Abstract
Calorie restriction extends lifespan in organisms ranging from yeast to mammals1. In yeast, the SIR2 gene mediates the life-extending effects of calorie restriction2. Here we show that the mammalian SIR2 orthologue, Sirt1 (sirtuin 1), activates a critical component of calorie restriction in mammals; that is, fat mobilization in white adipocytes. Upon food withdrawal Sirt1 protein binds to and represses genes controlled by the fat regulator PPAR-γ (peroxisome proliferator-activated receptor-γ), including genes mediating fat storage. Sirt1 represses PPAR-γ by docking with its cofactors NCoR (nuclear receptor co-repressor) and SMRT (silencing mediator of retinoid and thyroid hormone receptors). Mobilization of fatty acids from white adipocytes upon fasting is compromised in Sirt1+/− mice. Repression of PPAR-γ by Sirt1 is also evident in 3T3-L1 adipocytes, where overexpression of Sirt1 attenuates adipogenesis, and RNA interference of Sirt1 enhances it. In differentiated fat cells, upregulation of Sirt1 triggers lipolysis and loss of fat. As a reduction in fat is sufficient to extend murine lifespan3, our results provide a possible molecular pathway connecting calorie restriction to life extension in mammals.
Calorie restriction is the first regimen shown to extend mammalian lifespan. Calorie restriction induces a shedding of body fat from white adipose tissue (WAT), a decrease in body temperature and an increase in insulin sensitivity4. It has been suggested that calorie restriction functions in a passive way by lowering oxidative and other damage, thereby resulting in a longer lifespan. However, studies in yeast suggest that calorie restriction is a highly regulated response that requires a sensing step followed by the execution of a programme to extend lifespan5. The regulatory gene that mediates this programme is SIR2, which promotes survival in yeast6, Caenorhabditis elegans7 and, perhaps, higher eukaryotes in response to food scarcity. SIR2 is well suited for this function because it encodes an NAD-dependent protein deacetylase1, enabling it to monitor cellular metabolism and exert corresponding effects on gene expression.
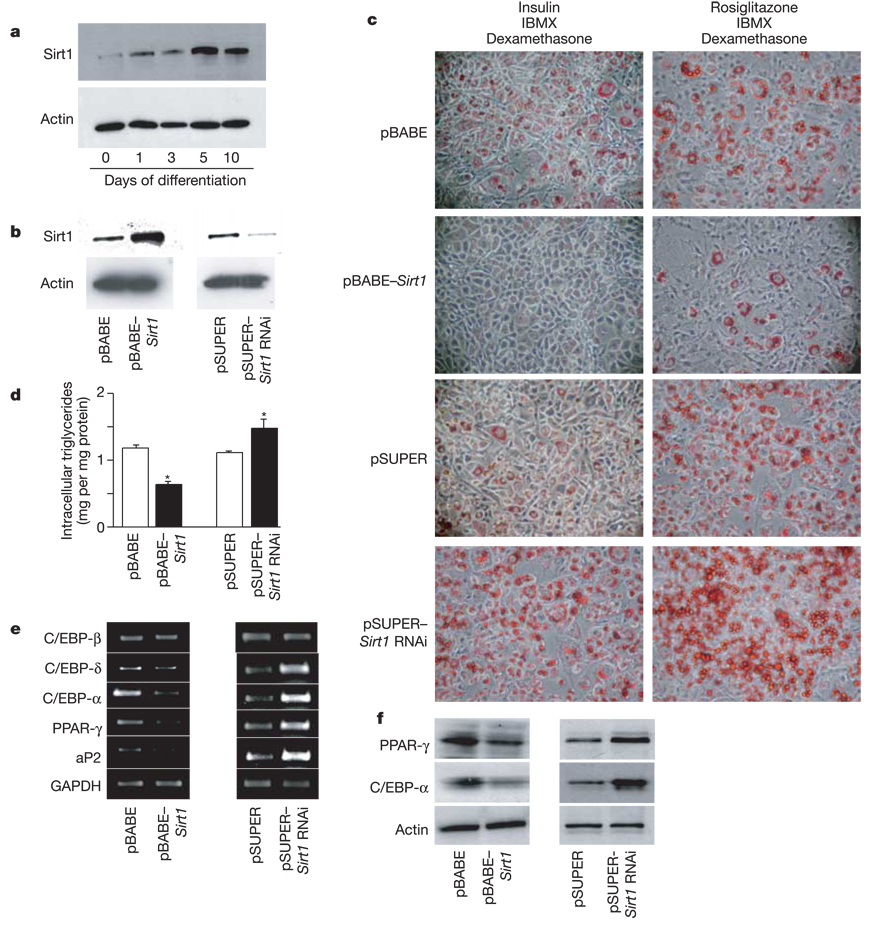
WAT seems to be a primary factor in longevity, as mice engineered to have reduced levels of it live longer3. This effect on lifespan may result from changes in hormones that are normally secreted from WAT in proportion to fat mass. Thus, we evaluated whether the mammalian Sir2 orthologue, Sirt1, senses nutrient availability in WAT and mediates corresponding effects on fat accumulation. To probe whether Sirt1 may actually modulate adipogenesis, we used mouse 3T3-L1 fibroblasts as an in vitro model8. Adipogenesis in these cells is promoted by the nuclear receptor PPAR-γ 9. Upon induction of adipogenesis by insulin, dexamethasone and isobutylmethylxanthine, we observed that Sirt1 protein levels increased and peaked at day 5 after hormonal stimulation (Fig. 1a). Sirt1 expression in 3T3-L1 cells was then modified through retroviral infection with either pBABE–Sirt1 or pSUPER–Sirt1 RNA interference (RNAi) for overexpression (tenfold) or downregulation (sevenfold) of the Sirt1 gene, respectively (Fig. 1b). 3T3-L1 cells undergo one or two mitotic divisions after induction as a prelude to terminal differentiation9,10. This occurred normally in cells that overexpressed or underexpressed Sirt1 as evaluated by 5-bromodeoxyuridine (BrdU) incorporation (data not shown). However, compared with cells infected with the control vector, stable 3T3-L1 cells overexpressing Sirt1 accumulated much less fat as determined by Oil red O staining after 7 days of differentiation (Fig. 1c) or direct measurement of intracellular triglyceride content (Fig. 1d). In contrast, downregulation of Sirt1 expression resulted in a significant increase in triglyceride accumulation after differentiation (Fig. 1c, d). These results indicate that Sirt1 acts a negative modulator of adipogenesis in the 3T3-L1 model.
Figure 1.
Sirt1 regulates adipogenesis and triglyceride accumulation in 3T3-L1 cells. a, Time course of Sirt1 protein expression on a western blot in 3T3-L1 cells upon induction of adipogenesis. b, Sirt1 protein levels in 3T3-L1 cells infected with control pBABE or pSUPER retroviral vectors or with pBABE–Sirt1 or pSUPER–Sirt1 RNAi vectors for overexpression or downregulation of Sirt1, respectively. c, Oil red O staining for triglycerides in infected 3T3-L1 cells differentiated for 7 days with IBMX and dexamethasone plus either insulin or rosiglitazone. d, Intracellular triglyceride levels in infected 3T3-L1 cells differentiated for 7 days with insulin, IBMX and dexamethasone. Data shown are means + s.e.m. and were obtained from two independent experiments carried out in triplicate. Asterisk indicates a statistical difference from respective control (P < 0.05). e, f, 3T3-L1 cells were infected and differentiated as in d. Protein and total RNA were extracted at day 7. e, Reverse transcription and PCRs were on complementary DNAs with gene-specific primers. One microgram of total RNA was used in each case. f, Protein levels of PPAR-γ and C/EBP-α in total cell extracts measured by western blotting. GAPDH and actin levels are shown as loading controls.
Insulin/insulin-like growth factor (IGF)-1 signalling helps to regulate adipogenesis11, and this pathway determines lifespan in nematode worms12. To evaluate the possible interaction of Sirt1 with the insulin/IGF-1 signalling cascade in adipocytes, we differentiated virally transduced 3T3-L1 cells in the absence of insulin but with rosiglitazone, a very potent, selective PPAR-γ agonist that acts downstream of the insulin pathway. Although rosiglitazone treatment promoted adipogenesis to a greater extent than the regular differentiation cocktail (Fig. 1c), it did not alter the phenotypes of cells with increased or decreased levels of Sirt1 (Fig. 1c), suggesting that Sirt1 affects regulators that act downstream of insulin/IGF-1 signalling.
Differentiation of 3T3-L1 fibroblasts increases levels of the transcription factor C/EBP-δ, which stimulates the expression of PPAR-γ and C/EBP-α10. PPAR-γ induces expression of target genes, such as the fatty-acid-binding protein aP2 (also known as adipose tissue-specific FABP)13. Moreover, PPAR-γ can maintain expression of itself, perhaps by binding to PPAR-γ sites in the promoter of the PPAR-γ gene (Pparg)9,14. To gain an insight into the mechanisms by which Sirt1 represses fat accretion, we measured protein and messenger RNA expression of key factors in the transcriptional programme in the different virus-infected 3T3-L1 adipocytes. We observed a reduction in C/EBP-α, C/EBP-δ and aP2 mRNA, but not C/EBP-β, upon Sirt1 overexpression (Fig. 1e). A reduction in PPAR-γ and C/EBP-α was also observed by western blotting (Fig. 1f). In contrast, cells in which Sirt1 had been downregulated showed higher levels of PPAR-γ, C/EBP-δ, C/EBP-α and aP2 (Fig. 1e, f). These findings are consistent with the model that Sirt1 functions as a repressor of genes that drive white adipocyte differentiation and fat storage.
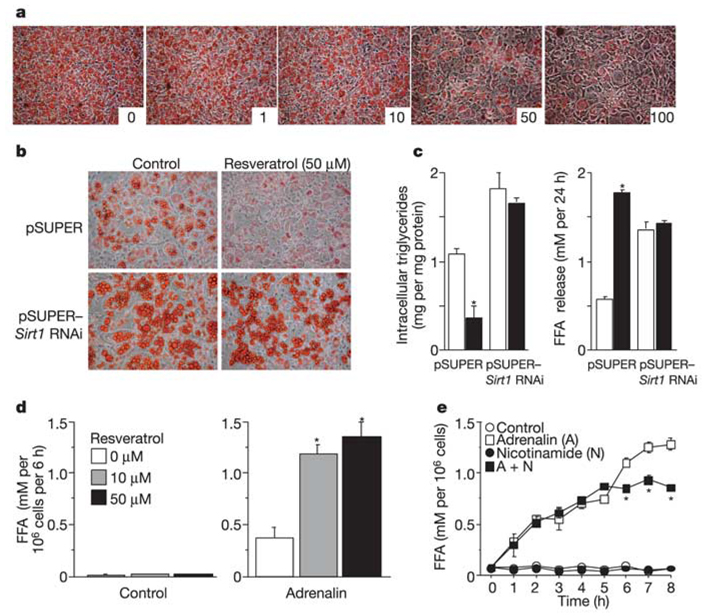
The above experiments do not address whether Sirt1 regulates fat accumulation in fully differentiated adipocytes. Thus, we fully differentiated 3T3-L1 cells and subsequently (12 days after induction) applied the Sirt1 activator resveratrol15 over a wide range of concentrations (Fig. 2a). After staining the cells for fat content, a strong reduction in fat was observed at 50 and 100 µM resveratrol. The loss of fat was due to activation of Sirt1, because there was no drug-mediated fat reduction in cells in which Sirt1 levels were knocked down (Fig. 2b). To validate further these visual results, we measured triglyceride content and free fatty acid (FFA) release in these cells. Triglyceride content was reduced and release of FFA was stimulated by resveratrol in the control cells, but not in the Sirt1 knockdown cells (Fig. 2c). These findings strongly suggest that upregulation of Sirt1 stimulates fat mobilization in fully differentiated 3T3-L1 adipocytes.
Figure 2.
Activation of Sirt1 by resveratrol decreases fat accumulation in differentiated adipocytes. a, Resveratrol addition to 3T3-L1 cells first fully differentiated into adipocytes. At day 12, resveratrol dissolved in DMSO was added in increasing concentrations (1, 10, 50 and 100 µM; as indicated) and Oil red O staining was performed 3 days later and compared with cells incubated with DMSO alone (resveratrol 0 µM). b, c, Effect of resveratrol on 3T3-L1 cells infected with the indicated viruses. Cells were differentiated into adipocytes as in Fig. 1d. At day 12, resveratrol (50 µM) was added and 3 days later, intracellular triglyceride content was evaluated by Oil red O staining (b) and in cell lysates (c). FFA released in the medium during the 3-day incubation period was also measured (c). Open bars, control; filled bars, resveratrol. d, e, Effect of resveratrol on primary adipocytes isolated from rat retroperitoneal WAT. Cells were incubated with or without adrenalin (10−7 and 10−5 M for d and e, respectively) in the presence of Sirt1 activator (resveratrol, d) or inhibitor (nicotinamide, e). FFA levels released in the medium were measured after 4 h (d) or at indicated time points (e). Data shown are means ± s.e.m. and were obtained from two independent experiments done in triplicate. Asterisk indicates a statistical difference from respective control (P < 0.05).
To determine whether Sirt1 stimulates fat mobilization in bona fide adipocytes, we cultured primary rat white adipocytes and activated fat mobilization with the known β-adrenergic inducer adrenalin. Addition of resveratrol greatly stimulated the release of FFA triggered by adrenalin (Fig. 2d), consistent with the findings in the 3T3-L1 cells. In a converse experiment using these rat adipocytes, we addressed whether inhibition of Sirt1 would blunt the mobilization of fat by adrenalin. Addition of the known Sirt1 inhibitor nicotinamide16 indeed reduced the release of FFA triggered by adrenalin (Fig. 2e). The above findings indicate that Sirt1 not only represses the differentiation programme of adipocytes, but also activates the mobilization of fat in fully differentiated cells.
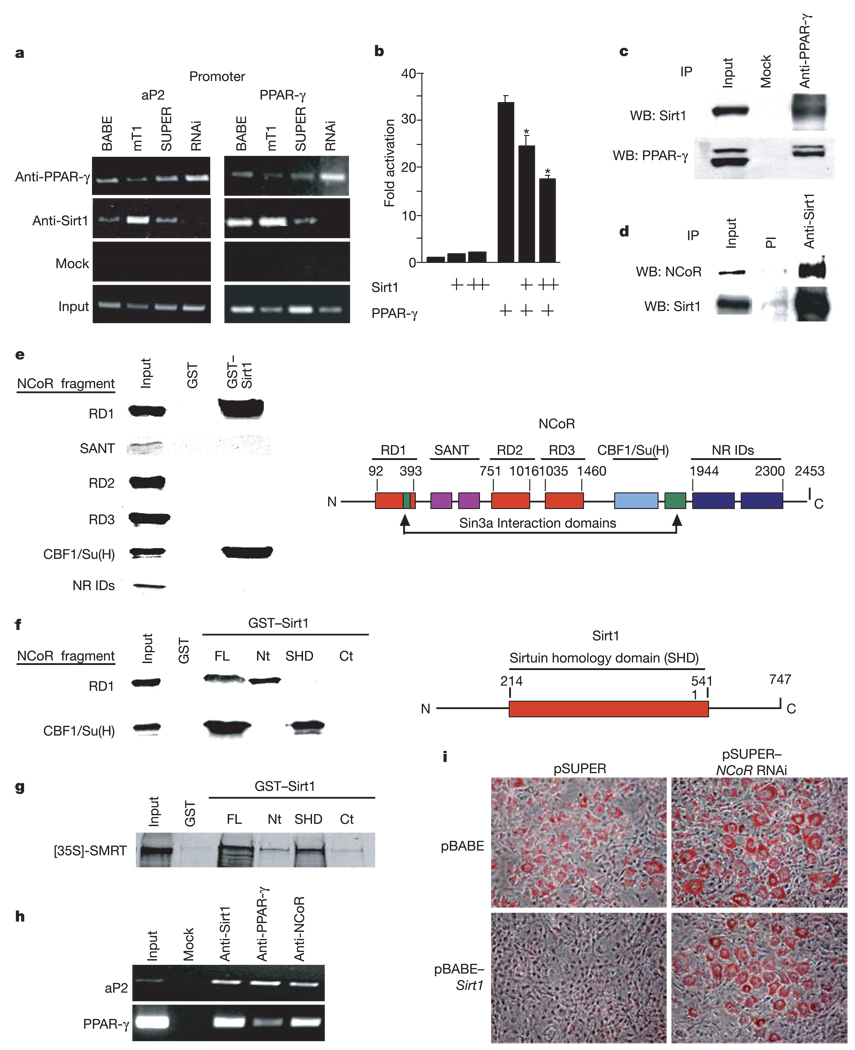
Sirt1 is an NAD-dependent deacetylase that can repress activity of p53 (ref. 1) and forkhead proteins17,18. Repression of adipogenesis and fat retention in 3T3-L1 cells by Sirt1 might be explained by inhibition of another transcription factor, PPAR-γ, as its activity is crucial for differentiation and maintenance of adipocytes9,14,19. Thus, we determined by chromatin immunoprecipitation (ChIP) assays whether Sirt1 binds to PPAR-γ sites in the promoters of the aP2 and Pparg genes. In 3T3-L1 cells, Sirt1 and PPAR-γ were both bound to similar promoter regions of aP2 and Pparg (Fig. 3a). The interaction of Sirt1 with both promoter sequences was stronger in Sirt1-overexpressing cells and was lost in Sirt1 knockdown cells (Fig. 3a). Binding of Sirt1 to a control DNA region 2.5 kilobases (kb) upstream of the PPAR-γ site near Pparg was not observed (data not shown). Furthermore, luciferase reporter assays showed that Sirt1 repressed transactivation by PPAR-γ (Fig. 3b). These findings indicate that Sirt1 and PPAR-γ bind to the same DNA sequences and suggest that Sirt1 is a co-repressor of PPAR-γ.
Figure 3.
Sirt1 reduces fat accumulation by repressing PPAR-γ activity through Sirt1–NCoR interactions. a, ChIP assays on 3T3-L1 cells infected as in Fig. 1c. Cells were differentiated for 7 days and assayed (Methods). b, Luciferase assays of 293T cells transfected with mouse PPAR-γ2 and pGL3–PPRE and with increasing concentrations of mouse Sirt1. Luciferase was measured 24 h later and was corrected for transfection efficiency. Data represent mean + s.e.m. of triplicate experiments carried out in duplicate. Asterisk indicates a statistical difference (P < 0.05). c, Co-immunoprecipitation of endogenous PPAR-γ and Sirt1 proteins in whole-cell extracts prepared from uninfected differentiated 3T3-L1 adipocytes. d, Co-immunoprecipitation of endogenous Sirt1 and NCoR proteins in HEK293 whole-cell extracts using pre-immune (PI) or anti-Sirt1 serum. e, Mapping the Sirt1 interaction interface of NCoR. Constructs encoding the indicated NCoR fragments were 35S-methionine-translated and used for GST pull-down experiments as previously described30. A schematic representation of NCoR functional domains is shown on the right. NR IDs, nuclear receptor interaction domains. SANT; Swi3, Ada2, NCoR, TF11B shared motif. f, Mapping the NCoR interaction interface of Sirt1. GST pull-down experiments were conducted using radiolabelled NCoR RD1 and CBF1/Su(H) interaction domains and the indicated fragments of Sirt1 fused to GST. A schematic representation of full-length Sirt1 is shown on the right. FL, full length; Nt, N-terminal; Ct, C-terminal. g, GST pull-down between the indicated fragments of Sirt1 fused to GST and 35S-methionine-translated SMRT. h, ChIP assay in uninfected 3T3-L1 cells differentiated for 7 days with insulin, IBMX and dexamethasone. i, RNAi of NCoR. 3T3-L1 cells were infected with either pBABE or pBABE–Sirt1 and co-infected with either pSUPER or pSUPER–NCoR RNAi. Puromycin-selected cells were then differentiated with insulin, IBMX and dexamethasone and stained with Oil red O 7 days later.
To gain further evidence that Sirt1 is a PPAR-γ co-repressor, we next determined whether the two proteins interact. Sirt1 was detected in PPAR-γ immunoprecipitates from differentiated 3T3-L1 adipocytes (Fig. 3c). The PPAR-γ cofactor NCoR20 was also co-immunoprecipitated by anti-Sirt1 antiserum but not by pre-immune serum (Fig. 3d). Glutathione S-transferase (GST) pull-down experiments revealed that two NCoR fragments interact with Sirt1: repression domain 1 (RD1) and the CBF/Su(H) interaction domain (Fig. 3e). Reciprocally, NCoR RD1 was found to interact with the amino-terminal region of Sirt1 (amino acids 1–214, GST–Sirt1(Nt); Fig. 3f), and the CBF1/Su(H) interaction domain of NCoR interacted with the homology domain of Sirt1 (amino acids 214–541, GST–Sirt1(SHD); Fig. 3f). In analogous experiments, GST pull-down assays revealed an interaction between the Sirt1 homology domain and SMRT (Fig. 3g).
The interaction between Sirt1 and NcoR (and SMRT) suggests that Sirt1 represses PPAR-γ activity by docking with the cofactors. Consistent with this, ChIP assays revealed that NCoR also binds to known PPAR-γ sites of promoters of adipogenic genes (Fig. 4h). To test the possibility that Sirt1 functionally represses PPAR-γ by means of NCoR, we infected Sirt1-overexpressing 3T3-L1 fibroblasts with an NCoR RNAi virus. Western blots showed that doubly infected cells overexpressed Sirt1 and underexpressed NCoR (not shown). As expected, overexpression of Sirt1 in the absence of the NCoR RNAi virus reduced fat accretion (Fig. 4i). In contrast, this reduction was largely prevented on simultaneous treatment with NCoR RNAi (Fig. 4i), demonstrating that NCoR is required for repression of fat accumulation by Sirt1.
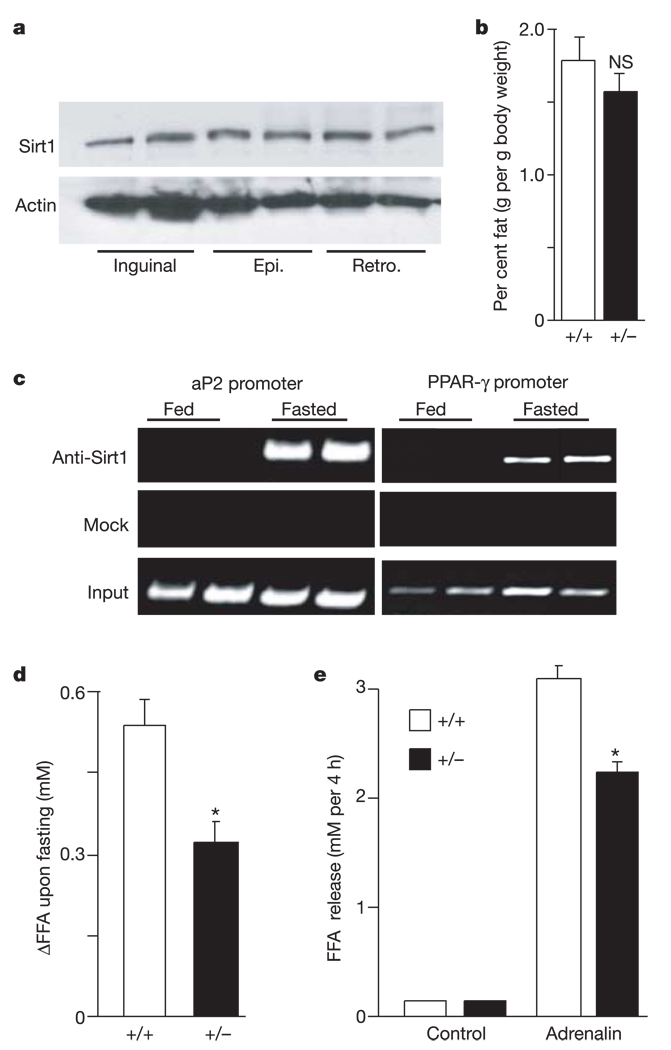
Figure 4.
Sirt1 promotes fat mobilization in vivo. a, Protein levels of Sirt1 in subcutaneous inguinal and visceral epididymal (Epi.) and retroperitoneal (Retro.) WAT depots in wild-type fed mice. Actin levels are shown as loading controls. Each lane represents an individual mouse. b, Weight of epididymal WAT depots in fed Sirt1 +/+ and Sirt1 +/− mice. Bars represent mean + s.e.m. (n = 10–12; NS, not significant). c, In vivo ChiP assay in FVB mice either fed or fasted overnight. Each lane represents an individual mouse. Findings represent five independent experiments done in duplicate. d, Change in FFA levels in blood from age-matched male Sirt1 +/+ and Sirt1 +/− mice between before and after overnight fasting. Bars represent mean + s.e.m. of 27–30 animals. Asterisk indicates a statistical difference from wild-type littermates (P < 0.05). e, Isolated adipocytes from Sirt1 +/+ and Sirt1 +/− mice incubated with or without adrenalin (10−5 M). After 4 h of incubation, FFA released in the medium were measured. Bars represent mean + s.e.m. of triplicate experiments. Asterisk indicates a statistical difference from Sirt1 +/+ adipocytes (P < 0.05).
To test for a role of Sirt1 in fat mobilization in vivo, we first determined whether Sirt1 was expressed in WAT in mice. Sirt1 was found in all white adipose depots examined, with no notable distribution differences (Fig. 4a). Next, to probe for an in vivo function, we used mice with germline mutations in the Sirt1 gene. Unfortunately, the total absence of Sirt1 in mice (Sirt1−/−) results in a high degree of post-natal lethality and other severe phenotypes, precluding their use in this study21. Therefore, we compared wild type to Sirt1+/− mice, which are phenotypically normal. No significant differences in epididymal WAT mass were observed between the wild type and heterozygous cohorts (Fig. 4b). As yeast Sir2 is important during calorie restriction, we assayed by ChIP the recruitment of Sirt1 to PPAR-γ-binding sites in the aP2 and PPAR-γ promoters in WAT of mice that were either fed or fasted. In mice fed ad libitum, Sirt1 was not bound to aP2 or PPAR-γ promoter sequences (Fig. 4c). However, Sirt1 was bound to these sequences after overnight food deprivation, showing that Sirt1 is recruited to PPAR-γ DNA-binding sites upon fasting.
As a test of the effect of Sirt1 on fatty acid mobilization in adipocytes, we next addressed whether fatty acid release from WAT upon fasting was altered in Sirt1+/− mice. Heterozygous gene ablation was associated with a 40–45% lowering in circulating FFA levels in the blood after overnight food deprivation compared with wild type (Fig. 4d, P < 0.05). To verify that these effects of Sirt1 genotype were due to differences in fat release from WAT and not reuptake of FFA from the blood by oxidative tissues, we cultured the same number of white adipocytes from Sirt1+/+ and Sirt1+/− mice, challenged them with adrenalin, and measured the release of FFA. Again, FFA release was reduced in the Sirt1+/− cells compared with the Sirt1+/+ cells (Fig. 4e).
Here we show that the mammalian Sir2 orthologue, Sirt1, is activated by food deprivation to trigger fat mobilization in WAT. The pharmacological activation of Sirt1 also elicits the lipolysis of triglycerides and the release of FFA. Because a reduction in fat storage in WAT is a primary way by which calorie restriction extends lifespan in mammals, our results provide a possible mechanism for understanding the regulation of mammalian lifespan by diet. Sirt1 represses WAT by inhibiting the nuclear receptor PPAR-γ 13,19. Starvation of animals causes Sirt1 to interact with PPAR-γ DNA-binding sites and thereby repress target genes that drive fat storage19. We do not yet know whether Sirt1 is activated upon fasting by a change in the NAD/NADH ratio, as has been proposed in yeast22, or by other metabolic changes. It is also uncertain whether Sirt1 deacetylates PPAR-γ, histones at target genes, or both.
The pathway of regulation described here may impact on age-related diseases. The accumulation of WAT during ageing is associated with several adverse complications, such as insulin resistance, type 2 diabetes and atherosclerosis23. Given the impact of Sirt1 on PPAR-γ activity and because PPAR-γ activity helps determine age-related insulin resistance24, Sirt1 may have an important role in metabolic diseases and link the effects of food consumption to body fat mass and diseases of ageing. It is likely that calorie restriction exerts other effects on mammals to increase longevity, besides reducing WAT, as longevity in mice with reduced fat is not as great as animals on a long-term calorie restriction regimen. Tissues that metabolize fat and carbohydrate may also be important in delivering some of the benefit of calorie restriction, and it will be of interest to determine whether Sirt1 upregulates metabolism upon food reduction to round out an optimal profile for long life.
Methods
Animal experimentation
Wild-type FVB male age-matched (12–16 weeks old) mice were used for the present studies. Sirt1+/+ and Sirt1+/− genotypes have been described previously21. All mice were housed under controlled temperature (25 ± 1 °C (s.d.)) and lighting conditions. Food provided was normal chow. All mice were cared for in accordance with the MIT animal care committee. Blood was collected from the retro-orbital sinus and kept on ice until centrifugation (1,500 g, 15min at 4 °C), and the plasma was stored at −20 °C until analysis. All animals were killed by decapitation. WAT depots were collected, weighed and quickly frozen in liquid nitrogen and stored at −70 °C until further processing by immunoblotting. Non-esterified free fatty acids were determined by enzymatic assays (Wako Pure Chemical Industries).
Cell culture, retroviral infection and transfection
3T3-L1, HEK293, 293T and Phoenix cells (ATCC, Rockville, MD) were cultured in Dulbecco’s modified Eagle’s medium with 10% FCS, and antibiotics. Primary adipocytes from Sprague–Dawley rats were prepared as described previously25. Transfections for luciferase assays were done as described9,13 using pSPORT6– PPAR-γ 2, pGL3–PPRE26 and pcDNA3–Sirt1. Data were corrected for transfection efficiency.
Retroviral infection was performed as described9. Phoenix cells were transfected with either pBABE, pBABE–Sirt1, pSUPERretro (Oligoengine), pSUPERretro–Sirt1 RNAi (5′-GATGAAGTTGACCTCCTCA-3′ ) or pSUPER–NcoR RNAi (5′ - GCTGCATCCAAGGGCCATG-3 ′ ) using Lipofectamine (Invitrogen). After 48 h of transfection, the medium containing retroviruses was collected, filtered, treated by polybrene (1 µg ml−1) and transferred to 3T3-L1 target cells. Infected cells were selected with puromycin (2.5 µg ml−1) for 7 days.
To stimulate adipogenesis and accumulation of lipids in cells, medium was supplemented to confluent cells (day 0) with 2 µM insulin or 10−7M rosiglitazone (Alexis Biochemicals), as stated in the text, 1 µM dexamethasone, and 0.25mM isobutylmethylxanthine (IBMX) for 2 days. The cells were then incubated with insulin or 10−7M rosiglitazone, changing the medium every second day. Fat accumulation was visualized by staining of lipids with Oil red O. Intracellular triglyceride levels were measured in cell lysates by an enzymatic method using a reagent kit from Boehringer Mannheim.
RNA and protein preparation and analysis
Total RNA from cultured cells were extracted (Qiagen) and analysed by semi-quantitative polymerase chain reaction with reverse transcription. GAPDH and 18S RNA levels were determined as a control for loading. Primer sequences have been described elsewhere19,27. Proteins from mouse tissues were extracted in a solution of pH 7.4 containing 20 mM HEPES, 250 mM sucrose, 4 mM EDTA, 1% Triton and protease inhibitor cocktail. 3T3-L1 cells were lysed in NET-N buffer (20 mM Tris-HCl, pH 8 containing 150 mM NaCl, 0.5% NP-40, 10% glycerol, 1 mM EDTA and a protease inhibitor cocktail).
Chromatin immunoprecipitation
For in vitro ChIP, infected 3T3-L1 cells were differentiated as mentioned above. For in vivo ChIP, epididymal WAT was dissected, minced and fixed overnight in PBS containing 1% formaldehyde and protease inhibitor cocktail. Tissues were then rinsed five times in PBS. Further ChIP assays were performed as described previously28, using 1:200 antibody dilutions to immunoprecipitate DNA–protein complexes. DNA was then purified using Qiagen PCR purification kit and PCR reaction was performed using primers for aP2 (5′-AAATTCAGAAGAAAGTAAACACATTATT-3′; 5′ -ATGCCCTGACCATGTGA-3′ ) and PPAR-γ proximal (amplifying a region at 0.3 kb upstream of ATG: 5′ -GAGCAAGGTCTTCATCATTACG-3′; 5′-CCCCTGGAGCTGGAGTTAC-3′ ) and distal (amplifying a region at 2.8 kb upstream of ATG: 5′-CTCTCCCACCCTCGCCATAC-3′ ; 5′ -TTGCCAGAGAAGCCAGTGACA-3 ′ ) promoters.
Pull-down and co-immunoprecipitation assays
Differentiated 3T3-L1 or HEK293 cells were lysed in NET-N buffer by agitation at 4 °C for 30 min. After a brief sonication, lysates were cleared by centrifugation and immunoprecipitated. Antibodies used were anti-Sirt1 (Upstate), anti-NCoR (Upstate), anti- PPAR-γ (SantaCruz), anti-C/EBP-α (SantaCruz) and anti-Sirt1 antiserum29 or preimmune serum. Immunoprecipitates were then analysed by immunoblotting. Equivalent amounts of GST or GST–Sirt1 fusion proteins were bound to glutathione-Sepharose (Pharmacia) and incubated with 35S-methionine-labelled NCoR or SMRT fragments prepared using TNT transcription-translation system (Promega). The reactions were washed five times with binding buffer (10mM Na-HEPES containing 10% glycerol, 1mM EDTA, 1mM DTT, 150mM NaCl and 0.05% NP-40) and bound proteins were eluted and resolved on denaturing SDS–polyacrylamide gel electrophoresis gels for analysis by autoradiography.
Statistical analysis
The main and interactive effects were analysed by analysis of variance (ANOVA) factorial or repeated measures when appropriate. When justified by the ANOVA analysis, differences between individual group means were analysed by Fisher’s PLSD test. Differences were considered statistically significant at P < 0.05.
Acknowledgements
We thank B. Spiegelman for the gift of constructs and for discussion; T. Kouzarides and T.Heinzel for Sirt1 and NCoR constructs, respectively; and R. Frye for the anti-Sirt1 antiserum. The Guarente laboratory was supported by grants from the NIH, the Ellison Medical Foundation, and the Howard and Linda Stern Fund. Studies in the Leid laboratory were supported by grants from the National Institutes of Health and by a NIEHS Center grant to the Oregon State University Environmental Health Sciences Center. T.S. was supported by a graduate fellowship from the Royal Thai Government. R.M.O. is supported by the Portuguese Foundation for Science and Technology. M.K. is a Howard Hughes undergraduate research fellow. F.P. was supported in part by a postdoctoral fellowship from the Canadian Institutes of Health Research.
Footnotes
Competing interests statement The authors declare competing financial interests: details accompany the paper on www.nature.com/nature.
References
- 1.Koubova J, Guarente L. How does calorie restriction work? Genes Dev. 2003;17:313–321. doi: 10.1101/gad.1052903. [DOI] [PubMed] [Google Scholar]
- 2.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 3.Blüher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 4.Weindruch R, Walford RL. The Retardation of Aging and Disease by Dietary Restriction. Springfield, Illinois: C. C. Thomas; 1988. [Google Scholar]
- 5.Lin SJ, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 6.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 8.Green H, Kehinde O. Sublines of mouse 3T3 cells that accumulate lipid. Cell. 1974;1:113–116. [Google Scholar]
- 9.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPARγ2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 10.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu. Rev. Cell Dev. Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 11.Nakae J, et al. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 12.Kenyon C. A conserved regulatory system for aging. Cell. 2001;105:165–168. doi: 10.1016/s0092-8674(01)00306-3. [DOI] [PubMed] [Google Scholar]
- 13.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPARγ2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 14.Ren D, Collingwood TN, Rebar EJ, Wolffe AP, Camp HS. PPARγ knockdown by engineered transcription factors: exogenous PPARγ2 but not PPARγ1 reactivates adipogenesis. Genes Dev. 2002;16:27–32. doi: 10.1101/gad.953802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howitz KT, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 16.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell. 2003;12:51–62. doi: 10.1016/s1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 17.Motta MC, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 18.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 19.Tamori Y, Masugi J, Nishino N, Kasuga M. Role of peroxysome proliferator-activated receptor-γ in maintenance of the characteristics of mature 3T3–L1 adipocytes. Diabetes. 2002;51:2045–2055. doi: 10.2337/diabetes.51.7.2045. [DOI] [PubMed] [Google Scholar]
- 20.Dowell P, et al. Identification of nuclear receptor corepressor as a peroxisome proliferator-activated receptor alpha interacting protein. J. Biol. Chem. 1999;274:15901–15907. doi: 10.1074/jbc.274.22.15901. [DOI] [PubMed] [Google Scholar]
- 21.McBurney MW, et al. The mammalian SIR2α protein has a role in embryogenesis and gametogenesis. Mol. Cell. Biol. 2003;23:38–54. doi: 10.1128/MCB.23.1.38-54.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S-J, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriely I, Barzilai N. The role of fat cell derived peptides in age-related metabolic alterations. Mech. Ageing Dev. 2001;122:1565–1576. doi: 10.1016/s0047-6374(01)00287-1. [DOI] [PubMed] [Google Scholar]
- 24.Miles PD, Barak Y, Evans RM, Olefsky JM. Effect of heterozygous PPARγ deficiency and TZD treatment on insulin resistance associated with age and high-fat feeding. Am. J. Physiol. 2003;284:E618–E626. doi: 10.1152/ajpendo.00312.2002. [DOI] [PubMed] [Google Scholar]
- 25.Marette A, Deshaies Y, Collet AJ, Tulp O, Bukowiecki LJ. Major thermogenic defect associated with insulin resistance in brown adipose tissue of obese diabetic SHR/N-cp rats. Am. J. Physiol. 1991;261:E204–E213. doi: 10.1152/ajpendo.1991.261.2.E204. [DOI] [PubMed] [Google Scholar]
- 26.Forman BM, et al. 15-Deoxy-Δ12,14 prostaglandin J2 is a ligand for the adipocyte determination factor PPARγ. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 27.Le Lay S, Lefrère I, Trautwein C, Dugail I, Krief S. Insulin and sterol-regulatory element-binding protein-1c (SREBP-1c) regulation of gene expression in 3T3-L1 adipocytes. J. Biol. Chem. 2002;277:35625–35634. doi: 10.1074/jbc.M203913200. [DOI] [PubMed] [Google Scholar]
- 28.Fajas L, et al. E2Fs regulate adipocyte differentiation. Dev. Cell. 2002;3:39–49. doi: 10.1016/s1534-5807(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 29.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dowell P, Peterson VJ, Zabriskie M, Leid M. Ligand-induced peroxisome proliferator-activated receptor alpha conformational change. J. Biol. Chem. 1997;272:2013–2020. doi: 10.1074/jbc.272.3.2013. [DOI] [PubMed] [Google Scholar]