Abstract
Gallibacterium anatis is a pathogen in chickens and other avian species where it is a significant cause of salpingitis and peritonitis. We found that bacterial cells and cell-free, filter-sterilised culture supernatant from the haemolytic G. anatis biovar haemolytica were highly cytotoxic towards avian-derived macrophage-like cells (HD11). We obtained the genome sequence of G. anatis 12656-12 and used a rational approach to identify a gene predicted to encode a 2026 amino acid RTX-toxin, which we named GtxA (Gallibacterium toxin). The construction of a gtxA knock-out mutant showed gtxA to be responsible for G. anatis’ haemolytic and leukotoxic activity. In addition, Escherichia coli expressing gtxA and an adjacent acyltransferase, gtxC, became cytolytic. GtxA was expressed during in vitro growth and was localised in the extracellular protein fraction in a growth phase dependent manner. GtxA had an unusual modular structure; the C-terminal 1000 amino acids of GtxA were homologous to the classical pore-forming RTX-toxins in other members of Pasteurellaceae. In contrast, the N-terminal approximately 950 amino acids had few significant matches in sequence databases. Expression of truncated GtxA proteins demonstrated that the C-terminal RTX-domain had a lower haemolytic activity than the full-length toxin, indicating that the N-terminal domain was required for maximal haemolytic activity. Cytotoxicity towards HD11 cells was not detected with the C-terminal alone, suggesting that the N-terminal domain plays a critical role for the leukotoxicity.
Keywords: RTX-toxin, leukotoxin, haemolysin, expression, mutagenesis
1. INTRODUCTION
Inflammation in the reproductive organs and peritoneum of egg-layers is a recurrent problem in commercial egg-layer flocks causing egg production drop, increased mortality and consequential economical losses and lowered animal welfare [4, 21]. Avian pathogenic Escherichia coli is often isolated from these lesions, but, several studies have demonstrated Gallibacterium anatis to be a frequent cause of oophoritis, salpingitis and peritonitis, either alone or as a co-pathogen [13, 25, 28, 29]. Moreover, G. anatis has been isolated from avian cases of septicaemia, hepatitis, enteritis and upper respiratory tract lesions [15, 17, 24, 25, 28]. G. anatis is a common part of the normal flora of both the upper respiratory tract and lower genital tract of chickens and other avian species [6], and can therefore be regarded as an opportunistic pathogen. Its pathogenesis has not been studied in depth, particularly not at the molecular level, and little is known about the genes and mechanisms behind G. anatis’ ability to cause disease. The activity of extracellular proteases and the ability to degrade of chicken IgG have been suggested to be involved [12], as has the products of a capsule biosynthesis locus [7]. G. anatis is divided into two biovars: the β-haemolytic biovar haemolytica and the non-haemolytic biovar anatis. The ability to lyse red blood cells is a prominent phenotype of pathogenic G. anatis isolates [9], and the produced haemolysin is a likely virulence factor. Gallibacterium is a Gram-negative genus belonging to the γ-proteobacterial family Pasteurellaceae [9], and various pathogenic members of Pasteurellaceae, e.g. the cause of periodontal disease in humans, Aggregatibacter actinomycemcomitans, the causative agent of bovine shipping fever Mannheimia haemolytica, and the swine pathogen Actinobacillus pleuropneumoniae produce haemolysins and leukotoxins belonging to the group of RTX-toxins (repeat in toxin). The RTX-toxins are important virulence factors and strains lacking these genes have reduced virulence [19, 39, 40]. A homologous toxin, HlyA, is produced by certain extra-intestinal strains of E. coli and has been the model toxin of this group. These pore-forming RTX cytotoxins share a number of common structural features. They are large (> 100 kDa), secreted proteins, containing varying numbers of nine amino acid (aa) glycine- and aspartate-rich calcium-binding repeats, and calcium is required for function. The proteins are exported by specific type-I secretion system (T1SS), most often encoded in the same transcriptional unit as the toxin [11]. A likewise co-transcribed acyltransferase posttranslationally adds acyl-groups to specific lysine residues in the toxin [37], these acyl-groups are essential for toxin function. The aim of this study was to examine G. anatis biovar haemolytica’s interactions with eukaryotic cells and to identify and characterize the genes and proteins responsible for the haemolytic phenotype. We found G. anatis to be highly cytotoxic towards avian macrophages, a trait likely to play a key role in pathogenesis. Furthermore, we identified and characterised a new type of RTX-toxin responsible for the leukotoxic and haemolytic activity in G. anatis biovar haemolytica.
2. MATERIALS AND METHODS
2.1. Bacterial strains and growth conditions
G. anatis biovar haemolytica strain 12656-12 Liver (referred to as 12656-12) was used in this study, this strain was originally isolated from the liver of a septicaemic chicken [5]. G. anatis 12656-12 was grown at 37 °C either on brain heart infusion (BHI) (Oxoid, Basingstoke, UK) agar supplemented with 5% citrated bovine blood in a closed plastic bag, or in BHI broth with aeration. Anaerogen (Oxoid) was used to produce anaerobic conditions in incubator jars. E. coli strains were grown in Luria–Bertani broth and agar, the medium was supplemented with 50 μg/mL kanamycin and 20 μg/mL chloramphenicol when appropriate. All chemicals were purchased from Sigma-Aldrich (St. Louis, USA).
2.2. Bioinformatics analyses
A draft version (115 contigs) of the genome sequence of G. anatis biovar haemolytica 12656-12 Liver1 was obtained from 454 Life Sciences (Branford, USA), using the pyrosequencing-based method [27]. Gene annotation was performed using Wasabi, a web-based annotation system for prokaryotic organisms provided by the Victorian Bioinfomatics Consortium, Monash University, Australia [8]. Sequence similarity searches were performed using BLASTP [1] (databases: non-redundant protein sequences (GenBank) and SwissProt), FASTA [30] and SSEARCH (databases: UniProtKB and SwissProt), and HHpred [36] (database: Interpro, 2009). All searches were performed in March 2009. Transterm [23] was used to predict transcriptional terminators.
2.3. Haemolysis assay
The haemolytic activity was assayed as previously described [33]; bovine blood was washed repeatedly in TN Buffer (10 mM Tris-HCl, 0.9% NaCl, pH 7.5) until the upper phase appeared colourless. A 2% (vol/vol) erythrocyte solution was prepared in TN-buffer supplemented with 10 mM CaCl2. Erythrocytes were incubated with filter-sterilised bacterial culture supernatant or bacteria in a 1:1 ratio at 37 °C for 1 h unless otherwise noted. Un-lysed erythrocytes and cell debris were collected by centrifugation and the amount of released haemoglobin was measured in a plate reader at 540 nm. 100% lysis was determined with 1% triton-X and background lysis was subtracted before calculation of haemolytic activity. The effect of heat was examined by incubating the supernatant at 60 °C for 30 min before the haemolysis assay. The effect of proteinase K was examined by incubating the supernatant with 4 μg/mL proteinase K at 37 °C for 30 min before haemolysis assay.
2.4. Culturing of HD11 cells and lactate dehydrogenase (LDH) cytotoxicity assay
The macrophage-like cell line HD11 derived from MC29 transformation of chicken bone marrow cells [3] was maintained in Roswell Park Memorial Institute (RPMI) 1640 medium + GlutaMAXTM-I + 25 mM HEPES (Gibco, Carlsbad, USA). The media was supplemented with 2.5% chicken serum, 7.5% foetal bovine serum (FBS), and 25 μg/mL gentamicin. The cells were cultured as an adherent cell line at 37 °C with an atmosphere of 5% CO2 and were sub-cultured every 2nd or 3rd day. For the cytotoxicity assays the cells were seeded in 96 well plates with 2 × 104 cells in RPMI added 5% FBS in a total volume of 100 μL. The cells were incubated overnight, and the media was changed. Ten μL filter-sterilised culture supernatant or bacteria resuspended in saline (0.9% NaCl) was added to the cells and incubated for 1 h. For E. coli, expression of recombinant proteins were induced as described in Section 2.7 and the OD600 (optical density at 600 nm) was adjusted to 1 corresponding to approximately 6 × 108 CFU/mL. G. anatis cells and supernatant was harvested in late exponential phase (OD600 = 1). The suspension of G. anatis was adjusted to OD600 of 1 corresponding to approximately 4 × 108 CFU/mL. Filter-sterilised culture supernatant was stored on ice and added to cells within 30 min after harvest. Cytotoxicity was determined with LDH cytotoxicity assay (Promega, Madison, USA) as described by the manufacturer. Each sample was assayed in triplicate wells and the experiments were repeated a minimum of three times.
2.5. Construction of a G. anatis gtxA mutant
A 1 509 bp fragment consisting of nucleotides 140 to 1648 of gtxA was PCR-amplified with primers 4240 and 4242, and a 1 484 bp fragment consisting of nucleotides 3995 to 5478 was amplified with primers 4243 and 4245 (primer sequences are listed in Tab. I). The two fragments were digested with restriction enzymes and ligated into the corresponding restriction sites in plasmid pWSK129 [42]. The gel-purified kanamycin-cassette (Tn903) from EcoRI-digested pUC4-KISS [2] was ligated into the EcoRI site between the two PCR-fragments. The kanamycin resistance gene was inserted in the same transcriptional direction as gtxA. The plasmid DNA was linearised by digestion with XhoI and SalI and column purified. The natural competence of G. anatis 12656-122 was induced by the MIV-method as previously described for Haemophilus influenzae [31]; G. anatis was grown in BHI to an OD600 of 0.2, washed once in MIV and incubated in MIV for 100 min. The linear DNA was added to the cells at a concentration of 0.5 μg DNA/mL. After 20 min, two volumes of BHI were added and the bacteria were incubated for 1 h before transformants were selected on blood agar plates with 5 μg/mL kanamycin. Colonies were re-streaked and the deletion was verified with primer pairs 39F + kanR, kanF + 5334R and 2871F + 3270R. The strain was designated ΔgtxA.
Table I.
Primer list. Primers used for construction and verification of gtxA mutants. Restriction sites are underlined.
| Primer name | Primer sequence (5′-3′) |
|---|---|
| 4240 | TATCGTCGACTATCCATCGCGGCATCAG |
| 4242 | AGCTGAATTCAAGCAAGTGCTATTGCTACCG |
| 4243 | AGCTGAATTCTTATGTCGGCGATCAAACAA |
| 4245 | TATGTCTAGAGGCGTTGGTGGATAAGAGAT |
| kanR | CGATAGATTGTCGCACCTGA |
| kanF | TATGGAACTGCCTCGGTGA |
| 39F | TGATGCAATCAAAGATAAAGTCG |
| 5734R | AATCGGCATTGGAGCTTTC |
| 2871F | AACCAAACCAATCCAAGGT |
| 3270R | ATTGCCGTCTTTGCCTACTG |
2.6. Construction of expression plasmids
Plasmids encoding full-length GtxA, the N-terminal domain of GtxA (amino acids 1–949 (Nterm)) and the RTX-domain of GtxA (amino acids 931–2026 (RTX)) with (+C) and without GtxC were constructed by the ligation of PCR-fragments into the expression vector pET28a (Novagen, EMD Biosciences, San Diego, USA). The PCR-fragments were amplified with pfx50 polymerase (Invitrogen, Carlsbad, USA), column purified, double digested with NcoI and XhoI and either column purified again or gel purified (fragments > 6 kb). The primers used for each construct are listed in Table II. The construct Nterm+C, nucleotides 1–2847 of gtxA in operon with gtxC, was made by the use of splicing by overlap extension [18], where the primers GtxUP-NcoI and gtxC-r-XhoI were used in the second round of PCR. Plasmid pET28a was double digested with NcoI and XhoI, dephosphorylated with Antarctic phosphatase (New England Biolabs, Ipswich, USA) and gel purified. Vector and PCR-fragments were ligated at a molar ratio of 1:3, transformed into chemically competent E. coli Top10F′ (Invitrogen) and selected on LB-agar plates with kanamycin. The sequence of the insert in each plasmid was verified by DNA-sequencing (Macrogen, Seoul, Korea). The plasmids were transformed into the E. coli expression strain ER2566 (New England Biolabs). Plasmid pLG575 encodes E. coli hlyB and hlyD, components of the T1SS secreting HlyA [26], and was introduced to promote secretion of the expressed proteins.
Table II.
Primer list. Primers used for constructs for expression in E. coli. Restriction sites are underlined. Bold indicates overlapping regions for splicing by overlap extension.
| Primer name | Primer sequence (5′-3′) | Construct | Primers used |
|---|---|---|---|
| GtxUP-NcoI | AGTCCCATGGGTCTTTCATTAAAAGAAAAAGTAACTGGAATA | gtxA+C | GtxUP-NcoI & gtxC-r-XhoI |
| gtxC-r-XhoI | CAGTCTCGAGTTATGAATTTTCTTCTATAAAAGCAGC | gtxA | GtxUP-NcoI & gtxA-r-XhoI |
| gtxA Cf NcoI | AGTCCCATGGCAATTGAATCTTTCAATTTAATCGCAA | RTX+C | gtxA Cf NcoI & gtxC-r-XhoI |
| gtxA-Nr-XhoI | CAGTCTCGAGTTAATTTAGGAAATCGGTCATTATGCCAT | RTX | gtxA Cf NcoI & gtxA-r-XhoI |
| gtxA-r-XhoI | CAGTCTCGAGTTAAACAAGATACATAGTGACCAGTTCAT | Nterm+C | GtxUP-NcoI & SOErev1 |
| SOErev1 | GTTATCCATAATAATTAATTTAGGAAATCGGTCATTATG | SOEfor2 & gtxC-r-XhoI | |
| SOEfor2 | TTCCTAAATTAATTATGGATAACTTCTCAACTTTAGG | Nterm | GtxUP-NcoI & gtxA-Nr-XhoI |
2.7. Expression of recombinant GtxA proteins in E. coli
Protein expression was induced on agar plates containing 0.1 mM IPTG incubated at 30 °C. For induction in broth, an overnight culture was diluted 1:50 and incubated at 37 °C with shaking until the culture reached an OD600 of 0.6. Then, IPTG (0.2 mM) was added and induction was maintained for 2 h at 30 °C. To release recombinant protein from the cells, cells were pelleted by centrifugation, resuspended in 0.1 M Tris/0.9% NaCl in 1/25 of the original volume, lysed by bead beating (FastPrep) for 45 s and spun down at 4 °C, and the supernatant was used immediately for liquid haemolysis assays and LDH cytotoxicity assay.
2.8. SDS-PAGE and Western blot analysis
Total cellular protein was obtained by harvesting 500 μL of culture and resuspending the cell pellets in 10 mM Tris, 500 μL/ per OD unit at the time of harvest. Extracellular proteins were prepared from filter-sterilised culture supernatant (low protein binding filter (0.22 μm) (Millex® GP, Millipore, Billerica, USA). Proteins were precipitated overnight by the addition of one volume ice-cold 96% ethanol, collected by centrifugation (13 000 g for 30 min at 0 °C), and resuspended in 10 mM Tris (1/100 of the original volume). Proteins were separated by SDS-PAGE in NuPAGE® Novex gels (Invitrogen). For Western blot analysis, proteins were transferred to polyvinylidene difluoride membranes (Invitrogen). The primary antibody, rabbit antiserum raised against ApxI from A. pleuropneumoniae [35], was used at a 1:1333 dilution and detected with Westernbreeze Chemiluminiscent Western Blot Immunodetection Kit (Invitrogen) as described by the manufacturer.
2.9. RNA purification
An overnight culture was diluted 1:100 in BHI and incubated at 37 °C with aeration. Cells were harvested at OD600 0.17, 0.6, 2, 3, as well as 1 h after growth had stopped and after 24 h of incubation. Total RNA was isolated with RNeasy protect Mini Kit (Qiagen, Hilden, Germany), on-column DNAse treatment was performed as described by the manufacturer (Qiagen).
2.10. Northern blotting
Blotting of RNA, probe labelling (with [α-32P]-dCTP) and hybridization was performed basically as described [10]. A 388 bp fragment within the RTX-half of gtxA was PCR-amplified with primers 3487F 5′-GCCTCTACCGCCGTTTCTG-3′ and 3874R 5′-GGCTGGCTAATAATTCATCACCTTG-3′ and used as template in the probe labelling reaction.
2.11. Nucleotide sequence accession number
The sequence corresponding to G. anatis gtxA and gtxC has been deposited in GenBank under accession number FJ917356.
3. RESULTS
3.1. Cytolytic activity of G. anatis
G. anatis biovar haemolytica is β-haemolytic on bovine-blood agar plates [9]. To test the target range we examined the haemolysis of strain 12656-12 on agar plates with blood from different species, and the bacterium produced clear zones of β-haemolysis when grown on agar plates containing horse, cow, swine or chicken blood (data not shown), confirming the broad target range reported by Greenham and Hill [14]. The bacterium was haemolytic under both aerobic and anaerobic culture conditions (data not shown).
This haemolytic activity has been suggested to originate from a secreted haemolysin [14]. To test this, liquid haemolysis assays were performed, where G. anatis 12656-12 cell-free culture supernatants, harvested in different phases of growth, were incubated with a suspension of bovine erythrocytes and the amount of released haemoglobin measured. G. anatis supernatant from mid to late exponential phase lysed the erythrocytes efficiently (Fig. 1A). This activity was inactivated by heat (60 °C) and proteinase K, and was reduced by the calcium chelater EGTA (data not shown), confirming that G. anatis produces a calcium-dependent secreted haemolytic protein.
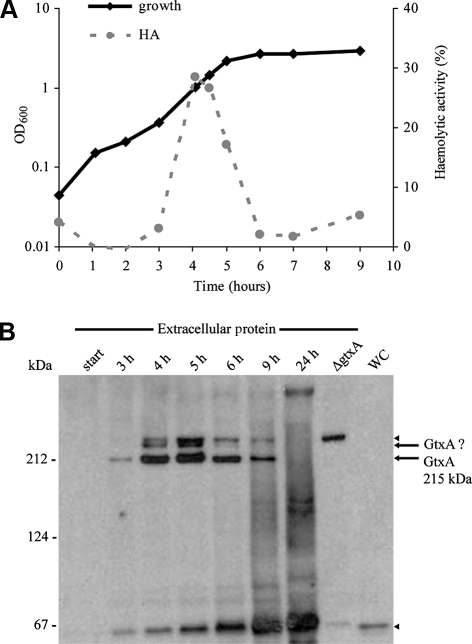
Figure 1.
Haemolytic activity of G. anatis culture supernatants and expression of GtxA. (A) Growth and haemolytic activity of cell-free culture supernatant of G. anatis 12656-12. An overnight culture was diluted 1:100 and growth (cell density measured by absorbance at 600 nm) and haemolytic activity (HA) in cell-free culture supernatant were recorded. The haemolytic activity of supernatant diluted 100-fold in BHI is shown. Extracellular proteins for Western blotting (Fig. 1B) were harvested in parallel. The experiment was repeated three times, the level of haemolytic activity varied but the relative pattern was consistent. (B) Levels of GtxA determined by Western blotting with ApxI-antiserum. Supernatants were harvested at the indicated time points, concentrated 100-fold as described in Materials and Methods and separated by SDS-PAGE in a 3–8% gel prior to blotting. Extracellular proteins from ΔgtxA were harvested at OD600 = 2 (lane marked ΔgtxA). WC = whole cell lysate from wild-type, the cells were harvested 5 h after inoculation. Size markers are indicated on the left. The experiment was repeated with the same result.
The lysis of erythrocytes may play a role in iron acquisition in the host, however, interactions with other types of cell, e.g. leukocytes, may play a more important role during natural infection. We therefore tested G. anatis’ cytotoxic activity towards leukocytes using the avian-derived macrophage-like cell line HD11. The HD11 cells showed rounding and detached from the surface after exposure to G. anatis (Fig. 2B). The cytotoxicity was quantified using the LDH cytotoxicity assay, which showed a pronounced cell death (Fig. 2C). This leukotoxic activity of G. anatis is likely to be essential in the pathogenesis of this bacterium and proteins responsible for the leukotoxic activity are thus expected to be important virulence factors.
Figure 2.
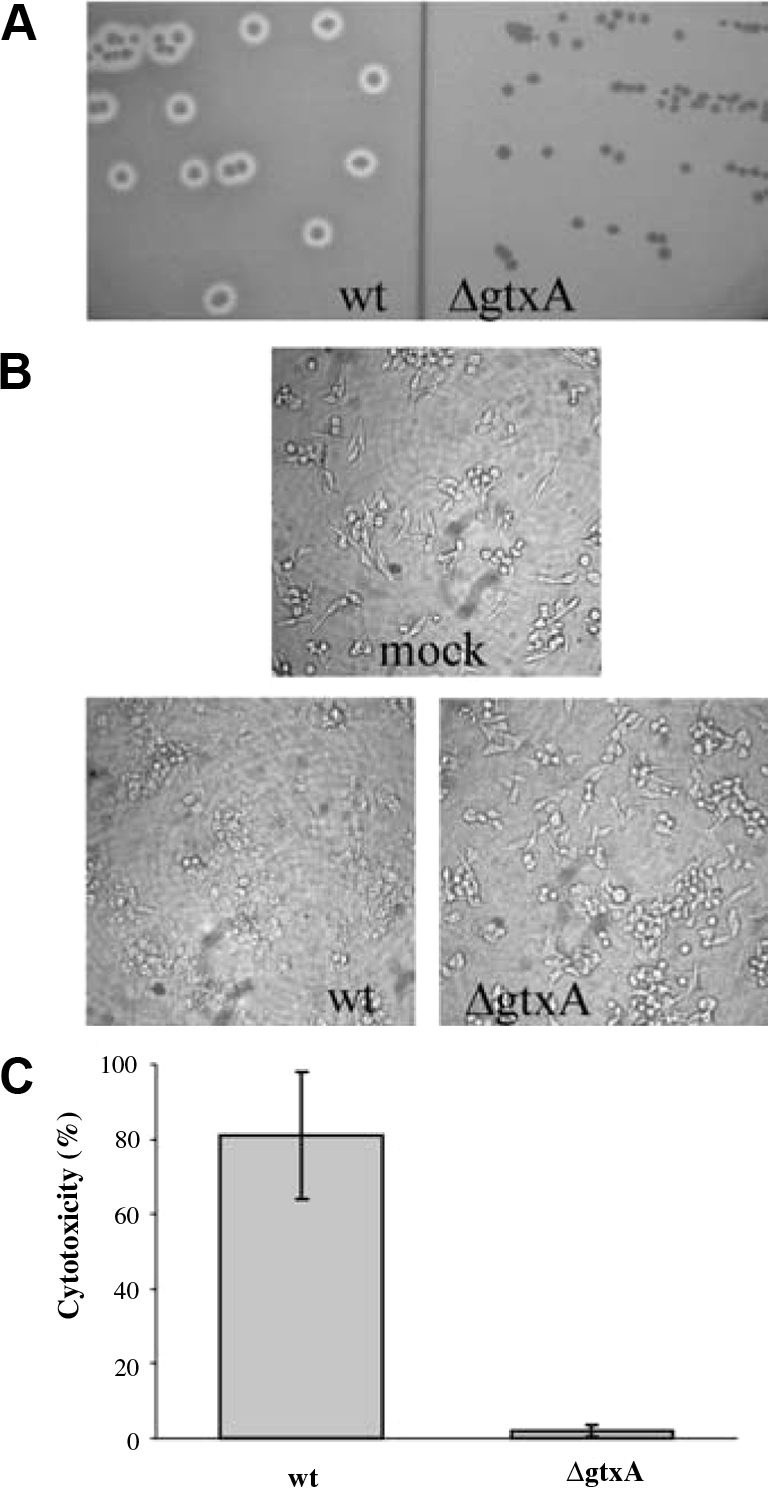
Haemolytic activity and cytotoxicity of G. anatis 12656-12 wild-type (wt) and the isogenic gtxA mutant (ΔgtxA). (A) β-haemolysis. The bacteria were streaked on BHI-agar plates with 5% bovine blood and incubated at 37 °C for 18 h. (B) Light microscopy (100 × magnifications) of HD11 cells after 1 h incubation with saline (mock), wt or ΔgtxA. Bacteria were harvested in late exponential phase (OD600 = 1) and added at an m.o.i. of 10. (C) Cytotoxicity quantified with LDH activity. HD11 cells were incubated with bacteria as described in 1B. The averages of three replicate wells are shown, bars represent the S.E.
3.2. Identification of an RTX-toxin in G. anatis’ genome sequence
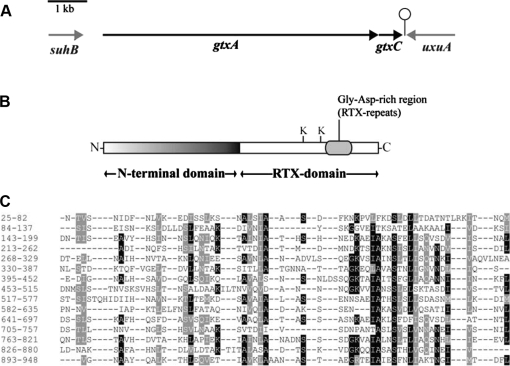
To identify a specific protein responsible for G. anatis’ cytotoxic phenotype, we obtained the genome sequence of G. anatis 12656-12 and searched for sequences encoding possible toxins. Proteins belonging the group of pore-forming RTX-toxins are important virulence factors and responsible for haemolytic and leukotoxic activity in bacteria related to Gallibacterium [11], making proteins of this type an obvious target to search for. BLAST searches with the amino acid sequences of different RTX-toxins (including ApxI and ApxII from A. pleuropneumoniae, LtxA from A. aggregatibacter and HlyA from E. coli) against the G. anatis 12656-12 genome sequence led to the identification of a putative G. anatis RTX-toxin of 2026 amino acids. The 6081 nucleotide (nt) open reading frame (ORF) encoding this protein was named gtxA: gtx for Gallibacterium toxin and A by analogy to the designation of toxin gene in other RTX operons [11]. gtxA is followed by a very short five nucleotides (nt) intergenic region and a 492 nt ORF (Fig. 3A) encoding a predicted protein of 163 amino acids. This protein has homology to acyltransferase proteins, which are required for activation of RTX-toxins, and showed 38% identity and 60% similarity to the acyltransferease HlyC from E. coli. By analogy, the gene was named gtxC. A rho-independent transcriptional terminator was found downstream of gtxC and probably marks the end of a transcriptional unit including both gtxA and gtxC. The genes flanking the gtxA-C operon were predicted to encode an inositol-1-monophosphatase (suhB upstream of gtxA), and a mannoate dehydratase gene (uxuA downstream of gtxC) (Fig. 3A), both of which are unlikely to be involved in GtxAC function.
Figure 3.
(A) The genetic organisation of gtxA, gtxC and their flanking genes in G. anatis 12656-12. Arrows indicate ORF. A predicted transcriptional terminator is indicated downstream of gtxC. (B) Organisation of GtxA. K indicates conserved lysine residues (Lys1484 and Lys1607). The glycine aspartate-rich region (position 1640–1830) is marked. (C) Alignment of the 15 repeats in the N-terminal domain of GtxA, the alignment was generated with Radar [16]. Numbers to the left indicate amino acid position in GtxA. Positions where the amino acid are identical (black) or similar (grey) in more than 50% of the repeats are marked.
Interestingly, GtxA (2026 aa) is twice as large as the “typical” RTX-toxins (approximately 1000 aa [11]) described from other members of the Pasteurellaceae family and HlyA from E. coli. The 1000 amino acids at the C-terminus of GtxA are homologous to these RTX-toxins, with whom the region shares approximately 20% sequence identity and 35% sequence similarity. This C-terminal region also contains several of the conserved features of RTX-toxins. HlyA from E. coli is acylated at Lys564 and Lys690 [37]; both these lysine and the preceding glycine residues are conserved in GtxA (Lys1484 and Lys1607 (Fig. 3B)), so these are likely acylation sites in GtxA mediated by GtxC. Downstream of the predicted acylation sites (aa 1640–1830), GtxA has a glycine and aspartate-rich region, which is also a conserved feature of the RTX-toxins.
In contrast, the N-terminal region (aa 1 to approximately 950) had limited similarity to available sequences, and no significant homologues were found by BLASTP searches against the GenBank database. However, the region from aa 520 to 879 had similarity (E-value 0.007) to a conserved domain (COG1511) of unknown function from predicted membrane proteins. Compared to the RTX-domain, the N-terminal domain is less acidic and contains a larger proportion of hydrophobic amino acids, particularly serine. The secondary structure was predicted to consist primarily of alpha-helices.
To get an idea of the function of the N-terminal domain, we performed a bioinformatic analysis of its amino acid sequence by the use of more sensitive search tools for sequence similarity and homology prediction. Homology searches using FASTA and SSearch found sequence similarity to the eukaryotic cytoskeletal proteins Talin-A and Talin-B from the amoeba Dictyostelium discoide (E-value 3.4 × 10−7 and 0.0061, respectively), and Talin from chickens (Gallus gallus) (E-value 0.0087). Furthermore, the homology detection server HHpred predicted homology to talin (probability = 100%). Talin binds to a range of other proteins, including actin, vinculin and the cytosolic part of integrins. Large proteins often consists of repeats arisen by duplications and examination of the N-terminal domain with the repeat finder Radar [16], found 15 repeats of 57 amino acids (Fig. 3C).
GtxA thus consists of two domains: an N-terminal repeat domain and a C-terminal RTX/cytolysin domain.
3.3. GtxA has cytolytic activity which is dependent on GtxC
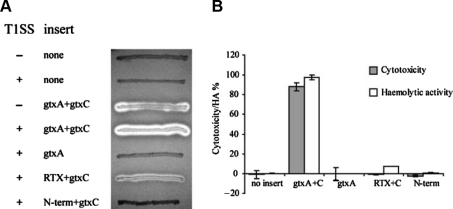
To examine if GtxA is a cytolytic protein, gtxA was cloned together with the predicted acyltransferase gene, gtxC, and introduced into the non-haemolytic expression strain E. coli ER2566. Upon expression of gtxA and gtxC, this strain exhibited a haemolytic phenotype on blood agar plates and in liquid haemolytic assays (Fig. 4), showing that GtxAC holds haemolytic activity. RTX-toxins are usually extracellular proteins exported by specific T1SS. Introduction of a plasmid (pLG575) expressing the T1SS encoded by E. coli hlyBD increased the haemolysis zone (Fig. 4A) and immunoblotting showed that a larger fraction of GtxA was present in the extracellular protein fraction (Fig. 5), demonstrating that the E. coli secretion system can secrete G. anatis GtxA. The cytotoxic activity of GtxA towards HD11 cells was assayed by LDH release assay and E. coli ER2566 expressing gtxAC was toxic to HD11 cells. E. coli containing vector with no insert (negative control) showed no toxicity after 1 h incubation (Fig. 4B).
Figure 4.
Cytotoxic activity of E. coli expressing GtxA. (A) β-haemolytic activity of E. coli ER2566 grown on LB-agar with 5% bovine blood and 0.1 mM IPTG, incubated at 30 °C. T1SS: + or − indicates the presence or absence of plasmid pLG575 which expresses the E. coli T1SS-components HlyB and HlyD. RTX = amino acids 931–2026 of GtxA, N-term = amino acids 1–949 of GtxA. (B) Liquid haemolysis assay and LDH cytotoxicity assay with E. coli ER2566/pLG575 expressing different versions of GtxA.
Figure 5.
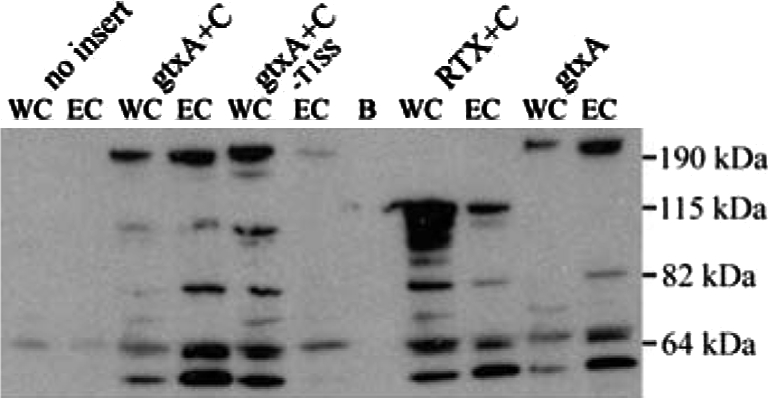
Expression of GtxA in E. coli. Western blot on whole cell lysate (WC) and extracellular protein (EC) from E. coli ER2566/pLG575 after induction with IPTG. Proteins were separated by SDS-PAGE in a 4–12% gel and blotted on a PVDF-membrane. The blot was probed with ApxI-antiserum. B = blank, -T1SS indicates the absence of plasmid pLG575. Size markers are indicated on the right (PageRuler Prestained Protein Ladder Plus, Fermentas, Burlington, Canada). The upper band in each lane has the expected size of full-length GtxA (215 kDa) or the RTX-domain (117 kDa). The smaller sized bands are likely degradation products.
The requirement of post translational acylation is one of the hallmarks of RTX-toxins. To assess whether this was also the case for the atypical GtxA, we examined the activity of GtxA expressed without its predicted acyltransferase GtxC. Figure 4 shows that when GtxA was expressed in the absence of GtxC it had no cytolytic activity against erythrocytes or leukocytes. Thus, the non-acylated protoxin is inactive, and posttranslational acylation is essential for both its haemolytic and leukotoxic activities. The secretion of GtxA was not hindered by the lack of acylation, as the non-acylated GtxA was detected in the culture supernatant in amounts similar to the acylated toxin (Fig. 5).
3.4. GtxA is responsible for G. anatis’ cytotoxic activity
To determine whether G. anatis’ haemolytic and leukotoxic activity originated from GtxA, we constructed a gtxA knock-out mutant. No molecular tools for genetic manipulation of Gallibacterium had previously been described, but, we found that G. anatis 12656-12 is naturally competent3, a trait we exploited in the construction of stable gtxA mutants by natural transformation. In the resulting mutants, the 2347 nucleotides between positions 1648 and 3995 in gtxA were deleted and replaced by a kanamycin resistance cassette.
In contrast to the wild-type, the ΔgtxA mutant was not haemolytic on blood agar plates (Fig. 2A) or in liquid haemolysis assay (data not shown). Furthermore, ΔgtxA showed no cytotoxicity towards HD11 cells (Figs. 2B and 2C). Identical results were obtained from two independently constructed gtxA mutants. Thus, gtxA is responsible for the haemolytic and leukotoxic activity of G. anatis.
3.5. Growth phase dependent levels of GtxA and its activity
The haemolytic activity of G. anatis supernatant was growth phase dependent: the activity peaked in late exponential phase, dropped at the transition to stationary phase, and was low in the supernatant from overnight cultures (Fig. 1A). This prompted us to hypothesize that the expression of GtxA was similarly growth phase dependent. To examine this and to establish GtxA’s localisation, we determined the amount of GtxA in the culture supernatant (extracellular proteins) and whole cell lysates at different times throughout growth using immunoblotting with ApxI-antiserum (Fig. 1B). The ApxI-antiserum recognised several proteins in the extracellular protein fraction including a band corresponding to the size of the predicted molecular mass of full-length GtxA (215 kDa). This band was absent in ΔgtxA, supporting that the band is GtxA. Like the haemolytic activity, the presence and amounts of GtxA in the supernatant were growth phase dependent (Fig. 1B): the amount of GtxA peaked at the transition to stationary phase, and the protein was not detected in day-old cultures (24 h) which is similar to the pattern reported for A. pleuropneumoniae ApxI and ApxII [20] and M. haemolytica LktA [38]. A second band (> 215 kDa) was also present in wild-type but absent in ΔgtxA suggesting that GtxA may exist in two different forms, possibly due to post translational modifications. Two further bands (65 kDa and > 215 kDa, respectively) were detected in both wild-type and mutant and are likely not related to GtxA. No protein of the size of GtxA was detected in whole cell lysates at any time point, consistent with the predicted extracellular localisation and in support of GtxA being secreted immediately after or in connection with synthesis. To examine transcription of gtxA, Northern blotting was performed with RNA from cells harvested throughout the growth phase. The blots showed gtxA to be transcribed during exponential growth and in the beginning of stationary phase, but no transcript was detected 2 h into stationary phase and in overnight cultures (data not shown), indicating the transcription of gtxA was shut down during stationary phase. In conclusion, GtxA is expressed during in vitro growth, it is a growth phase dependent extracellular protein and the growth phase dependence is influenced by transcriptional regulation, and the balance between accumulation of secreted GtxA and its subsequent degradation.
3.6. The N-terminal domain of GtxA is required for full cytolytic activity
The bioinformatical analysis showed that GtxA has an atypical organisation compared to other pore-forming RTX-toxins consisting of two parts, an RTX-domain and an N-terminal domain (Fig. 3B). To examine the contribution of the N-terminal domain to the cytolytic activity of GtxA, both the N-terminal domain (amino acids 1–949) and the RTX-domain (amino acids 931–2026) were expressed separately in E. coli and their haemolytic and leukotoxic activities examined and compared to those of the full-length protein (Fig. 4). E. coli expressing the RTX-domain together with GtxC showed haemolytic activity on blood agar plates and in liquid haemolysis assays, thus, the RTX-domain is a functional haemolytic protein by itself and the N-terminal domain is not essential for the lysis of red blood cells. However, the RTX-domain did exhibit a markedly lower haemolytic activity than the whole toxin, indicating that the N-terminal domain is required for the full haemolytic activity. No cytotoxic activity was detected from interactions between the RTX-domain and HD11 cells suggesting that the N-terminal domain plays an essential role for leukotoxicity. Immunoblotting (Fig. 5) showed that the RTX-domain was expressed and exported; therefore the differences in activity were not due to major differences in expression levels.
E. coli expressing only the N-terminal domain had no haemolytic or cytotoxic activity (Fig. 4). However, SDS-PAGE showed that this recombinant protein was primarily present in whole cell fraction and only in diminutive amounts in the extracellular protein fraction (data not shown). We therefore tested the activity of lysates (generated by FastPrep bead beating or lysozyme/sonication treatment) of cells expressing the N-terminal domain, and these did not show any activity of the in liquid haemolysis assays or towards avian macrophages, despite prolonged incubation. These results indicate that the N-terminal domain has no cytolytic activity by itself.
4. DISCUSSION
In this study, we report the first identification and characterisation of a specific virulence factor in Gallibacterium, named GtxA. GtxA is a new type of RTX-toxin and is responsible for G. anatis’ leukotoxic and haemolytic activity. GtxA shares sequence similarity with RTX-toxins from related members of Pasteurellaceae and the α-haemolysin HlyA from E. coli. Interestingly, GtxA deviates from these toxins and the group of pore-forming RTX-toxins in general because it contains an extra 950 amino acid N-terminal domain. We demonstrated that GtxA’s RTX-domain retained haemolytic activity in the absence of the N-terminal domain, confirming that it constitutes a functional unit by itself. The N-terminal domain was, however, required for full haemolytic activity of the toxin, and appeared essential for its leukotoxic activity (Fig. 4), suggesting a synergistic effect of the two domains.
The modular organisation of GtxA resembles the organisation of the adenylate cyclase toxin, CyaA, from Bordetella pertussis, which consists of two functional parts; an N-terminal adenylate cyclase domain and a C-terminal RTX-haemolysin domain [41]. However, GtxA’s N-terminal domain has no homology to the adenylate cyclase domain of CyaA, or any other type of RTX-toxin. The RTX-haemolysin domain of CyaA binds to integrin (CD11b/CD18) on the target cell, inserts into the membrane and forms a pore through which the adenylate cyclase domain is transported into the cytoplasm of the target cell. Once inside the target cell, the adenylate cyclase domain catalyzes the conversion of ATP to cyclic AMP (cAMP), a key secondary messenger. The increase in cAMP concentration affects signal transduction in the host cell [41]. The CyaA RTX-haemolysin domain also has pore-forming (haemolytic) activity on its own [34] similar to the RTX-domain of GtxA. We hypothesize that GtxA employs a similar mode of action, where the RTX-haemolysin domain is responsible for binding to target cells and the translocation of either the N-terminal domain alone or the whole toxin into the cytoplasm of the target cell. Both the predicted presence of sequence repeats (Fig. 3C) and the homology to eukaryotic cytoskeletal protein talin suggest a role for the N-terminal of GtxA in protein binding. We propose that the N-terminal domain of GtxA is involved in protein-protein interactions with intracellular host cell proteins, the cytoskeletal proteins actin and vinculin are potential candidates. The binding to cytoskeletal proteins may lead to direct morphological changes and interfere with signal transduction within the cell.
The genetic organisation of the gtxA locus differed from the canonical RTX-toxin operon structure which comprises 4 genes in the order rtxC, rtxA, rtxB, rtxD. The 4 genes encode an acyltransferase, the structural toxin, an ABC transporter protein, and an export channel protein, respectively. The two latter are components of a T1SS and are required for the secretion of the toxin. In G. anatis 12656-12, the acyltransferase gene (gtxC) was positioned downstream of gtxA, and a secretion system was not encoded adjacent to gtxA or gtxC. GtxA was exclusively detected in the extracellular protein fraction (Fig. 1B), and the E. coli T1SS facilitated export of GtxA (Fig. 5), therefore, G. anatis is likely to encode a T1SS responsible for the export of GtxA. The lack of a co-transcribed T1SS has also been described for ApxIIA from A. pleuropneumoniae, this protein is exported via a genetically unlinked T1SS, which also secretes ApxIA [32]. A BLAST search in the genome sequence of G. anatis 12656-12 with the E. coli secretion system components HlyB and HlyD as queries, identified three different predicted T1SS-encoding operons (data not shown). Two of these T1SS-operons additionally included each their gene encoding putative extracellular metalloproteases, which are likely to be their substrate proteins. The third T1SS-operon did not include any predicted substrate-encoding genes, and our preliminary data indicate that GtxA is secreted by this T1SS.
In G. anatis 12656-12, a single toxin, GtxA, is responsible for both the haemolytic and leukotoxic activity. Most A. pleuropneumoniae serotypes produce more than 1 of the 3 RTX-toxins ApxI, ApxII and ApxIII [22, 35], but this is seemingly not the case with G. anatis 12656-12. Genome searches supported this notion, as no other predicted proteins with homology to the pore-forming RTX-toxins were identified. However, the genome of 12656-12 did encode two large extracellular metalloproteases containing RTX-repeats.
Our characterisation of GtxA adds further variation to the theme of bacterial toxins in general and RTX-toxins in particular. GtxA contains a new type of domain and apparently uses yet undescribed mechanisms during the interaction with host cells and in the battle against the host immune system. We expect GtxA to be essential in the pathogenesis of G. anatis, and aim for further insight into the role of this toxin in the promotion of disease.
Acknowledgments
We thank Dr Rosemary Redfield and Dr Sunita Sinha, University of British Columbia, for valuable help with natural transformation of Gallibacterium. We thank Dr P. Kuhnert and Dr J. Frey, University of Bern, for providing plasmid pLG575 and ApxI-antiserum. This work was supported by the Danish Research Council for Technology and Production (23-03-0143). B.M. Kristensen was supported by the Faculty of Life Sciences, University of Copenhagen.
Footnotes
Bojesen A.M., unpublished data.
Kristensen B.M., unpublished data.
Kristensen B.M., unpublished results.
References
- 1.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J.H., Zhang Z., Miller W., Lipman D.J., Gapped BLAST and PSI-BLAST: a new generation of protein database search programs, Nucleic Acids Res. (1997) 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barany F., Two-codon insertion mutagenesis of plasmid genes by using single-stranded hexameric oligonucleotides, Proc. Natl. Acad. Sci. USA (1985) 82:4202–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beug H., Vonkirchbach A., Doderlein G., Conscience J.F., Graf T., Chicken hematopoietic-cells transformed by 7 strains of defective avian leukemia viruses display 3 distinct phenotypes of differentiation, Cell (1979) 18:375–390 [DOI] [PubMed] [Google Scholar]
- 4.Bisgaard M., Dam A., Salpingitis in poultry. II. Prevalence, bacteriology, and possible pathogenesis in egg-laying chickens, Nord. Vet. Med. (1981) 33:81–89 [PubMed] [Google Scholar]
- 5.Bojesen A.M., Torpdahl M., Christensen H., Olsen J.E., Bisgaard M., Genetic diversity of Gallibacterium anatis isolates from different chicken flocks, J. Clin. Microbiol. (2003) 41:2737–2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bojesen A.M., Nielsen S.S., Bisgaard M., Prevalence and transmission of haemolytic Gallibacterium species in chicken production systems with different biosecurity levels, Avian Pathol. (2003) 32:503–510 [DOI] [PubMed] [Google Scholar]
- 7.Bojesen A.M., Christensen J.P., Bisgaard M., Gallibacterium infections and other avian Pasteurellaceae, in: Pattison M., Mcmullin P.F., Bradbury J.M., Alexander D.J.(Eds.), Poultry diseases, Saunders Elsevier, Edinburg, 2008, pp. 160–163 [Google Scholar]
- 8.Bulach D.M., Zuerner R.L., Wilson P., Seemann T., McGrath A., Cullen P.A., et al. , Genome reduction in Leptospira borgpetersenii reflects limited transmission potential, Proc. Natl. Acad. Sci. USA (2006) 103:14560–14565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen H., Bisgaard M., Bojesen A.M., Mutters R., Olsen J.E., Genetic relationships among avian isolates classified as Pasteurella haemolytica, “Actinobacillus salpingitidis” or Pasteurella anatis with proposal of Gallibacterium anatis gen. nov., comb. nov. and description of additional genomospecles within Gallibacterium gen. nov, Int. J. Syst. Evol. Microbiol. (2003) 53:275–287 [DOI] [PubMed] [Google Scholar]
- 10.Frees D., Chastanet A., Qazi S., Sorensen K., Hill P., Msadek T., Ingmer H., Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus, Mol. Microbiol. (2004) 54:1445–1462 [DOI] [PubMed] [Google Scholar]
- 11.Frey J., Kuhnert P., RTX toxins in Pasteurellaceae, Int. J. Med. Microbiol. (2002) 292:149–158 [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Gomez E., Vaca S., Perez-Mendez A., Ibarra-Caballero J., Perez-Marquez V., Tenorio V.R., Negrete-Abascal E., Gallibacterium anatis-secreted metalloproteases degrade chicken IgG, Avian Pathol. (2005) 34:426–429 [DOI] [PubMed] [Google Scholar]
- 13.Gerlach H., Die Bedeutung von Pasteurella haemolytica in Hühnerbeständen, Prakt. Tierarzt (1977) 5:324–328 [Google Scholar]
- 14.Greenham L.W., Hill T.J., Observations on an avian strain of Pasteurella haemolytica, Vet. Rec. (1962) 74:861–863 [Google Scholar]
- 15.Harry E.G., A haemolytic coccobacillus recovered from poultry, Vet. Rec. (1962) 74:640 [Google Scholar]
- 16.Heger A., Holm L., Rapid automatic detection and alignment of repeats in protein sequences, Proteins (2000) 41:224–237 [DOI] [PubMed] [Google Scholar]
- 17.Hinz K.H., Bakteriologische Befunde bei Erkrankungen der Atmungsorgane von Junghennen, 9th Congress of the World Veterinary Poultry Association, Beograd, 1969, pp. 713–718 [Google Scholar]
- 18.Horton R.M., Cai Z.L., Ho S.N., Pease L.R., Gene-splicing by overlap extension: tailor-made genes using the polymerase chain-reaction, Biotechniques (1990) 8:528–535 [PubMed] [Google Scholar]
- 19.Jansen R., Briaire J., Smith H.E., Dom P., Haesebrouck F., Kamp E.M., et al. , Knockout mutants of Actinobacillus pleuropneumoniae serotype 1 that are devoid of Rtx toxins do not activate or kill porcine neutrophils, Infect. Immun. (1995) 63:27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarma E., Regassa L.B., Growth phase mediated regulation of the Actinobacillus pleuropneumoniae ApxI and ApxII toxins, Microb. Pathog. (2004) 36:197–203 [DOI] [PubMed] [Google Scholar]
- 21.Jordan F.T., Williams N.J., Wattret A., Jones T., Observations on salpingitis, peritonitis and salpingoperitonitis in a layer breeder flock, Vet. Rec. (2005) 157:573–577 [DOI] [PubMed] [Google Scholar]
- 22.Kamp E.M., Popma J.K., Anakotta J., Smits M.A., Identification of hemolytic and cytotoxic proteins of Actinobacillus pleuropneumoniae by use of monoclonal antibodies, Infect. Immun. (1991) 59:3079–3085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kingsford C.L., Ayanbule K., Salzberg S.L., Rapid, accurate, computational discovery of Rho-independent transcription terminators illuminates their relationship to DNA uptake, Genome Biol. (2007) 8:R22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kjos-Hanssen B., Egglederperitonitt forårsaket av patogen “kloakkbakterie” hos høns, Nord. Vet. Med. (1950) 2:523–531 [Google Scholar]
- 25.Kohlert R., Untersuchungen zur ätiologie der Eileiterentzündung beim Huhn, Monatsh. Veterinärmed. (1967) 23:392–395 [PubMed] [Google Scholar]
- 26.Mackman N., Nicaud J.M., Gray L., Holland I.B., Genetical and functional organisation of the Escherichia coli haemolysin determinant 2001, Mol. Gen. Genet. (1985) 201:282–288 [DOI] [PubMed] [Google Scholar]
- 27.Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., et al. , Genome sequencing in microfabricated high-density picolitre reactors, Nature (2005) 437:376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirle C., Schöngarth M., Meinhart H., Olm U., Untersuchungen zu Auftreten und Bedeutung von Pasteurella haemolytica-Infektionen bei Hennen unter besonderer Berücksichtigung von Erkrankungen des Legesapparates, Monatsh. Veterinärmed. (1991) 46:545–549 [Google Scholar]
- 29.Neubauer C., De Souza-Pilz M., Bojesen A.M., Bisgaard M., Hess M., Tissue distribution of haemolytic Gallibacterium anatis isolates in laying birds with reproductive disorders, Avian Pathol. (2008) 38:1–7 [DOI] [PubMed] [Google Scholar]
- 30.Pearson W.R., Lipman D.J., Improved tools for biological sequence comparison, Proc. Natl. Acad. Sci. USA (1988) 85:2444–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poje G., Redfield R.J., Transformation of Haemophilus influenzae, Methods Mol. Med. (2003) 71:57–70 [DOI] [PubMed] [Google Scholar]
- 32.Reimer D., Frey J., Jansen R., Veit H.P., Inzana T.J., Molecular investigation of the role of ApxI and ApxII in the virulence of Actinobacillus pleuropneumoniae serotype 5, Microb. Pathog. (1995) 18:197–209 [DOI] [PubMed] [Google Scholar]
- 33.Rowe G.E., Welch R.A., Assays of hemolytic toxins, Methods Enzymol. (1994) 235:657–667 [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto H., Bellalou J., Sebo P., Ladant D., Bordetella pertussis adenylate cyclase toxin. Structural and functional independence of the catalytic and hemolytic activities, J. Biol. Chem. (1992) 267:13598–13602 [PubMed] [Google Scholar]
- 35.Schaller A., Kuhn R., Kuhnert P., Nicolet J., Anderson T.J., MacInnes J.I., et al. , Characterization of apxIVA, a new RTX determinant of Actinobacillus pleuropneumoniae, Microbiology (1999) 145:2105–2116 [DOI] [PubMed] [Google Scholar]
- 36.Soding J., Biegert A., Lupas A.N., The HHpred interactive server for protein homology detection and structure prediction, Nucleic Acids Res. (2005) 33:W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanley P., Packman L.C., Koronakis V., Hughes C., Fatty acylation of 2 internal lysine residues required for the toxic activity of Escherichia coli hemolysin, Science (1994) 266:1992–1996 [DOI] [PubMed] [Google Scholar]
- 38.Strathdee C.A., Lo R.Y., Regulation of expression of the Pasteurella haemolytica leukotoxin determinant, J. Bacteriol. (1989) 171:5955–5962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tascon R.I., Vazquez-Boland J.A., Gutierrez-Martin C.B., Rodriguez-Barbosa I., Rodriguez-Ferri E.F., The RTX haemolysins ApxI and ApxII are major virulence factors of the swine pathogen Actinobacillus pleuropneumoniae: evidence from mutational analysis, Mol. Microbiol. (1994) 14:207–216 [DOI] [PubMed] [Google Scholar]
- 40.Tatum F.M., Briggs R.E., Sreevatsan S.S., Zehr E.S., Ling Hsuan S., Whiteley L.O., et al. , Construction of an isogenic leukotoxin deletion mutant of Pasteurella haemolytica serotype 1: characterization and virulence, Microb. Pathog. (1998) 24:37–46 [DOI] [PubMed] [Google Scholar]
- 41.Vojtova J., Kamanova J., Sebo P., Bordetella adenylate cyclase toxin: a swift saboteur of host defense, Curr. Opin. Microbiol. (2006) 9:69–75 [DOI] [PubMed] [Google Scholar]
- 42.Wang R.F., Kushner S.R., Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli, Gene (1991) 100:195–199 [PubMed] [Google Scholar]