Abstract
Clear cell renal cell carcinoma (ccRCC) is the most common form of adult kidney cancer, characterised by the presence of inactivating mutations in the VHL gene in the majority of cases1,2 and by infrequent somatic mutations in known cancer genes. To elucidate further the genetics of ccRCC, we have sequenced 101 cases through 3544 protein coding genes. Here we report the identification of inactivating mutations in two genes encoding enzymes involved in histone modification, SETD2, a histone H3 lysine 36 methyltransferase and JARID1C (KDM5C), a histone H3 lysine 4 demethylase in addition to mutations in the histone H3 lysine 27 demethylase, UTX (KMD6A), we recently reported3. The results highlight the role of mutations in components of the chromatin modification machinery in human cancer. Additionally, NF2 mutations were found in non-VHL mutated ccRCC and several other likely cancer genes were identified. These results indicate that substantial genetic heterogeneity exists in a cancer type dominated by mutations in a single gene and that systematic screens will be key to fully elucidating the somatic genetic architecture of cancer.
Renal cell carcinoma accounts for about 209,000 new cases per year worldwide and 102,000 deaths2. Compared to other adult carcinomas, the genetics of ccRCC are distinctive. The majority of ccRCC have either somatic or germline inactivating mutations in the VHL gene, which are absent in most other cancers. Known cancer genes that are frequently mutated in other adult epithelial cancers, for example the RAS genes, BRAF, TP53, RB, CDKN2A, PIK3CA, PTEN, EGFR and ERBB2, make only a small contribution to ccRCC (http://www.sanger.ac.uk/genetics/CGP/cosmic/). To further elucidate the somatic genetics of ccRCC, we sequenced the coding exons of 3544 genes ased sequencing in 101 ccRCC (Supplementary Table 1 for sample information) equating to approximately 745 Mb of cancer genome sequenced. A full list of genes is given in Supplementary Table 2 and available online (http://www.sanger.ac.uk/genetics/CGP/Studies/). Copy number analyses using high-density SNP array and genome-wide expression array analyses were also performed. The initial study was comprised of 96 primary pre-treatment tumours (Table 1) and 5 ccRCC cell lines for which there was a matching lymphoblastoid line. All somatic mutations were confirmed by sequencing of the relevant exons in normal DNAs from the same individuals.
Table 1.
Patient demographics and clinical characteristics of primary ccRCC screening set
| Sex | |
| Male | 56 |
| Female | 40 |
| Age, Years | |
| Median | 62 |
| Range | 32–85 |
| Stage at diagnosis | |
| I | 44 |
| II | 14 |
| III | 34 |
| IV | 2 |
| NA | 2 |
| Grade at diagnosis | |
| 1 | 4 |
| 2 | 35 |
| 3 | 39 |
| 4 | 16 |
| NA | 2 |
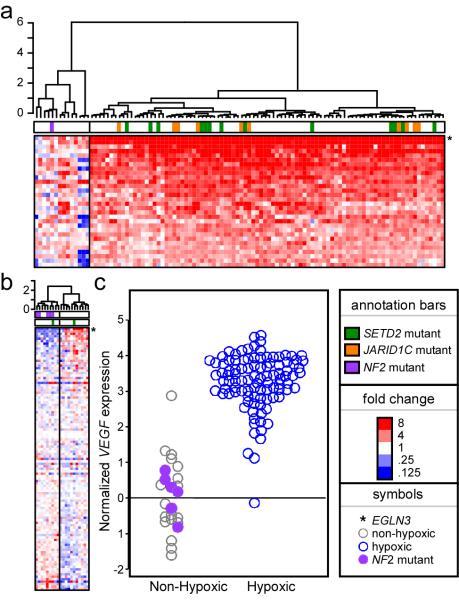
515 somatic base substitutions and small insertions/deletions were identified in the initial study (Supplementary Table 3). This included 56 cases (55%) with mutations in VHL, a prevalence in agreement with other reports 4. Evaluation of gene expression revealed two distinct phenotypes (Figure 1a). Seventy five out of 91 (82%) ccRCCs assessed for expression had up-regulation of genes associated with cellular hypoxia 5,6 with most (49/75, 65%) carrying VHL inactivating point mutations. Loss of 3p where VHL resides was the most frequent (88/101, 87%) copy number change seen on SNP array analyses. (Supplementary Figure 2; http://www.sanger.ac.uk/cgi-bin/genetics/CGP/cghviewer/CghHome.cgi). We identified a significantly (p<0.001, Supplemental Methods) higher proportion of small insertion/deletion mutations in ccRCC than seen in screens of the coding exons in pancreatic cancer7 and glioma 8 or several cancer types screened through all protein kinase genes 9 This may indicate an unidentified DNA repair defect, a common exposure or combination of these two. Average mutation prevalence was 0.75/Mb, somewhat lower than that observed for other adult cancers 9. The mutation spectrum in ccRCC was unremarkable, being dominated by C to T/G to A transitions (Supplementary Figure 1) as has been noted in several other adult cancers 9.
Figure 1. Gene expression analysis reveals two main classes of tumours - hypoxic and non-hypoxic.
A. Heatmap of hypoxia related gene expression (see methods for source of gene list) in primary ccRCC tumours. Red colour indicates a relative increase in gene expression while blue indicates decreased expression. Samples clustered to the left (highlighted with grey bar) do not show a hypoxic gene expression pattern while those to the right display the hypoxic expression pattern. EGLN3 is the most upregulated gene in the hypoxic group. JARID1C (orange bar) and SETD2 (green bar) mutant tumours are all clustered in the hypoxic group while the NF2 mutant tumour (purple bar) is in the non-hypoxic group. B. A similar pattern is observed in RCC cell lines with EGLN3 again being the most upregulated gene in the hypoxic group. Five NF2 mutant cell lines cluster in the non-hypoxic group. C. Clustering of NF2 mutant samples within low VEGF expression/non-hypoxic subgroup of ccRCC.
Genes with two or more non-synonymous mutations, a subset of those with at least one truncating mutation and/or identified as being of particular interest (Supplementary Table 5) were sequenced in a follow-up series of 311 primary RCC samples comprised of 246 ccRCC plus 65 additional samples of non-clear cell histology. Combined initial and follow-up screening data (Supplementary Tables 3 and 6) minus VHL mutations, were subjected to statistical analyses for the presence of positive selection, i.e. clustering of somatic mutations in a subset of genes consistent with a role in cancer development (Supplementary Methods). Five genes (SETD2, JARID1C, NF2, UTX, MLL2) have statistical support for being under selection at FDR<0.2, with all but MLL2 having strongest evidence for selection by truncating mutations (Supplementary Table 7).
Twelve of 407 (3%) ccRCC cases had somatic truncating mutations in SETD2, which encodes a histone H3K36 methyltransferase10, and 13/407 (3%) had truncating mutations in JARID1C, which encodes a histone H3K4 demethylase11 (Table 2, Supplementary Table 8). Screening of 779 cancer cell lines identified an additional SETD2 homozygous truncating mutation in the A498 ccRCC line. As assessment of homozygous deletions in primary tumour material is challenging, it is possible that we have underestimated the prevalence of inactivating mutations in these genes. No mutations were found in either SETD2 or JARID1C in the subset of non-clear cell cancers included in the follow-up screen and there was little evidence for involvement in other tumour types in cancer cell lines (Supplementary Table 9) in contrast to UTX 3. 88% (21/24) of samples with truncating SETD2 and JARID1C mutations had VHL mutations and/or the hypoxia expression phenotype (Figure 1a, b). One ccRCC cell line, LB996-RCC, was found to harbour (in addition to a NF2 truncating mutation) both a truncating UTX and SETD2 mutation, suggesting that these are not redundant in ccRCC development.
Table 2.
Mutation summary of highlighted genes in ccRCC
| Gene | Initial Screen Mutations |
Follow-up Screen Mutations |
Additional RCC cell line mutations* |
Total mutations |
|---|---|---|---|---|
| HIF1A | 1 nonsense | 1 splice/del, 1 frameshift | 3 | |
| JARID1C | 1 nonsense, 1 missense |
5 nonsense, 2 splice/del, 4 frameshift, 1 missense |
14 | |
| MLL2 | 1 nonsense, 2 missense |
9 missense, 1 nonsense, 4 silent | ND | 17 |
| NBN | 1 frameshift | 1 frameshift | ND | 2 |
| NF2 | 3 frameshift, 1 splice | 1 frameshift | 1 nonsense, 1 splice/del |
7 |
| PMS1 | 1 frameshift | 2 nonsense (Germline) | 3 | |
| SETD2 | 4 frameshift, 1 nonsense, 2 missense |
4 frameshift, 3 nonsense, 1 missense |
1 frameshift | 16 |
| UTX | 3 frameshift, 1 splice, 2 missense |
1 frameshift, 1 splice/del, 3 missense, 1 nonsense (Germline) |
12 | |
| WRN | 1 nonsense | 1 splice/frameshift, 1 missense | ND | 3 |
| ZUBR1 | 1 frameshift, 1 missense, 1 silent |
3 frameshift, 4 missense | ND | 10 |
no matching normal sequence available, presumptive somatic mutation. ND=not done. Detailed mutation annotation can be found in Supplementary Table 8.
Comparison of expression phenotypes of SETD2 and JARID1C mutated ccRCCs revealed a signature for both and marked difference between the two (Figure 2a, Supplemental Table 10a, b). Large scale transcriptional deregulation was noted in the SETD2 mutated subset with 298 genes showing significant differences (FDR<0.05, P=0.001 for association with SETD2 mutation) in expression relative to other cancers analysed (Figure 2b, Supplementary Table 10a). Nearly all of the significant expression changes were two-fold or less. In contrast, JARID1C mutant cancers revealed a much more restricted signature (Figure 2c, Supplementary Table 10b). Eighteen genes had significant changes in expression (FDR<0.05) in cancers with JARID1C mutations, including the metallothionien genes. Of note, those ccRCC with UTX mutations were also found to over-express metallothioniens (Figure 2c,d, Supplementary Table 10c) suggesting overlap in transcriptional deregulation caused by JARID1C and UTX loss (Figure 2e). Indeed, UTX and JARID1C are both implicated in H3K4 methylation status, JARID1C directly as a H3K4 demethylase11 and UTX as a component of the MLL2/3 H3K4 methylation complex12 13. In support of the importance of this axis, MLL2, an H3K4 methylase, was one of the other genes identified as a likely ccRCC cancer gene in our statistical analyses.
Figure 2. Gene deregulation in SETD2 and JARID1C/KDM5C mutant samples.
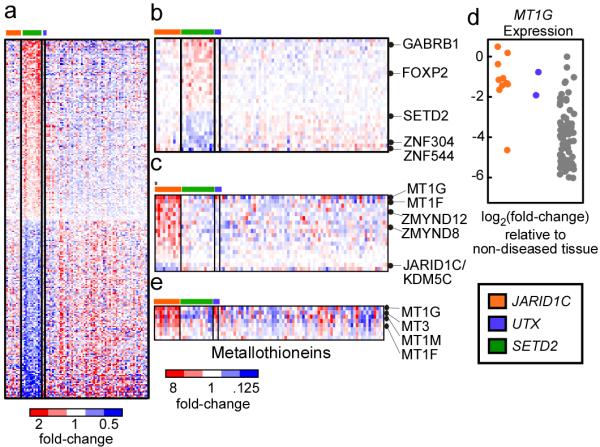
A. Genes (n=298) that are deregulated in tumor samples that contain non-synonymous SETD2 mutations (n=13) versus samples that lack such mutations (n=77) are plotted as a heatmap. Red color indicates increased gene expression compared to the average expression in the tumor samples, blue color indicates decreased gene expression. B. The most significantly deregulated genes in the SETD2 mutant samples. C. Heatmap of genes (n=18) that are deregulated in tumor samples that contain non-synonymous JARD1C/KDM5C mutations (n=10) versus samples that lack such mutations (n=80). The asterisks (*) highlights the sample containing the S1222P mutation. D. Expression of the MT1G gene in the tumor samples. Expression values are shown relative to non-diseased tissue and log2-transformed such that a log2-transformed value of −2 is equivalent to a 4-fold decrease in expression relative to non-diseased kidney. E. Metallothionein genes (n=8) were isolated examined for deregulated expression in JARID1C and UTX mutant samples. Significantly deregulated genes are indicated.
Five somatic truncating mutations in the NF2 gene were found in the full screen (Table 2, Supplementary Table 8). Germline NF2 truncating mutations predispose to neurofibromatosis II, characterised by predisposition to acoustic neuromas, meningiomas and schwannomas14. Somatic truncating mutations have been reported in these tumour types as well as mesothelioma (http://www.sanger.ac.uk/genetics/CGP/cosmic/). Sequencing in 779 cancer cell lines identified two truncating mutations in ccRCC cell lines SN12C and ACHN (http://www.sanger.ac.uk/genetics/CGP/CellLines/), supporting a novel role for NF2 in ccRCC. In contrast to JARID1C and SETD2, none of the NF2 mutant ccRCC samples harboured a VHL mutation or exhibited the hypoxia expression phenotype (Figure 1a, b). These data suggest that somatic NF2 mutations may account for a proportion of this subset of cases.
The screen identified a number of other potential new cancer genes in ccRCC (Table 2, Supplementary Table 8), including the identification of three samples with somatic HIF1a truncating mutations. Only these truncating mutations were found and two of the three samples had VHL point mutations. It has been shown that HIF1a and HIF2a have overlapping but non-identical targets and activities. HIF1a antagonises MYC function whilst HIF2a cooperates with MYC15,16. In VHL disease-associated ccRCC frequent absence of HIF1a staining with a preponderance of HIF2a positivity has been reported17, suggesting that there may be selection for loss of HIF1a during ccRCC progression. Three different truncating mutations (Table 2, Supplementary Table 8) were also identified in the DNA mismatch repair gene, PMS1. Notably, two truncating mutations found in the follow-up screen proved to be germline alleles. To our knowledge this is the first report of PMS1 mutations in ccRCC. Both germline cases were late onset (70 and 71 years old), without documented family history and none of the three mutated cancers were microsatellite unstable (data not shown). No truncating variants were detected in sequencing all coding exons of PMS1 in 528 normal controls indicating the germline alleles are not polymorphisms. Determining the extent truncating germline PMS1 alleles contribute to ccRCC will require larger cohort studies.
Somatic truncating mutations were detected in both initial and follow-up screens in WRN, NBN and ZUBR1(UBR4) (Table 2, Supplementary Table 8). WRN and NBN are both involved in DNA double strand break repair18 and recessive mutations in WRN and NBN give rise to Werner syndrome and Nijmegen breakage syndrome, respectively, both which predispose to cancer19,20 ZUBR1 encodes the p600 retinoblastoma associated protein and has been shown to be a cellular target for the bovine papilloma virus 21 and human papilloma type 16 E7 proteins22. Interaction of E7 with p600 has been shown to mediate cellular transformation independent of RB1 binding21,22 and knockdown of p600 in the absence of E7 induces anchorage independent growth22.
VHL inactivation alone induces senescence, suggesting a requirement for additional mutations to further drive ccRCC development in VHL mutant cases23. Conditional knockout of the VHL gene in renal epithelium does not generate any RCC phenotype, consistent with a need for additional hits (Teh, unpublished results). The mutations in the genes reported here likely contribute in this regard and the work further suggests that there are likely to be other mutated genes in ccRCC. As exemplified here, even in the context of a very prevalent driver mutation and dominant histological subtype, the numbers of cancers needed to adequately explore and capture the somatic genetic heterogeneity may be quite large, strongly supporting current efforts to expand mutational screening to large sample series with ultimately full genome sequencing of hundreds of cancers of all major subtypes (http://www.icgc.org/). For ccRCC, the data presented here will provide insights into its pathogenesis and the opportunity to understand the role of genetic subtypes in clinical behaviour and response to treatment.
Methods Summary
Genomic DNA samples were obtained from clinical tumour samples (>80% tumour cellularity), matching peripheral blood/adjacent normal kidney taken at nephrectomy and cancer cell lines as indicated utilizing standard protocols. Collection and use of patient samples were approved by the appropriate IRB of each Institution in addition to this study having LREC approval locally. RCC clinical samples and cell lines screened are given in Supplemental Table 1. SNPArray hybridization on the SNP6.0 platform was as per Affymetrix Protocols and as as described at http://www.sanger.ac.uk/cgi-bin/genetics/CGP/cghviewer/CghHome.cgi. PCR-based exon resequencing was performed and data analysed as previously described9 with sequencing traces being first analysed using a semiautomated system24 followed by manual inspection. PCR primer sequences are available for download at http://www.sanger.ac.uk/genetics/CGP/Studies/Renal/. Overall significance of an excess of non-silent mutations was determined using the methods previously described25 and is described in detail in the supplemental methods Gene expression profiling. RNA was harvested from fresh frozen patient tissue with Trizol according to manufacturer’s instructions (Invitrogen) and analyzed using human U133 Plus 2.0 Array probe sets according to manufacturer’s instructions (Affymetrix). Summarized expression values were computed using the robust multichip average (RMA) approach, corrected for batch effects, and used for clustering analysis and discriminate gene analysis using a moderated t-statistic. The patient and cell line expression data were deposited with Gene Expression Omnibus and Array Express under accession numbers GSE17895 and E-TABM-770, respectively. Further detail on analysis can be found in the Supplemental Methods.
Supplementary Material
Acknowledgements
We would like to acknowledge the Wellcome Trust for support under grant reference 077012/Z/05/Z and the Hauenstein and Gerber Foundations for support for the microarray expression work. We also thank Sancha Martin, Wendy McLaughlin and Sabrina Noyes for administrative support and Francis Brasseur for providing the matched-pair ccRCC cell lines.
References
- 1.Eble J, Epstein J, Sesterhann I. Pathology and Genetics of Tumours of the Urinary System and Male Genital Organs. IARC Press; Lyon, France: 2004. [Google Scholar]
- 2.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. The Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 3.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–3. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banks RE, et al. Genetic and Epigenetic Analysis of von Hippel-Lindau (VHL) Gene Alterations and Relationship with Clinical Variables in Sporadic Renal Cancer. Cancer Res. 2006;66:2000–2011. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- 5.Bommi-Reddy A, et al. Kinase requirements in human cells: III. Altered kinase requirements in VHLâ̂’/â̂’ cancer cells detected in a pilot synthetic lethal screen. Proc Natl Acad Sci U S A. 2008;105:16484–16489. doi: 10.1073/pnas.0806574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chi JT, et al. Gene Expression Programs in Response to Hypoxia: Cell Type Specificity and Prognostic Significance in Human Cancers. PLoS Med. 2006;3:e47. doi: 10.1371/journal.pmed.0030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–6. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun X-J, et al. Identification and Characterization of a Novel Human Histone H3 Lysine 36-specific Methyltransferase. J. Biol. Chem. 2005;280:35261–35271. doi: 10.1074/jbc.M504012200. [DOI] [PubMed] [Google Scholar]
- 11.Iwase S, et al. The X-Linked Mental Retardation Gene SMCX/JARID1C Defines a Family of Histone H3 Lysine 4 Demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Issaeva I, et al. Knockdown of ALR (MLL2) Reveals ALR Target Genes and Leads to Alterations in Cell Adhesion and Growth. Mol. Cell. Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MG, et al. Demethylation of H3K27 Regulates Polycomb Recruitment and H2A Ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 14.Ferner RE. Neurofibromatosis 1 and neurofibromatosis 2: a twenty first century perspective. Lancet Neurol. 2007;6:340–351. doi: 10.1016/S1474-4422(07)70075-3. [DOI] [PubMed] [Google Scholar]
- 15.Gordan JD, Bertout JA, Hu C-J, Diehl JA, Simon MC. HIF-2± Promotes Hypoxic Cell Proliferation by Enhancing c-Myc Transcriptional Activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, et al. HIF-1 Inhibits Mitochondrial Biogenesis and Cellular Respiration in VHL-Deficient Renal Cell Carcinoma by Repression of C-MYC Activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Mandriota SJ, et al. HIF activation identifies early lesions in VHL kidneys: Evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- 18.McKinnon PJ, Caldecott KW. DNA Strand Break Repair and Human Genetic Disease. Annl Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- 19.Kudlow BA, Kennedy BK, Monnat RJ. Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat Rev Mol Cell Biol. 2007;8:394–404. doi: 10.1038/nrm2161. [DOI] [PubMed] [Google Scholar]
- 20.Demuth I, Digweed M. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene. 2007;26:7792–7798. doi: 10.1038/sj.onc.1210876. [DOI] [PubMed] [Google Scholar]
- 21.DeMasi J, Huh K-W, Nakatani Y, Münger K, Howley PM. Bovine papillomavirus E7 transformation function correlates with cellular p600 protein binding. Proc Natl Acad Sci U S A. 2005;102:11486–11491. doi: 10.1073/pnas.0505322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huh K-W, et al. Association of the human papillomavirus type 16 E7 oncoprotein with the 600-kDa retinoblastoma protein-associated factor, p600. Proc Natl Acad Sci U S A. 2005;102:11492–11497. doi: 10.1073/pnas.0505337102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young AP, et al. VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat Cell Biol. 2008;10:361–369. doi: 10.1038/ncb1699. [DOI] [PubMed] [Google Scholar]
- 24.Dicks E, et al. AutoCSA, an algorithm for high throughput DNA sequence variant detection in cancer genomes. Bioinformatics. 2007;23:1689–1691. doi: 10.1093/bioinformatics/btm152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics. 2006;173:2187–98. doi: 10.1534/genetics.105.044677. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.