Abstract
The activating protein-1 (AP-1) transcription factor complex is a heterogeneous entity, composed in mammalian cells of dimers chosen from a group of at least eight proteins belonging to three families: jun, fos, and activating transcription factor (ATF). The AP-1 complexes participate in diverse biological processes that include cell proliferation, survival, and differentiation. These seemingly contrasting functions have been attributed to the intensity and duration of the signals provided by AP-1, but the biological consequences of changing composition of the AP-1 complex have not been fully explored. Here, we show that functional AP-1 is required for 1,25-dihydroxyvitamin D3 (1,25D)-induced monocytic differentiation, and that the composition of the AP-1 protein complex that binds TRE, its cognate DNA element, changes as cells differentiate. In HL60 cells in an early stage of differentiation, the principal AP-1 components detected by gel shift analysis include c-jun, ATF-2, fos-B, fra-1, and fra-2. In cells with a more established monocytic phenotype, the demonstrable AP-1 components are c-jun, ATF-2, jun-B, and fos-B. Following the addition of 1 nmol/L of 1,25D, the cellular content of each of these four proteins markedly increased in a sustained manner, whereas the increases in c-fos, fra-1, fra-2, and jun-D were minimal, if any. Small increases in mRNA levels encoding all AP-1 component proteins, except c-fos, were also noted. These findings provide a basis for the previously found participation of the c-Jun N-terminal kinase pathway in 1,25D-induced differentiation of myeloid leukemia cells, and direct attention to jun-B and fos-B as new cellular therapeutic targets, that may promote replicative quiescence associated with differentiation of malignant cells.
Introduction
Signaling of differentiation that is mediated by nuclear receptors such as the vitamin D receptor (VDR) is largely propagated by waves of transcription factor activation. The initial event that follows the entry into the cell of the physiologically active form of vitamin D, 1,25-dihydroxyvitamin D3 (1,25D), is its binding to VDR, which itself is a ligand-activated transcription factor. Liganded VDR forms heterodimers with members of the retinoid X receptor family of proteins, which then bind to vitamin D response elements in the promoter regions of the vitamin D primary response genes (reviewed in refs. 1–3).
The genes that are known to respond to vitamin D directly, primarily function in the metabolism of vitamin D itself, e.g., CYP-24, which hydroxylates 1,25D (4, 5), in calcium homeostasis, e.g., the calcium channel TRPV6 (6), or have still unknown functions (reviewed in refs. 7, 8). Curiously, thus far, none of the genes shown to be directly regulated by 1,25D have any known function in the induction of cell differentiation. However, in model systems of differentiation, secondary events that follow cell exposure to 1,25D have been shown to include major changes in cellular signaling networks (2, 9–12).
A well studied model of 1,25D-induced cell differentiation is provided by HL60 myeloid leukemia cells in culture (13, 14). In these, and other myeloid leukemia cells, 1,25D signaling includes the activation of the mitogen-activated protein kinase pathways, especially the Raf/MEK/ERK cascade, and the c-Jun-NH2-kinase (JNK) cascades (15–17). These kinases are believed to phosphorylate and thus activate nuclear transcription factors which then up-regulate the secondary and subsequent waves of 1,25D-induced gene expression that result in the mature monocyte/macrophage phenotype, and the cessation of cell proliferation (18, 19).
Several transcriptions factors have been clearly involved in 1,25D-induced monocytic differentiation of HL60 cells. These include Egr-1 (20), Sp-1 (21), and C/EBPβ (22). In addition, the involvement of activating protein-1 (AP-1) has been suggested by its participation in 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced macrophage differentiation (23, 24), and the need for the activation of JNK pathway in 1,25D-induced monocytic differentiation (16), because the principal target of JNK is c-jun, a component of the AP-1 complex. We therefore showed the requirement for the AP-1 DNA element (TRE)-mediated signaling in 1,25D-induced monocytic differentiation, and determined the constituent proteins that are the most likely to be important for the acquisition of the monocyte phenotype. Of the eight different proteins known to form the AP-1 transcription factor complex, the principal ones that are up-regulated by 1,25D and are present in the AP-1 complex during this process are c-jun, activating transcription factor 2 (ATF-2), jun-B, and fos-B.
Materials and Methods
Chemicals and antibodies
1,25D was a kind gift from Dr. Milan Uskokovic (BioXell, Nutley, NJ). [γ-32P]ATP was purchased from NEN Life Science Products, Inc. (Boston, MA). The antibodies against c-fos (4-10G and c-fos-C4), fra-1 (R-20), fra-2 (H-103), fos-B (H-75), c-jun (H-79), jun-B (N-17), jun-D (329), ATF-2 (N-96), and Crk-L (C-20) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The antibodies used for supershift analyses were purchased as concentrated (10×) preparations. Anti-CD14 (MY4-RD-1) and anti-CD11b (MO1-FITC) antibodies were obtained from Coulter Corporation (Miami, FL).
Cell culture
Human leukemia HL60-G cells (25), derived from a patient with promyeloblastic leukemia (26), were cultured in RPMI 1640 (Mediatech, Washington, DC) supplemented with 10% heat-inactivated, iron-enriched bovine calf serum (HyClone, Logan, UT) at 37°C in 5% CO2. The cell culture was passaged every 2 to 3 days and screened routinely for Mycoplasma contamination. For experiments, the cells were seeded at 3 × 105 cells/mL in the presence or absence of 1 nmol/L of 1,25D3 for the indicated times. Cell viability was determined using trypan blue (0.25%) exclusion.
Determination of markers of differentiation
Aliquots of 1 × 106 cells were harvested, washed twice with PBS (1× PBS), then the cell suspensions were incubated for 45 minutes at room temperature with 0.5 µL of MY4-RD-1 and 0.5 µL of MO1-FITC (Coulter, Miami, FL) to analyze the expression of surface cell markers, CD14 and CD11b, respectively. The cells were then washed thrice with ice-cold PBS, and resuspended in 1 mL 1× PBS. Two-parameter analysis was done using an Epics XL-MCL flow cytometer (Coulter, Fullerton, CA). Isotypic mouse IgG1 was used to set threshold variables. Monocyte specific esterase staining was preformed as described before (27).
Cell cycle distribution
HL60 cells (1 × 106) were washed twice with 1× PBS and fixed in 75% ethanol at −20°C for at least 2 hours. The fixed cells were spun down to remove fixative buffer and washed twice with 1× PBS, then the cell pellets were resuspended in 1 mL propidium iodide solution (10 µg/mL; RNase A, 10 µg/mL; Sigma, St. Louis, MO) at 37°C for 30 minutes. The cells were analyzed using an Epics XL-MCL instrument (Coulter, Fullerton, CA), and cell cycle distribution was determined by Multicycle Software Program (Phoenix Flow System, San Diego, CA).
Oligonucleotides
AP-1 decoy oligonucleotide (28) was used to study the role of AP-1 transcription factors in the monocytic differentiation of HL60 cells. The AP-1 decoy, and control oligonucleotides used in the experiments, were phosphorothioate oligos synthesized by the Molecular Resource Facility of the New Jersey Medical School. The HL60 cells were incubated with 5 µmol/L decoy oligos for 12 hours before being treated with 1,25D. The following are the sequences of synthetic decoy oligonucleotides that were used in the experiments (residues changed in the mutant oligonucleotides are indicated in lowercase type): AP-1, 5′-CGCTTCATGACTcaGCCGGAA-3′, mutant AP-1, 5-CGCTTCATGACTtgGCCGGAA-3′, AP-1 and mutant AP-1 double-stranded oligonucleotides were generated by annealing the synthetic oligonucleotides with the respective complimentary sequences.
Cellular uptake of oligonucleotides was checked by incubating 106 cells with 10 ng of 32P-labeled oligonucleotide in the growth medium for 4 or 24 hours. Cells were washed four times with 1× PBS, and the radioactivities in the cell pellets and culture medium were determined. The cell uptake of oligonucleotides was calculated as the percentage of radioactivity in the cell pellets versus the total radioactivity added into the cells.
RNA extraction and RT-PCR
RNA was extracted from cells using RNeasy kit (Qiagen, Carlsbad, CA), which is based on the selective binding properties of silica gel–based membrane with the microspin technique. GeneAmp RNA PCR Core Kits (Perkin-Elmer, Boston, MA) were used and procedures were done as previously described. The forward and reverse primer sequences used, respectively, were: c-Jun, 5′-AGGAGGAGCCTCAGACAGTG-3′ and 5′-TGTTTAAGCTGTGCCACCTG-3′; jun-B, 5′-ACTCTT TAGAGACTAAGTGCG-3′ and 5′-GAAACAGACTCGATTCATA-3′; jun-D, 5′-CGCAGCCTCAAACCCTGCCTTTCC-3′ and 5′-CAAACAGGAATGTGGACTCGTAGC-3′; c-fos, 5′-CCTGCCCCTTCTCAACGAC-3′ and 5′-GCTCCACGTTGCTGATGCT-3′; fos-B, 5′-ACCCTTTTCTGATCGTCTCG-3′ and 5′-CTGCTCACACTCACACTCG-3′; fra-1, 5′-GTCATTGCTAGGATACCAAAC-3′ and 5′-CACTGTCCAGCAAGGGTCTGT-3′; fra-2, 5′-CCCAGTGTGCAAGATTAGCC-3′ and 5′-CCCAGTGTGCAAGATTAGCC-3′; β-actin, 5′-CACTCTTCCAGCCTTCCTTCC-3′ and 5′-CGGACTCGTCATACTCCTGCTT-3′. For reverse transcription, samples were incubated in an Eppendorf PCR system at 42°C for 15 minutes, then 99°C for 5 minutes, and 5°C for 5 minutes. For PCR, samples were incubated in GeneAmp PCR System as follows: 95°C for 105 seconds, 35 cycles of 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 7 minutes. The RT-PCR products were separated in 1.5% agarose gels containing ethidium bromide (1 µg/mL). The intensities of the bands were measured using Image QuaNT program (Molecular Dynamics, Sunnyvale, CA). In preliminary experiments, we established that 35 cycles of amplification gave optimal signals for each of the products studied here and were in the linear range.
Cell extracts
Whole cell extracts were prepared as described previously. Cell pellets were washed twice with 1× PBS then resuspended with a lysis buffer containing 20 mmol/L Tris-HCl (pH 7.4), 150 mmol/L NaCl, 1 mmol/L EDTA, 1 mmol/L EGTA, 1% Triton X-100, 2.5 mmol/L sodium pyrophosphate, 1 mmol/L β-glycerophosphate, 1 mmol/L Na3VO4, 1 mmol/L phenylmethylsulfonyl fluoride, 1 µg/mL leupeptin, and 1 µg/mL aprotinin. The protein concentrations were determined by using Bio-Rad (Hercules, CA) protein assay kit. Equal amounts of 3× SDS sample buffer containing 150 mmol/L Tris-HCl (pH 6.8), 30% glycerol, 3% SDS, 1.5 mg/mL bromophenol blue dye, and 100 mmol/L DTT was then added to each sample. Nuclear extracts were prepared by a previously described procedure (29). Briefly, 107 cells were resuspended in 0.5 mL ice-cold hypotonic buffer [10 mmol/L HEPES-KOH (pH 7.9), 1.5 mmol/L MgCl2, 10 mmol/L KCl, 0.5 mmol/L DTT, and Complete protease inhibitor cocktail from Sigma]. Cells were kept on ice for 10 minutes to allow them to swell, vortexed for 10 seconds, and centrifuged at 16,000 × g for 30 seconds. Supernatant was saved as cytoplasmic extract and the pellet was resuspended in 50 µL of nuclear extraction buffer [20 mmol/L HEPES-KOH (pH 7.9), 25% glycerol, 420 mmol/L NaCl, 1.5 mmol/L MgCl2, 0.2 mmol/L EDTA, 0.5 mmol/L DTT, and Complete protease inhibitor cocktail], placed on ice for 20 minutes, and centrifuged at 16,000 × g for 15 minutes. The supernatant was saved as the nuclear extract and stored at −80°C.
Western blotting
Equal amounts of whole cell extracts (40 µg of protein) were separated on 10% SDS-PAGE gel and transferred to nitrocellulose membranes (Amersham Pharmacia Biotech, Piscataway, NJ). The membranes were blocked with 5% milk in 1× TBS/0.1% Tween 20 for 1 hour, subsequently blotted with primary antibodies, then the membranes were blotted with a horseradish-linked secondary antibody for 1 hour. The protein bands were visualized with a chemiluminescence assay system (Amersham). The protein loading of the gel and efficiency of the transfer were controlled by stripping the membrane and reprobing for Crk-L, a constitutively expressed adaptor protein in HL60 cells. The absorbance of each band was quantitated using an image quantitator (Molecular Dynamics).
Electrophoretic mobility shift assay
AP-1 gel shift assay was done as previously described (30). Double-stranded oligonucleotides from promoter regions of hVDR containing the proximal (−77 to −97 relative to the transcription start site) TRE, a binding site for AP-1 (5′-CTGGCAAGAGAGGACTGGACC-3′ hVDR-AP-1), and the mutated TRE (5′-CTGGCAAGAGAGtgCTGGACC-3′), were synthesized by Molecular Resource Facility of the New Jersey Medical School. Ten micrograms of the nuclear extracts were preincubated with binding buffer containing 50 mmol/L Tris-HCl (pH 7.9), 10 mmol/L MgCl2, 1 mmol/L EDTA, 100 mmol/L KCl, 2 mmol/L DTT, 20% (v/v) glycerol, 0.1% (v/v) Nonidet P-40, and 2 µg [d(I-C)] for 15 minutes at 4°C. The extracts were then incubated for an additional 30 minutes at room temperature with 50 pg of 32P-labeled double-stranded oligonucleotide. The specificity of the AP-1 binding was estimated by competition with either a 50× molar excess of the unlabeled double stranded hVDR-AP-1 nucleotide or mutant AP-1 double-stranded nucleotide added to parallel samples during the preincubation period. Supershift assay was done using antibody against components of AP-1 family proteins. The complexes were separated on 4% polyacrylamide gel under nondenaturing conditions with a constant current of 22 mA for 3 hours at 4°C. The gel was dried and set up for autoradiography.
Statistical analysis
All experiments were repeated at least thrice. The significance of the differences between the means of the various subgroups were assessed by two-tailed Student’s t test. The computations were done with an IBM-compatible personal computer using Microsoft EXCEL.
Results
Functional AP-1 complexes are required for 1,25D-induced monocytic differentiation of HL60 cells
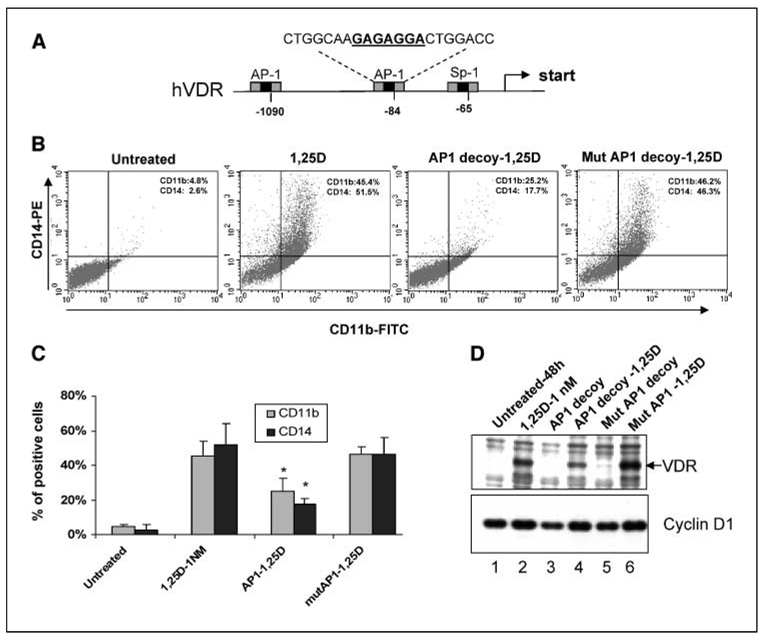
Because AP-1 is not a single entity, the currently popular approaches of gene silencing or knock-down are not feasible to study its function. We therefore used the transcription factor binding DNA element “decoy” strategy to reduce the availability of AP-1 complexes for binding to the same elements in promoters of AP-1 target genes (Fig. 1A; refs 28, 31). We chose a 1 nmol/L concentration of 1,25D because it is only moderately higher than the concentration of 1,25D in the plasma, and thus, may approximate its concentration in target tissues (32), and 48 hours as the principal time of exposure to 1,25D. At this time point, a large proportion of the treated cells exhibits markers of monocyte phenotype, CD11b and CD14 (Fig. 1B), as well as the cytoplasmic enzyme, monocyte-specific esterase (Fig. 2B). The illustrative primary data (Fig. 1B) are summarized in Fig. 1C, and show that a great excess of a synthetic double-stranded oligonucleotide (5 µmol/L) containing the AP-1 binding TRE element markedly inhibits 1,25D-induced differentiation. The decoy was shown to act by intracellular events, rather than having a direct effect on the cell surface, by demonstrating that the expression of VDR, one of the genes positively regulated by AP-1 (33, 34), is down-regulated by the decoy, but not by its mutated version (Fig. 1D). Thus, the binding of AP-1 complexes to TREs in gene promoters seems to be one of the requirements for the induction of the monocytic phenotype.
Figure 1.
AP-1 transcription factor is required for 1,25D-induced differentiation. A, schema of the promoter region of human VDR (hVDR) showing the location and sequence of the proximal TRE, which binds the AP-1 transcription factors, in the promoter region of human VDR. This sequence was used as the decoy, and as the probe in gelshif t analyses. B, HL60 cells were treated with 1,25D (1 nmol/L for 48 hours) in the absence or presence of TRE oligonucleotides decoy (5 mmol/L) to AP-1 binding element. The myelomonocytic markers CD11b and CD14 were determined by two-parameter flow cytometry. 1,25D-induced differentiation was markedly inhibited by promoter competition with AP-1 decoy oligo, whereas a TRE-containing oligo with a mutation in the AP-1 binding sequence had no effect on differentiation. C, summary of experiments in (B). Columns, mean; bars, ± SD (n = 3); compared with the mutated decoy treatment of the cells (*, P < 0.05). D, protein levels of VDR, encoded by an AP-1-dependent gene, and cyclin D, used here as a control, were determined by immunoblotting, and show the effect of the decoy on VDR but not on cyclin D1 expression in 1,25D-treated cells. Note also the unexpected increase in cyclin D1 in 1,25D-treated cells (lane 2), which may be due to a larger proportion of cells expressing cyclin D1, due to the increased accumulation of the treated cells in the G1 cell cycle compartment following 48 hours of treatment with 1,25D (Fig. 2C; refs. 50, 51).
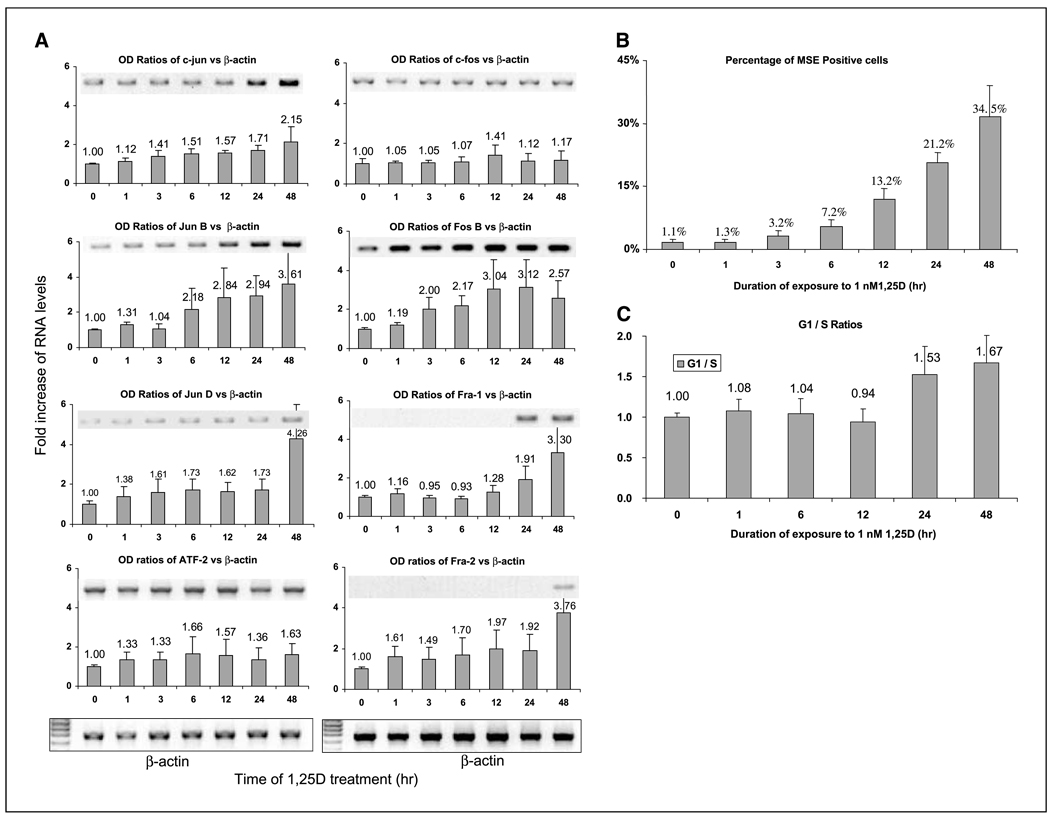
Figure 2.
1,25D increases mRNA levels of all members of the AP-1 family except c-fos. A, HL60 cells were treated with 1 nmol/L of 1,25D for the indicated times, then the mRNA level of each AP-1 complex member was determined by semiquantitative PCR. This experiment was repeated at least thrice, and the quantitated mRNA level of each member is shown by a bar chart, and illustrated by a representative gel. Columns, mean of three experiments (numbers above columns, mean values); bars, SD. B, monocytic differentiation of HL60 cells as determined by positivity for the monocyte-specific esterase, also known as “nonspecific esterase, ” staining. Differentiation was evident at 3 hours and continued to increase during the 1,25D treatment (MSE, monocyte-specific esterase). C, evidence of a G1 to S phase block, shown by an increasing ratio of cells in the G1 compartment as compared with cells in the S phase compartment, determined as described in Materials and Methods.
Potential components of the AP-1 complex are selectively up-regulated by 1,25D at both mRNA and protein levels
We hypothesized that the composition of AP-1 complexes in proliferating HL60 cells differs from the composition of these complexes in differentiating cells. The possibilities through which such changes can take place include altered rates of gene transcription and mRNA translation, as well as changes in mRNA and protein stability. We initiated the study of these mechanisms by a semiquantitative determination of the steady-state levels of the mRNA for eight potential members of the AP-1 complex. The increased abundance of mRNAs encoding all these proteins except c-fos was noted, but with different kinetics for individual mRNAs (Fig. 2A). In most cases, the elevated levels of mRNA were detected 1 hour after administration of 1,25D, preceding the earliest evidence of monocytic differentiation, which was apparent at 3 hours (Fig. 2B). The levels of mRNAs that encode c-jun and jun-B continued to increase during the period of observation (48 hours), whereas ATF-2 and fos-B mRNAs seemed to peak at ~ 6 to 12 hours. The levels of several mRNAs studied showed sharp increases at 24 to 48 hours. These included jun-D, fra-1, and fra-2 (Fig. 2A). Interestingly, none of the mRNAs declined in abundance, although the cells were beginning to approach replicative quiescence, as shown by increasing G1 to S phase block (Fig. 2C).
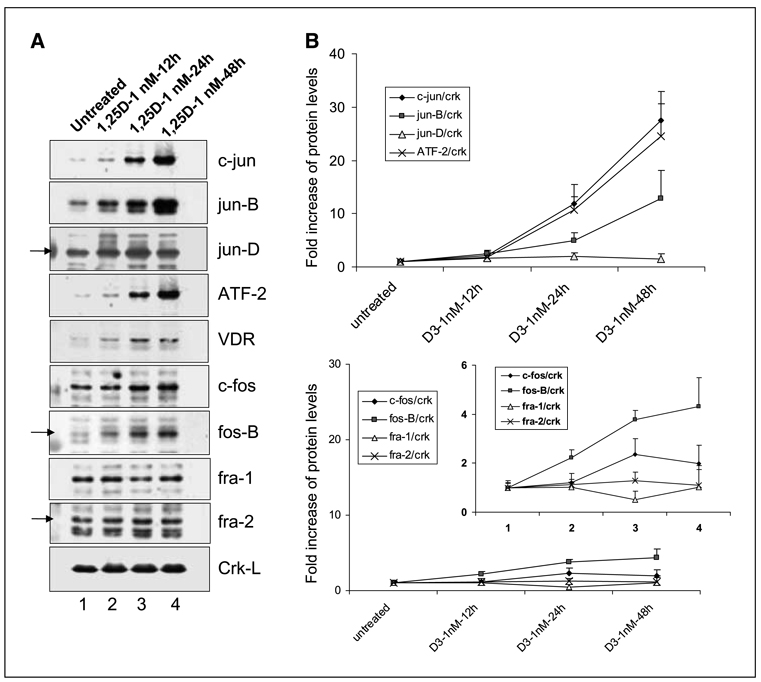
Similar to mRNA, protein levels of the potential AP-1 complex components, determined by Western blotting, showed no decrease following the exposure to 1,25D (Fig. 3). Remarkably, the up-regulation of several proteins (up to ~ 30-fold) far exceeded the relatively small (up to ~ 4-fold) increases in mRNAs. Also, there was greater selectivity in the up-regulation of the AP-1 component proteins; c-jun and ATF-2 showed the greatest, and approximately equal, increases, whereas jun-B and fos-B also showed significant up-regulation. The abundance of the other members of the fos family studied here (c-fos, fra-1, and fra-2) and jun-D was not detectably increased. This suggests that only four of the eight potential components of AP-1 complex are of special importance in the induction and/or maintenance of monocytic differentiation in this system.
Figure 3.
Increased expression of AP-1 proteins in HL60 cells treated with 1 nmol/L 1,25D. Immunoblot analysis of whole cell protein extracts in untreated and 1,25D-treated cells. A, a series of representative immunoblots for jun and fos family members and ATF-2. VDR was used as a control for a protein known to increase in abundance following 1,25D treatment, and Crk-L, a constitutively expressed protein was used here as loading/transfer control. B, points, mean obtained by OD quantitation of three similar experiments; bars, SD. Inset in the lower frame represents the same data at a higher magnification, designed to more clearly show the extent of the experimental variation (D3, 1,25D).
Evidence for the presence of c-Jun, jun-B, ATF-2, and fos-B proteins in complexes bound to TRE in differentiating cells
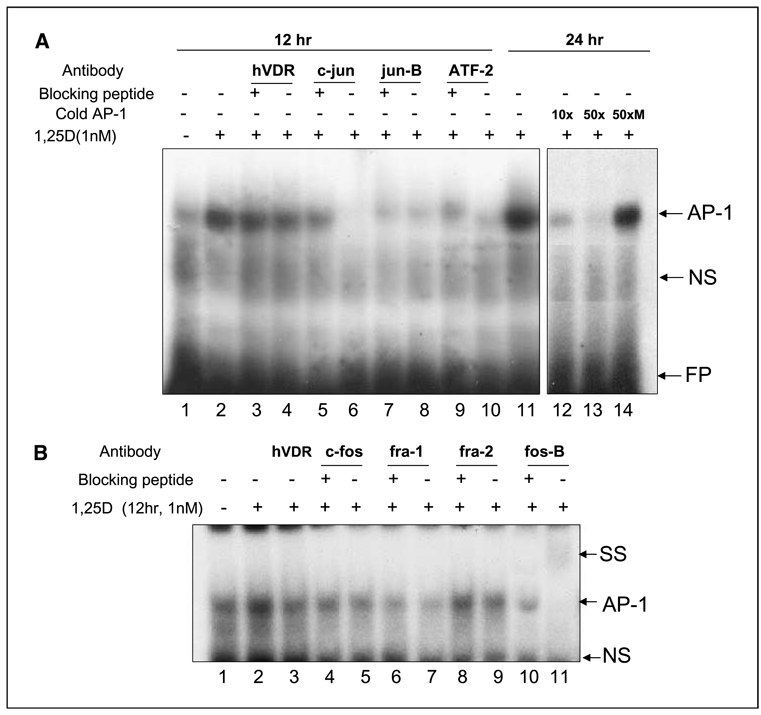
We then employed gel shift analysis to determine which proteins were detectable in the complexes binding TRE in intact cells differentiating on the presence of 1,25D. Because in untreated cells, the AP-1 binding is weak (see Fig. 4A, lane 1), and thus, difficult to analyze, we used the 12-hour time point as representative of the early situation. As Fig. 4 shows, binding of the AP-1 complex to TRE increases with time of treatment by 1,25D to at least 24 hours (lanes 1, 2, and 11, Fig. 4A; lanes 1 and 2, Fig. 4B). The binding is competed by the unlabeled “self” probe, but not a mutated AP-1 probe analyzed at 24 hours, as greater intensity of the complex at that time facilitated the analysis (Fig. 4A, lanes 12–14). Attempts to show supershifts at this time point were successful only with antibody to fos-B (Fig. 4B, lanes 10 and 11), showing the presence of this protein in the AP-1 complex. However, incubation with antibodies to the other potential components of the AP-1 complex showed that although a supershift could not be detected, the antibody to c-jun blocked binding to TRE, whereas partial blocking was observed by antibodies to ATF-2, fra-1, and fra-2. As controls in these experiments, the peptide used to generate the antibody was preincubated with the antibody to neutralize its action, and comparisons were made individually for each pair, i.e., with and without the blocking peptide (Fig. 4A, lanes 5–10; Fig. 4B, lanes 4–11). An antibody to VDR was used as an irrelevant antibody, and only marginally reduced the effect on AP-1 binding (Fig. 4A, lanes 3 and 4). No AP-1 binding block was observed by antibodies to c-fos (Fig. 4B, lanes 5 and 6) or to jun-D at this or later times of 1,25D exposure (data not shown).
Figure 4.
Gel-shift analysis at an early differentiation time point (12 hours) of the component proteins in AP-1 complexes in HL60 cells treated with 1 nmol/L 1,25D. A, representative blot showing the gradual and marked increase in the intensity of AP-1 binding to the proximal TRE sequence from the human VDR (hVDR) gene promoter (lanes 1, 2, and 11), and reduction in AP-1 complex binding when the indicated excess of unlabeled (“cold”), but not the mutated (M) sequence, was added prior to the binding reaction (lanes 11–14). In these experiments, preincubation of nuclear extracts with antibodies was used in the attempt to show a supershift (SS), or blocking of binding of the protein complex to the proximal TRE sequence in the hVDR promoter. The antibody to hVDR was used as an irrelevant antibody control, and as an additional control, the action of each antibody was neutralized by the addition of the peptide used to generate the antibody (e.g., lane 5 versus lane 6, etc.). Note the block of c-jun binding to TRE, and block of the upper part of the AP-1 complex by the antibody to ATF-2, a protein larger (70 kDa) than other potentialmembers of the AP-1 protein complex (lanes 9 and 10). There was no apparent change in the binding of jun-B to TRE at this time point. B, a similar analysis of the members of the fos family at this time point showed a slight reduction of binding by fra-1 and fra-2, a supershift with fos-B antibody, but little or no observable effect with c-fos antibody. This analysis is representative of three similar experiments. AP-1, complex binding to hVDR proximal TRE; SS, supershift; NS, nonspecific signal; FP, free probe.
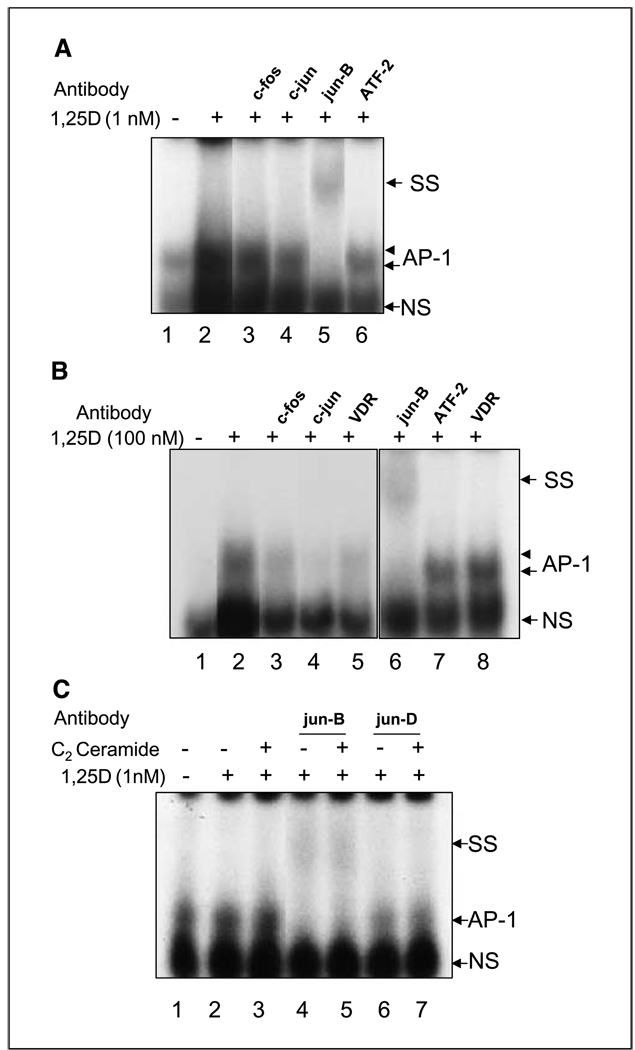
When nuclear extracts were prepared from cells in a later stage of 1,25D-induced differentiation (48 hours) the major difference noted was that a supershift was clearly seen with the antibody against jun-B (Fig. 5A and C), whereas the faint supershift seen at early differentiation time point with fos-B antibody was no longer detected. However, a strong block to TRE binding could still be noted with this antibody (data not shown). Interestingly, the ATF-2 antibody blocked only the more slowly migrating part of the AP-1 complex (arrowhead, lane 6, Fig. 5A) suggesting that AP-1 complexes contained the 70 kDa ATF-2 protein, as well as complexes of more rapidly migrating, smaller proteins, such as the jun proteins.
Figure 5.
Gel-shift analyses of AP-1 protein binding at 48 hours. A, a supershift was produced by jun-B antibody (lane 5) at this time point, and block of the upper part of the AP-1 complex by ATF-2 antibody (arrowhead), as was also observed at 12 hours (lane 6; Fig. 4A, lane 10). No specific effect of c-fos antibody could be seen (lane 3), and c-jun antibody produced only a minor effect. B, the AP-1 complex generated by 100 nmol/L 1,25D produced a more intense signal, which was easier to interpret, and showed that binding was blocked by c-jun antibody (lane 4), but c-fos antibody produced only the nonspecific effect similar to VDR antibody (lanes 3 and 5). The supershift with jun-B (lane 6), and the block of the upper part of the AP-1 complex by ATF-2 (lane 7), are also shown. C, in an experiment in which C2 ceramide was added to 1,25D in an attempt to increase AP-1 complex intensity, the jun-B supershift was again seen, although ceramide did not change its intensity (lanes 4 and 5). No specific effect was seen with jun-D antibody (lanes 6 and 7). SS, supershift; AP-1, complex binding to hVDR proximal TRE; NS, nonspecific signal.
The continued involvement of c-jun in AP-1 complex formation at this time point was also suggested by data shown in Fig. 5A (lane 4), but was more convincingly shown when the experiment was done using a higher concentration of 1,25D (100 nmol/L), which gives a stronger AP-1 signal at 48 hours of treatment (Fig. 5B). The blocking of the upper part of the AP-1 complex by ATF-2 antibody was also confirmed at the 100 nmol/L concentration (Fig. 5B). In this experiment, the AP-1 signal was blocked by c-jun antibody, but not by c-fos antibody, nor by VDR antibody, used here as a control. When the experiment was repeated with the addition of a cell-permeable form of ceramide (C2), reported to potentiate the effects of 1,25D on HL60 cells (35, 36), the gel shift with jun-B antibody showed the supershifted band under both experimental conditions. No effect was observed with an antibody to jun-D (Fig. 5C), other than the nonspecific alteration of the signal in the presence of antibody, as illustrated in Fig. 5B. Thus, the data show that only the proteins which exhibit increased abundance following the exposure to 1,25D, i.e., c-jun, ATF-2, jun-B, and fos-B, can be detected in the AP-1 complexes that bind to TRE. In contrast, proteins constitutionally present in HL60 cells, c-fos, fra-1, fra-2 and jun-D, could not be shown in AP-1 complexes of cells differentiated for 48 hours, although they were approximately equally abundant in these cells (Fig. 3A).
Discussion
In this study, we show for the first time that binding of the AP-1 transcription complex to its cognate DNA element is required for 1,25D-induced differentiation of HL60 cells. Also, the composition of the complex changes as the cells exit the proliferative state and acquire the monocytic phenotype. At this time, four proteins have markedly increased their cellular abundance, and can be shown to be present in complexes that bind to the TRE. Two of these, c-Jun and ATF-2, have been reported to be involved in diverse cellular processes including cell proliferation and various specialized cellular functions (37–39). In contrast, jun-B and fos-B seem to be characteristic of the differentiated state in this system, particularly jun-B, which at 48 hours of 1,25D exposure, became the most abundant of the AP-1 complex proteins, as judged by Western blotting (Fig. 3A, second panel from the top), and the ease of its detection by supershift analysis (Fig. 5A and C). This finding is in accord with the reports that jun-B impedes cell cycle progression (40, 41), as its abundance markedly increased at the time that 1,25D-treated cells exited the cell cycle (24–48 hours), and accumulated primarily in the G1/G0 cell compartment (Fig. 2C). It has been suggested that the ratio of jun-B to c-Jun may determine the difference in AP-1 transcriptional activity (38, 40). This does not seem to be the case in 1,25D-induced monocytic differentiation, as the rates of increase of jun-B protein paralleled the increase in c-jun protein, and the ratio remained ~ 1:2 (Fig. 3B). However, it is possible that as the levels of both proteins increased above some predetermined threshold levels, the negative effect of jun-B on cell proliferation became manifest, and contributed to the observed G1 block (Fig. 2C). This has important implications for increasing the understanding of the mechanistic basis of antiproliferative effects of 1,25D, as jun-B is known to be capable of repressing the expression of cyclin D1 (40), and of promoting the expression of p16/INK4a inhibitor of cyclin-dependent kinases (41). Indeed, mice lacking the expression of the jun-B gene in myeloid cells develop chronic myeloid leukemia, a disease in which the cells do not undergo terminal differentiation (42). The importance of jun-B expression for terminal differentiation of myeloid cells is further illustrated by the earlier findings that TPA-induced macrophage differentiation of HL60 cells is accompanied by increases in jun-B mRNA levels (43, 44), and that normal human monocytes exposed to the macrophage colony-stimulating factor express jun-B transcripts, as well as transcripts of several other potential components of AP-1 (45).
Fos-B is the fourth most prominently up-regulated potential AP-1 component protein that is induced by 1,25D in HL60 cells (Fig. 3). It is the only member of the fos family of proteins that is responsive to 1,25D in the system studied here, and could be shown to be present in the AP-1 complexes by the supershift analysis (Fig. 4B). However, blocking antibody analysis shows that at an early time after exposure to 1,25D (12 hours), fra-1 and fra-2 could also be detected in the AP-1 complexes, consistent with their constitutive presence in the cells (Fig. 3A). The significance for induction of differentiation of the fra-1 and fra-2 presence in AP-1 complexes at this early period is clouded by the fact that most cells in the culture are still proliferating at normal rates (Fig. 2C; data not shown). Therefore, it is likely that fra-1 and fra-2 participate in various housekeeping functions in cell growth and proliferation.
The lack of a clear link between c-fos and 1,25D-induced differentiation was somewhat surprising in view of its frequent participation in AP-1 complex formation as a heterodimeric partner for c-jun, the high levels of c-fos mRNA reported in other differentiation systems (e.g., ref. 46), and the relative abundance of c-fos protein in HL60 cells (Fig. 3A). A possible explanation is that in hematopoietic differentiation, c-fos is antagonistic to monocyte/macrophage differentiation in a lineage-specific manner because in c-fos-deficient mice, increased numbers of bone marrow macrophages were found (47). An alternative possibility is that this abundance is simply due to the lack of macrophage differentiation into osteoblasts, and that an overexpression of c-fos could also favor myeloid differentiation, as reported in another murine system (48).
It is important to stress that the ability to show the presence of proteins c-jun, ATF-2, jun-B, and fos-B in AP-1 complexes of differentiated cells correlated with their inducibility by 1,25D, not simply with their cellular abundance. For instance, jun-D and c-fos proteins were easily detected in both untreated and 1,25D-treated HL60 cells, yet they could not be detected by supershift or antibody block analysis. Thus, the simple presence of the proteins in the cell does not guarantee its inclusion in the AP-1 complex, perhaps because these proteins are sequestered in the cytoplasm, or in nuclear complexes other than those that bind to the proximal TRE present in the human VDR promoter, the probe used in these studies.
Also of interest is the finding that the induction by 1,25D of the proteins that participate in AP-1 complexes characteristic of differentiated cells occurs principally at the posttranscriptional level. The levels of mRNAs encoded by all genes (except c-fos) studied here increased slightly (2- to 4-fold), yet only those proteins that were identified as components of the AP-1 complex showed more dramatic increases (10- to 30-fold). It is not clear if the mRNA levels found here represent increased transcriptional rates due to the presence of 1,25D3 in the cells, or the stabilization of the mRNA, as was found for c-jun and jun-B mRNA in TPA treated HL60 cells (49). Likewise, the mechanisms responsible for the greater increases in protein abundance of c-jun, ATF-2, jun-B, and fos-B remain to be elucidated, but reduced protein turnover is a distinct possibility. Interestingly, the increases in both mRNA and protein levels, when they do occur, are particularly sharp at 24 to 48 hours of 1,25D exposure (Fig. 2A and Fig. 3A). This coincides with the appearance of G1 to S phase block, and reinforces the view that 1,25D-induced differentiation can be divided into at least two, partially overlapping, stages (11). The importance of the JNK pathway activation in 1,25D-induced differentiation, which was previously shown (16, 17), seems to be predominately directed to the second stage.
The identification of four proteins as components of transcription activating complex offers mechanistic insights and directs attention to further studies of differentiation of neoplastic cells. Of particular importance may be an evaluation of jun-B as a tumor suppressor protein, and as such, a potential target for discovery of new drugs for cancer therapy.
Acknowledgments
We are grateful to Dr. Milan Uskokovic (BioXell) for the gift of 1,25-dihydroxyvitamin D3.
References
- 1.DeLuca HF. New concepts of vitamin D functions. Ann N Y Acad Sci. 1992;669:59–69. doi: 10.1111/j.1749-6632.1992.tb17089.x. [DOI] [PubMed] [Google Scholar]
- 2.Studzinski GP, McLane JA, Uskokovic MR. Crit Rev Eukaryotic Gene Expr. 1993;3:279–312. [PubMed] [Google Scholar]
- 3.Rachez C, Freedman LP. Mechanisms of gene regulation by vitamin D3 receptor: a network of coactivator interactions. Gene. 2000;246:9–21. doi: 10.1016/s0378-1119(00)00052-4. [DOI] [PubMed] [Google Scholar]
- 4.Ohyama Y, Ozono K, Uchida M, et al. Identification of a vitamin D-responsive element in the 5′-flanking region of the rat 25-hydroxyvitamin D3 24-hydroxylase gene. J Biol Chem. 1994;269:10545–10550. [PubMed] [Google Scholar]
- 5.Chen KS, DeLuca HF. Cloning of the human 1 α,25-dihydroxyvitamin D3 24-hydroxylase gene promoter and identification of two vitamin D-responsive elements. Biochim Biophys Acta. 1995;1263:1–9. doi: 10.1016/0167-4781(95)00060-t. [DOI] [PubMed] [Google Scholar]
- 6.van de Graaf SF, Boullart I, Hoenderop JG, Bindels RJ. Regulation of the epithelial Ca2+ channels TRPV5 and TRPV6 by 1α,25-dihydroxy vitamin D3 and dietary Ca2+ J Steroid Biochem Mol Biol. 2004;89–90:303–308. doi: 10.1016/j.jsbmb.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 7.Wang TT, Tavera-Mendoza LE, Laperriere D, et al. Large-scale in silico and microarray-based identification of direct 1,25-dihydroxyvitamin D3 target genes. Mol Endocrinol. 2005;19:2685–2695. doi: 10.1210/me.2005-0106. [DOI] [PubMed] [Google Scholar]
- 8.Carlberg C. Current understanding of the function of the nuclear vitamin D receptor in response to its natural and synthetic ligands. Recent Results Cancer Res. 2003;164:29–42. doi: 10.1007/978-3-642-55580-0_2. [DOI] [PubMed] [Google Scholar]
- 9.Cowley S, Paterson H, Kemp P, Marshall CJ. Activation of MAP kinase kinase is necessary and sufficient for PC12 differentiation and for transformation of NIH 3T3 cells. Cell. 1994;77:841–852. doi: 10.1016/0092-8674(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 10.Song X, Bishop JE, Okamura WH, Norman AW. Stimulation of phosphorylation of mitogen-activated protein kinase by 1α,25-dihydroxyvitamin D3 in promyelocytic NB4 leukemia cells: a structure-function study. Endocrinology. 1998;139:457–465. doi: 10.1210/endo.139.2.5747. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Studzinski GP. Activation of extracellular signal-regulated kinases (ERKs) defines the first phase of 1,25-dihydroxyvitamin D3-induced differentiation of HL60 cells. J Cell Biochem. 2001;80:471–482. doi: 10.1002/1097-4644(20010315)80:4<471::aid-jcb1001>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 12.Marcinkowska E. Evidence that activation of MEK1,2/erk1,2 signal transduction pathway is necessary for calcitriol-induced differentiation of HL-60 cells. Anticancer Res. 2001;21:499–504. [PubMed] [Google Scholar]
- 13.Abe J, Moriya Y, Saito M, Sugawara Y, Suda T, Nishii Y. Modulation of cell growth, differentiation, and production of interleukin-3 by 1α,25-dihydroxyvitamin D3 in the murine myelomonocytic leukemia cell line WEHI-3. Cancer Res. 1986;46:6316–6321. [PubMed] [Google Scholar]
- 14.Studzinski GP, Bhandal AK, Brelvi ZS. A system for monocytic differentiation of leukemic cells HL 60 by a short exposure to 1,25-dihydroxycholecalciferol. Proc Soc Exp Biol Med. 1985;179:288–295. doi: 10.3181/00379727-179-42098. [DOI] [PubMed] [Google Scholar]
- 15.Dwivedi PP, Hii CS, Ferrante A, et al. Role of MAP kinases in the 1,25-dihydroxyvitamin D3-induced trans-activation of the rat cytochrome P450C24 (CYP24) promoter. Specific functions for ERK1/ERK2 and ERK5. J Biol Chem. 2002;33:29643–29653. doi: 10.1074/jbc.M204561200. [DOI] [PubMed] [Google Scholar]
- 16.Wang Q, Wang X, Studzinski GP. Jun N-terminal kinase pathway enhances signaling of monocytic differentiation of human leukemia cells induced by 1,25-dihydroxyvitamin D3. J Cell Biochem. 2003;89:1087–1101. doi: 10.1002/jcb.10595. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Salman H, Danilenko M, Studzinski GP. Cooperation between antioxidants and 1,25-dihydroxy-vitamin D3 in induction of leukemia HL60 cell differentiation through the JNK/AP-1/Egr-1 pathway. J Cell Physiol. 2005;204:964–974. doi: 10.1002/jcp.20355. [DOI] [PubMed] [Google Scholar]
- 18.Coffman FD, Studzinski GP. Differentiation-related mechanisms which suppress DNA replication. Exp Cell Res. 1999;248:58–73. doi: 10.1006/excr.1999.4457. [DOI] [PubMed] [Google Scholar]
- 19.Studzinski GP, Harrison LE. Differentiation-related changes in the cell cycle traverse. Int Rev Cytol. 1999;189:1–58. doi: 10.1016/s0074-7696(08)61384-4. [DOI] [PubMed] [Google Scholar]
- 20.Shafarenko M, Liebermann DA, Hoffman B. Egr-1 abrogates the block imparted by c-myc on terminal M1 myeloid differentiation. Blood. 2005;3:871–878. doi: 10.1182/blood-2004-08-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kolla SS, Studzinski GP. Constitutive DNA binding of the low mobility forms of the AP-1 and SP-1 transcription factors in HL60 cells resistant to 1-β-d-arabinofuranosylcytosine. Cancer Res. 1994;54:1418–1421. [PubMed] [Google Scholar]
- 22.Studzinski GP, Wang X, Ji Y, et al. The rationale for deltanoids in therapy for myeloid leukemia: role of KSR-MAPK-C/EBP pathway. J Steroid Biochem Mol Biol. 2005;97:47–55. doi: 10.1016/j.jsbmb.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis AF, Meighan-Mantha RL, Riegel AT. Effects of TPA, bryostatin 1, and retinoic acid on PO-B, AP-1, and AP-2 DNA binding during HL-60 differentiation. J Cell Biochem. 1997;65:308–324. [PubMed] [Google Scholar]
- 24.Wang Y, Prywes R. Activation of the c-fos enhancer by the erk MAP kinase pathway through two sequence elements: the c-fos AP-1 and p62TCF sites. Oncogene. 2000;19:1379–1385. doi: 10.1038/sj.onc.1203443. [DOI] [PubMed] [Google Scholar]
- 25.Wajchman HJ, Rathod B, Song S, et al. Loss of deoxycytidine kinase expression and tetraploidization of HL60 cells following long-term culture in 1,25-dihydroxyvitamin D3. Exp Cell Res. 1996;224:312–322. doi: 10.1006/excr.1996.0141. [DOI] [PubMed] [Google Scholar]
- 26.Gallagher R, Collins S, Trujillo J, et al. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood. 1979;54:713–733. [PubMed] [Google Scholar]
- 27.Wang X, Gardner JP, Kheir A, Uskokovic MR, Studzinski GP. Synergistic induction of HL60 cell differentiation by ketoconazole and 1-desoxy analogues of vitamin D3. J Natl Cancer Inst. 1997;89:1199–1206. doi: 10.1093/jnci/89.16.1199. [DOI] [PubMed] [Google Scholar]
- 28.Bielinska A, Shivdasani RA, Zhang LQ, Nabel GJ. Regulation of gene expression with double-stranded phosphorothioate oligonucleotides. Science. 1990;250:997–1000. doi: 10.1126/science.2237444. [DOI] [PubMed] [Google Scholar]
- 29.Andrews NC, Faller DV. Rapid micropreparation technique for extraction of DNA binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991;19:2499. doi: 10.1093/nar/19.9.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danilenko M, Studzinski GP. Enhancement by other compounds of the anti-cancer activity of vitamin D3 and its analogs. Exp Cell Res. 2004;298:339–358. doi: 10.1016/j.yexcr.2004.04.029. [DOI] [PubMed] [Google Scholar]
- 31.Kim CW, Suh SI, Sung SH, Lee IK, Lee KS. A transcriptional factor decoy against AP-1 suppresses TGF-β1-induced type I collagen gene expression in cultured keloid fibroblasts. J Dermatol Sci. 2005;37:49–51. doi: 10.1016/j.jdermsci.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Haussler MR, Baylink DJ, Hughes MR, et al. The assay of 1α,25-dihydroxyvitamin D3: physiologic and pathologic modulation of circulating hormone levels. Clin Endocrinol. 1976;5 Suppl:151S–165S. doi: 10.1111/j.1365-2265.1976.tb03823.x. [DOI] [PubMed] [Google Scholar]
- 33.Miyamoto K, Kesterson RA, Yamamoto H, et al. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol Endocrinol. 1997;11:1165–1179. doi: 10.1210/mend.11.8.9951. [DOI] [PubMed] [Google Scholar]
- 34.Qi X, Pramanik R, Wang J, et al. The p38 and JNK pathways cooperate to trans-activate vitamin D receptor via c-Jun/AP-1 and sensitize human breast cancer cells to vitamin D3-induced growth inhibition. J Biol Chem. 2002;277:25884–25892. doi: 10.1074/jbc.M203039200. [DOI] [PubMed] [Google Scholar]
- 35.Okazaki T, Bielawska A, Bell RM, Hannun YA. Role of ceramide as a lipid mediator of 1 α,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J Biol Chem. 1990;265:15823–15831. [PubMed] [Google Scholar]
- 36.Okazaki T, Bielawska A, Domae N, Bell RM, Hannun YA. Characteristics and partial purification of a novel cytosolic, magnesium-independent, neutral sphingomyelinase activated in the early signal transduction of 1α,25-dihydroxyvitamin D3-induced HL-60 cell differentiation. J Biol Chem. 1994;269:4070–4077. [PubMed] [Google Scholar]
- 37.van Dam H, Castellazzi M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 38.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat Cell Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 39.Wagner EF. Functions of AP1 (Fos/Jun) in bone development. Ann Rheum Dis. 2002;61 suppl 2:ii40–ii42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 2000;19:2056–2068. doi: 10.1093/emboj/19.9.2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Passegue E, Wagner EF. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J. 2000;19:2969–2979. doi: 10.1093/emboj/19.12.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Passegue E, Jochum W, Schorpp-Kistner M, Mohle-Steinlein U, Wagner EF. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking jun-B expression in the myeloid lineage. Cell. 2001;104:21–32. doi: 10.1016/s0092-8674(01)00188-x. [DOI] [PubMed] [Google Scholar]
- 43.Datta R, Sherman ML, Stone RM, Kufe D. Expression of the jun-B gene during induction of monocytic differentiation. Cell Growth Differ. 1991;2:43–49. [PubMed] [Google Scholar]
- 44.Mollinedo F, Naranjo JR. Uncoupled changes in the expression of the jun family members during myeloid cell differentiation. Eur J Biochem. 1991;200:483–486. doi: 10.1111/j.1432-1033.1991.tb16208.x. [DOI] [PubMed] [Google Scholar]
- 45.Nakamura T, Datta R, Kharbanda S, Kufe D. Regulation of jun and fos gene expression in human monocytes by the macrophage colony-stimulating factor. Cell Growth Differ. 1991;2:267–272. [PubMed] [Google Scholar]
- 46.McCabe LR, Kockx M, Lian J, Stein J, Stein G. Selective expression of fos- and jun-related genes during osteoblast proliferation and differentiation. Exp Cell Res. 1995;218:255–262. doi: 10.1006/excr.1995.1154. [DOI] [PubMed] [Google Scholar]
- 47.Grigoriadis AE, Wang ZQ, Cecchini MG, et al. c-Fos: a key regulator of osteoclast-macrophage lineage determination and bone remodeling. Science. 1994;226:443–448. doi: 10.1126/science.7939685. [DOI] [PubMed] [Google Scholar]
- 48.Shafarenko M, Amanullah A, Gregory B, Liebermann DA, Hoffman B. Fos modulates myeloid cell survival and differentiation and partially abrogates the c-myc block in terminal myeloid differentiation. Blood. 2004;103:4259–4267. doi: 10.1182/blood-2002-09-2704. [DOI] [PubMed] [Google Scholar]
- 49.Sherman ML, Stone RM, Datta R, Bernstein SH, Kufe DW. Transcriptional and post-transcriptional regulation of c-jun expression during monocytic differentiation of human myeloid leukemic cells. J Biol Chem. 1990;265:3320–3323. [PubMed] [Google Scholar]
- 50.Quelle DE, Ashmun RA, Shurtleff SA, et al. Over-expression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993;7:1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- 51.Resnitzky D, Reed SI. Different roles for cyclins D1 and E in regulation of the G1-to-S transition. Mol Cell Biol. 1995;15:3463–3469. doi: 10.1128/mcb.15.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]