Abstract
A widely accepted species concept for bacteria has yet to be established. As a result, species designations are inconsistently applied and tied to what can be considered arbitrary metrics. Increasing access to DNA sequence data and clear evidence that bacterial genomes are dynamic entities that include large numbers of horizontally acquired genes have added a new level of insight to the ongoing species concept debate. Despite uncertainties over how to apply species concepts to bacteria, there is clear evidence that sequence-based approaches can be used to resolve cohesive groups that maintain the properties of species. This cohesion is clearly evidenced in the genus Salinispora, where three species have been discerned despite very close relationships based on 16S rRNA sequence analysis. The major phenotypic differences among the three species are associated with secondary metabolite production, which occurs in species-specific patterns. These patterns are maintained on a global basis and provide evidence that secondary metabolites have important ecological functions. These patterns also suggest that an effective strategy for natural product discovery is to target the cultivation of new Salinispora taxa. Alternatively, bioinformatic analyses of biosynthetic genes provide opportunities to predict secondary metabolite novelty and reduce the redundant isolation of well-known metabolites. Although much remains to be learned about the evolutionary relationships among bacteria and how fundamental units of diversity can be resolved, genus and species descriptions remain the most effective method of scientific communication.
Keywords: Species concepts, Natural product discovery, Taxonomy
Introduction
A fundamental goal of biology is to study the diversity of living organisms. This activity is broadly encompassed within the discipline of biological systematics, which includes developing methods to place organisms into taxonomic groups that share common features and evolutionary histories as well as describing new organisms that fall outside of circumscribed taxonomic boundaries. The foundations for these activities are rooted in Latin-based genus and species binomial nomenclature, which provides a common language for scientific communication.
Although identifying organisms to the species level has long been a fundamental goal of biology, developing broadly accepted species concepts that capture the complex evolutionary forces driving biological diversification remains problematic and controversial [2]. Prokaryotes present a particular challenge in that they reproduce largely by binary fission, and thus traditional biological species concepts are not readily applicable. In light of new information provided by DNA sequencing, there is an ongoing and high profile debate over how to recognize fundamental units of prokaryotic diversity that maintain the properties associated with species [1, 16, 24]. This debate is fueled by the observation that bacteria are adept at exchanging genes horizontally both among individuals in the same population and, to a lesser extent, between more distantly related taxa [33]. The process of horizontal gene transfer (HGT) can occur at homologous loci (homologous recombination) or can result in the introduction of new genes. Homologous recombination has a homogenizing effect on genetic diversification that can blur species boundaries and create uncertainty over taxonomic assignments when different genes within an individual strain maintain different evolutionary relationships. Although methods such as multilocus sequence typing can reveal these inconsistencies [34], concatenated trees are often strongly supported and produce relationships consistent with 16S rRNA phylogenies, suggesting that highly probable evolutionary relationships among closely related bacteria are resolvable even in the presence of HGT. Despite the uncertainties over how to incorporate HGT into an effective species concept for bacteria [9], well-chosen phylogenetic markers are rarely subject to this process and continue to be used to effectively identify cohesive clusters of bacteria that share evolutionary histories consistent with what would be expected for species-level taxa.
There are a number of reasons why it is important to describe new bacterial species even if current methods are flawed and revisions to existing taxa ultimately need to be made once more meaningful species concepts have been developed. One primary reason is that species descriptions require the designation of a type strain and its deposit into a publicly accessible culture collection. Also required is the deposit of relevant sequence data into a public database. Given that 16S rRNA gene sequencing has become the primary tool for initial taxonomic assignment, the importance of having type strain sequences available for comparative purposes can’t be overstated. These comparisons help to prevent taxonomic drift, or the spread of taxonomic boundaries across increasingly diverse lineages, as can happen when species names are sequentially assigned to query sequences based on top database matches as opposed to the closest type strain. The use of type strains for taxonomic assignments provides a more accurate assessment of the taxonomic novelty of the query strain, whereas the analysis of non-type strain sequences reveals a more complete picture of the distribution of related strains in nature and the diversity of the lineage to which the type strain belongs. Fortunately, the NCBI RefSeq Targeted Loci Project (http://www.ncbi.nlm.nih.gov/genomes/static/refseqtarget.html) includes a curated database of prokaryotic type strain 16S rRNA sequences that, when completed, will facilitate future comparisons.
The application of DNA sequence-based approaches has provided unprecedented opportunities to assess bacterial diversity with a level of resolution that could not be obtained using traditional culture-based methods. Sequence data also provide new opportunities to re-formulate species concepts and to test past species descriptions for accuracy. The ability to sequence complete bacterial genomes has provided the most significant advance in this regard and at the same time has added a new and unexpected level of complexity to the interpretation of species concepts. This complexity is amply demonstrated by observations that even well-studied bacteria such as Escherichia coli can share as little as 39% total protein content among different strains [42]. However, the realization that bacterial genomes maintain a mosaic structure with large numbers of acquired genes may be more an indication of an effective method of adaptation than evidence that sequence-based phylogenies created using the appropriate markers cannot be used to re-create accurate evolutionary histories. Advances in genome sequencing include the genomic encyclopedia of Bacteria and Archaea (GEBA) project (http://www.jgi.doe.gov/programs/GEBA/), a collaborative effort between the Joint Genome Institute and the DSMZ, which aims to use the tree of life as a guide to select underrepresented taxa for sequencing. GEBA promises to add considerably to our understanding of species concepts and genome evolution by filling in major sequencing voids in the tree of life and by providing a vast amount of new information about the evolutionary history of individual genes and the various hosts in which they are observed.
HGT and secondary metabolism
The types of genes most susceptible to horizontal gene transfer are generally those associated with non-essential functions or adaptive traits. Included among these “auxiliary” genes are those involved in the biosynthesis of secondary metabolites [17]. Secondary metabolites are produced by large (in some cases exceeding 100 kb) gene collectives, which include genes encoding both biosynthetic and tailoring enzymes as well as mechanisms of resistance and transport. These biosynthetic pathways are responsible for the remarkable structural diversity observed among bacterial secondary metabolites [14], and their tight genetic clustering is readily amenable to HGT. There is ample evidence for the transfer of secondary metabolite biosynthetic genes among bacteria [28]. The horizontal exchange of these pathways provides a particularly rapid and effective method for individual cells to test new secondary metabolites for selective advantages as opposed to the more traditional concepts of mutation-driven evolution. The susceptibility of genes involved in secondary metabolism to HGT suggests they would not be linked to specific taxonomic groups, but instead should be strain-specific. Strain specificity creates a unique set of challenges for the effective discovery of new secondary metabolites as it necessitates the screening of a large number of strains, and a good dose of serendipity, to discover new products. It also suggests that secondary metabolite phenotypes are not reliable taxonomic markers.
Species concepts have not been at center stage in the search for new microbial secondary metabolites. Historically, the tendency to describe the producer of each new secondary metabolite as a new species led to an explosion of species descriptions and an over-classification of the genus Streptomyces [4]. Although revisions to this genus have since been made, it remains the most specious prokaryotic taxon and the source of nearly 50% of the biologically active bacterial secondary metabolites discovered from microbial sources as of 2002 [6]. Despite this remarkable productivity, it is predicted that only a small percentage of the total number of compounds that can be produced by this genus have been discovered [41]. These predictions are supported by genome analyses of even well-studied species [5] where it is clear that the products of the majority of the biosynthetic pathways maintained by individual strains have yet to be discovered [32].
The large number of Streptomyces species that have been discovered to date is linked to the pharmaceutical industries focused efforts on this taxon, which resulted in the isolation and screening of untold thousands of strains. In general, the consensus at the time when microbial drug discovery efforts were a high priority for the pharmaceutical industry was that secondary metabolite production was strain-specific [39], thus necessitating the incorporation of large numbers of strains into screening protocols. Strain specificity is what would be expected if biosynthetic pathways were being exchanged at random, and the products had little effect on the survival of the new host. Given the resolving power of the taxonomic tools available at the time, it can be understood why the perception of strain specificity may have been maintained, as many of what appeared to be the same Streptomyces species were undoubtedly found to produce different metabolites. It would be interesting to re-visit these strains using higher resolution molecular tools to document the extent to which pathways are maintained within closely related populations.
Diminishing returns on efforts to discover new secondary metabolites from soil streptomycetes contributed to a pharmaceutical industry-wide paradigm shift away from natural products towards other discovery platforms and the expansion of research programs to poorly studied environments in an effort to isolate new actinomycete taxa. The marine environment has become an important focal point in these efforts, and marine-derived actinomycetes are now recognized as an important source of new secondary metabolites [7, 11, 13, 26, 29]. Although only four new marine genera have been described to date [19, 30, 40, 43], there appears to be considerable diversity that is unique to the marine environment [18], especially when the phylum Actinobacteria is broadly considered [20, 36].
The genus Salinispora as a model for species concepts
Among the new actinomycetes that have been cultured from marine samples, the genus Salinispora has become an interesting model with which to address species concepts. To date, S. tropica and S. arenicola have been formally described [30], and the description of a third species (“S. pacifica”) is in preparation. These bacteria are widely distributed in marine sediments [21] and have also been reported from a marine sponge [23]. Salinispora spp. are a rich source of structurally unique secondary metabolites including salinosporamide A, which is currently in clinical trials for the treatment of cancer [12]. In addition to the small molecules themselves, the elucidation of the biosynthetic machinery involved in producing these unusual compounds has led to a number of interesting discoveries including a new chlorination mechanism [10] and a new series of extender units in polyketide biosynthesis [27].
The three Salinispora species are closely related, yet can be readily distinguished using all current metrics commonly applied to the delineation of bacterial species. The genus as a whole shares 99% 16S rRNA sequence identity, while the pair-wise species comparisons differ by as few as five nucleotides (Fig. 1). To date, no intraspecific 16S diversity has been detected for S. tropica, while only two single nucleotide changes have been observed in S. arenicola. Interestingly, both of these changes are restricted to populations recovered from the Sea of Cortex, providing the first evidence of biogeographical isolation (allopatric diversification) within a Salinispora species [21]. There is also evidence of geographical isolation between S. tropica (Bahamas only) and S. pacifica (yet to be recovered from the Atlantic). Although it is widely disseminated that the 16S rRNA gene evolves too slowly to be used for species-level determinations [37], this marker nonetheless has proven effective for the delineation of closely related species within the genus Salinispora.
Fig. 1.
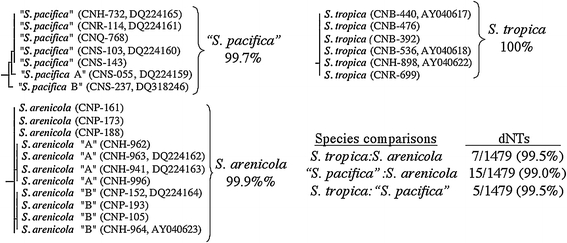
The three Salinispora species display little intra- (shown below species names) and inter-species 16S rRNA gene sequence diversity. dNTs = variable nucleotides
In contrast to the limited intraspecific Salinispora 16S sequence diversity that has been detected to date, most currently described bacterial species encompass considerably greater diversity. The inclusion of strains encompassing up to 3% 16S rRNA sequence divergence into a single bacterial species may in part be an artifact of trying to interpret sequence data in the context of past taxonomic assignments. This trend may have continued in part as a legacy of the report by Stackebrandt and Goebel [38], which stated that “species having 70% or greater DNA:DNA similarity usually have more than 97% 16S sequence identity.” Since a current requirement for the description of a bacterial species is that members generally share >70% DNA:DNA hybridization, it is possible that this statement has been interpreted to mean that 16S similarities as low as 97% can (or should) be used to circumscribe a bacterial species, although it is not clear that this was the authors' intent. In the case of some actinomycetes, it is now recognized that 16S values closer to 99% may be more realistic for species-level descriptions [31]. Although there is no reason to expect that one 16S value will be appropriate for all species-level units of diversity, or for that matter that this gene has sufficient resolution to delineate most species-level relationships, it is possible that many current species that encompass diverse 16S phylotypes may more appropriately represent genus level units of diversity or higher [25].
Despite the close phylogenetic relationships among the three Salinispora species, other metrics used to delineate bacterial species are also maintained when applied to the genus. In terms of DNA:DNA hybridization, the pair-wise comparisons of all three species yield values <70%. The average nucleotide identity (ANI) calculated from a comparison of 3,606 orthologous genes from the genome sequences of S. tropica (strain CNB-440) and S. arenicola (strain CNS-205) was 87% [35], which is well below the suggested cutoff value of 94% that was recently proposed to delineate genomic species [25]. Although these metrics are not based on ecological or evolutionary theory and therefore their use can be justifiably debated, they can be considered conservative, and in many cases will likely lead to the grouping together of lineages that should more accurately be considered independent.
In addition to being able to distinguish Salinispora species using sequence-based approaches, the three species can also be delineated by phenotypic traits. Surprisingly, the most dramatic of these is secondary metabolite production. Although species-specific secondary metabolite profiles have been reported for fungi [15], it is not clear that similar observations have been widely observed for actinomycetes. In the case of Salinispora species, the patterns are dramatic, with all S. arenicola strains examined to date producing compounds in the rifamycin and staurosporine classes, regardless of the global locations from which the strains were collected [22]. In the case of S. tropica, all strains produce compounds in the salinosporamide and sporolide classes, while none have been reported to produce the S. arenicola metabolites. Given that both of the classes of compounds observed in S. arenicola were originally reported from other bacteria and thus HGT can be inferred, these observations came as a surprise and contradict the concept that secondary metabolite production is strain specific. Clearly the fixation of specific pathways among globally distributed populations is powerful evidence of selection and implies that the products of these pathways have important ecological functions.
A comparison of the complete genome sequences of S. tropica and S. arenicola supports the species specificity of their core sets of secondary metabolites and provides new insight into the processes that drive diversification in the genus [35]. The most prominent feature emerging from the alignment of the two genome sequences is the concentration of species-specific genes in islands. Genomic islands are sites within which niche-specific genes are concentrated and ecological adaptations among closely related species resolved [8]. As might be expected, the biosynthetic genes linked to Salinispora species-specific secondary metabolite production are located on genomic islands. These biosynthetic operons are dynamic entities that hop among islands and display strong evidence of horizontal gene transfer suggesting that they are active sites of genome evolution and ecological adaptation [35]. Despite the relatively close relationship between the two Salinispora spp. for which genome sequences are available, the species specificity of certain pathways suggests that secondary metabolism can represent an important taxonomic feature for this group. Ongoing efforts by the J. Craig Venter Institute to sequence four additional Salinispora strains as well as large numbers of streptomycetes by the Broad Institute will provide a valuable new window within which to assess the distribution of biosynthetic pathways among actinomycetes.
Based on the results obtained to date from the three Salinispora species, it appears that a diversity-based approach targeting the cultivation of new actinomycete phylotypes provides an effective strategy for natural product discovery. However, there is no reason to expect that the compounds produced by a new phylotype will include new chemical structures as they may simply represent structures that are new relative to other closely related species. An alternative discovery approach is to evaluate the diversity of the biosynthetic genes maintained by individual strains prior to fermentation and chemical evaluation. Phylogenetic analyses of biosynthetic genes are providing new methods to predict secondary metabolite production and thus improve the discovery process. One of the best examples comes from analyses of ketosynthase domains derived from polyketide synthase genes. These domains tend to cluster together based on the compound produced [17], allowing sequence analyses of unknown strains to be used to predict the presence of specific biosynthetic pathways and their products (Gontang et al., submitted). Likewise, there is growing evidence that NRPS-derived condensation domains can be used for similar purposes. Thus, as more pathways are sequenced and experimentally characterized, it will become increasingly effective to use molecular screens to rapidly identify strains with the greatest genetic potential to produce new secondary metabolites or compounds within a targeted class of metabolites. This type of screening paradigm will allow more time to be spent varying fermentation conditions and monitoring pathway expression in a few high-quality strains as opposed to spending limited time on large numbers of strains that possess unknown biosynthetic potential.
Conclusions
Sequence-based approaches are providing new opportunities to assess bacterial diversity and resolve the relationships among groups of related strains that maintain the fundamental properties we expect from species-level taxa. Sequence-based approaches also have the capacity to provide an estimate of the genetic potential of strains to produce specific classes of secondary metabolites and to reveal the evolutionary relationships of these genes with respect to their hosts. Surprisingly, Salinispora species produce secondary metabolites in species-specific patterns. The ecological and evolutionary significance of this observation is not yet understood, nor is it known how broadly it may apply to other actinomycetes. If it is a general feature of these bacteria, it may be possible to develop a secondary metabolite-based chemotaxonomy for actinomycetes, as has been observed for fungi, plants and some marine invertebrates. However, the susceptibility of actinomycete biosynthetic pathways to horizontal gene transfer suggests there may be limits to this type of approach. The patterns of secondary metabolite production observed in the Salinispora example nonetheless suggest that the search for new actinomycete diversity represents a useful strategy for compound discovery, especially if this new diversity is derived from poorly explored environments where habitat-specific challenges to survival may select for the production of new metabolites. This strategy, coupled with the analysis of biosynthetic gene diversity, has the potential to reduce the redundant isolation of known compounds and allow more time to be devoted to those strains with the greatest potential to produce new secondary metabolites. What remains to be seen is which sequence-based approaches will ultimately prove most effective and become broadly employed by the natural products research community.
The field of microbiology is at a crossroads where the fundamental goal of assigning species names to bacteria is in danger of being abandoned. The growing acceptance of strain names instead of formal species descriptions portends a general lack of interest in taxonomy. If continued, this disinterest may leave large regions of taxonomic space filled with strain names but lacking the anchor points provided by formal species descriptions. Although efforts to create anchor points without formal species descriptions have been made [3], this is a precedent that will not necessarily be widely adapted and runs the risk of being applied without a standardized format. Assigning strain names is an easy solution to the growing problem that few are interested or being trained in bacterial systematics. It also avoids the very real problem that current methods to delineate species boundaries are insufficient and most likely inaccurate. A re-invigoration of the field in ways that make it less onerous to describe new species, possibly by the elimination of procedures that bog down the descriptive process in favor of increased reliance on methods that are generally available in most microbiology labs, may reduce this problem. At the same time, a working species concept that can be broadly applied to bacteria is needed. If these challenges can be met, there is reason to believe that the vast majority of bacterial diversity, possibly including that which has yet to be cultured, could be described using genus and species nomenclature over the next few generations. This is a goal that should be readily embraced as opposed to settling for strain names that lack taxonomic precision and will create communication challenges that will haunt future generations of microbiologists.
Acknowledgments
This work was supported by the California Sea Grant Program (R/NMP-98), NOAA Grant NAO80AR4170669 and the National Institutes of Health Grant GM085770.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 2.Agapow PM, Bininda-Emonds ORP, Crandall KA, Gittleman JL, Mace GM, et al. The impact of species concepts on biodiversity studies. Quart Rev Biol. 2004;79:161–179. doi: 10.1086/383542. [DOI] [PubMed] [Google Scholar]
- 3.Ahlgren NA, Rocap G, Chisholm SW. Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol. 2006;8:441–454. doi: 10.1111/j.1462-2920.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- 4.Anderson AS, Wellington EMH. The taxonomy of Streptomyces and related genera. Int J Syst Evol Microbiol. 2001;51:797–814. doi: 10.1099/00207713-51-3-797. [DOI] [PubMed] [Google Scholar]
- 5.Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 6.Berdy J. Bioactive microbial metabolites: a personal view. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 7.Bull AT, Stach JEM. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol. 2007;15:491–499. doi: 10.1016/j.tim.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Coleman ML, Sullivan MB, Martiny AC, Steglich C, Barry K, et al. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Res. 2009;19:744–756. doi: 10.1101/gr.086645.108. [DOI] [PubMed] [Google Scholar]
- 10.Eustaquio AS, Pojer F, Noel JP, Moore BS. Discovery and characterization of a marine bacterial SAM-dependent chlorinase. Nature Chem Biol. 2008;4:69–74. doi: 10.1038/nchembio.2007.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenical W, Jensen PR. Developing a new resource for drug discovery: marine actinomycete bacteria. Nature Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]
- 12.Fenical W, Jensen PR, Palladino MA, Lam KS, Lloyd GK, Potts BC. Discovery and development of the anticancer agent salinosporamide A (NPI-0052) Bioorganic & Medicinal Chem. 2009;17:2175–2180. doi: 10.1016/j.bmc.2008.10.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fiedler HP, Bruntner C, Bull AT, Ward AC, Goodfellow M, et al. Marine actinomycetes as a source of novel secondary metabolites. Antonie Van Leeuwenhoek. 2005;87:37–42. doi: 10.1007/s10482-004-6538-8. [DOI] [PubMed] [Google Scholar]
- 14.Fischbach MA, Walsh CT. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev. 2006;106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 15.Frisvad JC, Andersen B, Thrane U. The use of secondary metabolite profiling in chemotaxonomy of filamentous fungi. Mycol Res. 2008;112:231–240. doi: 10.1016/j.mycres.2007.08.018. [DOI] [PubMed] [Google Scholar]
- 16.Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, et al. Re-evaluating prokaryotic species. Nature Rev Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. [DOI] [PubMed] [Google Scholar]
- 17.Ginolhac A, Jarrin C, Robe P, Perriere G, Vogel T, et al. Type I polyketide synthases may have evolved through horizontal gene transfer. J Mol Evol. 2005;60:716–725. doi: 10.1007/s00239-004-0161-1. [DOI] [PubMed] [Google Scholar]
- 18.Gontang EA, Fenical W, Jensen PR. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl Environ Microbiol. 2007;73:3272–3282. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han SK, Nedashkovskaya OI, Mikhailov VV, Kim SB, Bae KS. Salinibacterium amurskyense gen. nov., sp. nov., a novel genus of the family Microbacteriaceae from the marine environment. Int J Syst Evol Microb. 2003;53:2061–2066. doi: 10.1099/ijs.0.02627-0. [DOI] [PubMed] [Google Scholar]
- 20.Jensen PR, Lauro FM. An assessment of actinobacterial diversity in the marine environment. Antonie Van Leeuwenhoek. 2008;94:51–62. doi: 10.1007/s10482-008-9239-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen PR, Mafnas C. Biogeography of the marine actinomycete Salinispora. Environ Microbiol. 2006;8:1881–1888. doi: 10.1111/j.1462-2920.2006.01093.x. [DOI] [PubMed] [Google Scholar]
- 22.Jensen PR, Williams PG, Oh D-C, Zeigler L, Fenical W. Species-specific secondary metabolite production in marine actinomycetes of the genus Salinispora. Appl Environ Microbiol. 2007;73:1146–1152. doi: 10.1128/AEM.01891-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim TK, Hewavitharana AK, Shaw PN, Fuerst JA. Discovery of a new source of rifamycin antibiotics in marine sponge actinobacteria by phylogenetic prediction. Appl Environ Microbiol. 2006;72:2118–2125. doi: 10.1128/AEM.72.3.2118-2125.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koeppel A, Perry EB, Sikorski J, Krizanc D, Warner A, et al. Identifying the fundamental units of bacterial diversity: a paradigm shift to incorporate ecology into bacterial systematics. Proc Nat Acad Sci. 2008;105:2504–2509. doi: 10.1073/pnas.0712205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Nat Acad Sci. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lam KS. Discovery of novel metabolites from marine actinomycetes. Curr Opin Microbiol. 2006;9:245–251. doi: 10.1016/j.mib.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Hazzard C, Eustaquio AS, Reynolds KA, Moore BS. Biosynthesis of salinosporamides from alpha beta-unsaturated fatty acids: implications for extending polyketide synthase diversity. J Amer Chem Soc. 2009;131:10376–10377. doi: 10.1021/ja9042824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez JV. Naturally mosaic operons for secondary metabolite biosynthesis: variability and putative horizontal transfer of discrete catalytic domains of the epothilone polyketide synthase locus. Mol Gen Genomics. 2004;270:420–431. doi: 10.1007/s00438-003-0937-9. [DOI] [PubMed] [Google Scholar]
- 29.Magarvey NA, Keller JM, Bernan V, Dworkin M, Sherman DH. Isolation and characterization of novel marine-derived actinomycete taxa rich in bioactive metabolites. Appl Environ Microbiol. 2004;70:7520–7529. doi: 10.1128/AEM.70.12.7520-7529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maldonado LA, Fenical W, Jensen PR, Kauffman CA, Mincer TJ, et al. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int J Syst Evol Microbiol. 2005;55:1759–1766. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- 31.Maldonado LA, Stach JEM, Ward AC, Bull AT, Goodfellow M. Characterisation of micromonosporae from aquatic environments using molecular taxonomic methods. Antonie Van Leeuwenhoek. 2008;94:289–298. doi: 10.1007/s10482-008-9244-0. [DOI] [PubMed] [Google Scholar]
- 32.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochman H, Lerat E, Daublin V. Examining bacterial species under the specter of gene transfer and exchange. Proc Nat Acad Sci. 2005;102:6595–6599. doi: 10.1073/pnas.0502035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papke RT, Zhaxybayeva O, Feil EJ, Sommerfeld K, Muise D, Doolittle WF. Searching for species in haloarchaea. Proc Nat Acad Sci. 2007;104:14092–14097. doi: 10.1073/pnas.0706358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, et al. Genomic islands link secondary metabolism to functional adaptation in marine actinobacteria. ISMEJ. 2009;3:1193–1203. doi: 10.1038/ismej.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rappe MS, Gordon DA, Vergin KL, Giovannoni SJ. Phylogeny of actinobacteria small subunit (SSU) rRNA gene clones recovered from marine bacterioplankton. Syst Appl Microbiol. 1999;22:106–112. [Google Scholar]
- 37.Rossello-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol Rev. 2001;25:39–67. doi: 10.1016/S0168-6445(00)00040-1. [DOI] [PubMed] [Google Scholar]
- 38.Stackebrandt E, Goebel BM. A place for DNA-DNA re-association and 16S Ribosomal-RNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 39.Strohl WR. Antimicrobials. In: Bull A, editor. Microbial Diversity and Bioprospecting. Washington DC: ASM Press; 2004. pp. 36–355. [Google Scholar]
- 40.Tian XP, Zhi XY, Qiu YQ, Zhang YQ, Tang SK, et al. Sciscionella marina gen. nov., sp. nov., a marine actinomycete isolated from a sediment in the northern South China Sea. Int J Syst Evol Microbiol. 2009;59:222–228. doi: 10.1099/ijs.0.001982-0. [DOI] [PubMed] [Google Scholar]
- 41.Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by the genus Streptomyces? Arch Microbiol. 2001;176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 42.Welch RA, Burland V, Plunkett G, Redford P, Roesch P, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Nat Acad Sci. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi H, Schumann P, Sohn K, Chun J. Serinicoccus marinus gen. nov., sp. nov., a novel actinomycete with L-ornithine and L-serine in the peptidoglycan. Int J Syst Evol Microbiol. 2004;54:1585–1589. doi: 10.1099/ijs.0.03036-0. [DOI] [PubMed] [Google Scholar]


