). In all proteins in the GHL family, ATP binding and/or hydrolysis appears to induce large conformational changes which are purported to be involved in the signaling of cellular processes (Ali et al., 2006; Ban et al., 1999; Chu et al., 2006; Corbett and Berger, 2003, 2005; Dollins et al., 2005; Dutta and Inouye, 2000; Immormino et al., 2004; Shiau et al., 2006).
The conformational changes observed in E. coli MutL in response to adenine nucleotides have been previously explored indirectly by size-exclusion chromatography and directly by crystallographic structures of the isolated N-terminal domain. Size-exclusion chromatography has shown that the full length E. coli MutL adopts a more compact size in the presence of a non-hydrolyzable ATP analog 5′-adenylyl-beta-gamma-imidodiphosphate (AMPPNP), which the authors attribute to N-terminal dimerization (Ban et al., 1999). Similarly, crystal structures of the N-terminal domain of E. coli MutL show that AMPPNP binding results in the dimerization of the N-terminal domains, which is distinct from the monomeric structure of the apo N-terminal domain (Ban et al., 1999). The crystal structure of the AMPPNP bound form of E. coli MutL also shows that upon AMPPNP binding, the ATP lid of the Bergerat fold folds over and makes contacts with the AMPPNP.
In stark contrast to E. coli MutL, the N-terminal fragment of hPms2, one of two subunits in eukaryotic MutLα, is a monomer in the crystal structure, even in the presence of an ATP analog. Unlike E.coli MutL, the N-terminal fragment of hPms2 is hydrolytically proficient, and dimerization does not appear to be a requirement for ATP hydrolysis (Guarne et al., 2001). Additionally, the ATP lid of the Bergerat fold becomes more disordered, and fewer residues of this lid are seen in the N-terminal Pms2 structure with ATPγS bound than in the apo structure. Partial proteolysis experiments show that ATP binding to MutLα causes a reduction in digestion by trypsin, and yeast two-hybrid experiments suggest that ATP promotes an interaction between the N-terminal domains of yMlh1 and yPms1 (Raschle et al., 2002; Tran and Liskay, 2000). These data suggest that ATP induces conformational changes in MutLα; however, to date, there are no data on the full-length structures of either MutL or MutLα.
In this paper we present an analysis of the full-length structure of MutLα at the level of individual molecules. Using AFM, coupled with biochemical analyses, we have examined nucleotide-induced changes in the quaternary structure of human and yeast MutLα. Our AFM images reveal that MutLα can adopt four distinct conformations. Based on the observed conformational changes, we propose a model for how ATP binding and hydrolysis drives conformational changes in MutLα. These observed conformational changes are likely coupled to the signaling of downstream events in mismatch repair.
Results
MutLα Exists in Four Different Conformational States
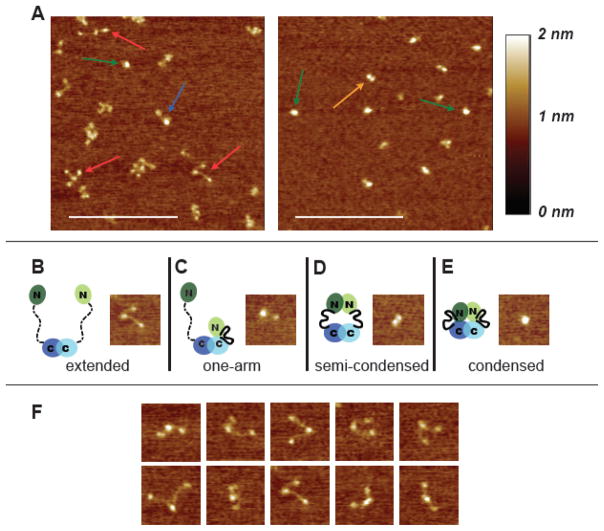
Inspection of AFM images such as those in Figure 1A reveals that yMutLα can adopt four different conformational states. In the absence of additional nucleotide cofactors, MutLα exists predominately in an open and “v-shaped” extended conformation, in which a large compact central domain is connected to two smaller domains by flexible arms (Figure 1B, 1F, and S1). Because it is known that yMlh1 and yPms1 form the heterodimeric complex via their C-termini (Pang et al., 1997), the larger structure found at the vertex of the V is undoubtedly the dimerized C-termini, while the two domains located at the ends of the elongated linker arms are the N-termini. A statistical analysis revealed that one arm is approximately 40% longer than the other and that there is a broad distribution of angles between the two arms (Figure S1 and Table S1). These results are consistent with secondary structure predictions made using PsiPred and PONDR (Bryson et al., 2005; Jones, 1999; McGuffin et al., 2000), which suggest that the N- and C- terminal domains of yMlh1 and yPms1 have well defined secondary structures that are connected by disordered regions of ~160 and ~300 amino acids, respectively. In addition, the flexible linker arms have a smaller cross-section than the coiled-coil domains of Rad50 as visualized by AFM (van Noort et al., 2003), further reinforcing the idea that the arms that connect the N- and C-termini lack secondary structure. Finally, the volume of proteins in AFM images has been shown to depend linearly on molecular weight (Ratcliff and Erie, 2001; Yang et al., 2005; Yang et al., 2003), and the measured volumes of the three observed domains are consistent with the predicted molecular weights of the structured N-termini and dimerized C-terminal domains (Tables S2 and S3).
Figure 1.
AFM images of yMutLα. (A) 500 nm × 500 nm images of MutLα deposited in the absence of nucleotide (left) and in the presence of 5 mM ADP (right). Arrows point to examples of the four different conformational states seen in the images: red = extended; blue = one armed; orange = semi-condensed; green = condensed. Scale bar (white) is 250 nm. (B–E) 100 nm × 100 nm images of the four states accompanied by cartoons of MutLα in the different conformational states. In the cartoons, domains are indicated by ovals, connected by a flexible linker. Disordered linker is shown as a dashed line; ordered linker is shown as a solid heavy line. Mlh1 is shown in light green and blue, and Pms2 (yPms1) is shown in dark green and blue. (F) A selection of 100 nm × 100 nm images of the extended state to show the flexibility of the linker arms.
In addition to the extended conformation, three other minor populations of conformations are seen in the AFM images (Figures 1 & 2). Approximately 20% of the molecules exhibit a “one-armed” conformation, with a large domain connected to a smaller one by a flexible linker arm (Figure 1C). Measurement of the volume of the domains (Ratcliff and Erie, 2001; Yang et al., 2005; Yang et al., 2003) reveals that the large structure in this state is larger than the central structure in the extended conformation (Table S3). These results suggest that in these one-armed conformations, one of the N-terminal domains may be either interacting directly with the dimerized C-terminal region and/or that one linker arm may have folded (“condensed”) to create a globular domain consisting of the two C-termini, one linker arm, and one N-terminus. Roughly 10% of the molecules are in a semi-condensed state, with two domains similar in size (Figure 1D). In this state, the linker arms are not visible and the centers of the two domains are separated by 15 ± 4 nm. These results suggest that in this state, the two N-terminal domains have associated with one another but not with the C-terminal domains. The semi-condensed state is distinguished from the one-armed state by the size of the protein domains and their relative orientations to each other. Finally, ~ 15% of the molecules exhibit a compact structure with no protrusions (condensed state, Figure 1E). In this condensed state, the N- and C-terminal domains of both proteins in the heterodimer appear to be interacting to form a single compact molecule with no separate domains or linker arms visible.
Figure 2.
Distribution of the four conformational states of MutLα under varying conditions: apo (n=727), 0.1 mM ADP (n=912), 0.1 mM ATP (n=391), 5 mM ADP (n=559), and 5 mM ATP (n=1021). Red = extended; Blue = one-arm; Orange = semi-condensed; Green = condensed. Data were tallied by hand from 1 × 1 μm AFM images taken from multiple experiments. Error bars represent the variation in percentages of the different states from randomly selected subsets of the images.
Adenine nucleotides induce large conformational changes in MutLα
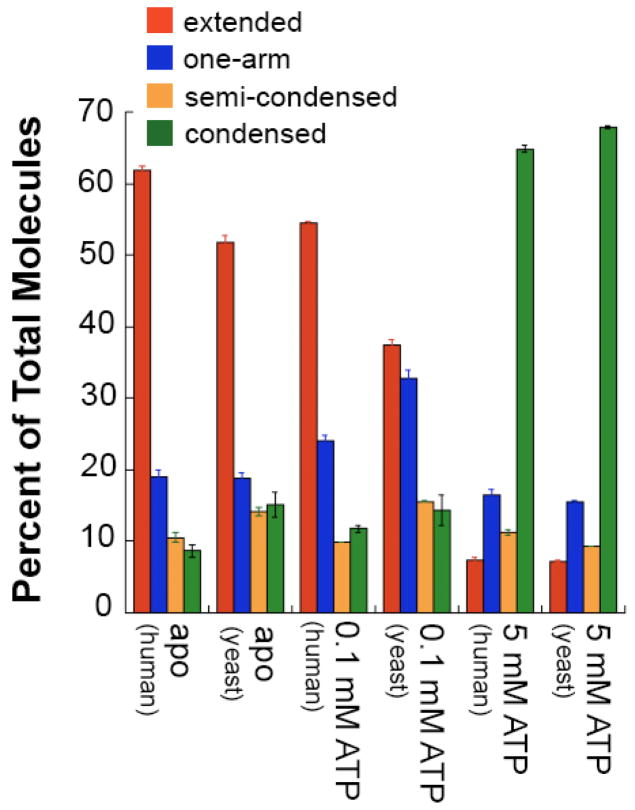
Genetic studies have shown that mutations that impair the binding or hydrolysis of ATP in either yPms1 or yMlh1 result in defective MMR in vivo (Hall et al., 2002; Tran and Liskay, 2000). In addition, biochemical studies suggest that ATP binding and hydrolysis induce conformational changes in MutL homologs (Ban and Yang, 1998; Hall et al., 2002; Raschle et al., 2002; Tran and Liskay, 2000). To assess the role of adenine nucleotides in controlling the conformations of MutLα, we imaged MutLα under a variety of nucleotide conditions (Figure 2). Based on previous studies of the ATPase activities of the N-terminal domains of yMlh1 and yPms1 (Hall et al., 2002), we chose conditions in which we expected either only the ATPase site of Mlh1 to be occupied (0.1 mM) or both sites to be occupied (5 mM).
Addition of either 0.1 mM ATP or ADP results in a decrease in the population of molecules in the extended conformation and increase in the population of the one-armed state (Figure 2). These results suggest that binding of ADP or ATP promotes a conformation of MutLα in which the N-terminal domain and linker arm of Mlh1 or Pms1 are interacting with or folded onto the C-terminal dimerization domains of Mlh1 and/or Pms1. Because Mlh1 appears to have a higher affinity for ATP (Hall et al., 2002), it is likely that these conformational changes are occurring in Mlh1 rather than Pms1.
At high concentrations of ATP (5 mM) where both yMlh1 and yPms1 should have nucleotide bound, MutLα exists predominately in the condensed conformation (70%) (Figure 2). Similar results were obtained with 1 mM ATP (data not shown). These results indicate that binding and/or hydrolysis of ATP drives the formation of this highly compact state in which the N-termini are interacting with one another and with the linker arms and/or the C-terminal dimerization domains. Interestingly, high concentrations of ADP (5 mM) also cause a significant increase in the condensed state (40% versus 15% without nucleotide). These results suggest that occupancy of both of the ATPase sites with either ATP or ADP facilitates the formation of the condensed state, although ATP is more efficient than ADP.
Proteolysis Protection reveals ATP-induced structural changes in solution
To further assess these conformational changes, we used partial proteolysis to probe MutLα conformations in solution (Figure 3). Consistent with our AFM results and previous experiments using yMutLα and hMutLα (Raschle et al., 2002, Tran and Liskay, 2000), yMutLα is readily digested by trypsin in the absence of nucleotide cofactor. The addition of 5 mM ATP results in a dramatic protection of MutLα from protease digestion (Figure 3). In addition, 0.1 mM ATP causes a slight protection of MutLα relative to no added nucleotide, with Pms1 being ~ 2-fold more sensitive to digestion than Mlh1 (Figure 3 and data not shown). These results indicate that ATP binding induces a conformational change that masks the trypsin protease sites and supports the idea that the conformational changes observed by AFM also occur in solution.
Figure 3.
Partial trypsin proteolysis of yMutLα. From left to right: undigested yMutLα, digested yMutLα, digested yMutLα + 0.1 mM ATP, digested yMutLα + 5 mM ATP. The lower amount of protein in the lane containing digested MutLα was reproducible in multiple experiments, indicating that this difference is not simply due to less protein being loaded into this lane. Because there are many trypsin sites that could produce very short peptides, which would not be visible on the gel, the lower intensity is probably due to apo MutLα having an increased sensitivity to trypsin relative to MutLα in the presence of 0.1 mM ATP.
ATP binding causes an increase in secondary structure
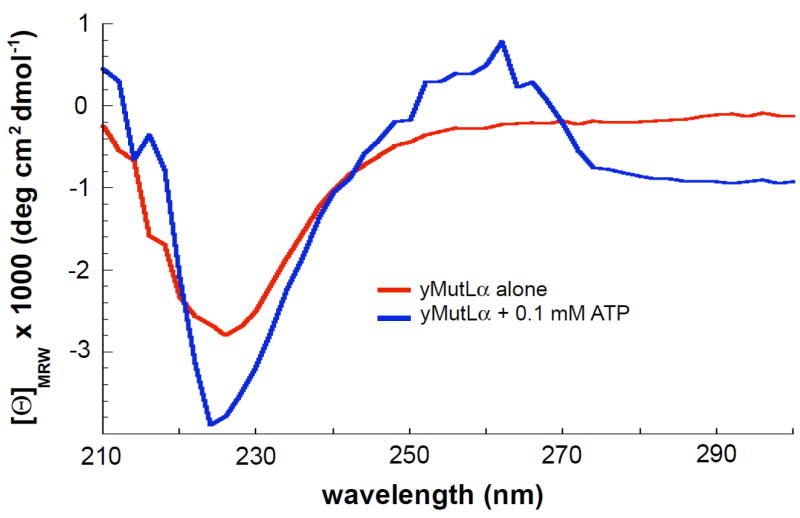
Although the AFM data reveal that ATP can induce dramatic changes in the conformation of MutLα and the partial proteolysis data suggest conformational changes, neither technique provides direct information about secondary structure formation. To determine if ATP binding promotes the formation of structure in the flexible arms, we examined secondary structure formation using circular dichroism spectroscopy (CD). Due to the high extinction coefficient of ATP, we were able to only obtain CD spectra at 0.1 mM ATP (but not at 5 mM ATP). At this concentration of ATP, there is a significant increase in the one-armed state in the AFM images and a slight increase in the amount of protein protected from trypsin digestion (Figures 2 and 3). Comparison of the CD spectra in the absence and presence of 0.1 mM ATP (Figure 4) shows that the addition of 0.1 mM ATP causes a significant increase in the CD signal in the region centered at 225 nm. This negative peak, centered at 225 nm, is characteristic of α-helices and/or β-sheet secondary structures in proteins (Kelly et al., 2005). There is also a positive peak, centered at 260 nm, that is consistent with a rigid environment for aromatic amino acid residues (Kelly et al., 2005). These data, taken together with our AFM data and partial proteolysis results, suggest that binding of ATP to one subunit of MutLα (probably Mlh1) induces the formation of secondary structure, likely in the flexible linker arm, and promotes the interaction between a linker arm and the N- and C- termini.
Figure 4.
CD spectra of 0.4 μM yMutLα in the absence (red) and presence (blue) of 0.1 mM ATP. The signal differences seen at 220–230 nm demonstrate that in the presence of ATP, the protein adopts more secondary structure (either α-helix or β-sheet). Plotted curves are the average of 3 experiments, with the background spectra (either imaging buffer alone or imaging buffer + 0.1 mM ATP) subtracted.
Human MutLα displays similar conformational changes
To determine if the results with yeast MutLα are general, we examined the conformations of human MutLα (hMlh1-hPms2) alone, with 0.1 mM ATP and with 5 mM ATP. As expected, we observe the same four states for hMutLα that we see for yMutLα. In addition, the distributions of the states under the same conditions are similar for hMutLα and yMutLα (Figure 5). Specifically, in the absence of ATP, both yeast and human MutLα exist predominantly in the extended state with defined N- and C- termini and disordered linker arms; whereas, in the presence of 5 mM ATP, both human and yeast MutLα are primarily condensed. There are some differences in the distributions in the presence of 0.1 mM ATP, which may result from differences in the ATP binding affinity of the yeast and human proteins (Figure 5), since very little is known about the binding affinity of any full-length eukaryotic MutLα.
Figure 5.
Comparison of the distributions of conformational states of hMutLα and yMutLα molecules. hMutLα apo (n=1253), hMutLα + 0.1 mM ATP (n=979), hMutLα + 5 mM ATP (n=302). Red = extended; blue = one-arm; orange = semi-condensed; green = condensed. Data were tallied by hand from 1μm × 1 μm AFM images taken from multiple experiments. Error bars represent the variation in percentages of the different states from randomly selected subsets of the images.
Hydrolysis is not required for the formation of the condensed state
To determine whether or not hydrolysis of ATP is required for the formation of the condensed state, we examined the conformational changes of a mutant of hMutLα in which the hydrolysis activity of both subunits is severely impaired (hMlh1•E34A-Pms2•E41A (hMutLα-EA)). In this mutant, the conserved glutamate residues in the active sites of both subunits have been changed to alanines, inhibiting hydrolysis of ATP (Raschle et al., 2002; Tomer et al., 2002). Images of hMutLα-EA in the absence of adenine nucleotide appear similar to images of the wild-type human and yeast MutLα, with the extended conformation dominating the population (Figure 6a). In addition, like wild-type MutLα, high concentrations of ATP (5mM) cause hMutLα-EA to condense (Figure 6b and 6c).
Figure 6.
Hydrolysis is not required for the formation of the condensed state. Images (500 nm × 500 nm) of hMutLα-EA (30 nM) in the absence (A) and presence (B) of 5 mM ATP. Arrows point out individual molecules in the v-shaped (A) and condensed (B) state. (C) Distribution of the four conformational states for apo yMutLα (n=727), yMutLα + 5 mM ATP (n=1021), yMutLα + Na•AMPPNP (n=213), and hMutLα•EA + 5mM ATP (n=161). Red = extended; blue = one-arm; orange = semi-condensed; green = condensed. Data were tallied by hand from 1 μm × 1 μm AFM images taken from multiple experiments. Error bars represent the variation in percentages of the different states from randomly selected subsets of the images.
The hMutLα-EA mutant is still capable of limited ATP hydrolysis, with a rate of ~0.2 min−1 (Raschle et al., 2002; F.K and P.M. unpublished results). Because MutLα and ATP are incubated together for less than a minute prior to being deposited on the surface, less than 20% of the hMutLα-EA molecules will have hydrolyzed ATP at the time of deposition. Given that ~70% of the molecules are in the condensed state for both the wild-type hMutLα and hMutLα-EA, it is highly unlikely that hydrolysis is required for formation of the condensed state (Figure 6c).
To further investigate the role of ATP hydrolysis, we imaged yMutLα in the presence of the non-hydrolyzable ATP analog AMPPNP. In our initial experiments, we used AMPPNP as purchased from Sigma, which comes as a tetralithium salt, and we found that it did not promote the condensation of MutLα, which is in direct contrast to the results obtained using the hMutLα-EA hydrolysis mutant. Because LiCl, in the absence of other salts, has been shown to inhibit the ATPase activity of GHL ATPases (Hu et al., 2003), we added 20 mM LiCl to our 5 mM ATP reactions and found that it inhibited the formation of the condensed state (data not shown). Consequently, we purified the commercially available AMPPNP using an ion exchange column and eluted it using NaCl (see Materials & Methods) to form the sodium salt (Na•AMPPNP). Depositions of yMutLα in the presence of Na•AMPPNP show that ~55% of the molecules are in the condensed state (Figure 6c). These results, taken together with the data on hMutLα-EA (Figure 6) and the observation that ADP also promotes the formation of the condensed state (Figure 2), strongly suggest that adenosine nucleotide binding, but not ATP hydrolysis, is required to induce the formation of the fully condensed state.
Discussion
X-ray crystallography is a good technique for characterization of the structure of ordered proteins and protein domains. However, there are no full-length structures of MutL or MutLα, likely because of the intrinsic disorder of the linker region connecting the N- and C- termini as well as the large conformational changes that take place upon adenine nucleotide binding. Analysis of populations of individual MutLα molecules in AFM images has allowed us to not only visualize the protein in four different conformational states but also to visualize the linker arm, which is unlikely to be detected with other techniques.
Nucleotide binding induces conformational changes and disorder-order transitions in MutLα
Our experiments show that in the absence of added adenine nucleotide, MutLα exists predominantly in an extended conformation (Figures 1 & 2). In this extended conformation, there are no visible interactions between the N- and C- terminal domains or between the two N-terminal domains of the heterodimer. There is likely no nucleotide bound to this extended state, nor to the other, lesser-populated states. The binding of one ATP to MutLα, likely to Mlh1 (KM = 69 μM in an N-terminal construct of yMlh1 vs. KM = 1500 μM in an N-terminal construct of yPms1) (Hall et al., 2002), drives the formation of the one-armed state. The one-armed state shows an increase in secondary structure compared to the extended state, as observed by CD in the absence (extended) and presence of 0.1 mM ATP (one-arm) (Figure 4). In addition, yMlh1 is 1.5 to 2 times less sensitive than yPms1 to trypsin under all conditions (Figure 3 and data not shown). Taken together, these results suggest that binding of ATP to Mlh1 promotes secondary structure formation in its linker arm, condensing the N-terminus and linker region of Mlh1 onto the dimerized C-terminal domains.
We also show that at 5 mM ATP (or ADP), where both nucleotide-binding sites should be occupied, both subunits in the heterodimer are condensed. In the condensed state, the interactions between linker regions and the N- and C-termini likely persist, with the addition of interactions between the two N-termini. The observations that the hydrolysis mutant, hMutLα-EA, condenses in the presence of ATP and that AMPPNP and ADP also promote the condensed state strongly suggests that condensation is linked to binding of adenine nucleotide but not to ATP hydrolysis.
The semi-condensed state is likely a conformational rearrangement of the condensed state, with the contacts between the two N-termini remaining, but with the linker regions partially unfolded and clearly no interaction between the N-termini and C-termini. This state suggests direct interaction between the two N-termini in MutLα, which is consistent with yeast two-hybrid data showing an interaction of the N-terminal domains of yPms1 and yMlh1 (Tran and Liskay, 2000). The presence of all four of these states in the absence of adenine nucleotide suggests that the four protein states are in dynamic equilibrium and that binding and hydrolysis of ATP followed by ADP release cycle MutLα through the different conformational states.
The observed ordering of the disordered regions in MutLα upon ADP or ATP binding is analogous to the ordering that happens after ligand binding in intrinsically disordered proteins (Dunker et al., 2001; Xie et al., 2007a; Xie et al., 2007b). Although ATP binding alone is unlikely to induce order in the ~160 and ~300 disordered amino acids in the linker arms, it probably promotes interactions of the linker regions with the other regions of MutLα, such as the N- and C-terminal domains, and these interactions, together with the ATP binding may lead to the significant increase in secondary structure that is observed in the CD spectra. As such, this disorder-order transition adds another class of such transitions to natively disordered proteins (Dunker et al., 2001; Xie et al., 2007a; Xie et al., 2007b), where binding of a small molecule to one region of a protein, which is predominately ordered, allosterically facilitates the folding of an adjacent region of the protein.
Comparison of MutLα with other GHL ATPases
The members of the GHL ATPase family are diverse in their function. Despite this diversity of function, there remains a common theme: the binding and subsequent hydrolysis of ATP drives protein conformational changes that are linked to the timing of their mechanistic cycles (Ali et al., 2006; Ban et al., 1999; Chu et al., 2006; Corbett and Berger, 2003, 2005; Dollins et al., 2005; Dutta and Inouye, 2000; Immormino et al., 2004; Shiau et al., 2006). Reflective of this common theme, the overall architectures of proteins in the GHL ATPase family are similar; the N-terminus contains a conserved ATPase binding domain and the C-terminus contains a variable dimerization domain (Dutta and Inouye, 2000). One of the sources of variation between GHL proteins is in the connection between the N- and C- domains. Most GHL proteins have a structured middle domain; whereas, MutLα contains large disordered regions between the N- and C- terminal domains, which were previously predicted (Guarne et al., 2004) and confirmed by this study.
In spite of the functional differences between the MutLα and the other GHL family members, the four states observed here show similarity to conformational states of Grp94, HtpG (both members of the hsp90 family of chaperones) and TopoVI (a type II topoisomerases), which have been studied using electron microscopy (EM), small angle x-ray scattering (SAXS) and/or x-ray crystallography (Corbett et al., 2007; Shiau et al., 2006; Wearsch and Nicchitta, 1996). Grp94, HtpG and TopoVI have ordered middle domains (in comparison with the disordered linker of MutLα) and no one-armed state has been observed for Grp94, HtpG or TopoVI.
In the absence of adenine nucleotide, both Grp94 and HtpG are seen by EM in an extended, v-shaped, conformation similar to the extended conformation seen here for MutLα by AFM. SAXS data reveal that TopoVI also exists in an open, v-shaped conformation (Corbett et al., 2007). The difference between the extended MutLα and extended Grp94, HtpG and TopoVI is the absence of the disordered linker arms connecting N- and C-termini in Grp94, HtpG and TopoVI. In the presence of the non-hydrolyzable ATP analog AMPPNP, HtpG undergoes a large conformational change from the open, extended state to a more compact state (Shiau et al., 2006). This state appears similar to the semi-condensed state of MutLα seen by AFM (Figure 1D), with the N-termini dimerized, but not collapsed upon the C-termini. When visualized by EM in the presence of ADP, HtpG becomes even more condensed than it is in the presence of AMPPNP (Shiau et al., 2006).
Examination of the crystal structures of HtpG in the absence and the presence of ADP along with the structure of yHsp82, the yeast homolog of Hsp90, shows that the Hsp90 proteins also exist in an extended conformation in the absence of ATP (Ali et al., 2006; Shiau et al., 2006). The addition of ADP causes dimerization of the N-terminal domains (seen with HtpG), while the addition of ATP causes a twisted collapse around the dimerized N-terminal domains (seen with yHsp82) and a 20 Å movement of the N-terminal domains towards each other (Ali et al., 2006). Perhaps the semi-condensed structure we visualize by AFM is an “untwisted” version of the condensed state.
ATPase cycle for MutLα
Each of the four observed states of MutLα represents a snapshot of the conformational changes involved in the process of ATP binding, hydrolysis and ADP release. Based on these four observed states, one possible model for an ATPase cycle suggests that adenine nucleotides bind sequentially to MutLα (Figure 6). The model is consistent with different KMs of the N-terminal domains of yMlh1 and yPms1 (Hall et al., 2002). The extended state corresponds to MutLα with no nucleotide bound (apo). The binding of the first ATP condenses one “arm”, likely Mlh1, forming the one-armed state; the binding of a second ATP condenses the other arm, forming the condensed state.
The one-armed state is formed by the ordering of the linker region of one protein in MutLα, causing it to condense onto the already dimerized C-termini. The occurrence of this state suggests that there may be contacts formed between the N- and C- terminal domains in MutLα after ATP binds to the protein. Partial proteolysis suggests that these interactions are dynamic, because the digestion patterns of yMutLα alone and yMutLα plus 0.1 mM ATP are similar (Figure 3).
The hydrolysis and subsequent release of both bound adenine nucleotides, either sequentially (following the one-armed pathway) or at the same time (following the semi-condensed pathway), returns the protein to the original extended conformation. Because low ADP concentration (0.1 mM ADP) increases the one-armed state, and high ADP concentration (5 mM ADP) can drive the formation of the condensed state, it is likely that ADP release, not ATP hydrolysis, causes the cycling of the protein through the four states. As the cycle progresses, conformational changes expose different surfaces and new structured regions to solution, making them available for binding by other proteins.
Our model contrasts with a previously proposed ATPase cycle for MutL that was based upon mutational studies alongside biochemical and crystallographic data (Ban et al., 1999). In the E. coli MutL model, both MutL monomers bind ATP at the same time, followed by N-terminal dimerization. Each MutL monomer then simultaneously hydrolyzes ATP, the N-terminal domains dissociate and ADP is released. The extended state we observe for the eukaryotic MutLα correlates well with the N-terminal dissociated state seen for MutL, while the semi-condensed state correlates with the dimerized N-terminal state. However, MutLα is a heterodimer, and each protein contained therein has differing ATP affinities and rates of ATP hydrolysis. These important differences make simultaneous ATP binding and hydrolysis unlikely for the subunits of MutLα. As we have observed in the one-armed state, the condensation of one protein subunit (after the binding of one ATP) occurs in the absence of the condensation of the other.
MutLα conformational changes in the context of MMR and cellular activities
It has long been suggested that MutL and MutL homologs function as a switch or molecular matchmaker, changing conformation to coordinate the functions of the mismatch repair machinery. In this paper, we report direct observations of the conformational changes of MutLα in response to adenine nucleotide. ATP hydrolysis is required for repair (Hall et al., 2001; Pang et al., 1997; Raschle et al., 2002; Spampinato and Modrich, 2000; Tran and Liskay, 2000) as well as the endonucleolytic activity of MutLα (Kadyrov et al., 2006). Since MutLα also interacts with PCNA and ExoI in the MMR pathway, perhaps our observed conformational changes mediate the timing of these interactions as well as the enzymatic function of MutLα.
The in vivo concentrations of ADP and ATP (~1 mM and ~3 mM, respectively) (Williams et al., 1993) suggest that MutLα would rarely exist in the apo form unless this form is stabilized by other proteins. It has been demonstrated that interactions between MutSα and MutLα are mediated by the N-terminal domain of Mlh1 alone and are not linked to the ATPase activity of MutLα (Plotz et al., 2003; Raschle et al., 2002). Since the ATPase activity of MutLα does not affect interactions with MutSα, MutSα may be able to interact with MutLα in any conformational state, but to do so, MutSα must be in the appropriate state.
Stable complexes involving MutSα, MutLα, PCNA, mismatched DNA and ATP have been observed in vitro (Constantin et al., 2005; Dzantiev et al., 2004; Hidaka et al., 2005; Lee and Alani, 2006; Zhang et al., 2005). Based on observed in vitro interactions and on in vivo ATP concentrations, MutLα is likely to be in the condensed state when interacting with MutSα and PCNA. With the subsequent hydrolysis of one or both bound ATP molecules, the protein likely changes from the condensed state to the semi-condensed, one-armed, and/or apo state, changing the surface of MutLα and possibly hiding the surface with which PCNA interacts, exposing the ExoI interaction surface. Once ADP and Pi are released into solution, MutLα will be “reset” and ready to participate in another round of mismatch repair. The time required to hydrolyze ATP and then release ADP and Pi, as well as the time required for conformational changes likely causes a “pause” in the process of mismatch repair, allowing for the proteins currently interacting with MutLα to complete their intended processes.
As mentioned in the introduction, the proteins that make up MutLα also participate in other DNA transactions. The conformational changes reported here may mediate protein-protein or protein-DNA interactions in those cellular activities as well.
Materials & Methods
Chemicals
ATP, ADP and AMPPNP were purchased from Sigma. All other materials were purchased from Fisher.
Protein Expression and Purification
yMutLα was purified as previously described (Hall and Kunkel, 2001), with the exception of HEPES, pH 7.3 being used in the dialysis buffer instead of Tris, pH 7.3. hMutLα and hMutLα-EA were purified as described (Kadyrov et al., 2006).
Atomic Force Microscopy
For AFM imaging, yMutLα, hMutLα or hMutLα-EA was diluted to a final concentration of 30 nM in imaging buffer (20 mM HEPES, pH 7.3, 5 mM MgOAc, 25mM NaOAc) at room temperature. A volume of 10 μL of the diluted protein was deposited onto freshly cleaved ruby mica (Spruce Pine Mica Company, Spruce Pine, NC). The sample was rinsed immediately with nanopure water; excess water was blotted from the surface and the surface was then dried using a stream of nitrogen.
For experiments where ATP, ADP or AMPPNP (Na•AMPPNP) were present, the adenine nucleotide and MutLα were mixed and then diluted in imaging buffer to a final adenine nucleotide concentration of 0.1 mM, 1 mM or 5 mM and protein concentration of 30 nM. The sample was then deposited as for MutLα protein alone. The sample preparation and deposition process (from addition of adenine nucleotide to drying of sample) take <1 minute.
AFM images were captured in air using either a Nanoscope III or IIIa (Digital Instruments, Santa Barbara, CA) microscope in tapping mode. Pointprobe Plus tapping mode silicon probes (Agilent Technology, Tempe, AZ) with resonance frequencies of approximately 170 kHz were used for imaging. Images were collected at a speed of 2–3 Hz with an image size of 1 μm at 512 × 512 pixel resolution.
Images were analyzed using Nanoscope III v5.12r3 software (Veeco, Santa Barbara, CA), Image SxM, v 1.69 (Barrett, 2006) and NIH ImageJ (Rasband, 1997–2006) with OpenAFM and Cell Counter plug-ins. Volume analysis was performed as previously described (Ratcliff and Erie, 2001; Yang et al., 2003). Statistical plots were generated with Kaleidagraph (Synergy Software, Reading, PA).
Purification of AMPPNP
A 1 mL HiTrap DEAE column (GE Biosciences) was prepared according to manufacturers instructions. AMPPNP was dissolved in ddH2O (0.5 mL; final concentration 0.1 M) and loaded onto the column by syringe. The column was washed with ten column volumes of ddH2O, and the AMPPNP was eluted with 0.25 M NaCl (~100 μL fraction size). The AMPPNP eluted in the first three fractions (after the 1 mL column volume) and pooled.
Circular Dichroism Spectropolarimetry
CD spectra (λ = 210–300 nm) of 0.4 μM yMutLα in imaging buffer in the presence and absence of 0.1 mM ATP were acquired on an AVIV spectropolarimeter at 10°C, with 2 nm resolution and an averaging time of 3 sec per point, in 1-cm quartz cuvettes. Plotted curves are the average of 3 experiments, with the background spectra (either imaging buffer alone or imaging buffer + 0.1 mM ATP) subtracted.
Partial Proteolysis
In experiments testing for protease protection, 5 ng of trypsin was added to reactions containing 8 μg of yMutLα alone and with 0.1 mM ATP or 5 mM ATP in imaging buffer (total reaction size was 20 μL). The reaction was allowed to proceed for 5 minutes at room temperature before stopping the reaction by the addition of SDS load and incubation at 95°C for 10 minutes. All reactions were then run on a 10% SDS-PAGE gel. Protein bands were visualized by staining with Coomassie Brilliant Blue R-250.
Supplementary Material
Figure 7.
ATPase cycle for MutLα. The extended state corresponds to MutLα with no nucleotide bound (apo). The binding of the first ATP condenses one “arm” (probably Mlh1), forming the one-armed state; the binding of a second ATP condenses the other arm (Pms2), forming the condensed state (or the semi-condensed state, which is related to the condensed state by a conformational rearrangement of the protein). The hydrolysis and release of both bound adenine nucleotides, either sequentially (following the one-armed pathway) or at the same time (following the semi-condensed pathway), returns the protein to the extended conformation.
Acknowledgments
We thank Gary Pielak & Lisa Charlton for assistance with and discussion about the CD experiments, Shannon Holmes, Mercedes Arana for critical evaluation of the manuscript, and John Fortune, for help with purification of yMutLα. We thank A Keith Dunker and Vladimir Uversky for discussion and help with analysis of disorder to order transitions. We also thank Lauryn Sass, Erika Pearson and Robert Immormino for critical evaluation of the manuscript and many thought provoking discussions.
This work was supported in part by grant number RSG-03-047 from the American Cancer Society (D.A.E), grant number GM45190 (P.M.) from NIH and the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (T.K.). P.M. is an investigator of the Howard Hughes Medical Institute.
References
- Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, Piper PW, Prodromou C, Pearl LH. Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature. 2006;440:1013–1017. doi: 10.1038/nature04716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- Barrett SD. Image SXM 2006 [Google Scholar]
- Bellacosa A. Functional interactions and signaling properties of mammalian DNA mismatch repair proteins. Cell Death Differ. 2001;8:1076–1092. doi: 10.1038/sj.cdd.4400948. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic acids research. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Maynard JC, Chiosis G, Nicchitta CV, Burlingame AL. Identification of novel quaternary domain interactions in the Hsp90 chaperone, GRP94. Protein Sci. 2006;15:1260–1269. doi: 10.1110/ps.052065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005 doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Benedetti P, Berger JM. Holoenzyme assembly and ATP-mediated conformational dynamics of topoisomerase VI. Nat Struct Mol Biol. 2007;14:611–619. doi: 10.1038/nsmb1264. [DOI] [PubMed] [Google Scholar]
- Corbett KD, Berger JM. Structure of the topoisomerase VI-B subunit: implications for type II topoisomerase mechanism and evolution. Embo J. 2003;22:151–163. doi: 10.1093/emboj/cdg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Berger JM. Structural dissection of ATP turnover in the prototypical GHL ATPase TopoVI. Structure (Camb) 2005;13:873–882. doi: 10.1016/j.str.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Dollins DE, Immormino RM, Gewirth DT. Structure of unliganded GRP94, the endoplasmic reticulum Hsp90. Basis for nucleotide-induced conformational change. J Biol Chem. 2005;280:30438–30447. doi: 10.1074/jbc.M503761200. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- Dzantiev L, Constantin N, Genschel J, Iyer RR, Burgers PM, Modrich P. A defined human system that supports bidirectional mismatch-provoked excision. Molecular cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Molecular cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- Guarne A, Junop MS, Yang W. Structure and function of the N-terminal 40 kDa fragment of human PMS2: a monomeric GHL ATPase. Embo J. 2001;20:5521–5531. doi: 10.1093/emboj/20.19.5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarne A, Ramon-Maiques S, Wolff EM, Ghirlando R, Hu X, Miller JH, Yang W. Structure of the MutL C-terminal domain: a model of intact MutL and its roles in mismatch repair. Embo J. 2004;23:4134–4145. doi: 10.1038/sj.emboj.7600412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall MC, Kunkel TA. Purification of eukaryotic MutL homologs from Saccharomyces cerevisiae using self-affinity technology. Protein Expr Purif. 2001;21:333–342. doi: 10.1006/prep.2000.1379. [DOI] [PubMed] [Google Scholar]
- Hall MC, Shcherbakova PV, Kunkel TA. Differential ATP binding and intrinsic ATP hydrolysis by amino-terminal domains of the yeast Mlh1 and Pms1 proteins. J Biol Chem. 2002;277:3673–3679. doi: 10.1074/jbc.M106120200. [DOI] [PubMed] [Google Scholar]
- Hall MC, Wang H, Erie DA, Kunkel TA. High affinity cooperative DNA binding by the yeast Mlh1-Pms1 heterodimer. J Mol Biol. 2001;312:637–647. doi: 10.1006/jmbi.2001.4958. [DOI] [PubMed] [Google Scholar]
- Hidaka M, Takagi Y, Takano TY, Sekiguchi M. PCNA-MutSalpha-mediated binding of MutLalpha to replicative DNA with mismatched bases to induce apoptosis in human cells. Nucleic acids research. 2005;33:5703–5712. doi: 10.1093/nar/gki878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Machius M, Yang W. Monovalent cation dependence and preference of GHKL ATPases and kinases. FEBS Lett. 2003;544:268–273. doi: 10.1016/s0014-5793(03)00519-2. [DOI] [PubMed] [Google Scholar]
- Immormino RM, Dollins DE, Shaffer PL, Soldano KL, Walker MA, Gewirth DT. Ligand-induced conformational shift in the N-terminal domain of GRP94, an Hsp90 chaperone. J Biol Chem. 2004;279:46162–46171. doi: 10.1074/jbc.M405253200. [DOI] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Jager AC, Rasmussen M, Bisgaard HC, Singh KK, Nielsen FC, Rasmussen LJ. HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLalpha and hMLH1-hEXO1 complexes. Oncogene. 2001;20:3590–3595. doi: 10.1038/sj.onc.1204467. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim Biophys Acta. 2005;1751:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Kosinski J, Steindorf I, Bujnicki JM, Giron-Monzon L, Friedhoff P. Analysis of the quaternary structure of the MutL C-terminal domain. J Mol Biol. 2005;351:895–909. doi: 10.1016/j.jmb.2005.06.044. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Lee SD, Alani E. Analysis of interactions between mismatch repair initiation factors and the replication processivity factor PCNA. J Mol Biol. 2006;355:175–184. doi: 10.1016/j.jmb.2005.10.059. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- Mendillo ML, Mazur DJ, Kolodner RD. Analysis of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 and MLH1-PMS1 complexes with DNA using a reversible DNA end-blocking system. J Biol Chem. 2005;280:22245–22257. doi: 10.1074/jbc.M407545200. [DOI] [PubMed] [Google Scholar]
- Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- Nielsen FC, Jager AC, Lutzen A, Bundgaard JR, Rasmussen LJ. Characterization of human exonuclease 1 in complex with mismatch repair proteins, subcellular localization and association with PCNA. Oncogene. 2004;23:1457–1468. doi: 10.1038/sj.onc.1207265. [DOI] [PubMed] [Google Scholar]
- O’Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–692. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- Pang Q, Prolla TA, Liskay RM. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to human hereditary nonpolyposis colorectal cancer-associated mutations. Mol Cell Biol. 1997;17:4465–4473. doi: 10.1128/mcb.17.8.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotz G, Raedle J, Brieger A, Trojan J, Zeuzem S. N-terminus of hMLH1 confers interaction of hMutLalpha and hMutLbeta with hMutSalpha. Nucleic acids research. 2003;31:3217–3226. doi: 10.1093/nar/gkg420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasband WS. ImageJ. US National Institutes of Health; Bethesda, Maryland, USA: 1997–2006. [Google Scholar]
- Raschle M, Dufner P, Marra G, Jiricny J. Mutations within the hMLH1 and hPMS2 subunits of the human MutLalpha mismatch repair factor affect its ATPase activity, but not its ability to interact with hMutSalpha. J Biol Chem. 2002;277:21810–21820. doi: 10.1074/jbc.M108787200. [DOI] [PubMed] [Google Scholar]
- Ratcliff GC, Erie DA. A novel single-molecule study to determine protein--protein association constants. J Am Chem Soc. 2001;123:5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- Schofield MJ, Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- Shiau AK, Harris SF, Southworth DR, Agard DA. Structural Analysis of E. coli hsp90 Reveals Dramatic Nucleotide-Dependent Conformational Rearrangements. Cell. 2006;127:329–340. doi: 10.1016/j.cell.2006.09.027. [DOI] [PubMed] [Google Scholar]
- Spampinato C, Modrich P. The MutL ATPase is required for mismatch repair. J Biol Chem. 2000;275:9863–9869. doi: 10.1074/jbc.275.13.9863. [DOI] [PubMed] [Google Scholar]
- Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair (Amst) 2004;3:1091–1101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Tomer G, Buermeyer AB, Nguyen MM, Liskay RM. Contribution of human mlh1 and pms2 ATPase activities to DNA mismatch repair. J Biol Chem. 2002;277:21801–21809. doi: 10.1074/jbc.M111342200. [DOI] [PubMed] [Google Scholar]
- Tran PT, Liskay RM. Functional studies on the candidate ATPase domains of Saccharomyces cerevisiae MutLalpha. Mol Cell Biol. 2000;20:6390–6398. doi: 10.1128/mcb.20.17.6390-6398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, Kunkel TA. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- van Noort J, van Der Heijden T, de Jager M, Wyman C, Kanaar R, Dekker C. The coiled-coil of the human Rad50 DNA repair protein contains specific segments of increased flexibility. Proc Natl Acad Sci U S A. 2003;100:7581–7586. doi: 10.1073/pnas.1330706100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wearsch PA, Nicchitta CV. Endoplasmic reticulum chaperone GRP94 subunit assembly is regulated through a defined oligomerization domain. Biochemistry. 1996;35:16760–16769. doi: 10.1021/bi962068q. [DOI] [PubMed] [Google Scholar]
- Williams SP, Fulton AM, Brindle KM. Estimation of the intracellular free ADP concentration by 19F NMR studies of fluorine-labeled yeast phosphoglycerate kinase in vivo. Biochemistry. 1993;32:4895–4902. doi: 10.1021/bi00069a026. [DOI] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional Anthology of Intrinsic Disorder. 3. Ligands, Post-Translational Modifications, and Diseases Associated with Intrinsically Disordered Proteins. J Proteome Res. 2007a doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional Anthology of Intrinsic Disorder. 1. Biological Processes and Functions of Proteins with Long Disordered Regions. J Proteome Res. 2007b doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sass LE, Du C, Hsieh P, Erie DA. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic acids research. 2005;33:4322–4334. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang H, Erie DA. Quantitative characterization of biomolecular assemblies and interactions using atomic force microscopy. Methods. 2003;29:175–187. doi: 10.1016/s1046-2023(02)00308-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li GM. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.