Summary
p53 stability and localization is essential for its tumor suppressor function. Ubiquitination by the E3 ubiquitin ligase Mdm2 is the major regulatory mechanism of p53, which induces p53 nuclear export and degradation. However, it is unclear whether ubiquitinated cytoplasmic p53 can be recycled. Here we report that USP10, a cytoplasmic ubiquitin-specific protease, deubiquitinates p53, reversing Mdm2-induced p53 nuclear export and degradation. Following DNA damage, USP10 is stabilized and a fraction of USP10 translocates to the nucleus to activate p53. The translocation and stabilization of USP10 is regulated by ATM -mediated phosphorylation of USP10 at Thr42 and Ser337. Finally, USP10 suppresses tumor cell growth in cells with wild-type p53, with USP10 expression downregulated in a high percentage of clear cell carcinomas, known to have few p53 mutations. These findings reveal USP10 to be a novel regulator of p53, providing an alternative mechanism of p53 inhibition in cancers with wild-type p53.
Introduction
p53 is an crucial tumor suppressor that is mutated in more than 50% of human cancers and whose major function is regulating cell fate following cellular stress and repressing the propagation of damaged cells (Lane, 1992; Riley et al., 2008; Vogelstein et al., 2000). p53 functions as a transcription factor, and through its target genes regulates a variety of cellular functions, including cellular senescence, energy metabolism, DNA repair, cell differentiation, cell cycle progression and apoptosis.
p53 activity, expression and localization are mainly regulated by posttranslational modifications, such as phosphorylation, acetylation, and ubiquitination (Appella and Anderson, 2001). Mdm2 is an E3 ubiquitin ligase that plays a major role in regulating p53. Mdm2-mediated ubiquitination of p53 induces p53 nuclear export and degradation (Boyd et al., 2000; Freedman and Levine, 1998; Geyer et al., 2000; Haupt et al., 1997; Honda et al., 1997; Kubbutat and Vousden, 1997; Stommel et al., 1999). Although nuclear export was considered to be required for p53 degradation, later studies demonstrated that p53 degradation could also occur in the nucleus (Geyer et al., 2000; Lohrum et al., 2001; Shirangi et al., 2002; Stommel and Wahl, 2004; Xirodimas et al., 2001). The extent of p53 ubiquitination was suggested to determine p53 localization, with monoubiquitination inducing p53 nuclear export and polyubiquitination inducing nuclear degradation of p53 (Li et al., 2003). Other E3 ubiquitin ligases have also been shown to regulate p53 stability and/or localization, such as COP1, Pirh2, ARF-BP1, MSL2 and Parc (Brooks and Gu, 2006).
It is well documented that the ubiquitination of many proteins can be reversed by deubiquitinases (DUBs). In the human proteome, there are about 90 DUBs, however the mechanisms regulating p53 deubiquitination remain enigmatic. The ubiquitin-specific protease HAUSP has been shown to deubiquitinate p53, with overexpression of HAUSP resulting in p53 stabilization (Li et al., 2002). However depletion of HAUSP in cells does not decrease p53 levels as predicted, but rather increases p53 levels (Cummins et al., 2004), apparently due to HAUSP's ability to bind and deubiquitinate Mdm2 (Cummins et al., 2004; Li et al., 2004). In fact, Mdm2 seems to be a better binding partner of HAUSP than p53 (Hu et al., 2006; Sheng et al., 2006). Thus it would appear that the regulation of p53 by HAUSP is a rather complex process. It is likely that Mdm2 is the preferred substrate for HAUSP in unstressed cells, while genotoxic stress decreases HAUSP's binding to Mdm2/MdmX through ATM-dependent phosphorylation (Meulmeester et al., 2005), tilting the balance toward p53 stabilization (Brooks et al., 2007). In addition, HAUSP mainly localizes in the nucleus (Everett et al., 1997; Song et al., 2008), although a fraction of HAUSP has been shown to be present in the cytoplasm and mitochondria. One pertinent question arising from these studies pertains to the fate of ubiquitinated cytoplasmic p53 and the pliability of this fate. It is possible that there is a cytoplasmic counterpart of HAUSP responsible for the deubiquitination of cytoplasmic p53, resulting in p53 stabilization and translocation back to the nucleus. Here we identify USP10 as a p53 DUB. In unstressed cells, USP10 mainly localizes in the cytoplasm and regulates p53 homeostasis. Following DNA damage, USP10 also translocates to the nucleus and contributes to p53 activation. These results reveal an important missing piece in the regulation of p53 localization and stability.
Results
USP10 interacts with p53 and stabilizes p53
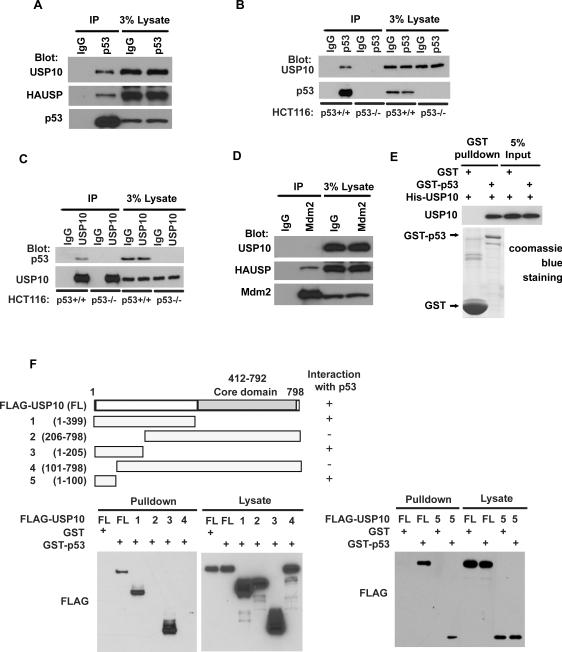
By serendipity, we found that the ubiquitin specific protease USP10 interacts with p53. As shown in Figures 1A and 1B, USP10 coimmunoprecipitated with p53 in HCT116 p53+/+ cells, but not HCT116 p53−/− cells. HAUSP also coimmunoprecipitated with p53 (Figure 1A), as previously reported (Li et al., 2002). Reciprocal immunoprecipitation with anti-USP10 also brought down p53 in HCT116 p53+/+ and U2OS cells but not in HCT116 p53−/− cells (Figure 1C and data not shown). Unlike HAUSP, USP10 did not interact with Mdm2 (Figure 1D). These results suggest a specific interaction between USP10 and p53 in vivo. To determine whether the USP10-p53 interaction is direct, we generated and purified recombinant USP10 and p53. Purified His-USP10 was able to interact with GST-p53 under cell-free conditions, suggesting a direct interaction between USP10 and p53 (Figure 1E). Mapping the region of USP10 required for p53 binding revealed that the N-terminal region (AA1–AA100) is critical for the interaction between USP10 and p53 (Figure 1F). Furthermore, FLAG-USP10 (1–100) itself could interact with p53, suggesting that AA1–100 of USP10 is both required and sufficient for p53 interaction.
Figure 1. USP10 interacts with p53.
(A–D) HCT116 p53+/+ and p53−/− cell lysates were subject to immunoprecipitation with control IgG, anti-p53 (A–B), anti-USP10 (C) or anti-Mdm2 (D) antibodies. The immunoprecipitates were then blotted with indicated antibodies. (E) Purified USP10 was incubated with GST or GST-p53 coupled to GSH-Sepharose. Proteins retained on Sepharose were then blotted with indicated antibodies. (F) H1299 cells transfected with indicated constructs were lysed and lysates incubated with GST or GST-p53-GSH-Sepharose. Proteins retained on Sepharose were blotted with the indicated antibody.
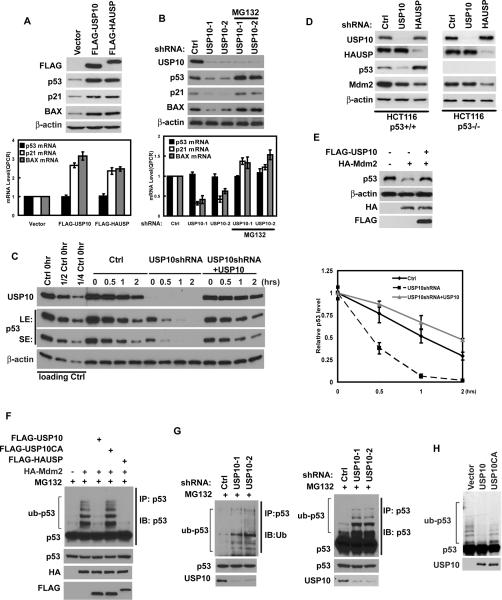
Since USP10 is an ubiquitin-specific protease, it is possible that USP10 could function to stabilize p53. Overexpression of USP10 significantly increased the levels of endogenous p53, with no effect on p53 mRNA levels (Figure 2A). We also observed increased mRNA and protein levels of p21 and Bax, two p53 target genes. On the other hand, overexpression of USP10 deleted of the p53 binding region (Δ1–100) did not change the level of endogenous p53 compared to vector transfected cells (Figure S1A). Consistent with previous results (Li et al., 2002), overexpression of HAUSP also increased p53 levels (Figure 2A). To confirm the role of USP10 in regulating p53 levels, we depleted USP10 using USP10 specific shRNA, and found that downregulation of USP10 decreased p53 protein levels with no effect on p53 mRNA levels (Figure 2B). While the mRNA and protein levels of p21 and Bax were also decreased. The decrease in p53 levels could be reversed by the addition of proteasome inhibitor MG132, suggesting that USP10 regulates p53 levels in a proteasome-dependent manner. A second USP10 shRNA behaved similarly (Figure 2B). These results suggest that USP10 can upregulate p53 levels, most likely by deubiquitinating and consequently stabilizing p53. To prove that USP10 affects p53 stability per se, we treated control cells or cells stably expressing USP10 shRNA with cycloheximide (CHX), and examined p53 stability. p53 stability was decreased in cells stably expressing USP10 shRNA; while reconstitution with shRNA-resistant USP10 restored p53 stability (Figure 2C). These results confirmed the specificity of our shRNA and also demonstrate that USP10 stabilizes p53 in cells.
Figure 2. USP10 stabilizes and deubiquitinates p53.
(A) HCT116 cells were transfected with indicated constructs. 48hrs later, proteins and mRNA were extracted and subjected to Western Blot or QRT-PCR. (B) HCT116 cells infected with lentivirus encoding indicated shRNAs were left untreated or treated with MG132 for 4hrs, then proteins and mRNA were extracted and subjected to Western Blot or QRT-PCR. (A and B) Lower panel: quantification of p53, p21 or BAX transcript levels. (C) HCT116 cells stably expressing control shRNA, USP10 shRNA, or USP10 shRNA together with shRNA-resistant USP10 were treated with cycloheximide (0.1 mg/ml), and harvested at the indicated times. The left panels show immunoblots of p53 and USP10. (LE: long exposure; SE: short exposure) Right panel: quantification of the p53 protein levels relative to β-actin. (A–C) Error bars represent SEM of triplicate experiments. (D) HCT116 p53+/+ and p53−/− cells infected with lentivirus encoding indicated shRNAs were lysed and lysates blotted with indicated antibody. (E) HCT116 cells transfected with indicated constructs were lysed and lysates blotted with indicated antibody. (F–G) Regulation of p53 ubiquitination levels in vivo by USP10. HCT116 cells transfected with indicated constructs (F) or stably expressing Ctrl or USP10 shRNA (G) were treated with MG132 for 4hrs before harvest. p53 was immunoprecipitated with anti-p53 polyclonal antibodies and immunoblotted with monoclonal anti-p53(DO-1) antibodies or anti-ub antibodies. (H) Deubiquitination of p53 in vitro by USP10. Ubiquitinated p53 was incubated with purified USP10 or USP10CA in vitro, and then blotted with anti-p53 antibodies. “See also Figure S1”
HAUSP has also been shown to stabilize Mdm2 (Cummins et al., 2004; Li et al., 2004) therefore we examined how USP10 would affect Mdm2 levels. Consistent with the previous report, downregulation of HAUSP decreased Mdm2 levels, which resulted in an increase in p53 levels (Figure 2D). Although downregulation of USP10 also resulted in decreased Mdm2 levels, we believe this is secondary to a decrease in p53 levels. Indeed, downregulation of USP10 did not affect Mdm2 levels in p53-deficient cells (Figure 2D and Figure S1B–C), while downregulation of HAUSP still decreased Mdm2 levels in p53-deficient cells. These results suggest that USP10 and HAUSP have differing roles in p53 homeostasis.
USP10 deubiquitinates p53
It is likely that USP10 functions to deubiquitinate p53 to counteract the action of E3 ubiquitin ligases such as Mdm2. Indeed, as shown in Figure 2E, although overexpression of Mdm2 significantly induced the degradation of p53, coexpression of USP10 effectively rescued p53 from Mdm2-induced degradation. We next examined whether USP10 regulates the levels of p53 ubiquitination in cells. To observe p53 ubiquitination, the proteasome inhibitor MG132 was used in Figures 2F–G. As shown in Figure 2F, Mdm2 induced the ubiquitination of p53; however, p53 ubiquitination was significantly diminished by coexpression of USP10 or HAUSP. On the other hand, USP10C488A (USP10CA), the catalytic-inactive USP10 mutant with a mutation at the core enzymatic domain (Soncini et al., 2001), lost the ability to reverse p53 ubiquitination induced by Mdm2 (Figure 2F and Figure S1D). Conversely, downregulation of USP10 increased p53 ubiquitination (Figure 2G). These results suggest that USP10 negatively regulates p53 ubiquitination in cells. In order to directly examine the deubiquitination activity of USP10 toward p53, we utilized a cell free system. We purified USP10 and USP10CA from bacteria, as well as ubiquitinated p53 from cells expressing FLAG-p53, pCMV-Mdm2 and HA-ub. We then incubated USP10 and ubiquitinated p53 in a cell-free system. As shown in Figure 2H, purified WT USP10, but not catalytically inactive USP10 effectively deubiquitinated p53 in vitro. These results demonstrate that USP10 deubiquitinates p53 both in vitro and in vivo.
USP10 localizes in the cytoplasm and counteracts Mdm2 action
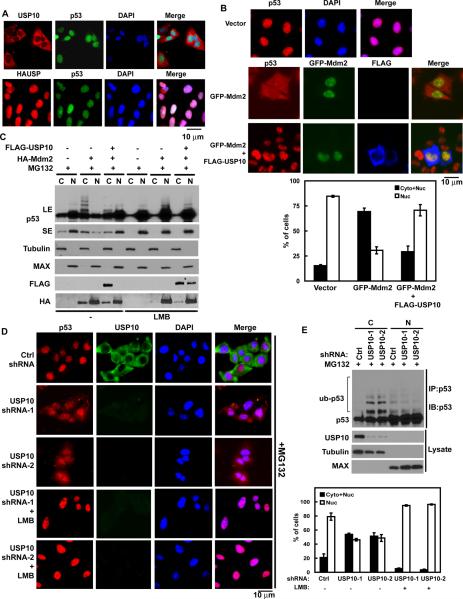
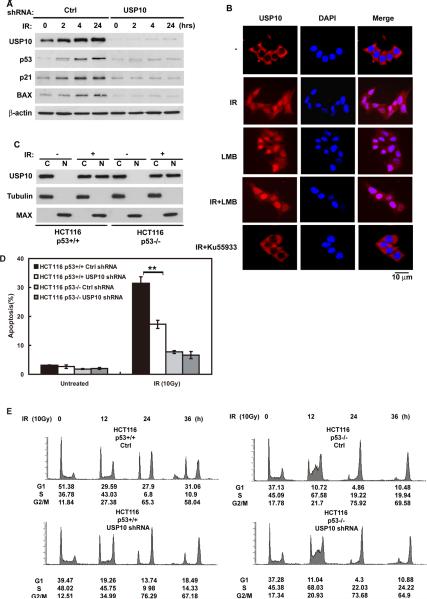
Previous studies suggest that ubiquitination of p53 by Mdm2 could induce p53 translocation from nucleus to cytoplasm (Boyd et al., 2000; Geyer et al., 2000; Li et al., 2003; Stommel et al., 1999). In addition, the cytoplasmic ubiquitin ligase Parc can ubiquitinate p53 and trap p53 in the cytoplasm (Nikolaev et al., 2003). However, it is not clear whether cytoplasmic p53 can be deubiquitinated and returned to the nucleus. The majority of HAUSP is localized in the nucleus (Marchenko and Moll, 2007; Song et al., 2008), USP10 however, is predominantly localized to the cytoplasm in both p53-proficient and p53-deficient cells (Figure 3A and Figure S2A). We also confirmed the localization of HAUSP and USP10 by cellular fractionation. As shown in Figure S2B, HAUSP is predominantly localized in the nucleus, while USP10 is localized in the cytoplasm. This led us to hypothesize that USP10 is the cytoplasmic p53 deubiquitinase that reverses Mdm2-induced nuclear export of p53. To test this, we performed immunofluorescence assays to detect the subcellular localization of p53. In control cells, p53 was readily detected in the nucleus. As previously demonstrated (Boyd et al., 2000; Geyer et al., 2000; Li et al., 2003; Stommel et al., 1999), in cells transfected with Mdm2, we observed nuclear export of p53 (Figure 3B). Coexpression of WT USP10 with Mdm2 reversed Mdm2-induced cytoplasmic translocation of p53. These results suggest that USP10 counteracts Mdm2's action and induce p53 translocation from the cytoplasm back to the nucleus. To confirm this result, we performed cell fractionation experiments and found that expression of Mdm2 induced ubiquitination and nuclear export of p53 (Figure 3C). The nuclear export of p53 was reversed by USP10 coexpression or LMB treatment. Therefore, a balance between USP10 and Mdm2 could determine p53 localization. If so, downregulation of USP10 could have similar effect to Mdm2 overexpression. Consistent with our prediction, downregulation of USP10 itself induced nuclear export of endogenous p53 (Figure 3D), which could be blocked by LMB. Similar results were obtained using a second USP10 shRNA (Figure 3D). Furthermore, downregulation of USP10 increased the ubiquitination of p53 in the cytoplasm, but not in the nucleus (Figure 3E). Although p53 could be downregulated by the ubiquitin pathway in both cytoplasm and nucleus (Figure S2C), the stability of cytoplasmic p53, but not that of nuclear p53, was decreased in cells depleted of USP10 (Figure S2D). Overall, our results from Figures 2 and 3 establish USP10 as the cytoplasmic deubiquitinase for p53 and the important role of USP10 in regulating homeostasis of p53 in cells.
Figure 3. Regulation of subcellular localization of p53 by USP10.
(A) Subcellular localization of USP10 and HAUSP. U2OS cells were fixed and stained as indicated. Bars, 10 μm. (B) HCT116 cells transfected with indicated constructs were treated with MG132 for 4hrs, then fixed and stained as indicated. Bars, 10μm. (C) HCT116 cells transfected with indicated constructs were treated with MG132 and then left untreated or treated with 25ng/ml leptomycin B (LMB), after 4hrs, cells were harvested and fractionated as described in Methods. Cellular fractions were then blotted with indicated antibodies. (C, cytoplasmic; N, nuclear). A cytoplasmic marker protein, Tubulin and a nuclear marker protein, MAX were used as controls to confirm the quality of fractions. LE: long exposure; SE: short exposure. (D) HCT116 cells infected with lentivirus encoding indicated shRNA were treated with MG132 and then left untreated or treated with LMB. Additional 4hrs later, cells were fixed and stained as indicated. Bars, 10μm. (B&D) Lower and Right panels: Quantification of cells with different p53 subcellular localization. Nuc: Nucleus only; Cyto + Nuc: both cytoplasm and nucleus. The data represent the average of three experiments and 150 cells were monitored in each experiment. (E) HCT116 cells stably expressing indicated shRNA were treated with MG132 for 4 hrs. Then cells were harvested and fractionated, cytoplasmic or nuclear p53 was immunoprecipitated with anti-p53 polyclonal antibodies and immunoblotted with monoclonal anti-p53 (DO-1) antibodies. “See also Figure S2”
USP10 regulates p53 function
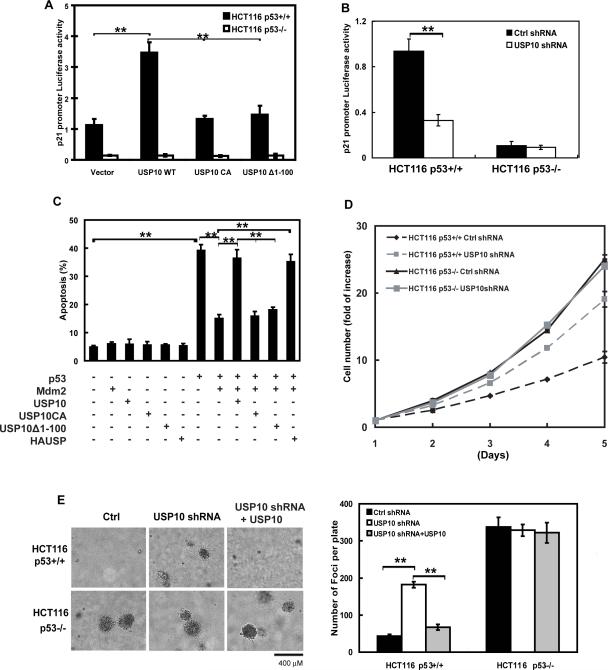
USP10's effects on p53 stabilization and nuclear import raised the possibility that USP10 regulates p53-dependent transcriptional activity, cell transformation and apoptosis. As shown in Figure 4A, overexpression of WT USP10 increased p21 promoter activity in HCT116 p53+/+ cells, but not in HCT116 p53−/− cells. In contrast, catalytically-inactive USP10 (USP10 CA) or USP10 deleted of the p53 binding region (Δ1–100) could not affect p53 transcriptional activity. Stable knockdown of USP10 by shRNA inhibited p21 promoter activity in HCT116 p53+/+ cells, but had little effect in HCT116 p53−/− cells (Figure 4B). These results suggest that USP10 regulates p53-dependent transcription activity. Furthermore, we tested whether USP10 directly affects p53-dependent apoptosis. As shown in Figure 4C, overexpression of p53 induced apoptosis, while Mdm2 strongly reduced p53-dependent apoptosis. However, coexpression of USP10 or HAUSP, but not USP10 CA or USP10Δ1–100, significantly reversed the inhibitory effect of Mdm2 on p53-mediated apoptosis. We further investigated how USP10 affects cell proliferation. As shown in Figure 4D, downregulation of USP10 increased cancer cell proliferation in p53+/+ cells, while USP10 expression levels have no apparent effect on proliferation in cells lacking p53. A similar effect was observed when cancer cells were culture in soft agar (Figure 4E). On the other hand, reconstitution of USP10 in cells with USP10 downregulation inhibited cancer cell proliferation (Figure 4E), suggesting the effect of USP10 knockdown is specific. Overall, these results suggest that USP10 potentiates p53 function in cells.
Figure 4. Effects of USP10 on p53-mediated transcriptional activity, cell growth repression and apoptosis.
(A) p53 reporter constructs for the p21 promoter were co-transfected with indicated constructs into HCT116 p53+/+ and HCT116 p53−/− cells. Reporter activity was then determined as described in materials and methods. (B) p53 reporter assay was performed in HCT116 p53+/+ and HCT116 p53−/− cells stably expressing indicated shRNA. (C) H1299 cells were transfected with indicated constructs. 48hrs later, apoptotic cells were determined. (D) HCT116 p53+/+ and HCT116 p53−/− cells stably expressing indicated shRNA were plated and cell proliferation was then quantified at indicated time. (E) Soft agar colony-formation assay was performed using HCT116 p53+/+ and HCT116 p53−/− cells stably expressing control shRNA, USP10shRNA or USP10shRNA together with shRNA-resistant USP10. Right panel: quantification of colonies formed in soft agar. Bars, 400μm. (A–E) Error bars represent SEM of triplicate experiments. **, P < 0.01 two tailed student's t test.
USP10 is upregulated and translocates to the nucleus following DNA damage and regulates p53-dependent DNA damage response
We have shown that USP10 can regulate p53 homeostasis in unstressed cells. Since p53 plays an important role in DNA damage response and becomes stabilized following DNA damage, we next asked whether USP10 is important for p53 stabilization and activation after DNA damage. Interestingly, downregulation of USP10 significantly decreased p53 stabilization and the expression of p53 target genes p21 and Bax after DNA damage (Figure 5A). We also observed increased p53 ubiquitination in cells depleted of USP10 following DNA damage (Figure S3A). Upstream signaling events such as ATM phosphorylation were unaffected by USP10 downregulation (Figure S3B). These results suggest that USP10 regulates p53 stabilization after DNA damage. We also observed that the expression of USP10 itself was increased after DNA damage. These results are rather surprising, since most DNA damage signaling is thought to occur in the nucleus. How is it that USP10, a largely cytoplasmic protein, affects p53 stabilization during DNA damage response? It is possible that p53 is still actively exported out of the nucleus and gets degraded in the cytoplasm during DNA damage response. Alternatively, USP10 could translocate into the nucleus to participate in DNA damage response. Indeed, we found by immunofluorescence that a portion of USP10 appears to relocalize to the nucleus following DNA damage (Figure 5B). LMB treatment also induced USP10 nuclear localization, suggesting that USP10 is actively exported out of the nuclear in unstressed cells. The translocation of USP10 could be blocked by the ATM inhibitor Ku55933 (Hickson et al., 2004), suggesting that USP10 translocation after DNA damage is ATM-dependent. To confirm the translocation of USP10, we performed cell fractionation assays. As shown in Figure 5C, increased amounts of USP10 were detected in the nucleus following DNA damage, and this translocation of USP10 also occurred in p53-null cells, suggesting the translocation of USP10 is p53 independent.
Figure 5. USP10 translocates into the nucleus and regulates p53 activity following DNA damage.
(A) HCT116 cells stably expressing indicated shRNA were irradiated (10Gy) and harvested at indicated time, cell lysates were then blotted with indicated antibodies. (B) HCT116 cells were left untreated, treated with 10Gy radiation, with LMB, with both or treated with Ku55933 (ATM specific inhibitor) for 1hr then irradiated. An additional 1hr later cells were fixed and stained as indicated. Bars, 10μm. (C) HCT116 p53+/+ or HCT116 p53−/− cells were irradiated (10Gy) or left untreated, after 1hr cells were harvested and fractionated. Cellular fractions were then blotted with indicated antibodies. (D) HCT116 p53+/+ or p53−/− cells stably expressing indicated shRNA were left untreated or treated with 10Gy radiation, after 48hrs, apoptotic cells were determined. Error bar represent SEM of triplicate experiments. **, P < 0.01 two tailed student's t test. (E) The same cells in (D) were treated with 10Gy radiation, then harvested at indicated time. Cell cycle profiles were determined by FACS. “See also Figure S3”
To confirm that translocated USP10 is functional, we fractionated cells treated with IR. As shown in Figure S3C–D, nuclear USP10 is still able to bind and deubiquitinate p53. These results imply that USP10 translocation could contribute to p53 activation in the nucleus. To test this notion, we further examined whether USP10 is required for p53-dependent function during DNA damage response. As shown in Figure 5D and Figure S3E, downregulation of USP10 inhibited IR-induced apoptosis in HCT116 p53+/+ cells. The IR-induced apoptosis in HCT116 p53−/− cells was attenuated, however downregulation of USP10 did not have any further effect. Furthermore, knockdown of USP10 in HCT116 p53+/+ cells resulted in defective DNA damage-induced G1 arrest (Figure 5E). These results are consistent with decreased Bax and p21 expression in cells with USP10 downregulation (Figure 5A), and suggest that USP10 is required for p53 activation following DNA damage.
USP10 phosphorylation by ATM is required for its stabilization and translocation following DNA damage
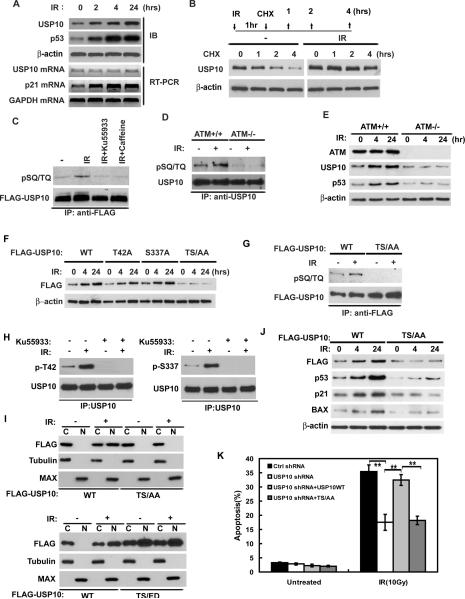
We next sought to determine the molecular mechanisms that regulate USP10 upregulation and translocation. Initial experiments indicated that the upregulation of USP10 occurs both in p53-WT and p53-null cells (Figure S3F). Furthermore, unlike p21, the upregulation of USP10 occurred without any change in USP10 mRNA (Figure 6A). These results suggest that DNA damage induced upregulation of USP10 is not regulated at the transcriptional level, and might be regulated at the posttranslational levels. To examine whether USP10 protein becomes stabilized, we irradiated and then treated cells with CHX. As shown in Figure 6B, USP10 accumulation in irradiated cells is due to increased stability.
Figure 6. USP10 phosphorylation by ATM regulates USP10 stabilization, translocation, and p53 activation following DNA damage.
(A) HCT116 cells were irradiated (10Gy) and harvested at indicated times. Protein and mRNA were extracted and analyzed by Western blot or QRT-PCR respectively. (B) HCT116 cells left untreated or irradiated were treated with CHX and harvested at indicated time. Cell lysates were then blotted with indicated antibodies. (C) HCT116 cells transfected with FLAG-USP10 were pretreated with DMSO, 25 mM Ku55933 or 3mM caffeine. After 2hr incubation, cells were left untreated or irradiated (10Gy). After additional 1hr, FLAG-USP10 was immunoprecipitated with anti-FLAG antibody and immunoblotted with phospho-SQ/TQ (pSQ/TQ) antibody. (D) ATM +/+ or ATM −/− cells were irradiated (10 Gy) or left untreated. After 1hr, USP10 was immunoprecipitated and immunoblot with indicated antibody. (E) ATM+/+ and ATM−/− cells were irradiated (10Gy) and harvested at indicated time and lysates blotted with indicated antibodies. (F) HCT116 cells stably expressing USP10 shRNA were reconstituted with shRNA resistant FLAG-USP10 WT, T42A, S337A or TS/AA (T42A and S337A double mutant). Cells were irradiated (10Gy) and harvested at indicated time and lysates blotted with indicated antibodies. (G) HCT116 cells stably expressing USP10 shRNA were reconstituted with shRNA resistant FLAG-USP10 WT or TS/AA. Cells were left untreated or irradiated (10Gy), USP10 phosphorylation was examined with pSQ/TQ antibody. (H) HCT116 cells were left untreated or treated with Ku55933 for 2hr then irradiated. The phosphorylation of USP10 was examined by site specific phospho antibodies:p-T42 and p-S337.(I) Cells as in (G) and cells expressing a phospho-mimetic mutant of USP10-T42E/S337D (TS/ED), lower panel, were irradiated (10Gy) or left untreated, and then harvested and fractionated. Fractions were then blotted with indicated antibodies. (J) Cells as in (G) were irradiated (10Gy) and harvested at indicated time, cell lysates were blotted with indicated antibodies. (K) Cells as in (G) were left untreated or irradiated, apoptotic cells were determined 48hrs later. Error bar represent SEM of triplicate experiments. **, P < 0.01 two tailed student's t test. “See also Figure S4”
Phosphorylation is a major posttranslational modification of the DNA damage response pathway, and has been shown to enhance protein stability and activity. For example, p53 is phosphorylated at Ser20 by the checkpoint kinase Chk2 after IR, which results in p53's dissociation from Mdm2 and its subsequent stabilization (Chehab et al., 2000; Hirao et al., 2000; Shieh et al., 2000). Therefore, we examined whether USP10 stabilization is regulated by ATM. As shown in Figure S4A–B, USP10 is less stable in ATM-deficient cells, suggesting that USP10 stability is regulated by ATM. We also observed DNA damage inducible interaction between USP10 and ATM (Figure S4C). These results implying that USP10 might be phosphorylated by ATM following DNA damage, which results in USP10 stabilization and relocalization. Indeed, following various genotoxic stresses (IR, UV or etoposide treatment), USP10 became phosphorylated at SQ/TQ motifs (Figure 6C and S4D, USP10 protein levels were equalized to specifically examine USP10 phosphorylation in experiments of Figure 6C–D, 6G–H, S4D and S4F–G), which are consensus phosphorylation sites for PI3-kinase like kinases (PIKKs), such as ATM, ATR, and DNA-PK (Abraham, 2001). Both pan-PIKK inhibitor caffeine and ATM inhibitor Ku55933 inhibited USP10 phosphorylation after DNA damage (Figure 6C). These results suggest that ATM regulates USP10 phosphorylation after DNA damage. We further confirmed the role of ATM in USP10 phosphorylation using ATM+/+ or ATM−/− cells. As shown in Figure 6D, USP10 failed to be phosphorylated at the SQ/TQ motifs in ATM−/− cells. Furthermore, USP10 levels did not increase following DNA damage in ATM−/− cells (Figure 6E). These results suggest that USP10 is phosphorylated by ATM following DNA damage, which contributes to its stabilization.
We next set out to determine the ATM phosphorylation sites of USP10. ATM specifically phosphorylates SQ/TQ motifs, of which there are two candidate sites in USP10: T42Q and S337Q. We found that mutation at either T42 or S337 partially affects USP10 stabilization and mutating both T42 and S337 (TS/AA) abolished USP10 stabilization following DNA damage (Figure 6F and S4E). Mutation of both T42 and S337 (TS/AA) also abolished USP10 phosphorylation by ATM (Figure 6G). To confirm these results, we generated phosphorylation site-specific antibodies against p-T42 and p-S337. As shown in Figure 6H and S4F–G, both T42 and S337 sites were phosphorylated following DNA damage and Ku55933 or T42/S337 mutations inhibited USP10 phosphorylation after DNA damage. These results suggest that T42/S337 are physiological sites for ATM in vivo. In addition, the TS/AA mutant failed to translocate into the nucleus following DNA damage (Figure 6I and Figure S4H). On the other hand, a phospho-mimetic mutant of USP10-T42E/S337D (TS/ED) translocated to the nucleus in the absence of genotoxic stress (Figure 6I and Figure S4I), suggesting phosphorylation of USP10 by ATM is sufficient to induce USP10 translocation. These results suggest that ATM-mediated phosphorylation of USP10 is required for USP10 translocation and stabilization.
We further examined the functional significance of USP10 phosphorylation by ATM. HCT116 cells stably expressing USP10 shRNA were reconstituted with shRNA-resistant WT USP10 or USP10 TS/AA. As shown in Figure 6J, cells expressing the USP10 TS/AA mutant show defective p53 stabilization and poor induction of Bax and p21 after DNA damage. In addition, reconstitution with WT USP10, but not the USP10 phospho-mutant, restored DNA damage-induced apoptosis (Figure 6K). The defect of the TS/AA mutant is not due to an intrinsic defect in its ability to bind and deubiquitinate p53, since the TS/AA mutant is still able to bind p53 (Figure S4J–M) and can deubiquitinate p53 (Figure S4N). Overexpression of the TS/AA mutant in cells is also able to reverse Mdm2-induced p53 ubiquitination (Figure S4O). Therefore, failure to translocate to the nucleus and be stabilized is the likely underlying mechanism for USP10 TS/AA's failure to activate p53 in response to DNA damage. Overall, our results establish the important role of USP10 phosphorylation in p53 activation following DNA damage.
USP10 is downregulated in renal cell carcinoma
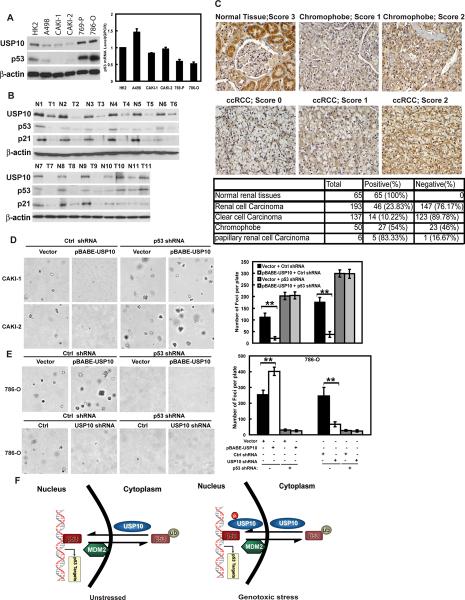
Since p53 is an important tumor suppressor that regulates cell proliferation and USP10 potentiates p53 function by deubiquitinating p53, it is possible that USP10 also acts as a tumor suppressor. Our results shown in Figures 4D–E demonstrate USP10's ability to inhibit cancer cell proliferation and lend support to the hypothesis that USP10 functions as a tumor suppressor in vivo. To further test this hypothesis, we examined the expression of USP10 in a panel of renal cell carcinoma (RCC) cell lines. We chose RCC to study USP10 expression because a very low percentage of RCC cases has been found to have p53 mutations (Soussi et al., 2000) and IARC p53 Database: www.IARC.fr/p53/index.html). Given the important function of p53 in tumor suppression, it is likely that the p53 pathway is compromised in RCC through other mechanisms, such as the downregulation of USP10. Indeed, we found that USP10 expression is significantly decreased in several RCC cell lines including A498, Caki-1 and Caki-2 cells, all of which contain WT p53 (Figure 7A). p53 expression was also lower in these cells than that of normal renal cells. This decreased p53 expression is not due to decreased p53 transcript (Figure 7A), and could be increased by MG132 treatment (Figure S5A), suggesting that p53 levels are regulated by the proteasome pathway in these cells. Interestingly, in RCC cell lines with mutant p53 (769-P, 786-O), USP10 as well as p53 levels were increased (Figure 7A). USP10 levels were also decreased in majority of fresh frozen RCC tissues compared to corresponding normal tissues (Figure 7B). The RCC samples with USP10 downregulation all contain WT p53 gene (T1–T9), although p53 levels were decreased. The decreased p53 expression is not due to Mdm2/Mdmx overexpression (Figure S5B). The Arf gene also does not show mutations in these tumor samples (data not shown). These results suggest that downregulation of USP10 might be an alternative way to suppress p53 activity in RCC. Interestingly, USP10 is overexpressed in some RCC tissues, and these tissues also contain mutant p53 (T10, T11). These results suggest that increased USP10 level in mutant p53 background might be beneficial to tumor growth. We further examined the expression of USP10 using RCC tissue microarray. The specificity of the USP10 antibody for immunohistochemistry was confirmed in Figure S5C. We scored the staining of USP10 from 0–3, and arbitrarily designated staining score 0–1 as negative and 2–3 as positive. Representative staining and scores were shown in Figure 7C. To confirm these results, we also used another USP10 antibody (Figure S5D). Strikingly, both two USP10 antibodies staining showed that close to 90% of clear cell carcinoma show negative staining of USP10. p53 staining was mostly negative in clear cell carcinoma (Figure S5D). About 50% of chromophobe and 20% of papillary RCC show negative USP10 staining (Figure 7C and S5D). These results suggest that USP10 is downregulated in RCC cases, especially clear cell carcinoma.
Figure 7. USP10 is downregulated in renal cell carcinoma.
(A) Expression of USP10 and p53 in Human renal tubular epithelial cell line (HK-2) and renal cell carcinoma (RCC) cell lines. Right panel shows p53 transcript level of HK-2 and RCC cells. Error bar represent SEM of triplicate experiments. (B) 11 pairs of fresh frozen RCC tissues and corresponding normal tissues were lysed, and cell lysates were blotted with indicated antibodies. (N: normal tissue; T: tumor tissue) (C) Immunohistochemical staining of USP10 in normal renal tissues and renal cell carcinoma. Lower table: quantification of USP10-positive or USP10-negative renal cell carcinoma cases. (ccRCC: clear cell Renal Cell Carcinoma). (D–E) Soft agar colony-formation assay was performed using CAKI-1 CAKI-2, or 786-O cells stably expressing indicated constructs. Right panels: quantification of colonies formed in soft agar. Error bar represent SEM of triplicate experiments. **, P < 0.01 two tailed student's t test. (F) The working model of p53 regulation by USP10. “See also Figure S5”
To confirm the role of USP10 in tumor suppression, we reconstituted USP10 in RCC cells with USP10 downregulation, and examined tumor cell growth using soft agar assay. Reconstitution of USP10 in CAKI-1 and CAKI-2 (WT p53 cell lines) restored p53 expression and increased p21 and BAX expression (Figure S5E). Furthermore, colony formation and cell proliferation were inhibited with USP10 reconstitution (Figure 7D and data not shown). Downregulation of p53 abolished the inhibitory effect of USP10 on tumor growth (Figure 7D and data not shown), confirming that USP10 inhibits cancer cell growth by stabilizing p53.
USP10 is overexpressed in RCC cell lines and tissues with mutant p53, correlating with increased p53 levels (Figure 7A–B). This is consistent with the observation that mutant p53 is often overexpressed in many cancers. To confirm that the mutant p53 is still subjected to Mdm2 and USP10 regulation, we investigated mutant p53 ubiquitination. As shown in Figure S5F, expression of Mdm2 enhanced p53 ubiquitination in 786O cells, suggesting Mdm2 targets mutant p53 for degradation. This is consistent with an recent interesting report that mutant p53 level is regulated by Mdm2 (Terzian et al., 2008). Coexpression of USP10 together with Mdm2 decreased mutant p53 ubiquitination (Figure S5F), while downregulation of USP10 increased mutant p53 ubiquitination (Figure S5G), suggesting that USP10 also regulates mutant p53 ubiquitination. Furthermore, expression of USP10 increased mutant p53 stability (Figure S5H), while downregulation of USP10 decreased mutant p53 stability (Figure S5I), suggesting that USP10 regulates mutant p53 stability.
Since mutant p53 is often dominant and displays gain of function, increased p53 levels could be advantageous to cancer. We reasoned that in contrast to cells with WT p53, increased expression of USP10 in mutant p53 background would be beneficial to cancer cell proliferation. Indeed, increased expression of USP10 in 786-O cells, which contain mutant p53, resulted in increased colony formation and cell proliferation (Figure 7E and data not shown), which could be reversed by p53 depletion. Although the levels of mutant p53 increased in cells ectopically expressing USP10, the expression of normal p53 target genes was not affected (Figure S5E). These results suggest that p53 is not functioning normally and has oncogenic gain-of-function in 786O cells. Conversely, downregulation of USP10 inhibited cell proliferation (Figure 7E and data not shown). Again, mutant p53 level was decreased when USP10 was downregulated, but the expression of p21 and Bax was not affected (Figure S5J). These results suggest that USP10 regulates p53 and cancer cell proliferation in a context-dependent manner. In cancer cells with WT p53, USP10 acts as a tumor suppressor and activating USP10 might be a sensible therapeutic strategy. On the other hand, in cancer cells with mutant p53, USP10 could promote cancer cell growth, and inhibition of USP10 is called for.
Overall, our studies suggest that USP10 is a deubiquitinase for p53. In unstressed cells, USP10 localizes in the cytoplasm and regulates p53 homeostasis. Following DNA damage, a fraction of USP10 translocates to the nucleus and contributes to p53 activation (Figure 7F). USP10, through its regulation of p53, appears to play an important role in cancer biology.
Discussion
The current study identifies USP10 as a p53 deubiquitinase. Unlike HAUSP, a previously identified deubiquitinase for p53 and Mdm2, USP10 only interacts with p53, but not Mdm2. In addition, while the majority of HAUSP localizes in the nucleus, USP10 seems to be predominantly cytoplasmic in unstressed cells. Although ubiquitination of p53 has long been known to induce its nuclear export, whether or not the exported p53 could be recycled back to the nucleus was unknown. Here we demonstrate that USP10 is able to reverse Mdm2-mediated p53 nuclear export. This is the first time that a deubiquitinase has been demonstrated to deubiquitinate cytoplasmic p53. Therefore, Mdm2 induced p53 ubiquitination could be counteracted at two locations: either by HAUSP in the nucleus or USP10 in the cytoplasm.
We also show that following genotoxic stress, a fraction of USP10 translocates to the nucleus, and this translocation is required for the stabilization and activation of p53. Mechanistically, the phosphorylation of Thr42 and Ser337 of USP10 by ATM is required for USP10 translocation and stabilization. Currently, we do not know how the phosphorylation of USP10 affects its stability and localization. Neither do we know the relationship between USP10 translocation and stabilization. USP10 contains a potential nuclear localization sequence (NLS) and several Leu/Ile rich regions, which are potential nuclear export signals (NES). Treatment of cells with LMB induced USP10 accumulation in the nucleus, suggesting that USP10 is actively exported out of nucleus in unstressed cells. It is possible that USP10 phosphorylation by ATM hinders USP10 nuclear export and shields it from degradation in the cytoplasm. We will examine this further in the future.
There are several mechanisms have been shown to stabilize and activate p53 following DNA damage. The phosphorylation of p53 by ATM or Chk2 has been shown to inhibit Mdm2 binding (Banin et al., 1998; Chehab et al., 2000; Shieh et al., 2000; Shieh et al., 1997). Furthermore, ATM-dependent phosphorylation of Mdm2 and Mdmx could destabilize these proteins and compromise their activity (Khosravi et al., 1999; Maya et al., 2001; Meulmeester et al., 2005; Pereg et al., 2005; Stommel and Wahl, 2004). Our results that USP10 is important for p53 stabilization and activation following DNA damage provide another regulatory route. Our results suggest that p53 is still ubiquitinated by residual Mdm2/Mdmx activity following DNA damage (Figure S3A), and the nuclear translocation of USP10 could further boost p53 activation. Since p53 is such an important factor in the determination of cell fate, such multiple regulatory mechanisms ensure optimal stress response.
Through its action toward p53, USP10 could function as a tumor suppressor. We show downregulation of USP10 in a high percentage of renal cell carcinoma samples. Furthermore, we provide evidence that USP10 inhibits cancer cell proliferation in cells with WT p53. These results suggest that USP10 could suppress tumorigenesis. Since very low percentage of RCC cases are found to have p53 mutations (Soussi et al., 2000) and IARC p53 Database: www.IARC.fr/p53/index.html), decreased expression of USP10 could be another mechanism to inhibit p53 function in RCC.
Paradoxically, USP10 could promote cancer cell proliferation in mutant p53 background. We found USP10 is overexpressed in RCC cells with mutant p53. Recent papers also suggest that increased USP10 expression in some breast cancer and glioblastoma samples (Deng et al., 2007; Grunda et al., 2006), although it is not clear whether p53 is mutated in these samples. One mystery in the p53 field of research is that mutant p53 is usually overexpressed in tumor samples, however, the mechanism responsible for this phenotype is not clear. Although it was thought that mutant p53 is intrinsically stable, a recent paper suggest that downregulation of Mdm2 might be one mechanism (Terzian et al., 2008). In contrast to mice with WT p53 background, loss of the Mdm2 in mutant p53 background increases p53 expression and promotes tumorigenesis. The explanation is that mutant p53 has a gain of function, and loss of Mdm2 increases mutant p53 levels and further promotes tumor development. Consistent with this study, we show that increased USP10 expression in mutant p53 background increases p53 levels and promotes cancer cell proliferation, while downregulation of USP10 inhibits cancer cell growth. Therefore, increased expression of USP10 could be another mechanism responsible for increased mutant p53 expression in human cancers. Future studies, for example generating USP10 knockout mice, are needed to establish the physiological role of USP10 in tumorigenesis.
Experimental Procedures
Deubiquitination of p53 in vivo and in vitro
Ubiquitination of p53 was detected as described previously (Li et al., 2002). For the in vivo deubiquitination assay, transfected HCT116 cells were treated for 4hr with a proteasome inhibitor MG132 (50μM) before being harvested. The cell extracts were subjected to immunoprecipitation and western blot.
To prepare ubiquitinated p53 as the substrate for the in vitro deubiquitination assay, HEK293 cells were transfected with both HA-UB and FLAG-p53, or pCMV-Mdm2. After treatment as described above, ubiquitinated p53 was purified from the cell extracts with anti-FLAG-affinity column in FLAG-lysis buffer. Following extensive washing with the FLAG-lysis buffer, the proteins were eluted with FLAG-peptides (Sigma). The recombinant His-USP10 and USP10CA were expressed in BL21 cells and purified with the His-tag purification column (Novagen). For the in vitro deubiquitination assay, ubiquitinated p53 protein was incubated with recombinant USP10 in a deubiquitination buffer for 2 h at 37°C.
Apoptosis Assay
Cells were washed with PBS and fixed in 4% paraformaldehyde at room temperature and then stained with DAPI. The number of cells with nuclear morphology typical of apoptosis were analyzed by fluorescence microscopy and scored in at least 400 cells per sample by an analyst blinded to the sample groups.
Colony and Soft Agar Colony-Formation Assays
The soft agar colony-formation assay was performed as described (Shim et al., 1997). Cells were plated in 0.3% top agarose in 35mm dishes and cultured for two weeks. Colonies were counted at room temperature under a light microscope.
Tissue Microarray
The tissue arrays of Kidney cancer samples were purchased from US Biomax (KD 2083, KD991t, KD804, KD241, KD208t). Immunohistochemical staining of USP10 (dilution 1:500) was carried out using IHC Select® HRP/DAB kit (Cat. DAB50, Millipore). The immunostaining was scored by pathologists in a blinded manner. A four-tier grading system (0 = negative, 1 = weak, 2 = moderate and 3 = strong staining intensity) was used. The normal proximal convoluted tubule show strong cytoplasmic staining and served as an internal control for strong staining (score 3) (Figure 7C, normal tissue). The score of tumor tissue was determined as compared to the staining intensity of normal tubules on the same slide.
Statistical Analysis
Results are reported as mean ± SEM of three independent experiments. Comparisons were performed with a two-tailed paired Student's t test (**, p < 0.01).
Supplementary Material
Acknowledgement
We thank Drs Bert Vogelstein (Johns Hopkins University), Thomas Roberts (Dana-Farber Cancer Institute and Harvard Medical School), Peter Howley (National Cancer Institute), Tyler Jacks (MIT Center for Cancer Research) for providing reagents used in this study. This work was supported in part by grants from the Richard Schulze Family Foundation and Research Grant and NIH (CA130996).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL DATA Supplemental Data include Supplemental Experimental Procedures and 5 figures.
Reference
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15:2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S, Taya Y, Anderson CW, Chessa L, Smorodinsky NI, Prives C, Reiss Y, Shiloh Y, et al. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol. 2000;2:563–568. doi: 10.1038/35023500. [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks CL, Li M, Hu M, Shi Y, Gu W. The p53--Mdm2--HAUSP complex is involved in p53 stabilization by HAUSP. Oncogene. 2007;26:7262–7266. doi: 10.1038/sj.onc.1210531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G(1) by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. p following 486. [DOI] [PubMed] [Google Scholar]
- Deng S, Zhou H, Xiong R, Lu Y, Yan D, Xing T, Dong L, Tang E, Yang H. Over-expression of genes and proteins of ubiquitin specific peptidases (USPs) and proteasome subunits (PSs) in breast cancer tissue observed by the methods of RFDD-PCR and proteomics. Breast Cancer Res Treat. 2007;104:21–30. doi: 10.1007/s10549-006-9393-7. [DOI] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. Embo J. 1997;16:1519–1530. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman DA, Levine AJ. Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- Grunda JM, Nabors LB, Palmer CA, Chhieng DC, Steg A, Mikkelsen T, Diasio RB, Zhang K, Allison D, Grizzle WE, et al. Increased expression of thymidylate synthetase (TS), ubiquitin specific protease 10 (USP10) and survivin is associated with poor survival in glioblastoma multiforme (GBM) J Neurooncol. 2006;80:261–274. doi: 10.1007/s11060-006-9191-4. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, Liu D, Elledge SJ, Mak TW. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science. 2000;287:1824–1827. doi: 10.1126/science.287.5459.1824. [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci U S A. 1999;96:14973–14977. doi: 10.1073/pnas.96.26.14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Vousden KH. Proteolytic cleavage of human p53 by calpain: a potential regulator of protein stability. Mol Cell Biol. 1997;17:460–468. doi: 10.1128/mcb.17.1.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono-versus polyubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Lohrum MA, Woods DB, Ludwig RL, Balint E, Vousden KH. C-terminal ubiquitination of p53 contributes to nuclear export. Mol Cell Biol. 2001;21:8521–8532. doi: 10.1128/MCB.21.24.8521-8532.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchenko ND, Moll UM. The role of ubiquitination in the direct mitochondrial death program of p53. Cell Cycle. 2007;6:1718–1723. doi: 10.4161/cc.6.14.4503. [DOI] [PubMed] [Google Scholar]
- Maya R, Balass M, Kim ST, Shkedy D, Leal JF, Shifman O, Moas M, Buschmann T, Ronai Z, Shiloh Y, et al. ATM-dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev. 2001;15:1067–1077. doi: 10.1101/gad.886901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, Dirks RW, Jochemsen AG. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Nikolaev AY, Li M, Puskas N, Qin J, Gu W. Parc: a cytoplasmic anchor for p53. Cell. 2003;112:29–40. doi: 10.1016/s0092-8674(02)01255-2. [DOI] [PubMed] [Google Scholar]
- Pereg Y, Shkedy D, de Graaf P, Meulmeester E, Edelson-Averbukh M, Salek M, Biton S, Teunisse AF, Lehmann WD, Jochemsen AG, et al. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc Natl Acad Sci U S A. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–291. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, Dalla-Favera R, Dang CV. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94:6658–6663. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi TR, Zaika A, Moll UM. Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. Faseb J. 2002;16:420–422. doi: 10.1096/fj.01-0617fje. [DOI] [PubMed] [Google Scholar]
- Soncini C, Berdo I, Draetta G. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene. 2001;20:3869–3879. doi: 10.1038/sj.onc.1204553. [DOI] [PubMed] [Google Scholar]
- Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Dehouche K, Beroud C. p53 website and analysis of p53 gene mutations in human cancer: forging a link between epidemiology and carcinogenesis. Hum Mutat. 2000;15:105–113. doi: 10.1002/(SICI)1098-1004(200001)15:1<105::AID-HUMU19>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. Embo J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. Embo J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Stephen CW, Lane DP. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp Cell Res. 2001;270:66–77. doi: 10.1006/excr.2001.5314. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.