Abstract
The influenza A nucleoprotein (NP) is an attractive target for avian flu vaccine development because of its high conversancy in the evolutionary chain of the virus. Here we identified two novel HLA-A*0201 restricted NP epitopes, named H5N1 NP373-381 AMDSNTLEL (NP373) and NP458-466 FQGRGVFEL (NP458), using computational bioinformatic analysis. The NP peptides showed a high binding affinity to HLA-A*0201 on T2 cells, and were able to induce the activation of the cytotoxic T cells in the human peripheral blood mononuclear cells. We examined the potential of using NP373 and NP458 peptide sequences supplemented with a single-chain trimer as potential DNA vaccine candidates in an HHD transgenic mouse model. A gene gun delivery system was used for administrating the vaccine candidates into the animals. The results from cytotoxicity and ELISPOT assays indicated that a significant amount of IFN-γ was secreted by the T cells of the vaccinated mice, and the T cells were able to eliminate the corresponding peptide-loaded T2 cells. The discovery of these novel immunogenic NP peptides provides valuable information for avian flu vaccine design and construction.
Keywords: influenza A virus, H5N1 nucleoprotein, HLA-A*0201 T-cell epitope, single-chain trimer, HHD transgenic mouse model
1. INTRODUCTION
The transmission of a pathogenic avian H5N1 virus from poultry to humans in Hong Kong and other Asian countries has caused a number of human deaths [13, 17, 19]. Confirmed cases of H5N1 transmission have also been reported in various African and European countries1 and all incidences have caused an enormous economic loss in the poultry industries worldwide. The threat of an H5N1 pandemic is becoming a major problem not only in Asian countries but countries worldwide. The urgency of developing a strategy to combat the H5N1 virus is an imperative task for most health authorities, and one possible solution is to develop a human vaccine against the H5N1 virus.
The production of a neutralizing antibody is the primary step to combat viral infection. Another important mechanism for virus elimination is the initiation of a cell-mediated immune response involving the induction of effector CD8+ cytotoxic T lymphocytes (CTL). Conserved components within the influenza A viral genome are good targets for inducing CTL responses because they can withstand viral mutations and have the capability to induce cross-reactive immune response among different subtypes and variations [14, 15, 24].
Nucleoprotein (NP) generally binds to the viral RNA and is synthesized in large amounts during viral replication within the infected cells [26]. In the influenza A virus, NP is a comparatively conserved protein, and there is no recorded evolution in the viral strains of most birds over the past 60 years. [26]. Therefore, because of its high conservancy during the evolution of the virus, NP is an attractive target for inducing T-cell immunity [10, 26]. One of the promising approaches involves a direct DNA vaccination to stimulate T-cell immunity against NP. Some DNA vaccines and recombinant viral vaccines such as those constituting an adenovirus encoding the whole NP gene, are found effective in eliciting the T-cell immunity [6, 7, 14, 23].
We previously showed that a single-chain trimer (SCT) system is a promising system for use in the construction of DNA vaccines [2]. Here, we identified two novel HLA-A*0201-restricted H5N1-NP-protein peptides and a modified SCT system was used in vaccine construction. The efficacy of the constructed vaccine candidates was investigated in a special genetically modified HHD transgenic mouse model. Our results suggest that the H5N1 NP373-381 AMDSNTLEL and NP458-466 FQGRGVFEL peptides might be promising candidates for use in H5N1 vaccine construction.
2. MATERIALS AND METHODS
2.1. Peptide construction
H5N1 NP peptides from the H5N1-Thailand-human-2004 strain were predicted by the HLA-peptide-binding prediction program, SYFPEITHI2. Nine 9-mer potential peptides were synthesized by solid-phase strategies. They were named NP48 KLSDYEGRL, NP55 RLIQNSITI, NP158 GMDPRMCSL, NP189 MVMELIRMI, NP256 LIFLARSAL, NP275 CLPACVYGL, NP357 QLSTRGVQI, NP373 AMDSNTLEL, and NP458 FQGRGVFEL. A peptide previously identified (N220 LALLLLDRL) was used as a positive control [2].
2.2. Animal model
HHD transgenic H-2Db-/- β2m-/- double knockout mice (6–8 weeks old) purchased from the Institute Pasteur in France were bred and maintained under pathogen-free conditions (Animal & Plant Care Facility, HKUST, Hong Kong, China). HHD mice were originally derived from C57BL mice with the mouse β2-microglobulin and MHC I knock-out, and HLA-A*0201 knock-in expressing the chimeric HLA class-I molecules, which were composed of the human β2-microglobulin, HLA-A*0201 α-1 and α-2 domains and the α-3 domain of H-2Db [21].
2.3. Production of recombinant NP
Recombinant NP was expressed from pET16b in BL21-condon plus (DE3)-RIL cells (Stratagene, La Jolla, CA, USA). Soluble NP was purified by a His-Bind Kit (Novagen, San Diego, CA, USA) and purified proteins were analyzed and confirmed by SDS-PAGE. Western blotting analysis was performed using an anti-influenza A NP antibody (Chemicon, Billerica, MA, USA).
2.4. T2-cell binding experiments
T2 cells (174 × CEM.T2, ATCC, Manassas, VA, USA) were identified as HLA-A*0201 positive, and they were transporters for the antigen-processing-deficient cells. The cells were maintained in IMDM medium (Invitrogen, Carlsbad, CA, USA) supplemented with 5% FBS (Invitrogen), 100 U/mL penicillin and 100 μg/mL streptomycin (Invitrogen) at 37 °C with 5% carbon dioxide. In T2-cell binding assays, the cells (1 × 105) were pulsed with each of the target NP peptides (10 μg/mL), respectively, in serum-free medium containing the human β2-microglobulin (5 μg/mL) for 2 h. The cells were washed with 1× PBS, stained with an FITC-conjugated anti-human HLA-A2 antibody (BD Pharmingen, San Diego, CA, USA), and finally subjected to flow cytometrical analysis (BD FASC Aria II, BD Bioscience, San Jose, CA, USA). The fluorescent index was calculated by the formula as follows: FI = [MFI (T2 + peptide)/MFI (T2 only)] − 1. The flu peptide, GILGFVFTL, was used as a positive control [1].
2.5. In vitro stimulation of human CD8+ T cells
Fresh human HLA-A2+ peripheral blood mononuclear cells (PBMC) (Hong Kong Red Cross, Hong Kong, China) were isolated by Ficoll-Hypaque density gradient centrafugation (GE Healthcare, Piscataway, NJ, USA). CD8+ T cells were purified using human CD8 microbeads (MACS, Miltenyi Biotec, Bergisch Gladbach, Germany) and primed with autologous dendritic cells (DC) loaded with NP [2, 16]. Briefly, PBMC were seeded on 60 mm dishes (BD Bioscience) for 2 h. The process for DC culture requires 7 days. Adherent monocytes were cultured in an AIM-V medium (Invitrogen) containing GM-CSF (800 U/mL, Amoytop Biotech, Xiamen, Fujian, China) and IL-4 (1 000 U/mL, Amoytop Biotech) for 4 days. On day 5 from the cell culturing process, 20 μg/mL of NP was added. On day 6, maturation of DC was carried out by adding TNF-α (10 ng/mL) and PGE2 (1 μg/mL, Sigma-Aldrich Corp, St. Louis, MO, USA) to the culture medium. On day 7, mature DC were co-cultured with the purified CD8+ T cells in AIM-V medium (Invitrogen) supplemented with 5% human AB serum (Sigma-Aldrich Corp), IL-2 (20 U/mL, Amoytop Biotech), and IL7 (10 ng/mL, Miltenyi Biotec) for 7 days. The CD8+ T cells were stimulated three times with protein-loaded DC, respectively.
2.6. Staining of intracellular cytokines
Intracellular cytokine staining was performed as previously described with minor modifications [28]. Briefly, primed T cells were incubated with peptide-loaded T2 cells in the presence of Brefeldin A (eBioscience, San Diego, CA, USA) for 8 h. The cells were stained with a PE-conjugated anti-human CD8 antibody (eBioscience) at 4 °C for 30 min. After washing with 1× PBS, the cells were fixed and permeabilized with a permeabilization buffer (eBioscience) according to the manufacturer’s instructions. The cells were finally stained with an FITC-conjugated anti-human IFN-γ antibody at 4 °C for 30 min in the dark, and the fluorescent signals were detected by flow cytometrical analysis (BD FASC Aria II, BD Bioscience). T2 cells loaded with previously identified non-responsive peptide (N160 LQLPQGTTL), were used as a negative experimental control [2].
2.7. Construction of plasmids
Plasmids including NP158HHDpBudCE4.1, NP189HHDpBudCE4.1, NP373HHDpBudCE4.1, and NP458HHDpBudCE4.1 were prepared as follows. In brief, the single-chain trimer gene (HHD) was constructed by connecting the leader sequence, the human β2-microglobulin, the human HLA-A*0201 α-1, α-2 domains, and the mouse H-2Db α-3 domain. It was subsequently cloned into a pBudCE4.1 vector (Invitrogen). Plasmids encoding different peptide sequences were constructed by adding the corresponding DNA fragments after the leader sequence of the HHD using PCR with the following primers: sense (NP158: 5′-ATCGACCGGTCTATATGCTGGAATGGACCCTCGGATGTGTTCCCTGGGAGGAGGTGCTAG-3′, NP189: 5′-ATCGACCGGTCTATATGCTATGGTGATGGAGCTGATCAGAATGATCGGAGGAGGTGCTAG-3′, NP373: 5′-ATCGACCGGTCTATATGCTGCCATGGACAGCAACACCCTGGAGCTGGGAGGAGGTGCTAG-3′, NP458: 5′-ATCGACCGGTCTATATGCTTTCCAGGGACGGGGAGTGTTCGAGCTGGGAGGAGGTGCTAG-3′) and antisense (5′-ATCGCTCGAGTCACGCTTTACAATC-3′) primers. The constructed fragments were cloned into HHDpBudCE4.1 at Age I and Xho I sites. The N220HHDpBudCE4.1 expressing an identified HLA-A*0201 epitope (N220) was used as a control plasmid [2].
2.8. Animal vaccination
Animal vaccination was carried out by a gene gun delivery system and the preparation of DNA-coated gold particle cartridges was performed as previously described [2]. Briefly, cartridges were prepared by precipitating the plasmid DNA on 1 μm gold particles (Bio-Rad, Hercules, CA, USA) in 0.05 M spermidine (Sigma-Aldrich Corp) and 1 M CaCl2. The microcarrier/DNA suspension mixed with polyvinylpyrrolidone (5 mg/mL, Sigma-Aldrich Corp) was coated into the plastic tubing. The coated tubing was cut into 0.5 inch cartridges, and each cartridge contained 1 μg of DNA. DNA-coated particles were delivered to the shaved abdominal region of the HHD mice using a Helios gene gun (Bio-Rad, Hercules, CA, USA) with a discharge pressure of 400 psi. The mice received DNA (1 μg) for each vaccination. All animal groups (three mice per group) received a total of three vaccinations at 1-week intervals.
2.9. Cytotoxicity assay
The cytotoxicity assay was performed using a Delfia EuTDA cytotoxicity detection kit (PerkinElmer, Waltham, MA, USA). Mice were sacrificed 1 week after the last vaccination for splenocyte extraction. Splenocytes were cultured in an IMDM medium (Invitrogen) supplemented with 5% FBS (Invitrogen), the corresponding target peptides (10 μg/mL), and IL-2 (10 U/mL, eBioscience) in 12-well plates (BD Bioscience) at 37 °C for 5 days. They were seeded with the TDA-labeled, peptide-loaded T2 cells in ratios of 50:1, 10:1, and 5:1, respectively. The mice immunized with N220HHDpBudCE4.1 were used as an experiment control. The splenocytes seeded with N220-loaded T2 cells were used as a positive control while those seeded with N160-loaded T2 cells served as a negative control [2]. After incubating at 37 °C for 1 h, the culture medium was collected and the fluorescent signals emitted by the dead peptide-loaded T2 cells were detected with the cytotoxicity reagents according to the procedures stated by the manufacturer. The cytotoxicity was determined by the following equation:
Two controls were set up for the cytotoxicity assay: a maximum release was measured from the peptide-loaded T2 cells lyzed by a lysis buffer, and the spontaneous release was measured as a natural release from the peptide-loaded T2 cells.
2.10. ELISPOT assay
An ELISPOT assay was performed as previously described with minor modifications [2]. Briefly, a 96-well nitrocellulose plate (MultiScreen-HA, Millipore, Billerica, MA, USA) was coated with an anti-mouse IFN-γ antibody (5 μg/mL, eBioscience) at 4 °C for 20 h followed by blocking with an IMDM medium (Invitrogen) supplemented with 5% FBS (Invitrogen) at 37 °C for 2 h. Splenocytes (1 × 106) were seeded into the four wells of a 96-well nitrocellulose plate, and the corresponding target peptides (10 μg/mL) were added. The plate was incubated at 37 °C for 24 h. The mice immunized with N220HHDpBudCE4.1 were used as experiment controls. The splenocytes with N220 peptide added were used as positive controls while those added with N160 peptide served as negative controls [2]. Subsequently, the plate was washed with 1× PBS containing 0.01% Tween 20 and incubated with a biotinylated anti-mouse IFN-γ antibody (0.5 μg/mL, eBioscience) at room temperature for 1 h. After washing, a streptavidin-AP solution (1 μg/mL, Sigma-Aldrich Corp) was added and the plate was incubated at room temperature for 30 min. After a final wash, spots were developed by adding a BCIP/NBT solution (Invitrogen) and the visible spots were counted by an ELISPOT reader (Thermo Labsystems, Franklin, MA, USA).
Two controls for each group of splenocytes were set up for the ELISPOT assay. Splenocytes without the addition of peptides were used as a negative control while the splenocytes with staphylococcal enterotoxin B (Sigma-Aldrich Corp) added were used as a positive control.
2.11. Statistical analysis
Statistical analysis was expressed as means ± standard error (SE). A comparison between the individual readings was performed using the student’s t-test. p values < 0.05 were regarded as statistically significant.
3. RESULTS
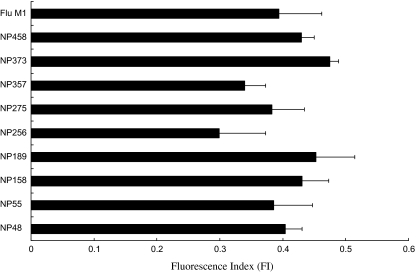
In order to search for the immunogenic NP peptide sequence, nine potential peptides were predicted using SYFPEITHI software. Various assays were used to determine the immunogenicity of the peptides and the desirable ones (i.e., those with both the highest binding affinity to HLA-A*0201 positive cells and the ability to stimulate human CD8+ T cells) were selected for further in vivo experimentation. In order to determine their binding affinity to HLA-A*0201 positive cells, the peptides were synthesized and a T2-cell binding assay was performed. T2 cells normally express unstable HLA molecules on the cell surface and therefore if a peptide is able to fit into the peptide-binding groove of an HLA molecule, it will form a more stable structural complex with the HLA molecule in the presence of β2-microglobulin. This complex can be detected with an anti-human HLA-A2 antibody via flow cytometry. In our assay, a flu peptide (GILGFVFTL), which is an HLA-A*0201 epitope, was used as a positive control for the experiment. The results indicate that the NP373 (AMDSNTLEL), NP189 (MVMELIRMI), and NP458 (FQGRGVFEL) showed the highest scores in the T2-cell binding assay (Fig. 1).
Figure 1.
A graph showing the results of the T2-cell binding assay. The binding affinity of the NP peptides to HLA-A2 in the presence of additional β2-microglobulin was detected by a T2-cell binding assay. T2 cells were pulsed with the target NP peptides, respectively, in serum-free medium supplemented with the β2-microglobulin for 2 h. The peptide-HLA-A2 complex was probed by an FITC-conjugated anti-human HLA-A2 antibody and the fluorescent signals were detected via flow cytometry. The binding affinity was presented as the fluorescent index that was calculated by the following formula: FI = [MFI (T2 + peptide)/MFI (T2 only)] − 1. The flu peptide (GILGFVFTL) was used as a positive control. The results represent the mean ± SE (n = 3).
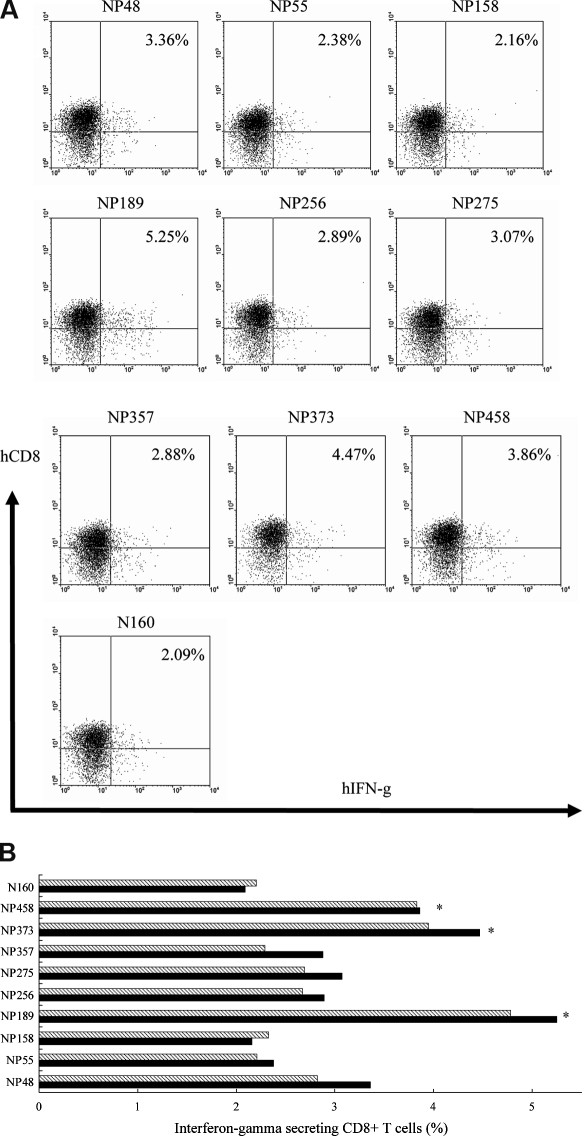
The potential of the nine peptides in stimulating the human CD8+ T cells was also determined. Two groups of CD8+ T cells isolated from PBMC of two healthy donors, respectively, were primed with autologous mature NP-loaded DC three times. Intracellular cytokine staining was performed to detect the release of IFN-γ from the CD8+ T cells, and the peptide-loaded T2 cells were used as the target cells. NP once processed by DC, will combine with the HLA molecules and eventually become presented on the cell surface for the priming of the CD8+ T cells. The primed CD8+ T cells could therefore recognize the peptide-MHC complex on the T2 cells if the target peptides are processed and presented on the DC surface. The results from intracellular cytokine staining showed that the highest number of CD8+ IFN-γ-secreting T cells was produced after the stimulation with the NP189 (MVMELIRMI), NP373 (AMDSNTLEL), and NP458 (FQGRGVFEL) peptides, and the lowest number of the stimulated T cells was observed after stimulation with the NP158 (GMDPRMCSL) peptide. The N160 (LQLPQGTTL) peptide-loaded T2 cells used as a negative control did not show any significant changes (Fig. 2) [2].
Figure 2.
An illustration of intracellular cytokine staining due to the secretion of IFN-γ from human CD8+ T cells. CD8+ T cells were primed with autologous NP-loaded DC three times at 1-week intervals. Primed CD8+ T cells were cultured with different target peptide-loaded T2 cells for 8 h and the number of IFN-γ-secreting CD8+ T cells was stained with PE-conjugated anti-human CD8 and an FITC-conjugated anti-human IFN-γ antibodies, and subsequently subjected to flow cytometry. A negative control was set up using T2 cells added with previously identified non-responsive peptide (N160). (A) Representative data from flow cytometrical analysis of one of the two experiments performed for the first donor. (B) A graph showing the data from two individual experiments for the first donor only. (C) Representative data from flow cytometrical analysis of one of the two experiments performed for the second donor. (D) A graph showing the data from two individual experiments for the second donor only. The p values were calculated by the student’s t-test (*p < 0.05) on comparisons between sample data points and the negative control data point.
From the results of the two assays, we selected four target peptides, NP158 (GMDPRMCSL), NP189 (MVMELIRMI), NP373 (AMDSNTLEL), and NP458 (FQGRGVFEL) for animal experimentation. Based on our findings, three of the peptides showed the highest binding affinity to the HLA-A2 molecules and they were able to activate the highest number of CD8+ T cells. Since NP158 (GMDPRMCSL) exhibited a comparatively low ability to stimulate T cells against the peptide-loaded T2 cells and induced only a small amount of the IFN-γ release, it was included in the sample peptides as a control for checking the consistency of the results previously shown by the in vitro T-cell stimulation experiment.
An animal experiment was performed to test the immunogenicity of the target peptides in vivo. DNA corresponding to the target peptides was cloned into the HHD gene to produce a DNA vaccine candidate containing the peptide-β2-microglobulin-MHC class-I heavy chain complex that could be expressed on the cell surface and trigger the immune system. Mice received a total of three administrations and were sacrificed 1 week after the last injection. Splenocytes were collected for the cytotoxicity and ELISPOT assays.
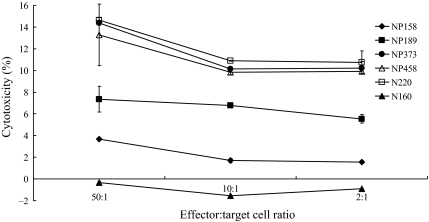
Cytotoxicity was measured by the amount of TDA released from the target cells. The target cells (T2 cells) were loaded with the target peptides. Therefore, the peptide-loaded T2 cells would be killed if the corresponding peptides were immunogenic to the mice. The results from the cytotoxicity assay showed that the highest percentage of cytotoxic response was triggered in mice immunized with plasmids encoding NP373 and NP458, and the effect was similar to that produced by the positive control (N220). During an 1 h incubation period, NP373 exhibited a 14% and 10% killing efficiency with the effector cell-to-target cell ratios of 50:1 and 10:1, respectively, and NP458 exhibited a 13% and 9% killing efficiency with the effector cell-to-target cell ratios of 50:1 and 10:1, respectively. However, the killing efficiency of T cells triggered by the NP189 was comparatively low (7%) with the effector cell-to-target cell ratios of 50:1. The result produced by NP158 was significantly lower, with only a 3.7% killing efficiency at an effector cell-to-target cell ratio of 50:1 (Fig. 3).
Figure 3.
A graph showing the percentage of specific killings of the target peptide-loaded T2 cells after immunization. Mice were immunized with NP158HHDpBudCE4.1, NP189HHDpBudCE4.1, NP373HHDpBudCE4.1, NP458HHDpBudCE4.1, and N220HHDpBudCE4.1, respectively, by gene gun delivery for three times at 1-week intervals. One week after the last immunization, splenocytes were collected and cultured with the corresponding peptides for 5 days. The splenocytes were subsequently incubated with the corresponding TDA-labeled peptide-loaded T2 cells at ratios of 50:1, 10:1, and 2:1, respectively, for 1 h. The culture media were collected and the fluorescent signals from the dead peptide-loaded T2 cells were detected by a Delfia EuTDA cytotoxicity detection kit and the cytotoxicity was calculated. The x-axis indicates the different ratios of the effector cells (splenocytes) to the target cells (TDA-labeled peptide-loaded T2 cells). The y-axis indicates the percentage of cell cytotoxicity. The cytotoxicity for the six groups of splenocytes vaccinated with the various plasmids was indicated as follows: NP158HHDpBudCE4.1 (-♦-), NP189HHDpBudCE4.1 (-■-), NP373HHDpBudCE4.1 (-●-), and NP458HHDpBudCE4.1 (-△-). N220HHDpBudCE4.1 (-□-) was used as a positive control. Splenocytes obtained from the mice vaccinated with the plasmid, N220HHDpBudCE4.1, but tested with N160-loaded T2 cells (-▲-), was used as a negative control. The results represent the mean ± SE (n = 3).
An ELISPOT assay was used to determine the splenocyte recognition of the corresponding peptides via IFN-γ release after animal immunization. The largest number of splenocytes (appeared as spots) was produced in mice after immunization with NP373 and NP458 containing the SCT-DNA constructs (79 and 72 spots, respectively). In contrast, only 14 and 4 spots were observed in animals immunized with NP189 and NP158 containing the SCT-DNA constructs, respectively. The results were very similar to the negative control (5 spots only). The immunogenicity triggered by NP189 in mice was comparatively low when compared with the results of the T2-cell binding assay and the intracellular cytokine staining. Nonetheless, the results indicate that the immunogenicity of NP373 and NP458 was more than 18 times higher than that induced by NP158 (Fig. 4).
Figure 4.
A graph presenting the results of the ELISPOT assay. Mice were immunized with NP158HHDpBudCE4.1, NP189HHDpBudCE4.1, NP373HHDpBudCE4.1, and NP458HHDpBudCE4.1 by gene gun delivery three times at 1-week intervals. N220HHDpBudCE4.1 was used as a positive control. One week after the last immunization, the splenocytes were collected and cultured with corresponding peptides for 24 h. The secretion of IFN-γ was measured by the ELISPOT assay. The N160 peptide (LQLPQGTTL) was used as a negative control for the assay with the mice vaccinated with N220HHDpBudCE4.1. The results represent the mean ± SE (n = 4). The p values were calculated by the student’s t-test (*p < 0.05) on comparisons between sample data points and the negative control data point.
The prediction process was repeated and the prediction score indicated that NP373 and NP485 may be able to bind to a variety of other HLA types, such as HLA-A1, A11, A24, A26, B7, B8, B14, B15, B27, B37, B38, B39, B40, B44, and B47 (Tab. I).
Table I.
Prediction scores of the NP373 and NP485 peptides for the binding to other HLA types via a HLA-peptide-binding prediction program, SYFPEITHI2.
| HLA Type | Score |
||
|---|---|---|---|
| NP373 AMDSNTLEL | NP485 FQGRGVFEL | Flu peptidea GILGFVFTL | |
| A*01 | 15 | 3 | 1 |
| A*0201 | 25 | 19 | 30 |
| A*1101 | 15 | 2 | 13 |
| A*2402 | 10 | 12 | 14 |
| A*26 | 11 | 12 | 17 |
| B*0702 | 16 | 12 | 13 |
| B*08 | 11 | 13 | 16 |
| B*1402 | 17 | 16 | 18 |
| B*1501 | 2 | 12 | 7 |
| B*1510 | 12 | 13 | 14 |
| B*2705 | 13 | 14 | 19 |
| B*2709 | 13 | 11 | 16 |
| B*37 | 13 | 12 | 17 |
| B*3801 | 16 | 13 | 13 |
| B*3901 | 19 | 13 | 15 |
| B*3902 | 12 | 19 | 13 |
| B*4001 | 17 | 13 | 13 |
| B*4402 | 17 | 12 | 14 |
| B*4701 | 10 | 10 | 13 |
| B*4901 | 11 | 7 | 9 |
The flu peptide (an epitope of HLA-A*0201) is used as a reference for the comparison of the binding scores.
4. DISCUSSION
HLA-A*0201 is a global common HLA type, which occurs at high frequencies in populations of northern and southeast Asia and North America [18]. In this study, we selected nine peptides that might be potential vaccine candidates for human influenza A using the HLA-peptide-binding prediction program. Two novel epitopes for HLA-A2 specific H5N1 NP were found, namely NP373-381 AMDSNTLEL (NP373) and NP458-466 FQGRGVFEL (NP458). We further repeated the prediction process to investigate the binding of the two identified epitopes to other HLA types (Tab. I). We found that these two epitopes might be able to bind to other HLA types, and therefore further experimentation may be required to confirm the prediction.
An ideal vaccine capable of counteracting infectious viruses should be able to trigger both neutralizing antibody production and the cytotoxic T-cell response which are critical processes in eliminating the virus during a viral pathogenesis. As previously mentioned, current studies of H5N1 vaccines have mainly focused on the generation of neutralizing antibodies [3, 11]. Another research stream involves the study of cell-mediated immunity against H5N1. Epitopes have been found in different internal proteins of influenza virus and with different HLA types [9, 20, 22, 25].
Amongst the identified epitopes, all NP epitopes are categorized as HLA-B types. We decided to examine the NP epitope binding specificity of HLA-A*0201 to CD8+ T cells. A desirable peptide sequence for inducing cytotoxic T-cell response should be presented as an endogenous peptide epitope through proteasome digestion and have a high binding affinity towards human MHC class-I molecules. Therefore, apart from the T2-cell binding assay, the stimulating ability of the predicted peptides was also accessed via an in vitro stimulation using human PBMC. In vitro stimulation was carried out by priming the T cells with the naturally processed NP peptides from autologous DC. Our results showed that the NP189 peptide stimulated the strongest immune response amongst the four peptides used. The binding affinity of NP158 in the T2-cell binding assay was relatively high but no significant change in the immune response was observed during in vitro administration of human PBMC, and the result produced was similar to the negative control. This suggests that the peptide prediction software can only give a computational estimation of immunogenicity of potential vaccine candidates. Hence, the predicted epitopes may not truly reflect experimental conditions in vitro nor mimic the real state in vivo.
Natural processing and peptide presentation on the HLA molecule significantly contribute to the magnitude of the immune response [5, 8]. In our experiment, the number of CD8+ T cells detected via intracellular cytokine staining was not as high as expected compared to the whole pool of CD8+ T cells. In normal natural processing, all epitopes on NP-loaded DC are presented to the CD8+ T cells, based on the cross-presentation characteristics of the DC [4, 16, 27]. Therefore all activated CD8+ T cells should be responsive to the immunogenic epitopes presented on NP. In our experiment, our target cells were loaded with only a single type of the target peptide which may explain why such a low percentage of cell activation caused by the corresponding peptide-specific CD8+ T cells is observed.
We used an SCT approach to construct our peptide vaccine candidates. With this method, peptides are able to specifically bind to the peptide-binding cleft created by the HLA-A*0201 α-1 and α-2 domains, and this phenomenon can imitate the natural binding of peptides onto the MHC class-I molecules during normal antigen processing [12]. Also the SCT system ensures the stability of the MHC I-peptide complex after translation [29], and a more precise and direct priming of the professional antigen presenting cells to the cytotoxic T cells is achieved. Hence the stimulating efficiency of the peptide vaccine can be greatly maximized [2].
The selected peptides were tested for their cytotoxic T cell stimulating ability in vivo using a HHD transgenic mouse model. HHD is a special mouse type which lacks mouse β2-microglobulin and MHC I, but contains human β2-microglobulin and HLA-A*0201 α-1, α-2, as well as a mouse α-3 fusion gene. This genetically modified mouse immune system can impersonate a human system’s immune response to stimulation restricted to the reaction between HLA-A*0201 and T-cell receptors only, eliminating the background caused by mouse MHC class-I molecules [21]. In our experiment, cytotoxicity and ELISPOT assays showed that the NP373-381 AMDSNTLEL and NP458-466 FQGRGVFEL are the most immunogenic peptides. The reactivity of the two peptides was consistent with all the tests during the process of epitope identification. Another target peptide, NP189, although it showed good responses in the T2-cell binding assay and in vitro stimulation of human CD8+ T cells, it induced a weak response in animal experiments. This is another indication that the in vitro system does not reflect the in vivo situation. Nonetheless, in vitro experiments are essential for acquiring basic information to verify the characteristics of these potential peptides and discovered immunogenic peptides, since the testing of the peptides cannot be performed in clinical trials without extensive investigation. The T2-cell binding assay is a good methodology for examining the potential binding affinity of the target peptides to the groove of HLA-A*0201 molecules. The antigen processing and presentation, and the magnitude of the CD8+ T-cell responses corresponding to the stimulants can be investigated via in vitro stimulation of the CD8+ T cells. We believe that the HHD transgenic mouse model is an appropriate animal model for investigating the stimulating effects of the peptide-HLA complex to the corresponding T-cell responses.
In summary, we identified two novel HLA-A*0201 T-cell epitopes, NP373 and NP458, in the H5N1 viral NP. Our results indicate that these two epitopes are capable of inducing a strong immune response, and that the SCT system can improve the immune responses. The finding of these novel cytotoxic T-cell epitopes presented provides significant information for the development of a human H5N1 vaccine. In the future, experiments using a mixture of SCT-DNA constructs encoding H5N1 NP epitopes instead of using one type of SCT-DNA construct encoding a single peptide are necessary to investigate whether there are any improvements for the immunogenicity.
Acknowledgments
We thank the Hong Kong Red Cross (Hong Kong, China) for supplying the blood samples, and Mr William Chau and his team for taking care of the experimental animals (Animal & Plant Care Facility, HKUST, Hong Kong, China). The study was supported by the Research Fund for the Control of Infectious Diseases (RFCID), Food and Health Bureau, Hong Kong SAR government.
Footnotes
References
- 1.Bednarek M.A., Sauma S.Y., Gammon M.C., Porter G., Tamhankar S., Williamson A.R., Zweerink H.J., The minimum peptide epitope from the influenza virus matrix protein: extra and intracellular loading of HLA-A2, J. Immunol. (1991) 147:4047–4053 [PubMed] [Google Scholar]
- 2.Cheung Y.K., Cheng S.C.S., Sin F.W.Y., Chan K.T., Xie Y., Induction of T-cell response by a DNA vaccine encoding a novel HLA-A*0201 severe acute respiratory syndrome coronavirus epitope, Vaccine (2007) 25:6070–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cinatl J. Jr., Michaelis M., Doerr H.W., The threat of avian influenza A (H5N1) part IV: Development of vaccines, Med. Microbiol. Immunol. (2007) 196:213–225 [DOI] [PubMed] [Google Scholar]
- 4.Cresswell P., Ackerman A.L., Giodini A., Peaper D.R., Wearsch P.A., Mechanisms of MHC class I-restricted antigen processing and cross-presentation, Immunol. Rev. (2005) 207:145–157 [DOI] [PubMed] [Google Scholar]
- 5.Dutoit V., Taub R.N., Papadopoulos K.P., Talbot S., Keohan M.L., Brehm M., et al., Multiepitope CD8(+) T cell response to a NY-ESO-1 peptide vaccine results in imprecise tumor targeting, J. Clin. Invest. (2002) 110:1813–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epstein S.L., Tumpey T.M., Misplon J.A., Lo C.Y., Cooper L.A., Subbarao K., et al., DNA vaccine expressing conserved influenza virus proteins protective against H5N1 challenge infection in mice, Emerg. Infect. Dis. (2002) 8:796–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epstein S.L., Kong W.P., Misplon J.A., Lo C.Y., Tumpey T.M., Xu L., Nabel G.J., Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein, Vaccine (2005) 23:5404–5410 [DOI] [PubMed] [Google Scholar]
- 8.Gnjatic S., Atanackovic D., Matsuo M., Jäger E., Lee S.Y., Valmori D., et al., Cross-presentation of HLA class IU epitopes from exogenous NY-ESO-1 polypeptides by nonprofessional APC, J. Immunol. (2003) 170:1191–1196 [DOI] [PubMed] [Google Scholar]
- 9.Heiny A.T., Miotto O., Srinivasan K.N., Khan A.M., Zhang G.L., Brusic V., et al., Evolutionarily conserved protein sequences of influenza a viruses, avian and human, as vaccine targets, PLoS ONE (2007) 2:e1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiromoto Y., Yamazaki Y., Fukushima T., Saito T., Lindstrom S.E., Omoe K., et al., Evolutionary characterization of the six internal genes of H5N1 human influenza A virus, J. Gen. Virol. (2000) 81:1293–1303 [DOI] [PubMed] [Google Scholar]
- 11.Horimoto T., Kawaoka Y., Strategies for developing vaccines against H5N1 influenza A viruses, Trends Mol. Med. (2006) 12:506–514 [DOI] [PubMed] [Google Scholar]
- 12.Huang C.H., Peng S., He L., Tsai Y.C., Boyd D.A., Hansen T.H., et al., Cancer immunotherapy using a DNA vaccine encoding a single-chain trimer of MHC class I linked to an HPV-16 E6 immunodominant CTL epitope, Gene Ther. (2005) 12:1180–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kodihalli S., Goto H., Kobasa D.L., Krauss S., Kawaoka Y., Webster R.G., DNA vaccine encoding hemagglutinin provides protective immunity against H5N1 influenza virus infection in mice, J. Virol. (1999) 73:2094–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kodihalli S., Kobasa D.L., Webster R.G., Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines, Vaccine (2000) 18:2592–2599 [DOI] [PubMed] [Google Scholar]
- 15.Laddy D.J., Yan J., Corbitt N., Kobasa D., Kobinger G.P., Weiner D.B., Immunogenicity of novel consensus-based DNA vaccines against avian influenza, Vaccine (2007) 25:2984–2989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leclerc D., Beauseigle D., Denis J., Morin H., Paré C., Lamarre A., Lapointe R., Proteasome-independent major histocompatibility complex class I cross-presentation mediated by papaya mosaic virus-like particles leads to expansion of specific human T cells, J. Virol. (2007) 81:1319–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li K.S., Guan Y., Wang J., Smith G.J., Xu K.M., Duan L., et al., Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia, Nature (2004) 430:2009–2013 [DOI] [PubMed] [Google Scholar]
- 18.Middleton D., Menchaca L., Rood H., Komerofsky R., New allele frequency database: http://www.allelefrequencies.net, Tissue Antigens (2003) 61:403–407 [DOI] [PubMed] [Google Scholar]
- 19.Nicholson K.G., Wood J.M., Zambon M., Influenza, Lancet (2003) 362:1733–1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parida R., Shaila M.S., Mukherjee S., Chandra N.R., Nayak R., Computational analysis of proteome of H5N1 avian influenza virus to define T cell epitopes with vaccine potential, Vaccine (2007) 25:7530–7539 [DOI] [PubMed] [Google Scholar]
- 21.Pascolo S., Bervas N., Ure J.M., Smith A.G., Lemonnier F.A., Pérarnau B., HLA-A2.1-restricted education and cytolytic activity of CD8(+) T lymphocytes from beta2 microglobulin (beta2m) HLA-A2.1 monochain transgenic H-2Db beta2m double knockout mice, J. Exp. Med. (1997) 185:2043–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rimmelzwaan G.F., Fouchier R.A., Osterhaus A.D., Influenza virus-specific cytotoxic T lymphocytes: a correlate of protection and a basis for vaccine development, Curr. Opin. Biotechnol. (2007) 18:529–536 [DOI] [PubMed] [Google Scholar]
- 23.Roy S., Kobinger G.P., Lin J., Figueredo J., Calcedo R., Kobasa D., Wilson J.M., Partial protection against H5N1 influenza in mice with a single dose of a chimpanzee adenovirus vector expressing nucleoprotein, Vaccine (2007) 25:6845–6851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ulmer J.B., Donnelly J.J., Parker S.E., Rhodes G.H., Felgner P.L., Dwarki V.J., et al., Heterologous protection against influenza by injection of DNA encoding a viral protein, Science (1993) 259:1745–1749 [DOI] [PubMed] [Google Scholar]
- 25.Wang M., Lamberth K., Harndahl M., Røder G., Stryhn A., Larsen M.V., et al., CTL epitopes for influenza A including the H5N1 bird flu: genome-pathogen-, and HLA-wide screening, Vaccine (2007) 25:2823–2831 [DOI] [PubMed] [Google Scholar]
- 26.Webster R.G., Bean W.J., Gorman O.T., Chambers T.M., Kawaoka Y., Evolution and ecology of influenza A viruses, Microbiol. Rev. (1992) 56:152–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson N.S., Villadangos J.A., Regulation of antigen presentation and cross-presentation in the dendritic cell network: facts, hypothesis, and immunological implications, Adv. Immunol. (2005) 86:241–305 [DOI] [PubMed] [Google Scholar]
- 28.Xie L.H., Sin F.W.Y., Cheng S.C.S., Cheung Y.K., Chan K.T., Xie Y., Xie Y., Activation of cytotoxic T lymphocytes against CML28-bearing tumors by dendritic cells transduced with a recombinant adeno-associated virus encoding the CML28 gene, Cancer Immunol. Immunother. (2008) 57:1029–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Y.Y., Netuschi N., Lybarger L., Connolly J.M., Hansen T.H., Cutting edge: single-chain trimers of MHC class I molecules form stable structures that potently stimulate antigen-specific T cells and B cells, J. Immunol. (2002) 168:3145–3149 [DOI] [PubMed] [Google Scholar]