Abstract
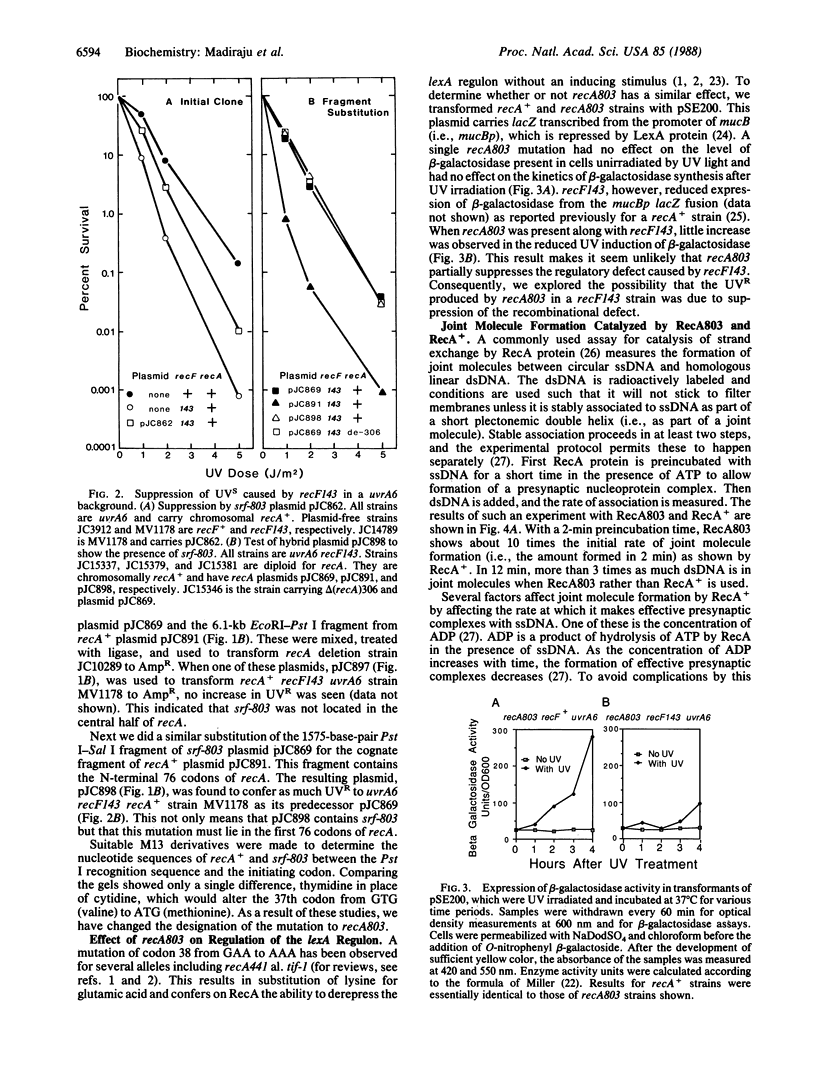
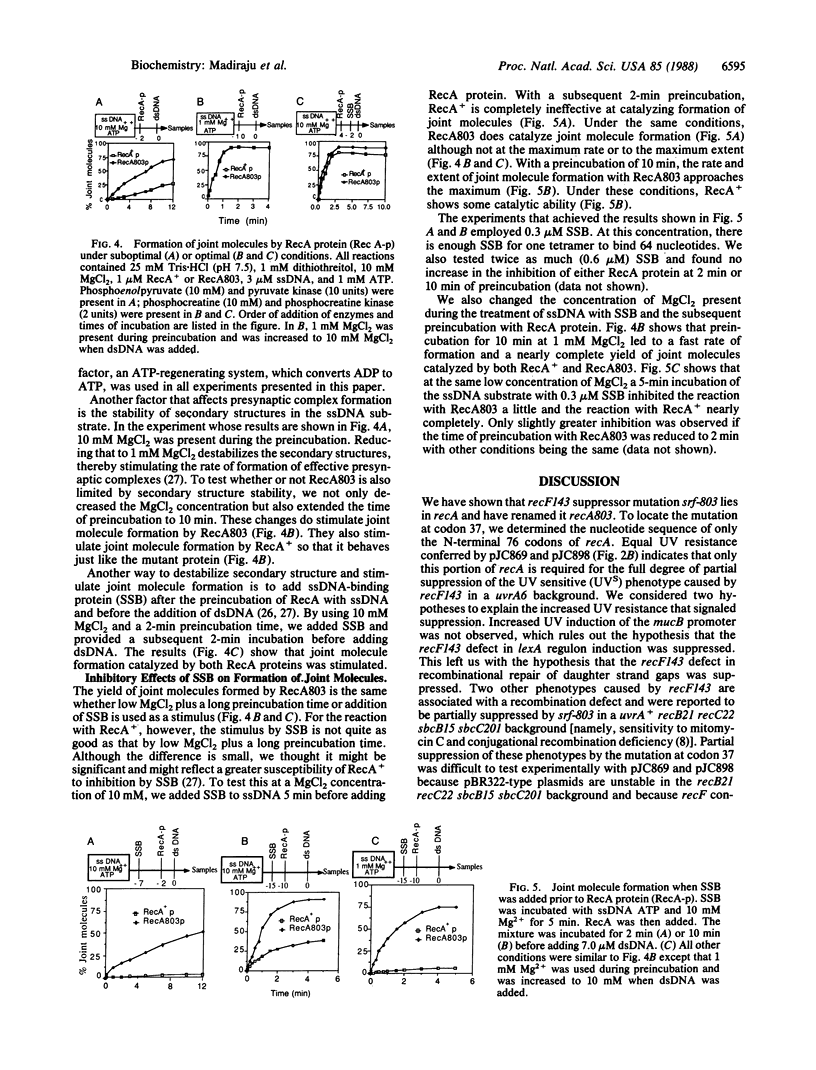
A mutation partially suppressing the UV sensitivity caused by recF143 in a uvrA6 background was located at codon 37 of recA where GTG (valine) became ATG (methionine). This mutation, originally named srf-803, was renamed recA803. Little if any suppression of the recF143 defect in UV induction of a lexA regulon promoter was detected. This led to the hypothesis that a defect in recombination repair of UV damage was suppressed by recA803. The mutant RecA protein (RecA803) was purified and compared with wild-type protein (RecA+) as a catalyst of formation of joint molecules. Under suboptimal conditions, RecA803 produces both a higher rate of formation and a higher yield of joint molecules. The suboptimal conditions tested included addition of single-stranded DNA binding protein to single-stranded DNA prior to addition of RecA. We hypothesize that the ability of RecA803 to overcome interference by single-stranded DNA binding protein is the property that allows recA803 to suppress partially the deficiency in repair caused by recF mutations in the uvrA6 background. Implications of this hypothesis for the function of RecF protein in recombination are discussed.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanar M. A., Sandler S. J., Armengod M. E., Ream L. W., Clark A. J. Molecular analysis of the recF gene of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Aug;81(15):4622–4626. doi: 10.1073/pnas.81.15.4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla Z., O'Brien P., Clark A. J. Genetic analysis of UV mutagenesis of the Escherichia coli glyU gene. Mol Gen Genet. 1987 Apr;207(1):1–8. doi: 10.1007/BF00331483. [DOI] [PubMed] [Google Scholar]
- Clark A. J., Sandler S. J., Willis D. K., Chu C. C., Blanar M. A., Lovett S. T. Genes of the RecE and RecF pathways of conjugational recombination in Escherichia coli. Cold Spring Harb Symp Quant Biol. 1984;49:453–462. doi: 10.1101/sqb.1984.049.01.051. [DOI] [PubMed] [Google Scholar]
- Cohen A., Clark A. J. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986 Jul;167(1):327–335. doi: 10.1128/jb.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M. M., Lehman I. R. Enzymes of general recombination. Annu Rev Biochem. 1987;56:229–262. doi: 10.1146/annurev.bi.56.070187.001305. [DOI] [PubMed] [Google Scholar]
- Cunningham R. P., DasGupta C., Shibata T., Radding C. M. Homologous pairing in genetic recombination: recA protein makes joint molecules of gapped circular DNA and closed circular DNA. Cell. 1980 May;20(1):223–235. doi: 10.1016/0092-8674(80)90250-0. [DOI] [PubMed] [Google Scholar]
- Davison J., Heusterspreute M., Merchez M., Brunel F. Vectors with restriction-site banks. I. pJRD158, a 3903-bp plasmid containing 28 unique cloning sites. Gene. 1984 Jun;28(3):311–318. doi: 10.1016/0378-1119(84)90148-3. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Walker G. C. The muc genes of pKM101 are induced by DNA damage. J Bacteriol. 1983 Sep;155(3):1306–1315. doi: 10.1128/jb.155.3.1306-1315.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn R., Radding C. M. Separation of the presynaptic and synaptic phases of homologous pairing promoted by recA protein. J Biol Chem. 1984 Jun 25;259(12):7495–7503. [PubMed] [Google Scholar]
- Kato T., Rothman R. H., Clark A. J. Analysis of the role of recombination and repair in mutagenesis of Escherichia coli by UV irradiation. Genetics. 1977 Sep;87(1):1–18. doi: 10.1093/genetics/87.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moreau P. L. Effects of overproduction of single-stranded DNA-binding protein on RecA protein-dependent processes in Escherichia coli. J Mol Biol. 1987 Apr 20;194(4):621–634. doi: 10.1016/0022-2836(87)90239-7. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Rothman R. H., Clark A. J. Defective excision and postreplication repair of UV-damaged DNA in a recL mutant strain of E. coli K-12. Mol Gen Genet. 1977 Oct 24;155(3):267–277. doi: 10.1007/BF00272805. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Amundsen S. K., Chaudhury A. M., Cheng K. C., Ponticelli A. S., Roberts C. M., Schultz D. W., Taylor A. F. Roles of RecBC enzyme and chi sites in homologous recombination. Cold Spring Harb Symp Quant Biol. 1984;49:485–495. doi: 10.1101/sqb.1984.049.01.055. [DOI] [PubMed] [Google Scholar]
- Tessman E. S., Peterson P. Plaque color method for rapid isolation of novel recA mutants of Escherichia coli K-12: new classes of protease-constitutive recA mutants. J Bacteriol. 1985 Aug;163(2):677–687. doi: 10.1128/jb.163.2.677-687.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms B., Wackernagel W. Regulatory role of recF in the SOS response of Escherichia coli: impaired induction of SOS genes by UV irradiation and nalidixic acid in a recF mutant. J Bacteriol. 1987 Apr;169(4):1731–1736. doi: 10.1128/jb.169.4.1731-1736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Clark A. J. Overproduction of the Escherichia coli recA protein without stimulation of its proteolytic activity. J Bacteriol. 1981 Oct;148(1):386–390. doi: 10.1128/jb.148.1.386-390.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Molin S., Gustafsson P., Nordström K. Plasmids with temperature-dependent copy number for amplification of cloned genes and their products. Gene. 1979 Jun;6(2):91–106. doi: 10.1016/0378-1119(79)90065-9. [DOI] [PubMed] [Google Scholar]
- Volkert M. R., Hartke M. A. Effects of the Escherichia coli recF suppressor mutation, recA801, on recF-dependent DNA-repair associated phenomena. Mutat Res. 1987 Nov;184(3):181–186. doi: 10.1016/0167-8817(87)90015-0. [DOI] [PubMed] [Google Scholar]
- Volkert M. R., Hartke M. A. Suppression of Escherichia coli recF mutations by recA-linked srfA mutations. J Bacteriol. 1984 Feb;157(2):498–506. doi: 10.1128/jb.157.2.498-506.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkert M. R., Margossian L. J., Clark A. J. Two-component suppression of recF143 by recA441 in Escherichia coli K-12. J Bacteriol. 1984 Nov;160(2):702–705. doi: 10.1128/jb.160.2.702-705.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T. C., Smith K. C. Mechanisms for recF-dependent and recB-dependent pathways of postreplication repair in UV-irradiated Escherichia coli uvrB. J Bacteriol. 1983 Dec;156(3):1093–1098. doi: 10.1128/jb.156.3.1093-1098.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis D. K., Uhlin B. E., Amini K. S., Clark A. J. Physical mapping of the srl recA region of Escherichia coli: analysis of Tn10 generated insertions and deletions. Mol Gen Genet. 1981;183(3):497–504. doi: 10.1007/BF00268771. [DOI] [PubMed] [Google Scholar]


