Abstract
TMPRSS2-ERG fusion is the most common oncogenic rearrangement in prostate cancer (CaP). Due to this chromosomal rearrangement one TMPRSS2 allele loses its promoter, and one of the ERG alleles gains that promoter leading to its overexpression in these tumor cells. Some studies suggest that TMPRSS2, an androgen regulated type II trans-membrane serine protease, may have an effect on CaP progression. We hypothesized that a difference in TMPRSS2 expression may be present in vivo between CaP cells with and without TMPRSS2-ERG fusion, or a compensatory mechanism for the allelic loss of TMPRSS2 may balance that expression difference. Therefore, TMPRSS2 mRNA expression was evaluated in micro-dissected CaP cells with and without TMPRSS2-ERG fusion in 132 CaP patients and analyzed for its correlation with other androgen receptor (AR) regulated genes and clinico-pathologic features. In vivo TMPRSS2 expression correlated with that of other AR-regulated genes, including PSA/KLK3 and PMEPA1, offering potential as AR surrogates. A significantly reduced expression of TMPRSS2 was evident in malignant cells harboring TMPRSS2-ERG fusion, but not in CaP cells without TMPRSS2-ERG fusion, further defining these two genetically distinct types of CaP.
Keywords: prostate cancer, TMPRSS2, quantitative expression, TMPRSS2-ERG fusion
Introduction
Tumor markers are widely used for the early diagnosis, staging and management of several malignancies. In prostate cancer (CaP), prostate specific antigen (PSA) is the most widely used tumor marker, leading to the diagnosis of clinically localized disease in the vast majority of cases, since its use became widespread in the early 1990s (1). The focus of tumor marker research in CaP is now on altered genes, particularly those involved in androgen receptor (AR) signaling.
The TMPRSS2 gene is located on chromosome 21 and is a type II transmembrane protease expressed in prostate secretory epithelium, as well as in other parts of the urogenital, gastrointestinal and respiratory tracts (2,3). TMPRSS2 is regulated by androgens with some alternative signaling pathways noted in cases of androgen ablation. Overexpression of this serine protease has been demonstrated in neoplastic prostate epithelium, with significant increase in the TMPRSS2 mRNA level demonstrated in poorly differentiated CaP (3–6). The ETS-related gene (ERG), an oncogene and member of the ETS transcription family, has been identified as the most consistently overexpressed proto-oncogene in the transcriptome of malignant prostate epithelial cells (7). Recurrent fusions of the 5’ untranslated region of TMPRSS2 to ERG were discovered in the majority of CaP tissue specimens exhibiting ERG overexpression (8). Several other studies have confirmed these findings, but few studies have investigated the role of wild type TMPRSS2, also an androgen-inducible gene, in CaP cells. As the genomic rearrangements leading to TMPRSS2-ERG fusion separate one copy of TMPRSS2 from its promoter, and TMPRSS2 may play a role in CaP development (9), we were interested in defining TMPRSS2 expression and regulation in CaP cells with TMPRSS2-ERG fusion. In these CaP cells the promoter-less copy of TMPRSS2 is silenced, which may lead either to lower TMPRSS2 expression, or the dosage loss may be compensated as it has been shown for numerous genes, some in this region of chromosome 21 (10–11).
In this study we compared TMPRSS2 mRNA expression in laser-capture microdissected malignant and benign prostate epithelial cells of CaP patients with and without the TMPRSS2-ERG fusion product. The relationship between TMPRSS2 expression and that of other androgen-regulated genes was also determined.
Materials and Methods
The prostate tissue specimens used in this study were obtained under an IRB-approved protocol at Walter Reed Army Medical Center. Microdissected cells were evaluated from 132 men who had undergone RP as primary therapy for CaP. All patients were hormone naïve.
Benign and malignant cells were laser capture microdissected (LCM) by a pathologist from optimum cutting temperature (OCT) medium-embedded and H&E stained frozen prostate sections of RP specimens (2000 laser shots per sample). Total RNA was isolated from the LCM specimens using the MicroRNA kit (Stratagene, La Jolla, CA). The RNA was quantified with RiboGreen dye (Molecular Probes, Eugene, OR) and a VersaFluor fluorimeter (BioRad, Herculese, CA). Real time quantitative reverse transcriptase polymerase chain reaction (RT-PCR) was then performed (7). The isolated total RNA was converted to cDNA (Sensiscript, Qiagen, Valencia, CA). Quantitative gene expression analysis was performed by TaqMan-based QRT-PCR on ABI 7700 (Applied Biosystems, Foster City, CA). The TaqMan primers and probe for TMPRSS2 were: forward primer: CAC GGA CTG GAT TTA TCG ACA A; reverse primer: CGT CAA GGA CGA AGA CCA TGT, probe: TGA GGG CAG ACG GCT A; for PSA/KLK3 forward primer: CCC ACT GCA TCA GGA ACA AA, reverse primer: GAG CGG GTG TGG GAA GCT, probe: ACA CAG GCC AGG TAT TTC AGG TCA GCC; for AR forward primer: GGT GTC ACT ATG GAG CTC TCA CAT, reverse primer: GCA ATC ATT TCT GCT GGC G, probe: CTT CAA AAG AGC CGCTGA AGG GAA ACA G; for PMEPA1 forward primer: CAT GAT CCC CGA GCT GCT, reverse primer: TGA TCT GAA CAA ACT CCA GCT CC, probe: AGG CGG ACA GTC TCC TGC GAA AC; for PCA3 forward primer: CAC ATT TCC AGC CCC TTT AAA TA, reverse primer: GGG CGA GGC TCA TCG AT, probe: GGA AGC ACA GAG ATC CCT GGG AGA AAT G; for ERG forward primer: CAG GTC CTT CTT GCC TCC C, reverse primer: TAT GGA GGC TCC AAT TGA AAC C, probe: TGT CTT TTA TTT CTA GCC CCT TTT GGA ACA GGA; for TMPRSS2-ERG A-type fusion forward primer: CGC GGC AGG AAG CCT TAT, reverse primer: CGC GGT CAT CTC TGT CTT AGC, probe: GCC TAC GGA ACG CCA CAC; for LTF forward primer: AGA GAC TCC CCC ATC CAG TGT AT, reverse primer: CTG TCT TTC GGT CCC GTA GAC TT, probe: CAT TGC GGA AAA CAG; for AMACR: “assay-on-demand” Hs00204885 (Invitrogen, Carlsbad, CA). The expression of GAPDH was analyzed as endogenous control (Human GAPDH TaqMan mix, Applied Biosystems, Foster City, CA). Target gene expression in each sample was normalized to GAPDH, with negative control provided by RNA samples without reverse transcription. Standard thermal cycling conditions were 95 °C for 10min, 50 cycles at 95 °C for 15 sec, and 60 °C for 1min. Gene expression results were obtained as average CT (threshold cycle) values of duplicate samples.
Patient characteristics examined included diagnostic serum PSA (ng/mL), race (African American versus Caucasian and other), pathologic T stage (pT2, pT3), Gleason sum (6, 7, 8–9), margin status (positive versus negative), and PSA recurrence. Recurrence was defined as a single post-operative PSA of ≥ 0.2ng/mL two months after surgery. Paired t test was applied to compare the difference of TMPRSS2 expression levels between tumor tissue and benign glands, using log2 transformed expression values due to their not normal distribution. Spearman correlation analysis was used to determine correlation between the mRNA expression levels in tumor cells of TMPRSS2 and other genes: ERG, PSA/KLK3, AR, PMEPA1, PCA3, LTF and AMACR. Wilcoxon or Kruskal-Wallis tests were used to examine the association of TMPRSS2 ratio (N/T) with clinical-pathological characteristics as well as TMPRSS2-ERG fusion status. Continuous gene expression was separated into quartiles to perform Kaplan-Meier estimation of time to PSA recurrence. P value of 0.05 was adopted as statistically significant. All statistical analysis was performed with the SAS software package (version 9.1, SAS Institute Inc., Cary, NC).
Results
A total of 132 patients with frozen prostate tissue sections available in the CPDR Tissue Bank were included in this investigation. A summary of patient demographical characteristics, as well as clinico-pathological and molecular data are shown in Table 1 for categorical variables and Table 2 for continuous variables. This cohort is part of a CaP patient series undergoing radical prostatectomy at WRAMC and was selected based on the availability of sufficient amount of frozen tumor cells for optimal micro-dissection. Therefore the proportion of higher pathological stage (pT3) is increased from the overall 39% to 60% (Table 1). Of the 110 patients evaluated for TMPRSS2-ERG fusion status 72 (65.4%) was fusion positive based on the expression of fusion mRNA in their tumor cells (Table 1).
Table 1.
Descriptive statistics for categorical clinico-pathological and molecular variables (N=132)
| Variable | N | % |
|---|---|---|
| Race | ||
| Caucasian & other | 102 | 77.3 |
| African American | 30 | 22.7 |
| Pathologic T stage | ||
| pT2 | 50 | 40.0 |
| pT3 | 75 | 60.0 |
| Missing | 7 | |
| Post-operative Gleason sum | ||
| 6 | 40 | 30.8 |
| 7 | 65 | 50.0 |
| 8 – 9 | 25 | 19.2 |
| Missing | 2 | |
| Margin status | ||
| Negative | 89 | 67.9 |
| Positive | 42 | 32.1 |
| Missing | 1 | |
| ERG fusion | ||
| No | 38 | 34.6 |
| Yes | 72 | 65.4 |
| Missing | 22 | |
| PSA recurrence | ||
| No | 97 | 73.5 |
| Yes | 35 | 26.5 |
Table 2.
Descriptive statistics for continuous clinical and molecular variables (N=132)
| Variable | Median (range) |
|---|---|
| Age at surgery (year) | 61.6 (40.2–73.6) |
| Serum PSA (ng/ml) | 6.2 (1.1–38.9) |
| Follow up time post surgery (month) | 69.2 (2.7–138.7) |
| TMPRSS2 expression level in tumor tissue* | 5.9 (0.04–533.7) |
| TMPRSS2 expression level in benign tissue* | 11.4 (0.5–2530.1) |
| TMPRSS2 ratio (N/T) | 2.3 (0.02–249.9) |
TMPRSS2 expression level = 2−TMPRSS 2ΔCt
TMPRSS2 ^Ct = TMPRSS2 Ct value - GAPDH Ct value
In this cohort the median level of TMPRSS2 expression in CaP cells, normalized to house keeping gene GAPDH, was about half of that in the matched benign epithelium (Table 2). Paired t-test analysis (using mean log2 transformed expression values as described in the Methods) revealed that TMPRSS2 gene expression was significantly lower (3.6 fold) in malignant compared to matched benign prostate epithelial cells in TMPRSS2-ERG fusion positive patients (N=72, P<0.0001). In ERG fusion negative CaP patients (N=38), however, the mean difference in TMPRSS2 expression level between benign and tumor glands was not significant (1.5 fold, P=0.0536). The relationship between TMPRSS2 expression and TMPRSS2-ERG fusion status in CaP cells is also apparent when patients are grouped based on TMPRSS2 expression level. Of 15 patients with the highest TMPRSS2 expression in CaP cells (2-fold or higher than in matched benign cells), only 3 (20%) had TMPRSS2-ERG fusion. In contrast, of 29 patients with the lowest TMPRSS2 expression in their CaP cells (5-fold or lower than in matched benign cells) 23 (79%) had TMPRSS2-ERG fusion.
The ratio of TMPRSS2 expression in benign over tumor cells is significantly different between patients with and without TMPRSS2-ERG fusion (Table 3, P=0.006). In fusion positive patients (N=72) the median TMPRSS2 expression is 3.23 times lower in the CaP cells harboring the fusion compared to matched benign epithelial cells, in contrast in fusion negative patients (N=38) this ratio is 1.72. TMPRSS2 expression did not significantly associate with the clinico-pathological parameters analyzed (race, pathological T stage, post-operative Gleason sum, margin status) (Table 3) or with time to PSA recurrence (Kaplan-Meier analysis, data not shown).
Table 3.
Association of TMPRSS2 ratio (N/T) with ERG fusion and clinico-pathological features
| Variable | TMPRSS2 Ratio (N/T) | P | ||
|---|---|---|---|---|
| N | Median | Range | value | |
| Race | 0.4437 | |||
| Caucasian & other | 102 | 2.39 | 0.02–249.9 | |
| African American | 30 | 1.56 | 0.16–56.30 | |
| Pathologic T stage | 0.8900 | |||
| pT2 | 50 | 2.92 | 0.02–247.3 | |
| pT3 | 75 | 1.92 | 0.18–249.9 | |
| Post-operative Gleason sum | 0.5484 | |||
| 6 | 40 | 3.07 | 0.16–56.3 | |
| 7 | 65 | 2.07 | 0.12–247.3 | |
| 8 – 9 | 25 | 2.31 | 0.02–249.9 | |
| Margin status | 0.5699 | |||
| Negative | 89 | 2.44 | 0.02–247.3 | |
| Positive | 42 | 1.98 | 0.18–249.9 | |
| ERG fusion | 0.0060 | |||
| No | 38 | 1.72 | 0.12–37.01 | |
| Yes | 72 | 3.23 | 0.47–249.9 | |
The expression of TMPRSS2 mRNA was compared with that of other androgen regulated genes measured in the same CaP cell population. TMPRSS2 gene expression was found to correlate with tissue PSA/KLK3 (p=0.0005), PMEPA1 (p<0.0001), AR (p<0.0001), PCA3 (P=0.0187) and ERG (P=0.0158) (Table 4). In contrast, CaP related genes that are not androgen regulated, such as AMACR (P=0.934) and LTF (P=0.1466), did not correlate with TMPRSS2 expression (Table 4).
Table 4.
Spearman correlation between TMPRSS2 level and other gene levels in tumor gland
| TMPRSS2 | N | R* | P-value |
|---|---|---|---|
| ERG | 121 | 0.22 | 0.0158 |
| AR | 114 | 0.39 | < .0001 |
| PSA/KLK3 | 110 | 0.33 | 0.0005 |
| PCA3 | 99 | 0.24 | 0.0187 |
| PMEPA1 | 97 | 0.44 | <.0001 |
| AMACR | 80 | 0.009 | 0.934 |
| LTF | 86 | 0.16 | 0.1466 |
Discussion
Our evaluation of the quantitative expression of the TMPRSS2 transcript in prostate epithelial cells of 132 CaP patients revealed that TMPRSS2 transcript level in CaP cells reflects TMPRSS2-ERG fusion status. No significant association of TMPRSS2 expression with the clinico-pathological features analyzed was found.
Current research continues to elaborate the relevance of androgen-regulated genes, including TMPRSS2, in CaP development and progression. Previous studies examining TMPRSS2 regulation had found the gene to be overexpressed in primary CaP tissue compared to benign prostate tissue (3,4,6). However, TMPRSS2 was down-regulated in CaP bone metastasis-derived xenografts (5). One study concluded that TMPRSS2 was expressed in normal basal cells and in adenocarcinoma, but not in normal secretory cells or stroma (3). In contrast, TMPRSS2 expression was detected in secretory epithelium by in situ hybridization in another study, where 16 of 19 CaP cases (of which only 13 was untreated) had higher TMPRSS2 mRNA in tumor than in benign areas (4). Poorly differentiated tumors (N=7) had further elevated TMPRSS2 expression. An increased TMPRSS2 protein expression by immunohistochemistry (IHC) was reported in CaP and in BPH, and a further increased expression in high Gleason grade cancer, where the TMPRSS2 protein was mislocalized (12). A recent QRT-PCR analysis found that TMPRSS2 expression is higher in CaP tissue than in normal prostate, and it is increased with Gleason score, tumor grade and stage (13).
We hypothesized that due to the frequent genomic fusions between TMPRSS2 and ERG in CaP (8) in patients with the fusion the promoter-less copy of TMPRSS2 is silenced, which may lead either to lower TMPRSS2 expression in these CaP cells, or the dosage loss may be compensated as it has been shown for numerous genes, including some in this region of chromosome 21 (10–11). For the precise quantitative analysis of TMPRSS2 mRNA levels in CaP cells with or without the fusion, as well as in matched benign epithelium, we utilized laser micro-dissection and TaqMan QRT-PCR. Our analysis revealed that patients with TMPRSS2-ERG fusion (N=72) had significantly lower level of TMPRSS2 expression in their CaP cells compared to matched benign epithelium. In a subset of patients (N=15), most of whom (80%) did not have detectable TMPRSS2-ERG fusion transcripts, TMPRSS2 expression in CaP cells was significantly higher (over 2-fold) compared to matched benign cells. Although TMPRSS2 deficient mice did not have an apparent phenotype (14), TMPRSS2 may have a tumor progression related role in CaP cells through activating PAR2 (9).
An in vivo co-regulation of TMPRSS2 expression with other AR regulated genes, including PSA/KLK3 and PMEPA1, was demonstrated. A practical potential is that quantitative evaluation of the expression of these genes may serve as AR surrogate (15), although one must be aware of the TMPRSS2-ETS fusion status when considering TMPRSS2.
In summary, this manuscript describes, using carefully defined specimens and highly quantitative methodology, that CaP cells with TMPRSS2-ERG fusion have decreased expression of TMPRSS2. In contrast, CaP cells harboring no TMPRSS2-ERG fusion have higher expression of TMPRSS2, a gene implicated in signaling pathways that promote tumor progression, further defining these two genetically distinct types of CaP.
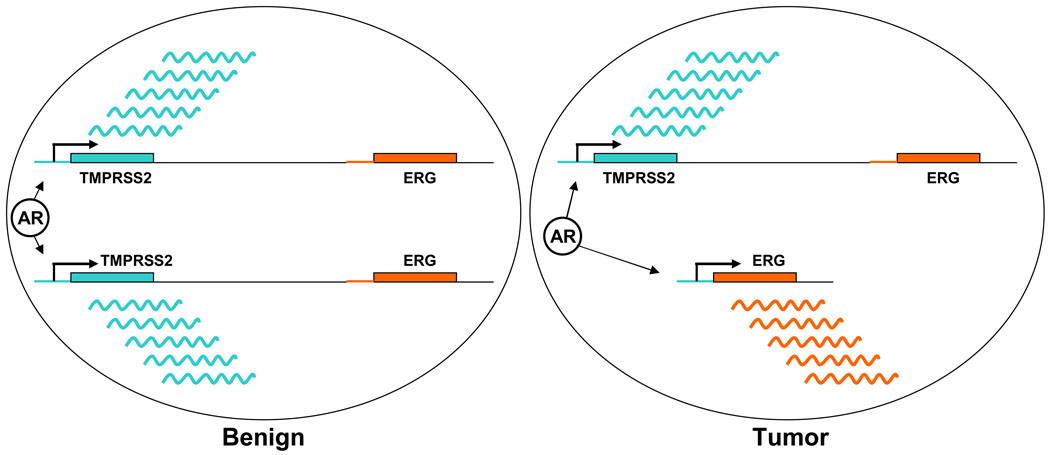
Figure 1. TMPRSS2 and ERG genes in tumor and benign prostate epithelial cells.
Schematic representation of TMPRSS2 and ERG genes on chromosome 21 in a diploid benign prostate epithelial cell (left panel), and in a prostate tumor cell with TMPRSS2-ERG fusion (right panel). AR in a circle represents androgen receptor protein. Wavy lines represent mRNA expressed from the TMPRSS2 gene (green) and the ERG gene (orange) fused to the AR-regulated TMPRSS2 promoter.
Acknowledgement
This work was supported in part by grant 5R01 DK065977 for S. S. and G. P. from the National Institutes of Health.
Footnotes
Note:
The views expressed in this manuscript are those of the authors and do not reflect the official policy of the Department of the Army, Department of Defense or the U.S. Government. None of the authors have competing financial interests.
References
- 1.Makarov DV, Loeb S, Getzenberg RH, Partin AW. Biomarkers for Prostate Cancer. Annu Rev Med. 2008;60:139–151. doi: 10.1146/annurev.med.60.042307.110714. [DOI] [PubMed] [Google Scholar]
- 2.Paoloni-Giacobino A, Chen H, Peitsch MC, Rossier C, Antonarakis SE. Cloning of the TMPRSS2 gene, which encodes a novel serine protease with transmembrane, LDLRA, and SRCR domains and maps to 21q22.3. Genomics. 1997;44:309–320. doi: 10.1006/geno.1997.4845. [DOI] [PubMed] [Google Scholar]
- 3.Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, et al. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59:4180–4184. [PubMed] [Google Scholar]
- 4.Vaarala MH, Porvari K, Kyllonen A, Lukkarinen O, Vihko P. The TMPRSS2 gene encoding transmembrane serine protease is overexpressed in a majority of prostate cancer patients: detection of mutated TMPRSS2 form in a case of aggressive disease. Int J Cancer. 2001;94:705–710. doi: 10.1002/ijc.1526. [DOI] [PubMed] [Google Scholar]
- 5.Afar DE, Vivanco I, Hubert RS, Kuo J, Chen E, Saffran DC, et al. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 2001;61:1686–1692. [PubMed] [Google Scholar]
- 6.Vaarala MH, Porvari KS, Kellokumpu S, Kyllönen AP, Vihko PT. Expression of transmembrane serine protease TMPRSS2 in mouse and human tissues. J Pathol. 2001;193:134–140. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH743>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 7.Petrovics G, Liu A, Shaheduzzaman S, Furusato B, Sun C, Chen Y, et al. Frequent overexpression of ETS-related gene-1 (ERG1) in prostate cancer transcriptome. Oncogene. 2005;24:3847–3852. doi: 10.1038/sj.onc.1208518. [DOI] [PubMed] [Google Scholar]
- 8.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 9.Wilson S, Greer B, Hooper J, Zijlstra A, Walker B, Quigley J, et al. The membrane-anchored serine protease, TMPRSS2, activates PAR-2 in prostate cancer cells. Biochem J. 2005;388:967–972. doi: 10.1042/BJ20041066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 11.Aït Yahya-Graison E, Aubert J, Dauphinot L, Rivals I, Prieur M, Golfier G, et al. Classification of human chromosome 21 gene-expression variations in Down syndrome: impact on disease phenotypes. Am J Hum Genet. 2007;81:475–491. doi: 10.1086/520000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucas JM, True L, Hawley S, Matsumura M, Morrissey C, Vessella R, et al. The androgen-regulated type II serine protease TMPRSS2 is differentially expressed and mislocalized in prostate adenocarcinoma. J Pathol. 2008;215:118–125. doi: 10.1002/path.2330. [DOI] [PubMed] [Google Scholar]
- 13.Bi X, He H, Ye Y, Dai Q, Han Z, Liang Y, et al. Association of TMPRSS2 and KLK11 gene expression levels with clinical progression of human prostate cancer. Med Oncol. 2009 Feb 26; doi: 10.1007/s12032-009-9185-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Kim TS, Heinlein C, Hackman RC, Nelson PS. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol Cell Biol. 2006;26:965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sterbis JR, Gao C, Furusato B, Chen Y, Shaheduzzaman S, Ravindranath L, et al. Higher expression of the androgen-regulated gene PSA/HK3 mRNA in prostate cancer tissues predicts biochemical recurrence-free survival. Clin Cancer Res. 2008;14:758–763. doi: 10.1158/1078-0432.CCR-07-1356. [DOI] [PubMed] [Google Scholar]