Abstract
Bone mineral density (BMD), a diagnostic parameter for osteoporosis and a clinical predictor of fracture, is a polygenic trait with high heritability. To identify genetic variants that influence BMD in different ethnic groups, we performed a genome-wide association study (GWAS) on 800 unrelated Southern Chinese women with extreme BMD and carried out follow-up replication studies in six independent study populations of European descent and Asian populations including 18,098 subjects. In the meta-analysis, rs2273061 of the Jagged1 (JAG1) gene was associated with high BMD (p = 5.27 × 10−8 for lumbar spine [LS] and p = 4.15 × 10−5 for femoral neck [FN], n = 18,898). This SNP was further found to be associated with the low risk of osteoporotic fracture (p = 0.009, OR = 0.7, 95% CI 0.57–0.93, n = 1881). Region-wide and haplotype analysis showed that the strongest association evidence was from the linkage disequilibrium block 5, which included rs2273061 of the JAG1 gene (p = 8.52 × 10−9 for LS and 3.47 × 10−5 at FN). To assess the function of identified variants, an electrophoretic mobility shift assay demonstrated the binding of c-Myc to the “G” but not “A” allele of rs2273061. A mRNA expression study in both human bone-derived cells and peripheral blood mononuclear cells confirmed association of the high BMD-related allele G of rs2273061 with higher JAG1 expression. Our results identify the JAG1 gene as a candidate for BMD regulation in different ethnic groups, and it is a potential key factor for fracture pathogenesis.
Main Text
Osteoporosis is a major public health problem, a disease characterized by low bone mass, poor bone quality, and an increased predisposition to fracture. BMD, an important clinical predictor of fracture, is a complex trait of strong genetic determination and is associated with a heritability estimate of 0.6 to 0.8.1–4 Some large cohort studies and meta-analyses have started to clearly delineate the association of certain polymorphisms.5–8 Several genome-wide association studies (GWAS) have recently confirmed some associated loci, although they demonstrate only a small effect size on BMD variation in the general population.9–13 It has also been suggested that genes predisposing to the risk of osteoporosis vary between different ethnic groups.14,15 Because BMD is a polygenic trait, it is probable that many additional associated variants or ethnic-specific loci remain to be identified. The identification of additional genes that determine BMD variation would therefore provide insight into the complex genetic architecture and pathogenetic mechanisms of osteoporosis and offer strategies for therapeutic development.
In this GWAS, a multistep genome-wide association strategy (Figure S1 available online) was adopted to identify additional and/or ethnic-specific genes and their variants underlying BMD variation. The association of genome-wide data was first studied in 800 unrelated Hong Kong Southern Chinese (HKSC) females with extreme BMD (discovery sample). The most promising single-nucleotide polymorphisms (SNPs) were then tested for replication in another 720 HKSC subjects with extreme BMD (first-step replication). An association between replicated SNPs and BMD variation was confirmed in five other independent cohorts including 17,378 subjects of European descent or Asian descent (second-step replication). Finally, a region-wide association study was conducted to detect the causal variant of identified gene in the pooled extreme sample of discovery cohort and first-step replication cohort (n = 1520), and the potential biological function of the gene variant was further validated.
The characteristics of population are shown in Table 1. In the discovery stage, 800 unrelated women with extreme high or low BMD were selected from a HKSC cohort with extreme BMD (n = 1520). These subjects were selected from a database (>7000 Southern Han Chinese volunteers) of the Osteoporosis Centre of the University of Hong Kong. The low-BMD subjects are defined as an individual having BMD Z-score ≤−1.28 at either lumbar spine (LS) or femoral neck (FN) (the lowest 10% of the total cohort), while high-BMD subjects are individuals with BMD Z-score ≥+1.0 at either site. Subjects who reported that they had diseases or environmental factors that may affect BMD and bone metabolism were excluded. The recruitment procedure and exclusion criteria have been detailed elsewhere.16 Subjects were excluded for having a history of chronic medical illness, premature menopause below age 40 years, malabsorption, major gastrointestinal operation, metabolic bone diseases, endocrine disorders including hyper- and hypothyroidism, or medications that may affect bone and calcium metabolism, as well as for taking hormonal replacement therapy, antiosteoporosis medications, and active vitamin D3 metabolites. BMD (g/cm2) at LS and FN was measured by dual-energy X-ray absorptiometry (DXA; Hologic QDR 4500 plus, Hologic Waltham, MA, USA) with standard protocol. The age-corrected and standardized BMD (mean 0, SD 1), termed BMD Z-scores, was generated for each gender. The in vivo precision of the machine was 1.2% and 1.5% for LS and FN BMD, respectively.17
Table 1.
Description of Studied Populations
|
Discovery Cohort |
Replication Cohorts |
|||||||
|---|---|---|---|---|---|---|---|---|
|
First Step |
Second Step |
|||||||
|
Hong Kong Southern Chinese Discovery Cohort |
Hong Kong Southern Chinese First-Step Replication Cohort |
Hong Kong Osteoporosis Study Prospective Cohort |
Northern Chinese Population |
|||||
| Group | High BMD | Low BMD | High BMD | Low BMD | BMD | Nonfracture | Fracture | BMD |
| Number | 376 | 424 | 264 (168) | 456 (124) | 1881 | 1637 | 244 | 1584 |
| Age (years) | 46.6 ± 14.8 | 51.1 ± 15.9 | 48.3 ± 16.3 | 49.0 ± 16.0 | 62.3 ± 8.9 | 60.7 ± 8.2 | 67.4 ± 9.9 | 56.2 ± 11.3 |
| Height (m) | 1.58 ± 0.06 | 1.53 ± 0.07 | 1.65 ± 0.09 | 1.57 ± 0.08 | 1.52 ± 0.06 | 1.53 ± 0.06 | 1.50 ± 0.06 | 1.60 ± 0.06 |
| Weight (kg) | 61.1 ± 9.9 | 49.1 ± 6.7 | 67.1 ± 11.4 | 52.1 ± 9.5 | 55.4 ± 9.5 | 55.7 ± 9.5 | 54.1 ± 9.3 | 59.5 ± 9.4 |
| BMD Z-Score | ||||||||
| Lumbar Spine | 1.05 ± 0.78 | −1.59 ± 0.53 | 1.36 ± 0.89 | −1.34 ± 0.72 | −0.43 ± 1.18 | −0.39 ± 1.18 | −0.72 ± 1.17 | −0.05 ± 1.24 |
| Femoral Neck | 1.15 ± 0.79 | −1.36 ± 0.60 | 1.01 ± 0.84 | −1.16 ± 0.69 | −0.35 ± 0.97 | −0.29 ± 0.97 | −0.78 ± 0.93 | 0.71 ± 0.92 |
Data are expressed as mean ± standard deviation. The subject number is total number (the number of male subjects, if applicable, is given in parentheses) in each cohort. A total of 1520 subjects with extreme BMD phenotype were selected from a growing database of Hong Kong Southern Chinese (more than 7000 volunteers). The low BMD subjects are defined as an individual having BMD Z-score ≤ −1.28 at either lumbar spine (LS) or femoral neck (FN), which is equivalent to the lowest 10% of the total cohort, while high BMD subjects are individuals with BMD Z-score ≥ +1.0 at either of the two skeletal sites. First, a subset of 800 women was selected from the Hong Kong Southern Chinese (n = 1520) as the discovery cohort, and then the remainder of individuals (n = 720, 59.4% women) of the extreme population were used for the first-step replication.
The discovery sample was genotyped via the Infinium assay (Illumina, San Diego, CA) with Human610-quad chip, including 564,214 SNPs. PLINK (version 1.04) was used for the data management and quality control statistics.18 785 individuals and 488,853 SNPs were retained for analysis after the exclusion based on strict quality-control criteria. Subjects were excluded according to the following criteria: (1) genotyping call rate less than 95% (n = 5); (2) autosomal heterozygosity less than 27% or more than 31% (the same five subjects with low genotyping call rate); (3) being related or identical to other individuals in the sample (n = 7); and (4) discordance of observed gender and estimated sex (n = 3). SNPs were excluded if (1) genotyping call rate was 95% or less (1158 SNPs); (2) Hardy-Weinberg equilibrium (HWE) p value was less than 1.0 × 10−4 (904 SNPs); (3) minor allele frequency (MAF) was less than 0.01 (73,589 SNPs). Finally, the average genotyping call rate of those retained SNPs was 99.91%.
In the genome-wide association (GWA) scan, the association tests of SNPs with standardized BMD were implemented via PLINK (version 1.04). For each SNP, the asymptotic p value for the relationship between the number of minor alleles and BMD was derived from a two-sided t-statistic assuming the minor allele having an additive effect. Each SNP was tested for its association with BMD at both LS and FN. We used the genomic control19 and EIGENSTRAT20 methods for detecting possible bias resulting from population substructure and stratification. Quantile-quantile plots were constructed via R-2.5.2 for Windows. The entire genome association data and the LD block structure of genomic region were plotted via Haploview v4.1.21
A two-step replication method was used in this study to validate the top SNPs identified from the GWA scan. SNPs from the GWA scan that had a false-positive report probability of less than 0.5 (corresponding to a p value threshold of 4.75 × 10−5 or less) as suggested by Wacholder et al. (2004)22 and the fact that one in 5000 SNPs is hypothesized as a true positive (according to results from previous GWAS10,23) were used for the first-step replication. Furthermore, SNPs with p value of less than 0.05 in the first-step replication and also those reaching genome-wide significance in the pooled analysis conducted in the 1520 HKSC subjects with extreme BMD were further validated in five independent cohorts, including the Hong Kong Osteoporosis Study prospective cohort (n = 1881 women), a Northern Chinese population (n = 1584 women), and three in silico replication cohorts: the Framingham Osteoporosis Study (FOS, n = 3569), the deCODE Genetics Study (dCG, n = 7610, ∼85.6% women), and the TwinsUK Study (TUK, n = 2734 women). In this GWA, the p value threshold for GWA significance was 1.02 × 10−7 or less (Bonferroni correction, 0.05 divided by 488,853 SNPs).
The first-step replication sample included the remainder subjects with extreme BMD (428 females and 292 males) from the same HKSC extreme cohort, who we did not included in the discovery phase. SNPs were genotyped with the high-throughput Sequenom genotyping platform (Sequenom, San Diego, CA). Genotyping was performed blind to the phenotype status of the samples and was repeated in 5% of the samples for verification and quality control. The association analyses were the same as those used for the discovery cohort. Then the joint analysis were conducted in the pooled subjects with extreme BMD including the HKSC discovery cohort and first-step replication cohort (n = 1520).
The three in silico replication cohorts have been well described previously. The FOS is a community-based, longitudinal, prospective cohort comprising three generations of individuals in multigenerational pedigrees and additional unrelated individuals.9 The dCG is a population-based sample for genetics study of complex diseases.11,12 The TUK is a population-based sample of twins from the UK studying the hereditary basis of a variety of age-related traits and diseases.10 BMD was measured in the three cohorts at LS and FN via DXA with standard protocols (Lunar DPX-L for FOS, Hologic QDR4500A for dCG, and Hologic QDR2000W for TUK). The three cohorts were genotyped with the Illumina Infinium HumanHap300 or HumanHap550 or HumanCNV370 Beadchip (TUK and dCG) or the Affymetrix 500K mapping array plus 50K supplemental array (FOS).
For the FOS cohort, sex- and cohort (Original and Offspring)-specific standardized residuals (mean 0, SD 1) of phenotypes were calculated. GWAS was performed with linear mixed effects regression models, with fixed SNP genotype effects, and random individual effects that correlate within pedigree according to kinship relationship.24 To account for the effect of population substructure, age- and weight-corrected BMD were also adjusted for the first four principal components from the EIGENSTRAT analysis. For the dCG and TUK cohorts, age- and weight-corrected BMD was computed for each sex separately to have a mean 0 and standard deviation 1. Single SNP association tests were performed with an additive genetic effect model that estimated the effect of one copy increment of the reference allele for all three cohorts. The p values were also corrected for genomic control in the dCG cohort.
Hong Kong Osteoporosis Study prospective cohort: This population is also a part of the HKSC database with both BMD and fracture data. A total of 1881 unrelated postmenopausal women were prospectively followed for incident fractures.25,26 The duration of follow-up corresponded to the time from baseline to the occurrence of fracture, death, or the last follow-up visit data. A low-traumatic fracture was defined as a fracture sustained after a fall from standing height or less. All nonosteoporotic site fractures (nose, toe, head, jaw, skull, and hands) and traumatic fractures were excluded. Radiographic morphometric vertebral fractures were defined by a reduction in vertebral height of at least 3 standard deviations in anterior, mid, or posterior ratios compared with normative means.27 Fractures were verified by retrieval of the X-ray report and hospital discharge summaries from the computerized patient record system of the Hospital Authority of the Hong Kong Government.
The Northern Chinese population: 1584 female Northern Chinese aged 20 years or above were recruited from three osteoporosis centers (two in Shanghai and one in Beijing) by posters. All the study subjects are residents of Shanghai or Beijing and are Chinese of Han origin. Patients with chronic diseases and conditions that might potentially affect bone mass and bone metabolism were excluded with the same exclusion criteria as the HKSC population.16,28,29 The DXA machine (GE-Lunar Corp. or Hologic Inc.) was used to measure BMD of the LS and FN via standard protocols. These two Chinese second-step replication cohorts were genotyped with the TaqMan system (Applied Biosystems, Foster City, CA). The statistical procedures of associations between SNP and BMD variation were the same as those used for the HKSC discovery cohort, with the addition of an “osteoporosis center” variable also included as a covariate in the association analysis of Northern Chinese population to eliminate the potential effect of subjects being recruited from different centers. The logistic regression model was used to examine the association of SNP with fracture risk by adjusting for covariates such as age and follow-up duration. Skeletal site-specific analyses were also conducted for spine and hip fractures. These statistical analyses were performed with SPSS v.16 (SPSS, Chicago, IL).
In all studies, all subjects gave informed consent and the study protocols were reviewed and approved by local institutional review boards.
The meta-analysis of SNP with BMD variation in (1) all of the Chinese populations used for the second-step replication (n = 3,465); (2) all of the second-step replication subjects of European descent (n = 13,913); and (3) all of the studied subjects from seven independent populations (n = 18,898) were performed via the inverse variance meta-analysis approach.
Furthermore, to study the association of the identified gene with BMD in more detail and to detect the functional variant, a region-wide and tagSNP-based method followed by imputation and sequencing was conducted.
Selected SNPs were genotyped with the high-throughput Sequenom genotyping platform (Sequenom, San Diego, CA) in all of the HKSC subjects with extreme BMD (n = 1520). SNPs with genotyping call rate < 90% and HWE p < 0.001 were excluded. Both single marker- and LD block-based haplotype association analyses with BMD were performed with the PLINK (version 1.04). The single SNP analysis was the same as that used for the HKSC discovery cohort. The conditional haplotype testing model was also used to assess the correlation structure and identify the most promising candidate causal variant(s).30
The imputation of genotypes for all untyped SNPs from HapMap in the studied genomic region was conducted with the IMPUTE program.31 We used the release 27 HapMap Chinese (CHB [Chinese Han Beijing] and CHD [Chinese in Denver]) data as the reference panel. The IGG program32 was used for the integration of HapMap data and the exportation of IMPUTE format.
Sequencing was conducted in the coding region of identified gene and its ∼500 bp flanking genomic sequence in 25 high-BMD subjects who carried the protective allele of the identified SNP and 25 low-BMD subjects who carried the risk allele.
In addition, to test the best-fitted genetic model for the identified SNP, the likelihood ratio test of the additive or dominant or recessive regression model (1 df) against the full 2 df model was performed. The interaction with sex, age (<50 and >50 years), and menopause was tested with regression models.
Finally, the potential biological function of the identified gene variant was studied. The FASTSNP program33 was used for predicting the function of SNPs of interest. An electrophoretic mobility shift assay (EMSA) was conducted to evaluate the predicted function. The gene expression in primary human bone-derived cell cultures (HBDCs) and human peripheral blood mononuclear cells (PBMCs) were compared among three genotype groups of identified SNP.
The results of GWA scan were as follows. 488,853 SNPs were studied for their associations with BMD at LS and FN in 800 unrelated HKSC women with extreme BMD. Quantile-quantile plots (Figure 1) revealed the presence of a number of SNPs associated with BMD at LS and FN. A substantial fraction of the most strongly associated SNPs could have true associations with BMD. There were many more significant associations than expected by chance. For example, we observed 110 signals with p value less than 1 × 10−4, where we would expect less than 50 signals under the null distribution. Population stratification was unlikely to be a key confounding factor influencing BMD association in this GWA scan analysis. First, the HKSC discovery subjects were selected from a relatively homogenous and large population belonging to the same geographical area. Second, the peak association signals persisted after the adjustment for residual population structure with EIGENSTRAT (Figure S2). Third, the respective genomic inflation factor (λ) was 1.007 and 1.003 for BMD at the LS and FN, indicating no significant population stratification.
Figure 1.
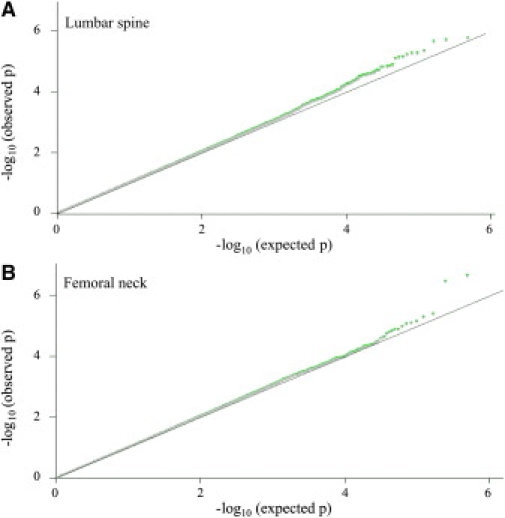
Quantile-Quantile Plots of Discovery Test Statistics for the Genome-wide Associations of Single Nucleotide Polymorphisms with Bone Mineral Density
y axis is the observed –log10 (p) values. x axis is the expected –log10 (p) values under the null distribution of no association for studied genome-wide SNPs, which would be expected to follow the solid black line. (A) BMD at lumbar spine (LS); (B) BMD at femoral neck (FN). The green and gray dots are p values without and with correction of the factor for chi-square statistics of the genomic control method (λ = 1.007 for LS BMD and λ = 1.003 for FN BMD), respectively.
In the GWA scan, a total of 62 SNPs with 42 for LS and 27 for FN (Figure S3) exceeded the false-positive report probability and were used for the first-step replication. Altogether these SNPs represented 40 independent signals (r2 < 0.2). Among these 62 SNPs, p values of 6 SNPs were less than 0.05 (with the consistent direction of effect as the GWA data) in the first-step replication (Table S1). Of these 62 SNPs, the association of rs2273061 in the Jagged1 (JAG1) gene (minor allele G) with high BMD reached the GWA significance with p value (β) being 1.06 × 10−8 (0.34) and 1.83 × 10−6 (0.25) for BMD at LS and FN, respectively, in the joint analysis of HKSC discovery and first-step replication subjects with extreme BMD (n = 1520). The SNP rs2273061 was then used for the second-step replication. Within the JAG1 gene region, several SNPs showed strong associations with BMD at LS and/or FN in the GWA scan (Figure S4). Two (rs2273061 and rs6133987) of them had p value less than 4.75 × 10−5 for BMD in the discovery cohort. These two SNPs were in a moderate LD (D′ = 0.8, r2 = 0.2).
Second-step replication studies verified the significant finding of rs2273061 of JAG1 gene with BMD in the initial GWA analysis (Table 2). The in silico replication in three independent cohorts of European descent consistently showed the same direction of the association with G of rs2273061 being associated with high BMD (all p < 0.05). The association of rs2273061 with BMD was also replicated in the Hong Kong Osteoporosis Study prospective cohort (p = 0.0025, β = 0.12 for LS and p = 0.0045, β = 0.09 for FN, n = 1881). Although the replication study in the Northern Chinese population (n = 1584) failed to reach statistical significance (p > 0.05), which is probably due to the small sample size, the results also suggested that the allele G was related to high BMD. So far, the loci or variant reported to be associated with osteoporosis generally have small effects on BMD variation.9–13 The regression coefficient estimated for BMD on genotype (β) is biased because of selection for extremes in BMD. To overcome this, we estimated the regression coefficient of genotype on BMD (β′) because selection on the independent variable does not bias the regression coefficient.34 To eliminate the potential effect of “winner's curse,” we used the data from the first-step replication cohort instead of the discovery cohort for estimation. The estimated β = β′ × variance (BMD Z-score)/2p(1 − p), where β′ is the regression coefficient of using phenotype as the covariate and genotype as the response, the variance of phenotype is from the total HKSC population and p is the frequency of reference allele. In our HKSC population, the allelic variance of JAG1 gene rs2273061 explained ∼1.3% of LS BMD variance and ∼0.5% of FN BMD variance. The smaller effect size of rs2273061 in populations of European descent may be due to a difference in allele frequency between Asians and Europeans, with the frequency of allele G of being approximately 0.31 and 0.60 in HKSC and cohorts of European descent, respectively. We believe that the functional genetic variation of rs2273061 might not be population specific but the difference in allele frequency of the variant allele might contribute to the observed racial differences in the prevalence of osteoporosis and osteoporotic fractures.
Table 2.
Summary of Association Results of Jagged 1 Gene rs2273061 in the Discovery Cohort and First- and Second-Step Replication Cohorts
|
BMD (Effect Estimated in SD) |
Fracturea |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Lumbar Spine |
Femoral Neck |
All |
Spine |
Hip |
|||||||||
| A1 | A2 | MAF | p | β | p | β | p | OR | p | OR | p | OR | |
| Discovery Stage | |||||||||||||
| Hong Kong Southern Chinese discovery cohort (n = 800) | Gb | A | 0.308 | 4.73E-05 | 0.320 | 5.36E-04 | 0.270 | NA | NA | NA | NA | NA | NA |
| First-Step Replication | |||||||||||||
| Hong Kong Southern Chinese first-step replication cohort (n = 720) | Gb | A | 0.319 | 9.06E-05 | 0.350 | 5.59E-03 | 0.180 | NA | NA | NA | NA | NA | NA |
| Second-Step Replicationc | |||||||||||||
| Hong Kong Osteoporosis Study prospective cohort (n = 1,881) | Gb | A | 0.307 | 2.50E-03 | 0.120 | 4.50E-03 | 0.090 | 9.00E-03 | 0.730 | 0.0630 | 0.780 | 1.60E-02 | 0.480 |
| Northern Chinese population (n = 1,584) | Gb | A | 0.346 | 0.1985 | 0.043 | 0.1180 | 0.043 | NA | NA | NA | NA | NA | NA |
| Framingham Study cohort (n = 3,569) | A | Gb | 0.405 | 7.02E-03 | 0.063 | 0.1050 | 0.032 | NA | NA | NA | NA | NA | NA |
| deCODE Genetics Study (n = 7,610) | A | Gb | 0.397 | 7.50E-03 | 0.051 | 4.30E-02 | 0.036 | NA | NA | NA | NA | NA | NA |
| TwinsUK Study cohort (n = 2,734) | A | Gb | 0.407 | 4.50E-02 | 0.052 | 0.0742 | 0.045 | ||||||
| Meta-analysis | |||||||||||||
| Total second-replication Chinese (n = 3,465) | 1.24E-02 | 0.088 | 1.49E-02 | 0.067 | NA | NA | NA | NA | NA | NA | |||
| Total second-replication subjects of European descent (n = 13,913) | 2.63E-04 | 0.055 | 2.14E-02 | 0.037 | NA | NA | NA | NA | NA | NA | |||
| Total subjects (n = 18,898) | 5.27E-08 | 0.072 | 4.15E-05 | 0.054 | NA | NA | NA | NA | NA | NA | |||
Abbreviations: A1, minor allele; A2, major allele; MAF, minor allele frequency; β, regression coefficient;
OR, odds ratio; NA, not available.
The results were adjusted for age and follow-up duration.
The reference allele. p values are two-sided.
p values of BMD listed in the second-step replication cohorts were one-sided p value.
Meta-analysis of rs2273061 in subgroups and all seven populations also supported the above association of the JAG1 gene with BMD variation (Table 2). The p values (β) for the association for all 18,898 subjects were 5.27 × 10−8 (0.07) for BMD at LS and 4.15 × 10−5 (0.05) for BMD at FN, respectively.
Furthermore, as shown in Table 2, the fracture trait results supported the associations of rs2273061 with osteoporotic fractures (p = 0.009, OR = 0.7, 95% CI 0.57–0.93), and the association remained significant even with site-specific analysis of fracture at the spine (p = 0.063, OR = 0.8, 95% CI 0.60–1.01) or at the hip (p = 0.016, OR = 0.5, 95% CI 0.26–0.87). The allele G was associated with lower risk of fracture, which is consistent with its association with higher BMD. The significance was partly but not completely attenuated if BMD was added as a covariate in the model of non-site-specific analysis (p = 0.031, OR = 0.8, 95% CI 0.58–0.97), suggesting that the effect of this variant on fracture risk is mediated through both BMD-dependent and -independent mechanisms. This finding is in agreement with the current understanding that bone strength is not being entirely explained by BMD and that other phenotypes such as bone size and geometry are important determinants of bone strength and fracture risk independently of BMD.35
Sequencing of the coding region of the JAG1 gene and its ∼500 bp flanking genomic sequence in 25 high-BMD and 25 low-BMD subjects carrying the variant allele of rs2273061 did not identify any new variants or mutations, although with this limited sample size we cannot exclude the existence of rare variants with large effects.
In the region-wide association analysis, a total of 48 SNPs were genotyped in the 20 kb flanking regions of JAG1 gene (Table S2), including 27 tagSNPs selected based on the HapMap Phase II CHB data with r2 > 0.8; 17 SNPs, potentially functional, or located in 2 kb flanking regions; 2 SNPs with p value less than 1.0 × 10−3 in this discovery phase; and 2 SNPs in high LD with rs2273061 (r2 > 0.8) based on the HapMap CHB data. Nine SNPs located in exons or within 2 kb flanking regions were monomorphic in the HKSC population. 39 SNPs with genotyping call rate > 90% and HWE p values > 0.001 were used for statistical analysis.
The single marker association analysis showed that the strongest association evidence was from the LD block 5 that included seven SNPs located from intron 3 to intron 4 having p values less than 1.0 × 10−4 (Figure 2). Three SNPs showed very strong significant associations, with two (rs2273061 and rs7265068) reaching GWA significance (p < 1.02 × 10−7) and the third (rs6514116) almost reaching the significance threshold (p = 1.53 × 10−7) for LS BMD (Table S2). These three SNPs are in high LD in individuals of both Chinese (r2 > 0.9) and European (r2 > 0.7, CEU) descent. In silico replication of rs6514116 and rs7265068 confirmed the same direction of association with BMD in the three replication cohorts of European descent (p < 0.05). In the haplotype analyses, the most significant association signals were detected in LD block 5 of the JAG1 gene with p value being 8.52 × 10−9 for LS and 3.47 × 10−5 for FN (Table S3). The haplotype data corresponded to the single marker analysis, but did not add further information to outcome. The conditional haplotype analysis revealed that the global association was largely attenuated (even disappeared) when conditioned on any of the most significant three SNPs (p = 0.0168 for LS and p = 0.2804 for FN), which may suggest that the significant haplotype associations were mostly derived from the effects of those three SNPs.
Figure 2.
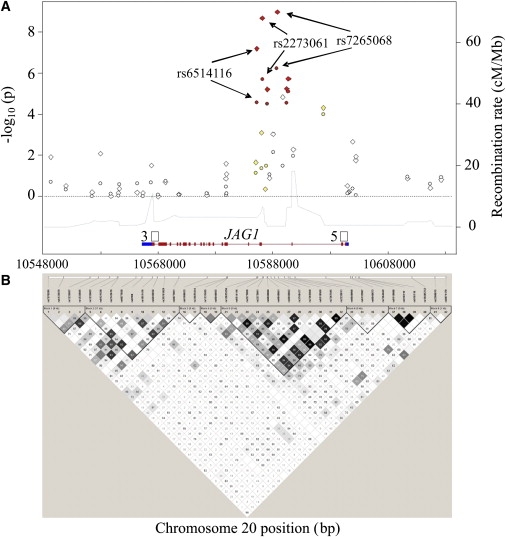
Characteristics for the 20 kb Downstream and Upstream Regions around the Jagged 1 Gene
(A) The fine scale recombination rate (right scale) across the studied region (HapMap rel22_B36); and the −log10 (p) values (left scale) plot for single marker association results of BMD at lumbar spine (LS, diamond dots) and femoral neck (FN, circle dots). The colors of dots are coded according to the degree of linkage disequilibrium (LD) with SNP rs2273061 identified originally in the genome-wide association discovery stage (r2 ≥ 0.8 red; 0.5 ≤ r2 < 0.8 orange; 0.2 ≤ r2 < 0.5 yellow; r2 < 0.2 white). The solid horizontal line at the bottom represents the JAG1 gene region (NCBI_B36) and its vertical bars represent exons.
(B) LD plot for the studied region based on the r2 statistic. Blocks connecting SNP pairs are shaded based on the LD strength between SNPs by using the disequilibrium coefficient r2, which are ranged from 0 (white) to 1.0 (black).
Subsequently, the imputation analysis further supported that the three SNPs rs6514116, rs2273061, and rs7265068 appeared consistently with the strongest association signals with BMD variation (Figure S5). All together, 82 SNPs in this studied region were included in the association analyses. Of these 82 SNPs, 43 were imputed SNPs that are extracted from the HapMap database with MAF ≥ 0.01 and 39 were genotyped SNPs in this study.
Additional analyses revealed that: (1) the additive model (1 df) best fitted the data for all of rs6514116, rs2273061, and rs7265068 (p > 0.8) and neither the dominant nor recessive (p < 0.01) model yielded an adequate fit when tested against the full 2 df model; (2) effects on BMD were not significantly different between females and males (p > 0.1); in subjects of age > 50 years compared to those aged < 50 years (p > 0.1); and for women before or after menopause (p > 0.1); (3) the associations of these SNPs with BMD persisted even with the adjustment of height and weight: the p values of rs6514116, rs2273061, and rs7265068 were 6.34 × 10−5, 5.76 × 10−6, and 2.41 × 10−6 for LS and 0.0270, 0.0053, and 0.0024 for FN, respectively.
The bioinformatics analyses suggested that the allele change (A to G) at rs2273061 located in intron 3 could generate one binding site for the transcription factor c-Myc (v-myc myelocytomatosis viral oncogene homolog [avian] [MIM 190080]; Figure S6). To confirm this, an electrophoretic mobility shift assay (EMSA) was performed with recombinant c-Myc protein (Abnova Corporation) and double-stranded oligonucleotide probes (Sigma-Aldrich Corp., St. Louis, MO). The EMSA results verified the specific binding of c-Myc to the oligonucleotide sequence containing G but not A nucleotide of rs2273061 (Figure 3). c-Myc is a helix-loop-helix (HLH) transcription factor that is also expressed in osteoblasts and chondrocytes.36,37 Studies found that the c-Myc transcript levels remained almost constant during the differentiation of murine preosteoblast MC3T3-E1 cells and that the forced expression of c-Myc can significantly increase the alkaline phosphatase expression and osteocalcin gene expression.38 Furthermore, we documented a significant relation between rs2273061 genotype and mRNA expression of JAG1 in human bone-derived cells (Figure 4A) and peripheral blood mononuclear cells (Figure 4B) with AA subjects having lower JAG1 mRNA expression than GG subjects. Quantitative real-time PCR studies from peripheral blood mononuclear cells revealed that the subjects with GG genotype had approximately 30-fold higher JAG1 mRNA expression levels than did subjects with AA genotype (p = 0.037, Figure 4C). However, the function of rs2273061 on JAG1 gene expression in osteoblasts will need to be further confirmed via chromatin immunoprecipitation (ChIP) assay and luciferase reporter assay. Further investigation including allelic-specific transcription study in heterozygotes for rs2273061 could help to confirm that the association observed is mediated by a difference in JAG1 expression.
Figure 3.
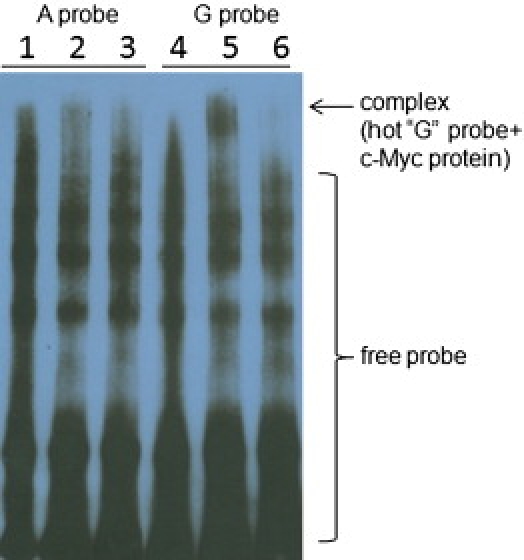
Electrophoretic Mobility Shift and Competition Assays with c-Myc Recombinant Protein and Allelic Variants of SNP rs2273061 in JAG1
The allele A probe, corresponding to JAG1 intron 3 sequences centering rs2273061 (underlined in the following sequences) was prepared by annealing of the Biotin-labeled oligonucleotide 5′-GACAACCTGTTACCACTTATTTACCTTCTTTA-3′ with the complementary sequence 5′-TAAAGAAGGTAAATAAGTGGTAACAGGTTGTC-3′; the allele G probe, prepared by annealing the Biotin-labeled oligonucleotide 5′-GACAACCTGTTACCACTTGTTTACCTTCTTTA-3′ with the complementary sequences 5′-TAAAGAAGGTAAACAAGTGGTAACAGGTTGTC-3′. Electrophoretic mobility shift assay (EMSA) was conducted with a commercial kit (Panomics, Fremont, CA). 1 ng of Biotin-labeled probe was incubated with 0.36 μg of c-Myc recombinant protein for 30 min at 15°C in a 10 μl reaction volume containing 2 μl 5 × binding buffer and 1 μg poly d(I-C). For competition reactions, the unlabeled probe was used at 660-fold molar excess of the labeled probe. After incubation, the samples were separated by electrophoresis on a 6% nondenaturing polyacrylamide gel with 0.5 × TBE buffer. DNA-protein complexes were electroblotted to Pall Biodyne B nylon membrane (Pall Corp., Pensacola, FL) and visualized by exposure to Chemiluminescent Detection Film (Agfa, Shanghai, China).
Lanes: 1, labeled A probe; 2, labeled A probe + c-Myc; 3, labeled A probe + c-Myc + unlabeled A probe; 4, labeled G probe; 5, labeled G probe + c-Myc; 6, labeled G probe + c-Myc + unlabeled G probe. Binding was not observed with oligonucleotide containing A allele (lane 2) but was present with oligonucleotide containing the G allele (lane 5). Binding to the G allele resulted in a complex that was specifically competed by unlabeled mutant probe containing the G allele (lane 6).
Figure 4.
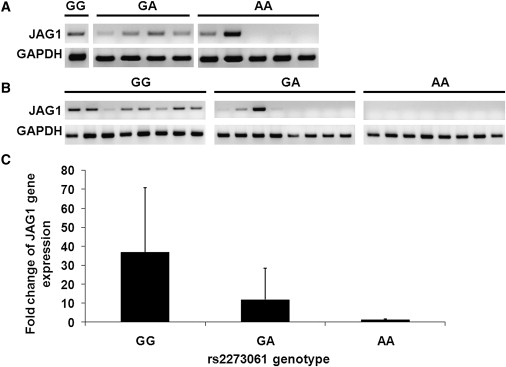
JAG1 Gene Expression in Subjects with Different rs2273061 Genotypes
(A and B) JAG1 mRNA expression normalized to GAPDH by reverse transcription PCR (RT-PCR) in human bone-derived cells (HBDCs, A) and peripheral blood mononuclear cells (PBMCs, B) from individual subjects with GG, GA, and AA genotype of rs2273061.
(C) The relative quantity of JAG1 mRNA expression normalized to ribosomal RNA by quantitative real-time PCR (q-PCR) in PBMCs from individuals with different rs2273061 genotypes (n = 20 per genotype, results are mean ± SD).
HBDCs and PBMCs from healthy subjects were isolated as described previously.46,47 Total RNA was extracted from HBDCs and PBMCs with Trizol reagent according to the manufacturer's instruction (Invitrogen, CA). 0.5 μg RNA was used for cDNA synthesis by M-MLV Reverse Transcriptase according to the manufacturer's protocol (Invitrogen, CA). (A) and (B) showed JAG1 gene expression with RT-PCR with primers (5′-TCGAGTTGGAGATCCTGTCC-3′) and (5′-GGGTGTGGGATGCACTTATC-3′) spanning exon 2 to 4 with annealing temperature at 59°C. Results were normalized to GAPDH gene expression with primers (5′-GCCTCCTGCACCACCACC-3′) and (5′-CCGTTCAGCTCAGGGATGA-3′). (C) showed q-PCR results with Taqman one-step PCR master mix kit with gene-specific target primers and 3-FAM probes of JAG1 gene (ABI gene expression assay ID number Hs00164982_m1). Each sample was simultaneously quantified with eukaryotic 18S primers and target probes (ABI gene expression assay 4319413E). Amplification was performed in an ABI Prism 7000 sequence detector, with standard settings of 40 cycles with an annealing temperature of 60°C. All samples were assayed in duplicates. Results were expressed as fold-change of JAG1 gene expression of GG and GA genotypes relative to AA genotype by comparative Ct method (−2ΔΔCT). q-PCR in PBMCs revealed the GG genotype group had approximately 30-fold higher JAG1 mRNA expression levels than the AA genotype group (p = 0.037, t test).
Recent studies have reported the involvement of the JAG1 gene in bone formation. The JAG1 gene encodes a cell surface protein—Jagged 1—belonging to the Delta/Serrate domain (DSL) family, which is the ligand binding to Notch receptors. Expression of JAG1 has been reported in osteoblastic cells in vivo and in vitro and during bone regeneration. Activation of JAG1 gene is associated with increased bone mineral deposition.38 Jagged1 protein was detected in trabecular osteoblasts and endosteal osteoblasts, but not in periosteal cortical osteoblasts, and its expression was increased with intermittent parathyroid hormone treatment.39
Mutations in the JAG1 gene are associated with the Alagille syndrome (AGS [MIM 118450]), a multisystem developmental disorder with autosomal dominant inheritance.40–42 Vertebral deformities and shortened vertebral body are reported in many patients with AGS.43 The association of AGS with vertebral deformities and the differential expression pattern of the JAG1 in trabecular rather than cortical osteoblasts39 is in line with the observed results in this study that the variants in JAG1 gene are more significantly associated with BMD variation at LS than FN, because spine comprises mostly trabecular bone whereas hip has 50% cortical and 50% trabecular bone.
Apart from JAG1, it is likely that many more sequence variations on BMD determination are yet to be identified. Besides the two SNPs in the JAG1 gene, four SNPs in two genomic regions were also replicated in the first-step replication population but did not reach a GWA significance of p < 1.02 × 10−7 in pooled analyses. These variants may account for some of the remaining heritability of BMD and deserve further investigation in a large population. Indeed, evidence for replication in some loci identified from the recently published large meta-analysis of GWAS of five populations of European descent44 was seen in our HKSC population, although they were not prominent in this GWA scan data. Of the reported 25 GWA significant SNPs in 20 loci, 14 SNPs in 10 loci, i.e., 50% of reported association signals observed in persons of European descent, were replicated in our HKSC discovery sample with a consistent direction of effect (p < 0.05; Table S4). With the most significant and independent SNP (r2 < 0.2) from these 10 replicated loci, the 10 SNPs combined explained ∼3.5% and ∼3.9% of the variance of BMD at LS and FN in the HKSC population, respectively. Together with the effect of JAG1 gene, ∼4.8% and ∼4.4% of the BMD variance at LS and FN was explained, respectively. Failure to replicate the other variants may have been due to differences in ethnicity or sampling methods or sample size. The selected extreme sampling strategy was adopted because it has a substantially greater statistical power over a random sample for detection of allelic association, but our discovery sample may have less power to detect variants with modest effect. Overall, the pooled discovery and first replication sample of 1520 subjects with extreme BMD have a 84% power to detect a variant with 1% effect on the BMD variation at a p < 1.0 × 10−7 significance level (assuming MAF = 0.3, LD = 1, Genetic power calculator).45
In conclusion, this study identified the JAG1 gene (20p11.23-p12.1) as a candidate for BMD regulation and a potential key factor for fracture pathogenesis. We also replicated some genomic loci/variants that have been reported to be associated with BMD variation. This study provides new insight into the complex genetic architecture of osteoporosis. The identified variants warrant further biological and clinical investigation.
Acknowledgments
This project is supported by Hong Kong Research Grant Council; The Bone Health Fund of HKU Foundation; and Matching Grant, CRCG Grant, and The Osteoporosis Research Fund of The University of Hong Kong. The Twins UK study was funded by the Wellcome Trust, European Community's Sixth and Seventh Framework Programmes (FP7/2007-2013), ENGAGE project HEALTH-F4-2007-201413, and the FP-5 GenomEUtwin Project (QLG2-CT-2002-01254). The study also receives support from the Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London. T.D.S. is an NIHR senior Investigator. The project also received support from a Biotechnology and Biological Sciences Research Council (BBSRC) project grant (G20234). The authors acknowledge the funding and support of the National Eye Institute via an NIH/CIDR genotyping project (PI, Terri Young). For genotyping of the Twins UK study, we thank the staff from the Genotyping Facilities at the Wellcome Trust Sanger Institute for sample preparation, Quality Control and Genotyping led by Leena Peltonen and Panos Deloukas; Le Centre National de Genotypage, France, led by Mark Lathrop; Duke University, North Carolina, USA, led by David Goldstein; and the Finnish Institute of Molecular Medicine, Finnish Genome Center, University of Helsinki, led by Aarno Palotie. The Framingham Osteoporosis Study (FOS) study was funded by grants from the US National Institute for Arthritis, Musculoskeletal and Skin Diseases and National Institute on Aging (R01 AR/AG 41398 to D.P.K. and R01 AR 050066 to D.K.). The Framingham Heart Study of the National Heart, Lung, and Blood Institute of the National Institutes of Health and Boston University School of Medicine were supported by the National Heart, Lung, and Blood Institute's Framingham Heart Study (N01-HC-25195) and its contract with Affymetrix, Inc. for genotyping services (N02-HL-6-4278). Analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. A portion of this research was conducted with the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The following are employees of deCODE Company, Iceland, and own stock or stock options of deCODE Company: U.S., B.V.H., U.T., and K.S.
Supplemental Data
Web Resources
The URLs for data presented herein are as follows:
Department of Twin Research and Genetic Epidemiology, http://www.twinsUK.ac.uk
EIGENSTRAT, http://genepath.med.harvard.edu/∼EIGENSTRAT.htm
FASTSNP program, http://fastsnp.ibms.sinica.edu.tw
Genetic power calculator, http://pngu.org/∼purcell/gpc/
Haploview program, http://www.broad.mit.edu/mpg/haploview
IGG program, http://bioinfo.hku.hk:13080/iggweb/
IMPUTE program, https://mathgen.stats.ox.ac.uk/impute/impute.html
The International HapMap project, http://www.hapmap.org
Online Mendelian Inheritance in Man (OMIM), http://www.nlm.nih.gov/Omim
References
- 1.Dequeker J., Nijs J., Verstraeten A., Geusens P., Gevers G. Genetic determinants of bone mineral content at the spine and radius: a twin study. Bone. 1987;8:207–209. doi: 10.1016/8756-3282(87)90166-9. [DOI] [PubMed] [Google Scholar]
- 2.Arden N.K., Baker J., Hogg C., Baan K., Spector T.D. The heritability of bone mineral density, ultrasound of the calcaneus and hip axis length: A study of postmenopausal twins. J. Bone Miner. Res. 1996;11:530–534. doi: 10.1002/jbmr.5650110414. [DOI] [PubMed] [Google Scholar]
- 3.Ng M.Y., Sham P.C., Paterson A.D., Chan V., Kung A.W. Effect of environmental factors and gender on the heritability of bone mineral density and bone size. Ann. Hum. Genet. 2006;70:428–438. doi: 10.1111/j.1469-1809.2005.00242.x. [DOI] [PubMed] [Google Scholar]
- 4.Ioannidis J.P., Ng M.Y., Sham P.C., Zintzaras E., Lewis C.M., Deng H.W., Econs M.J., Karasik D., Devoto M., Kammerer C.M. Meta-analysis of genome-wide scans provides evidence for sex- and site-specific regulation of bone mass. J. Bone Miner. Res. 2007;22:173–183. doi: 10.1359/jbmr.060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uitterlinden A.G., Ralston S.H., Brandi M.L., Carey A.H., Grinberg D., Langdahl B.L., Lips P., Lorenc R., Obermayer-Pietsch B., Reeve J., APOSS Investigators. EPOS Investigators. EPOLOS Investigators. FAMOS Investigators. LASA Investigators. Rotterdam Study Investigators. GENOMOS Study The association between common vitamin D receptor gene variations and osteoporosis: A participant-level meta-analysis. Ann. Intern. Med. 2006;145:255–264. doi: 10.7326/0003-4819-145-4-200608150-00005. [DOI] [PubMed] [Google Scholar]
- 6.Ralston S.H., Uitterlinden A.G., Brandi M.L., Balcells S., Langdahl B.L., Lips P., Lorenc R., Obermayer-Pietsch B., Scollen S., Bustamante M., GENOMOS Investigators Large-scale evidence for the effect of the COLIA1 Sp1 polymorphism on osteoporosis outcomes: The GENOMOS study. PLoS Med. 2006;3:e90. doi: 10.1371/journal.pmed.0030090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ioannidis J.P., Ralston S.H., Bennett S.T., Brandi M.L., Grinberg D., Karassa F.B., Langdahl B., van Meurs J.B., Mosekilde L., Scollen S., GENOMOS Study Differential genetic effects of ESR1 gene polymorphisms on osteoporosis outcomes. JAMA. 2004;292:2105–2114. doi: 10.1001/jama.292.17.2105. [DOI] [PubMed] [Google Scholar]
- 8.Tran B.N., Nguyen N.D., Eisman J.A., Nguyen T.V. Association between LRP5 polymorphism and bone mineral density: A Bayesian meta-analysis. BMC Med. Genet. 2008;9:55. doi: 10.1186/1471-2350-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiel D.P., Demissie S., Dupuis J., Lunetta K.L., Murabito J.M., Karasik D. Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med. Genet. 2007;8(Suppl 1):S14. doi: 10.1186/1471-2350-8-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richards J.B., Rivadeneira F., Inouye M., Pastinen T.M., Soranzo N., Wilson S.G., Andrew T., Falchi M., Gwilliam R., Ahmadi K.R. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Styrkarsdottir U., Halldorsson B.V., Gretarsdottir S., Gudbjartsson D.F., Walters G.B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Center J.R., Nguyen T.V. Multiple genetic loci for bone mineral density and fractures. N. Engl. J. Med. 2008;358:2355–2365. doi: 10.1056/NEJMoa0801197. [DOI] [PubMed] [Google Scholar]
- 12.Styrkarsdottir U., Halldorsson B.V., Gretarsdottir S., Gudbjartsson D.F., Walters G.B., Ingvarsson T., Jonsdottir T., Saemundsdottir J., Snorradóttir S., Center J.R. New sequence variants associated with bone mineral density. Nat. Genet. 2009;41:15–17. doi: 10.1038/ng.284. [DOI] [PubMed] [Google Scholar]
- 13.Xiong D.H., Liu X.G., Guo Y.F., Tan L.J., Wang L., Sha B.Y., Tang Z.H., Pan F., Yang T.L., Chen X.D. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am. J. Hum. Genet. 2009;84:388–398. doi: 10.1016/j.ajhg.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dvornyk V., Liu X.H., Shen H., Lei S.F., Zhao L.J., Huang Q.R., Qin Y.J., Jiang D.K., Long J.R., Zhang Y.Y. Differentiation of Caucasians and Chinese at bone mass candidate genes: Implication for ethnic difference of bone mass. Ann. Hum. Genet. 2003;67:216–227. doi: 10.1046/j.1469-1809.2003.00037.x. [DOI] [PubMed] [Google Scholar]
- 15.Lambrinoudaki I., Kung A.W. Absence of high-risk “s” allele associated with osteoporosis at the intronic SP1 binding-site of collagen Ialpha1 gene in Southern Chinese. J. Endocrinol. Invest. 2001;24:499–502. doi: 10.1007/BF03343882. [DOI] [PubMed] [Google Scholar]
- 16.Kung A.W., Lai B.M., Ng M.Y., Chan V., Sham P.C. T-1213C polymorphism of estrogen receptor beta is associated with low bone mineral density and osteoporotic fractures. Bone. 2006;39:1097–1106. doi: 10.1016/j.bone.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 17.Kung A.W., Yeung S.S., Lau K.S. Vitamin D receptor gene polymorphisms and peak bone mass in southern Chinese women. Bone. 1998;22:389–393. doi: 10.1016/s8756-3282(97)00301-3. [DOI] [PubMed] [Google Scholar]
- 18.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 20.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 21.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Wacholder S., Chanock S., Garcia-Closas M., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J. Natl. Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., Dixon R.J., Meitinger T., Braund P., Wichmann H.E., WTCCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Therneau T. Nineteenth Edition. R Foundation for Statistical Computing; Vienna, Austria: 2008. Kinship: Mixed Effects Cox Models, Sparse Matrices, and Modelling Data from Large Pedigrees. R Package Version 1.1.0. [Google Scholar]
- 25.Kung A.W., Luk K.D., Chu L.W., Tang G.W. Quantitative ultrasound and symptomatic vertebral fracture risk in Chinese women. Osteoporos. Int. 1999;10:456–461. doi: 10.1007/s001980050254. [DOI] [PubMed] [Google Scholar]
- 26.Kung A.W., Lee K.K., Ho A.Y., Tang G., Luk K.D. Ten-year risk of osteoporotic fractures in postmenopausal Chinese women according to clinical risk factors and BMD T-scores: A prospective study. J. Bone Miner. Res. 2007;22:1080–1087. doi: 10.1359/jbmr.070320. [DOI] [PubMed] [Google Scholar]
- 27.Black D.M., Cummings S.R., Stone K., Hudes E., Palermo L., Steiger P. A new approach to defining normal vertebral dimensions. J. Bone Miner. Res. 1991;6:883–892. doi: 10.1002/jbmr.5650060814. [DOI] [PubMed] [Google Scholar]
- 28.Liu J.M., Zhao H.Y., Ning G., Zhao Y.J., Zhang L.Z., Sun L.H., Xu M.Y., Chen J.L. Relationship between body composition and bone mineral density in healthy young and premenopausal Chinese women. Osteoporos. Int. 2004;15:238–242. doi: 10.1007/s00198-003-1536-7. [DOI] [PubMed] [Google Scholar]
- 29.Zhao H.Y., Liu J.M., Ning G., Zhao Y.J., Chen Y., Sun L.H., Zhang L.Z., Xu M.Y., Chen J.L. Relationships between insulin-like growth factor-I (IGF-I) and OPG, RANKL, bone mineral density in healthy Chinese women. Osteoporos. Int. 2008;19:221–226. doi: 10.1007/s00198-007-0440-y. [DOI] [PubMed] [Google Scholar]
- 30.Han S., Kim-Howard X., Deshmukh H., Kamatani Y., Viswanathan P., Guthridge J.M., Thomas K., Kaufman K.M., Ojwang J., Rojas-Villarraga A. Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE) Hum. Mol. Genet. 2009;18:1171–1180. doi: 10.1093/hmg/ddp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchini J., Howie B., Myers S., McVean G., Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 32.Li M.X., Jiang L., Ho S.L., Song Y.Q., Sham P.C. IGG: A tool to integrate GeneChips for genetic studies. Bioinformatics. 2007;23:3105–3107. doi: 10.1093/bioinformatics/btm458. [DOI] [PubMed] [Google Scholar]
- 33.Yuan H.Y., Chiou J.J., Tseng W.H., Liu C.H., Liu C.K., Lin Y.J., Wang H.H., Yao A., Chen Y.T., Hsu C.N. FASTSNP: An always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sham P.C., Purcell S., Cherny S.S., Abecasis G.R. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am. J. Hum. Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang Q.Y., Kung A.W. Genetics of osteoporosis. Mol. Genet. Metab. 2006;88:295–306. doi: 10.1016/j.ymgme.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y.W., Chang H.S., Lin C.H., Yu W.C. HPV-18 E7 conjugates to c-Myc and mediates its transcriptional activity. Int. J. Biochem. Cell Biol. 2007;39:402–412. doi: 10.1016/j.biocel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 37.Zhang Y., Hassan M.Q., Li Z.Y., Stein J.L., Lian J.B., van Wijnen A.J., Stein G.S. Intricate gene regulatory networks of helix-loop-helix (HLH) proteins support regulation of bone-tissue related genes during osteoblast differentiation. J. Cell. Biochem. 2008;105:487–496. doi: 10.1002/jcb.21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nobta M., Tsukazaki T., Shibata Y., Xin C., Moriishi T., Sakano S., Shindo H., Yamaguchi A. Critical regulation of bone morphogenetic protein-induced osteoblastic differentiation by Delta1/Jagged1-activated Notch1 signaling. J. Biol. Chem. 2005;280:15842–15848. doi: 10.1074/jbc.M412891200. [DOI] [PubMed] [Google Scholar]
- 39.Weber J.M., Forsythe S.R., Christianson C.A., Frisch B.J., Gigliotti B.J., Jordan C.T., Milner L.A., Guzman M.L., Calvi L.M. Parathyroid hormone stimulates expression of the Notch ligand Jagged1 in osteoblastic cells. Bone. 2006;39:485–493. doi: 10.1016/j.bone.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Alagille D., Estrada A., Hadchouel M., Gautier M., Odièvre M., Dommergues J.P. Syndromic paucity of interlobular bile ducts (Alagille syndrome or arteriohepatic dysplasia): Review of 80 cases. J. Pediatr. 1987;110:195–200. doi: 10.1016/s0022-3476(87)80153-1. [DOI] [PubMed] [Google Scholar]
- 41.Alagille D., Odièvre M., Gautier M., Dommergues J.P. Hepatic ductular hypoplasia associated with characteristic facies, vertebral malformations, retarded physical, mental, and sexual development, and cardiac murmur. J. Pediatr. 1975;86:63–71. doi: 10.1016/s0022-3476(75)80706-2. [DOI] [PubMed] [Google Scholar]
- 42.Krantz I.D., Piccoli D.A., Spinner N.B. Alagille syndrome. J. Med. Genet. 1997;34:152–157. doi: 10.1136/jmg.34.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krantz I.D., Rand E.B., Genin A., Hunt P., Jones M., Louis A.A., Graham J.M., Jr., Bhatt S., Piccoli D.A., Spinner N.B. Deletions of 20p12 in Alagille syndrome: Frequency and molecular characterization. Am. J. Med. Genet. 1997;70:80–86. [PubMed] [Google Scholar]
- 44.Rivadeneira F., Styrkársdottir U., Estrada K., Halldórsson B.V., Hsu Y.H., Richards J.B., Zillikens M.C., Kavvoura F.K., Amin N., Aulchenko Y.S., Genetic Factors for Osteoporosis (GEFOS) Consortium Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat. Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purcell S., Cherny S.S., Sham P.C. Genetic Power Calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19:149–150. doi: 10.1093/bioinformatics/19.1.149. [DOI] [PubMed] [Google Scholar]
- 46.Gartland A., Buckley K.A., Dillon J.P., Curran J.M., Hunt J.A., Gallagher J.A. Isolation and culture of human osteoblasts. Methods Mol. Med. 2005;107:29–54. doi: 10.1385/1-59259-861-7:029. [DOI] [PubMed] [Google Scholar]
- 47.Cheung C.L., Chan B.Y., Chan V., Ikegawa S., Kou I., Ngai H., Smith D., Luk K.D., Huang Q.Y., Mori S. Pre-B-cell leukemia homeobox 1 (PBX1) shows functional and possible genetic association with bone mineral density variation. Hum. Mol. Genet. 2009;18:679–687. doi: 10.1093/hmg/ddn397. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


