Abstract
Genomewide association studies to map common disease susceptibility loci have been hugely successful with over 300 reproducibly associated loci reported to date,1 but, perhaps surprisingly, have not yet provided convincing evidence for any susceptibility locus subject to parent of origin effects. We used imputation to extend existing genomewide association datasets2, 3, 4 and here report robust evidence, at rs941576, for paternally inherited risk of type 1 diabetes (T1D, ratio of allelic effects for paternal vs maternal transmissions = 0.75, 95%CI=0.71–0.79), in the imprinted region of chromosome 14q32.2, which contains a functional candidate gene, DLK1. Our meta-analysis also provided support at genomewide significance for a T1D locus at chromosome 19p13.2, with the highest association at marker rs2304256 (OR=0.86, 95%CI=0.82–0.90) in the TYK2 gene, which has previously associated with systemic lupus erythematosus.5
We used imputation to assess association with T1D across 2.6 million polymorphic SNPs from the International HapMap Project in a total of 7514 cases and 9405 controls of European ancestry from three existing genomewide association studies: WTCCC (UK)2, GAIN/NIMH (USA)3, T1DGC (UK)4 (supplementary table 1). The R package snpMatrix6 was used to conduct the imputation and calculate single SNP association score tests for each HapMap SNP. The score tests are based on the Cochran-Armitage test, with a Mantel extension to allow combination over different strata (UK region in the case of the WTCCC and T1DGC samples, and an estimated ancestry score derived from principal components in the case of the GoKinD/NIMH samples3). For imputed SNPs, the score statistics are calculated using the expected value of the imputed SNP, given observed SNPs, with the expectation calculated under the null hypothesis.
Overall, there was some over-dispersion of test statistics (λ = 1.14 and λ = 1.09 for 1 degree of freedom (df) and 2df tests respectively). This is consistent with the large sample size (almost 17,000 samples) and the over-dispersion observed in earlier analysis of these data without HapMap imputation4. Barrett et al argue that the greater contributor to over-dispersion in these data is bias (eg differential genotyping error) rather than population structure,4 and therefore cluster plots for all SNPs used to impute associated SNPs were examined carefully. Three loci showed suggestive evidence for association (p < 10−7) in regions not previously associated with T1D (supplementary figure 1; supplementary table 2). One SNP, rs229484, is proximal (30kb) to a nearby known T1D locus (rs2295413), also at 22q13.1, but is separated by two moderate recombination hotspots and there is low LD between the two markers (r2=0.1, D′=0.4).
In order to replicate these potential effects, we carried out direct genotyping of the three SNPs using TaqMan in a subset of the GWA samples, additional case-control and family samples and obtained evidence for association in two of the three loci (table 1, supplementary table 3). In these two loci, the overall levels of significance were < 10−8: rs2304256 p = 4.13 × 10−9, rs941576 p = 1.62 × 10−10.
Table 1.
Association testing of two SNPs using direct genotyping in case-control and family samples
| a. rs2304256 C>A on chromosome 19p13.2 | |||||
|---|---|---|---|---|---|
| Cohort | N | Fq (A) | Odds ratio (A:C) | (95% CI) | p value |
| WTCCC | 1766/1384 | 0.299 | 0.84 | (0.75–0.94) | 2.68 × 10−3 |
| T1DGC | 3838/3883 | 0.294 | 0.85 | (0.80–0.92) | 1.45 × 10−5 |
| Additional | 2686/4794 | 0.290 | 0.87 | (0.81–0.94) | 6.02 × 10−4 |
| Families | 3099 | 0.266 | 0.96 | (0.90–1.03) | 0.290 |
| Case-control combined | 8290/10061 | 0.293 | 0.86 | (0.82–0.90) | 1.43 × 10−10 |
| Families & case-control | (see above) | - | - | - | 4.13 × 10−9 |
| b. rs941576 A>G on chromosome 14q32.2 | |||||
|---|---|---|---|---|---|
| Cohort | N | Fq (G) | Odds ratio G:A | (95% CI) | p value |
| WTCCC | 1798/1406 | 0.43 | 0.90 | (0.81–1.00) | 0.049 |
| T1DGC | 3754/3736 | 0.43 | 0.88 | (0.82–0.94) | 9.3 × 10−5 |
| Additional | 2670/4840 | 0.43 | 0.92 | (0.86–0.99) | 0.030 |
| Families | 4057 | 0.45 | 0.87 | (0.82–0.93) | 1.8 × 10−5 |
| Case-control combined | 8222/9982 | 0.43 | 0.90 | (0.86–0.94) | 9.8 × 10−7 |
| Families & case-control | (see above) | - | - | - | 1.62 × 10−10 |
Association testing using observed (not imputed) genotypes in a subset of GWA samples, additional case control samples and family samples. SNP names are followed by alleles, ordered as major>minor. N is number of cases/controls, or number of informative transmissions. Fq is the frequency of the minor allele in controls or parents.
rs2304256 C>A (OR for A vs C = 0.86) is located within the TYK2 gene at chromosome 19p13.2, which is implicated in IFN-α, IL-6, IL-10 and IL-12 signalling. This is a region of wide LD containing several functional candidate genes (supplementary figure 2). rs2304256 is one of six SNPs in 1000 Genomes (pilot 1, April 2009) in mutual tight LD (r2 > 0.9); two are located within TYK2 (rs34725611 and rs11085725 in introns 6 and 23 respectively) and the remaining three (not yet in dbSNP) are downstream of TYK2 and upstream of ICAM3. No other SNPs had r2 > 0.62 with any of these six. rs2304256 itself is a non-synonymous SNP (Val362Phe) which has also been associated with systemic lupus erythematosus (SLE)5; in both T1D and SLE the minor (and inferred non-ancestral7) allele (A/Phe) appears protective5.
Most interestingly, the newly identified locus with the strongest association with T1D susceptibility occured in a well established imprinted region on chromosome 14q32.28 marked by SNP rs941576 A>G (OR for G vs A = 0.9). Beyond the insulin T1D susceptibility locus, marked by rs7111341 in Barrett et al,4 we do not know of any other T1D SNPs in established imprinted genes. Within this imprinted region of just over 1Mb, a mixture of paternally derived (DLK1, RTL1, DIO3) and maternally derived (MEG3, MEG8) genes are expressed8 (figure 1). Therefore, we tested for a parent of origin effect, expecting to see excess transmissions of the risk allele from either fathers or mothers (but not both) if the SNP was acting to influence one of these imprinted genes. A simple way to do this is to consider separately the paternal and maternal transmissions in a transmission disequilibrium testing (TDT) framework, and this showed strong evidence for reduced paternal transmission of the protective G allele (p=6.3 × 10−8). Although the maternal transmissions are distorted in the same direction and a small effect of the maternal copy cannot be discounted, there is no significant evidence for such an effect (p = 0.11; table 2). However, effects due to the action of maternal genotype in utero are confounded with imprinting effects9, so we fitted a model allowing for both maternal genotype and imprinting effects. This has been approached in case-parent trio data by log-linear modelling of counts of trios by parental and affected offspring genotype. We extended this method to allow for the fact that many of our families had multiple affected offspring (see supplementary methods) and found that the imprinting-only model was preferred (supplementary table 4); under that model, the imprinting effect was highly significant (p = 1.85 × 10−8) with the ratio of allelic effects for paternally to maternally inherited alleles equal to 0.75. This test gains power by using information on parental asymmetry induced by parent-of-origin effects. Asymmetry was clearly exhibited in our data: the protective allele (G) is less common amongst fathers of affected offspring than mothers (0.43 vs 0.47, p = 6.53 × 10−7). To reassure ourselves against a false positive result, driven by unusual patterns in a subset of the data, we reanalysed the families subdivided by broad geographical region, and found consistent effect estimates across all regions (table 3).
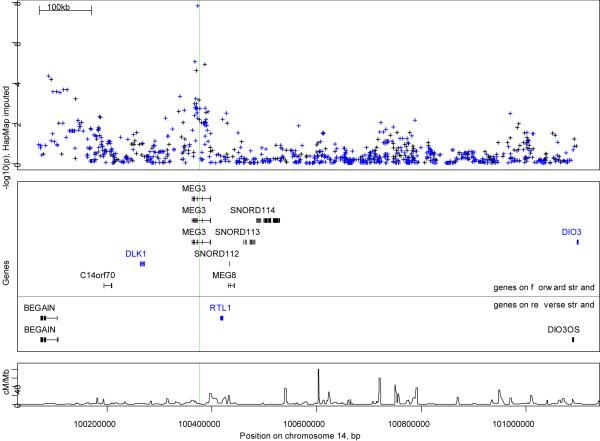
Figure 1.
The imprinted region on chromosome 14q32.2. The region shown is delimited by the most distant genes known to be imprinted 8 with positions according to Hs_NCBI36. The top panel shows -log 10(p) from 1 degree of freedom tests of association with SNPs across the region. SNPs which were directly genotyped are in black, SNPs imputed from HapMap in blue. The second panel shows the location and orientation of genes in the region. Paternally expressed genes are shown in blue, maternally expressed genes in black. The third panel shows recombination rates (cm/Mb) from HapMap. A solid green line shows the location of rs941576 in all panels for reference.
Table 2.
Transmission Disequilibrium Tests of rs941576 A>G
| Transmissions from | Fq | G Untransmitted | G Transmitted | p value |
|---|---|---|---|---|
| All parents | 0.45 | 2166 | 1891 | 1.6 × 10−5 |
| Fathers | 0.43 | 869 | 657 | 6.3 × 10−8 |
| Mothers | 0.47 | 793 | 730 | 0.11 |
Parental frequency (Fq) and transmissions of the rs941576 protective G allele, overall and separated by parent of origin. Frequencies are calculated using all parents. Note that because only transmissions from heterozygous (informative) parents are shown, transmission of a G allele implies non-transmission of A (and vice versa). The sum of maternal and paternal transmissions is less than the number of transmissions from all parents because it is not always possible to identify which parent transmitted which allele.
Table 3.
Imprinting analysis of rs941576 A>G
| Region | N | exp(-θ̂) | 95% CI | p |
|---|---|---|---|---|
| UK | 361 | 0.792 | 0.724 – 0.866 | 9.40 × 10−3 |
| Asia-Pacific | 32 | 0.88 | 0.656 – 1.18 | 0.662 |
| Other Europe | 257 | 0.725 | 0.644 – 0.815 | 6.08 × 10−3 |
| USA | 184 | 0.764 | 0.676 – 0.863 | 0.028 |
| Finland | 397 | 0.697 | 0.632 – 0.769 | 2.25 × 10−4 |
| Overall | 1231 | 0.749 | 0.712 – 0.789 | 1.85 × 10−8 |
Imprinting analysis using family data divided by broad geographical region. N is the number of informative families (which is less than the total number of families available, as only transmissions from asymmetric parents are informative). exp(-θ̂) is the ratio of the allelic effect for a paternally inherited risk allele compared to a maternally inherited allele.
The SNP rs941576 lies within intron 6 of the maternally expressed non-coding RNA gene, MEG3. However, our observation that only transmissions from fathers alter T1D risk suggest the causal variant influences one of the paternally expressed imprinted genes in its neighbourhood: DLK1, RTL1 or DIO3. rs941576 is between and downstream of both DLK1 and RTL1 and upstream of DIO3, at distances of 105kb, 41kb and 721kb respectively. Unusually for a locus identified from GWA data, the signal is restricted to rs941576 and there are no SNPs in HapMap or the current pre-release of 1000 Genomes Project (pilot 1, April 2009) which are in strong or moderate LD with rs941576 (all r2 < 0.5, data not shown). Although that does not preclude the existence of an as yet unknown variant (SNP or structural variant) in tighter LD, rs941576 lies within a region conserved across mammalian species, including opossum. This is interesting because the region is not imprinted in the opossum, there is no sequence homology to MEG3 and, while there is some sequence homology to mouse and human RTL1 gene, it appears to be extensively degraded in opossum10. Thus, if the region is conserved because it contains regulatory elements of nearby genes, these must regulate one of the genes common to all mammals, ie DLK1 or DIO3.
Although rs941576 lies some distance from the paternally expressed genes in the region, regulatory regions can lie >100kb from their target genes, particularly in imprinted regions11. This region is already subject to long-range cis-acting regulation from the intergenic differentially methylated region (DMR) located 12.5kb upstream of MEG3.12 Insertion of a transgene in the mouse downstream of this DMR causes loss of imprinting on the paternal chromosome, biallelic expression of the mouse homologue of MEG3, Gtl2 and reduced expression of Dlk113. Thus, it is plausible that this SNP (or another unknown variant nearby) could alter the regulation of the paternally expressed DLK1 or RTL1 genes.
Of the paternally expressed genes, only DLK1 has a strong functional candidacy. It is most strongly expressed in human heart, pancreatic islet cells, pituitary tissue, ovaries, placenta and testes (T1DBase, BioGPS), is related to members of the Notch-Delta family of signalling molecules and encodes a membrane bound protein, which can be cleaved to form fetal antigen 1 (FA1).14 FA1 is involved in differentiation of many cell types15 including pancreatic beta cells where FA1 immunoreactivity has been localised to glucagon-negative cells in the mature pancreas.16 FA1 is also involved in hematopoiesis including differentiation and function of B lymphocytes17,18 and has been shown to increase expression of pro-inflammatory cytokines in human bone marrow mesenchymal stem cells and promote B cell proliferation in human peripheral blood.19 Thus there are a number of ways in which variation in the expression of DLK1 could alter susceptibility to T1D, which is caused by autoimmune destruction of insulin-producing beta cells in the pancreas.
The mechanisms underlying imprinting are not yet fully understood, but are known to involve epigenetic processes including DNA methylation and histone acetylation. The causal variant underlying this association could be acting directly to alter the expression of the paternally inherited copy of a nearby gene (DLK1 appears to be the strongest candidate), or it could act by interfering subtly with the imprinting mechanism and in turn alter expression of either the paternally or maternally inherited copies of a target gene. Although rs941576 may be tagging an unknown causal variant, there is support for the hypothesis that this SNP is itself the causal variant, given its isolation from other SNPs in terms of linkage disequilibrium, and its location in a conserved and, presumably, regulatory region.
Rare disorders related to imprinting defects are known (eg Prader-Willi syndrome, OMIM 176270). For common complex diseases, over 300 reproducibly associated1 loci have been reported, but we are not aware of any convincing evidence for another susceptibility locus subject to parent of origin effects. At least one common disease locus overlaps a known imprinted region: the T1D associated region of chromosome 11p15 contains the insulin and IGF2 genes, but a previous report by our group of potential parent of origin effects at this locus in T1D20 has not yet been substantiated. We are aware of only one other report of a parent of origin effect, in basal cell carcinoma,21 although this was only demonstrated in a single population and at a relatively modest level of statistical significance (p ≈ 0.01).
Supplementary Material
Acknowledgments
This work was funded by the Juvenile Diabetes Research Foundation International, the Wellcome Trust and the National Institute for Health Research Cambridge Biomedical Centre. The Cambridge Institute for Medical Research (CIMR) is in receipt of a Wellcome Trust Strategic Award (079895). We thank all study participants and family members.
We acknowledge use of DNA from the 1958 British Birth Cohort collection, funded by the Medical Research Council (grant G0000934) and the Wellcome Trust (grant 068545/Z/02). We thank The Avon Longitudinal Study of Parents and Children laboratory in Bristol and the British 1958 Birth Cohort team, including S. Ring, R. Jones, M. Pembrey, W. McArdle, D. Strachan and P. Burton, for preparing and providing the control DNA samples.
We acknowledge use of DNA from The UK Blood Services collection of Common Controls (UKBS collection), funded by the Wellcome Trust grant 076113/C/04/Z, by the Wellcome Trust/Juvenile Diabetes Research Foundation grant 061858, and by the National Institute of Health Research of England. The collection was established as part of the Wellcome Trust Case-Control Consortium.
We thank David Dunger, Barry Widmer, and the British Society for Paediatric Endocrinology and Diabetes for the TID case collection.
We acknowledge use of DNA from the Human Biological Data Interchange and Diabetes UK for the USA and UK multiplex families, respectively; the Norwegian Study Group for Childhood Diabetes (D. Undlien and K. Ronningen) for the Norwegian families; D. Savage, C. Patterson, D. Carson and P. Maxwell for the Northern Irish families; the Genetics of Type 1 Diabetes in Finland (GET1FIN); J. Tuomilehto, L. Kinnunen, E. Tuomilehto-Wolf, V. Harjutsalo and T. Valle for the Finnish families; and C. Guja and C. Ionescu-Tirgoviste for the Romanian families.
This research utilizes resources provided by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Human Genome Research Institute (NHGRI), National Institute of Child Health and Human Development (NICHD), and Juvenile Diabetes Research Foundation International (JDRF) and supported by U01 DK062418.
This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data are available from http://www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113.
We gratefully acknowledge the National Institute of Mental Health for generously allowing the use of their control CEL and genotype data. Control subjects from the National Institute of Mental Health Schizophrenia Genetics Initiative (NIMH-GI), data and biomaterials are being collected by the “Molecular Genetics of Schizophrenia II” (MGS-2) collaboration. The investigators and co-investigators are as follows: P.V. Gejman (Collaboration coordinator) and A.R. Sanders (ENH/Northwestern University, MH059571); F. Amin (Emory University School of Medicine, MH59587); N. Buccola (Louisiana State University Health Sciences Center, MH067257); W. Byerley (University of California-Irvine,MH60870); C.R. Cloninger (Washington University, St. Louis, U01, MH060879); R. Crowe (PI) and D. Black (University of Iowa, MH59566); R. Freedman (University of Colorado, MH059565); D. Levinson (University of Pennsylvania, MH061675); B. Mowry (University of Queensland, MH059588); and J. Silverman (Mt. Sinai School of Medicine, MH59586). The samples were collected by V.L. Nimgaonkar’s group at the University of Pittsburgh as part of a multi-institutional collaborative research project with J. Smoller and P. Sklar (Massachusetts General Hospital) (grant MH 63420).
We acknowledge the National Institutes of Health for allowing the use of their control allele signal intensity and genotype data. The dataset(s) used for the analyses described in this manuscript were obtained from the GAIN Database, controlled through dbGaP accession number phs000018.v1.p1.
We also thank H. Stevens, P. Clarke, G. Coleman, S. Duley, D. Harrison, S. Hawkins, T. Mistry and N. Taylor for preparation of DNA samples.
We thank Anne Ferguson-Smith for helpful advice on the chromosome 14q32 region.
Appendix
Online Methods
Sample selection and genotyping
A total of 7514 cases and 9045 controls samples were included, from three GWA studies: WTCCC (UK), T1DGC (UK) GAIN/NIMH (USA); the samples, and their genotyping have been described previously.2,3,4 Numbers of samples from each study, and genotyping platform are given in supplementary table 1. SNP and sample exclusion criteria were as applied previously.4 Briefly, all subjects were of White European ancestry, and samples were excluded if they showed evidence of non-European ancestry, or of being duplicates of or closely related to another sample in the study. SNPs were excluded if the minor allele frequency (MAF) fell below 1% in cases or controls, if they deviated from Hardy-Weinberg equilibrium (p < 5.7 × 10−7), if the call rate fell below 95% (WTCCC and T1DGC) or if a genotype-calling metric indicated insufficient separation of the signal clouds (GoKinD/NIMH).22
SNPs showing suggestive association in the imputed analysis were genotyped directly using TaqMan (Applied Biosystems) on a subset of the GWA samples (the T1DGC, all WTCCC cases and about half the WTCCC controls were unavailable to us), additional case-control samples, and a set of family samples with T1D affected offspring (supplementary table 1). The additional case and control samples have also been described previously.4 The family samples were drawn from across Europe and America, and are predominantly of White European origin; we did not exclude subjects who self-reported a non-White European origin as testing for transmissions within families is equivalent to a pseudo case control approach, with ethnically matched controls. All Taqman genotyping data were scored twice to minimize error; the second operator was unaware of case-control status and family structure.
Imputation
For each of the three studies considered, we divided SNPs from HapMap version 2 (release 24) into two sets - those which were genotyped and passed quality control (QC) thresholds in the study (X) and those which were not genotyped or failed QC (Y). The R package snpMatrix6 from the Bioconductor project23 was to used calculate imputation “rules” for prediction of each SNP in Y from nearby SNPs in X using HapMap genotypes and to carry out association tests for the imputed SNPs. The algorithms used in snpMatrix, together with the parameter settings we used, are described below.
In regions of high LD, the genotype of one SNP can be related to the genotypes of others by a linear regression24,25,26. The first step in calculating an imputation rule is to select a set of “tag” SNPs by forward stepwise regression of the the Y SNP on the nearest 50 X SNPs (subject to a maximum missing data requirement). New SNPs are added to the regression until either (a) R2 > 0.95, (b) the change in R2 is < 0.05, or (c) the number of tag SNPs reaches four. Regression calculations are carried out at the genotype level, with each SNP genotype coded 0, 1, or 2. If a prediction R2 ≥ 0.95 cannot be achieved using this stepwise regression approach, then an alternative imputation rule is attempted using the set of tag SNPs selected by the forward stepwise procedure. Using the conventional EM algorithm, frequencies are estimated for the haplotypes of the Y SNP plus the selected tags. Conditional probabilites of the Y allele given the tag SNP haplotype are calculated and provide the imputation rule. This rule is used in preference to the regression rule if the improvement in R2 exceeds 0.1.
These imputation rules are then applied to the main study dataset to calculate the expectation of each Y SNP conditional on typed SNPs. Note that this expectation is not generally an integer and the Cochran-Armitage test then becomes a t-test comparing the mean imputation score in cases with that in controls. Extension to allow for stratified comparisons and to combine information from different studies is straightforward: differences between mean scores are simply averaged over strata (and studies), with weights inversely proportional to their variances. These procedures are all implemented in snpMatrix.
This imputation method is computationally faster than those based on hidden Markov models27 or on variable length Markov chains.28 For a subset of our data we compared our imputation results with those from IMPUTE27 and found them to be very similar. It has an additional advantage over such methods in that, since each imputation is based on a small number of tag SNPs, it is easier to differentiate between genuine associations and those caused by poor clustering and differential measurement error; for each putative association, allele signal plots for all tags were visually inspected.
Association analysis
Single SNP association score tests were performed for each HapMap SNP within each cohort using direct genotypes if available, or imputed genotypes if not. The score is
where Y i and Xi are the phenotypic (case/control) and genotype data respectively for subject i. When a SNP is not directly observed, Xi is replaced by its expected value calculated under the null hypothesis as described above. When it is poorly imputed, this expected value is shrunk towards X̅ and contributes little to the test statistic. The permutation variance (the variance under random permutation of Y ) is used to calculate the χ2 test. The score statistics were combined first across strata within cohorts and finally across cohorts using the method proposed by Mantel.29 The scores (Ui where i denotes cohort or stratum) and the variances (V i) are summed to form an overall test of association, (Σ Ui)T (Σ V i)>−1 (Σ Ui). Strata were defined by UK region in the case of the WTCCC and T1DGC samples, and an estimated ancestry score derived from principal components in the case of the GoKinD/NIMH samples.3 Testing for association with SNPs on the X chromosome was carried out using the method proposed by Clayton.30 Over-dispersion of the test statistics was calculated after removal of known T1D loci4 and these parameters used to calculate adjusted p values given in table 2.
SNPs showing overall association (p < 1 × 10−7) in regions not previously reported4 were subject to further screening. Cluster plots of each SNP used for imputation were examined manually, and the result discarded unless all cluster plots for all cohorts were considered clearly separated. One of the cohorts studied (USA) was not designed as a T1D case-control study, and was serendipitously assembled after cases and controls were genotyped on different versions of the Affymetrix 500K chip and to different protocols. This cohort was subject to greater differential bias than the other cohorts. As a result, many SNPs were found which showed (often extreme) association in the USA samples (p < 1 × 10−7) but no association in the T1DGC and WTCCC samples combined (p < 1 × 10−3); for these SNPs, only the data from T1DGC and WTCCC were combined.
Family data were analysed by transmission disequilibrium test, splitting mulitplex families into parent offspring trios and using a pseudo-case control framework to estimate allelic effects. A score statistic was also generated, and a score test for association in case-controls and families combined conducted by summing the scores and variances as described above.
Imprinting test
We use a logistic regression approach to test for imprinting/and or maternal genotype effects on risk in offspring. This approach was originally proposed by Weinberg9,31 for data consisting of trios of an affected case and both his or her parents, but required extension to deal with our data which included families with multiple affected offspring. Weinberg’s approach is to analyse counts of case-parent trios classified by genotype of mother, M, father, P, and the affected offspring, O, in a 3 × 3 × 3 table. Of the 15 cells in this table consistent with Mendelian transmission, five concern families in which the genotypes of the two parents are concordant; these are not informative in the analysis. The remaining ten cells can be organized by mating type and offspring genotype into five pairs in which the maternal and paternal genotypes are considered interchangable (Supplementary Table 5). In the absence of maternal genotype and imprinting effects, and assuming that, in the population from which families are drawn, the two possible parental genotype combinations within each mating type are equally frequent, then their frequencies in case-parent trios will also not differ systematically. However maternal genotype and imprinting effects will distort these ratios. In supplementary table 5, pairs of genotype configurations are set out with the configuration in which the mother carries more copies of the “2” allele than the father appearing first. The table also sets out the predictions of a multiplicative model for relative risk conditional upon genotype and upon parents; the genotype relative risk for the offspring (γ1/1, γ1/2, and γ2/2), are modified by multiplicative effects of the maternal genotype (φ1/2 and φ1/2, φ1/1 being taken as 1) and by a factor λ if a “2” allele was received from the mother rather than from the father. The ratio of these two risks for each mating type gives the ratio of expected frequencies in case-parent trios. This model can be fitted to the observed pairs of case-parent trio frequencies using any standard logistic regression program, thus allowing estimation and testing of maternal genotype and imprinting effects.
Extension of this method to deal with families in which there may be several affected offspring is relatively straightforward. Again we tabulate counts of families by genotype of mothers, father, and offspring, but there are now more possible cells in the tabulation. For example, with two affected offspring there are seven informative pairs of genotype configurations (Supplementary Table 6). Under the assumption that the SNP under observation is the sole causal variant or has r2 = 1 with a sole causal variant, disease occurrences in the offspring are conditionally independent given their genotypes and their parents, and the ratio of expected frequencies is given by the ratio of products of predicted relative risks for the two offspring. Extension to the case of more than two affected offspring follows similar principles. For families with three affected offspring there are nine informative pairs of genotype configuration, for four affected offspring, eleven, and so on. Logistic regression can then be used to estimate and test for effects of maternal genotype and imprinting in the general case where, as in our study, the data consist of families with varying numbers of affected offspring.
In the case where the SNP tested is not the sole causal variant (or in perfect LD with it), disease occurrences in offspring are not conditionally independent and there may be some bias. We would expect this to be small when the SNP has high r2 with the causal variant. We also note that type 1 error rate will be unaffected by departure from conditional independence when testing the hypothesis of no imprinting and no maternal genotype effect against presence of either (or both) effects, although the method may then not be fully efficient.
Footnotes
URLs
1000 Genomes http://www.1000genomes.org
BioGPS http://biogps.gnf.org
International HapMap Project http://www.hapmap.org
T1DBase http://www.t1dbase.org
References
- 1.Donnelly P. Progress and challenges in genome-wide association studies in humans. Nature. 2008;456:728–731. doi: 10.1038/nature07631. [DOI] [PubMed] [Google Scholar]
- 2.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper JD, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrett JC, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suarez-Gestal M, et al. Replication of recently identified systemic lupus erythematosus genetic associations: a case-control study. Arthritis Res Ther. 2009;11:R69. doi: 10.1186/ar2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clayton D, Leung HT. An R package for analysis of whole-genome association studies. Hum Hered. 2007;64:45–51. doi: 10.1159/000101422. [DOI] [PubMed] [Google Scholar]
- 7.Spencer CCA, et al. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagan JP, et al. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One. 2009;4:e4352. doi: 10.1371/journal.pone.0004352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg CR, et al. A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet. 1998;62:969–978. doi: 10.1086/301802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edwards CA, et al. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 2008;6:e135. doi: 10.1371/journal.pbio.0060135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arney KL. H19 and Igf2-enhancing the confusion? Trends Genet. 2003;19:17–23. doi: 10.1016/s0168-9525(02)00004-5. [DOI] [PubMed] [Google Scholar]
- 12.Lin SP, et al. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 13.Steshina EY, et al. Loss of imprinting at the Dlk1-Gtl2 locus caused by insertional mutagenesis in the gtl2 5′ region. BMC Genet. 2006;7:44. doi: 10.1186/1471-2156-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen CH, et al. Protein structure of fetal antigen 1 (FA1). a novel circulating human epidermal-growth-factor-like protein expressed in neuroendocrine tumors and its relation to the gene products of dlk and pG2. Eur J Biochem. 1994;225:83–92. doi: 10.1111/j.1432-1033.1994.00083.x. [DOI] [PubMed] [Google Scholar]
- 15.Laborda J. The role of the epidermal growth factor-like protein dlk in cell differentiation. Histol Histopathol. 2000;15:119–129. doi: 10.14670/HH-15.119. [DOI] [PubMed] [Google Scholar]
- 16.Tornehave D, et al. FA1 immunoreactivity in endocrine tumours and during development of the human fetal pancreas; negative correlation with glucagon expression. Histochem Cell Biol. 1996;106:535–542. doi: 10.1007/BF02473268. [DOI] [PubMed] [Google Scholar]
- 17.Sakajiri S, et al. Dlk1 in normal and abnormal hematopoiesis. Leukemia. 2005;19:1404–1410. doi: 10.1038/sj.leu.2403832. [DOI] [PubMed] [Google Scholar]
- 18.Raghunandan R, et al. Dlk1 influences differentiation and function of B lymphocytes. Stem Cells Dev. 2008;17:495–507. doi: 10.1089/scd.2007.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdallah BM, et al. dlk1/FA1 regulates the function of human bone marrow mesenchymal stem cells by modulating gene expression of pro-inflammatory cytokines and immune response-related factors. J Biol Chem. 2007;282:7339–7351. doi: 10.1074/jbc.M607530200. [DOI] [PubMed] [Google Scholar]
- 20.Bennett ST, et al. the IMDIAB group Insulin VNTR allele-specific effect in type 1 diabetes depends on identity of untransmitted paternal allele. Nat Genet. 1997;17:350–352. doi: 10.1038/ng1197-350. [DOI] [PubMed] [Google Scholar]
- 21.Stacey SN, et al. New common variants affecting susceptibility to basal cell carcinoma. Nat Genet. 2009;41:909–914. doi: 10.1038/ng.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plagnol V, et al. A method to address differential bias in genotyping in large-scale association studies. PLoS Genet. 2007;3:e74. doi: 10.1371/journal.pgen.0030074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapman JM, et al. Detecting disease associations due to linkage disequilibrium using haplotype tags: a class of tests and the determinants of statistical power. Hum Hered. 2003;56:18–31. doi: 10.1159/000073729. [DOI] [PubMed] [Google Scholar]
- 25.Clayton D, et al. Use of unphased multilocus genotype data in indirect association studies. Genet Epidemiol. 2004;27:415–28. doi: 10.1002/gepi.20032. [DOI] [PubMed] [Google Scholar]
- 26.Vella A, et al. Localization of a type 1 diabetes locus in the IL2RA/CD25 region by use of tag single-nucleotide polymorphisms. Am J Hum Genet. 2005;76:773–779. doi: 10.1086/429843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marchini J, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 28.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am J Hum Genet. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantel N. Chi-square tests with one degree of freedom: Extensions of the Mantel-Haenszel procedure. J Am Stat Assoc. 1963;58:690–700. [Google Scholar]
- 30.Clayton D. Testing for association on the X chromosome. Biostatistics. 2008;9:593–600. doi: 10.1093/biostatistics/kxn007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weinberg CR. Methods for detection of parent-of-origin effects in genetic studies of case-parents triads. Am J Hum Genet. 1999;65:229–235. doi: 10.1086/302466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.