Abstract
The fission yeast Schizosaccharomyces pombe is especially well-suited for both genetic and biochemical analysis of meiotic recombination. Recent studies have revealed ~50 gene products and two DNA intermediates central to recombination, which we place into a pathway from parental to recombinant DNA. We divide recombination into three stages – chromosome alignment accompanying nuclear “horsetail” movement, formation of DNA breaks, and repair of those breaks – and we discuss the roles of the identified gene products and DNA intermediates in these stages. Although some aspects of recombination are similar to those in the distantly related budding yeast Saccharomyces cerevisiae, other aspects are distinctly different. In particular, many proteins required for recombination in one species have no clear ortholog in the other, and the roles of identified orthologs in regulating recombination often differ. Furthermore, in S. pombe the dominant joint DNA molecule intermediates contain single Holliday junctions, and intersister joint molecules are more frequent than interhomolog types, whereas in S. cerevisiae interhomolog double Holliday junctions predominate. We speculate that meiotic recombination in other organisms shares features of each of these yeasts.
1 S. pombe: An Excellent Model Organism for Studying Meiotic Recombination
Homologous genetic recombination plays two important roles during meiosis, the special nuclear divisions during which chromosome number is reduced from two (diploid) to one (haploid). First, recombination provides the physical connection between homologs that aids their pairing and proper segregation at the first meiotic division (MI), and second, it increases genetic diversity that aids evolution (see chapter by Egel). Elucidating the molecular mechanism of meiotic recombination requires a combination of genetic and biochemical analysis. Fungi, such as yeasts, have been particularly useful in this regard, for they have the essential features of meiosis found in complex organisms yet are more tractable for genetics and biochemistry. Notably, in many fungi the haploid products (spores) from each meiosis are enclosed in an ascus. Analysis of the haploid progeny from one ascus reveals all of the products of a single meiotic recombination event at each locus analyzed.
Meiosis has been especially well-studied in the budding yeast Saccharomyces cerevisiae (see chapters by Keeney, Heyer, Lichten, and Hunter) and the distantly related fission yeast Schizosaccharomyces pombe discussed here (see also chapters by Hiraoka, Moreno and Martin-Castellanos, and Watanabe). Both haploid and diploid cells of these yeasts can be grown indefinitely by mitotic division; genetic analysis that uses recessive markers is simpler in haploids. Large cultures of cells can be synchronously induced for meiosis, facilitating biochemical analysis. The nucleotide sequences of their relatively small genomes, ~14 Mb, are essentially complete, permitting comprehensive genomic studies. In addition, S. pombe offers special advantages. The strongest meiotic recombination-deficient (Rec−) mutants of S. pombe produce many viable spores in part because this species has only three chromosomes, which, in the absence of recombination, would be still expected to segregate correctly and produce viable spores 12.5% of the time (2−3). S. pombe also has a mechanism for actively segregating non-recombinant (achiasmate) chromosomes at MI (Molnar et al. 2001; Davis and Smith 2005). Consequently, strong Rec− mutants that cannot initiate recombination are nevertheless able to produce ~10 – 25% as many viable spores as the wild type, an outcome that greatly aids analysis of such mutants (Ponticelli and Smith 1989; Young et al. 2004). All commonly used strains are derived from a single culture (Munz et al. 1989): their near isogenicity simplifies the use of strains and comparisons of results between labs. The M26 and closely related hotspots of recombination are exceptionally strong and are the best-defined meiotic hotspots in terms of nucleotide sequence (see section 6.1).
Some aspects of the molecular biology of S. pombe are more similar to those of humans than are those of S. cerevisiae. These aspects of S. pombe include more complex centromeres and origins of replication, the presence of RNAi and certain histone modifications, and the specifics of cell-cycle control. In contrast, S. pombe is unusual in not having a fully developed synaptonemal complex (SC; Olson et al. 1978; Bähler et al. 1993), a large meiosis-specific structure joining paired homologs (see chapters by Suja and McKim). The role of the SC is not clear, but its absence from S. pombe indicates that it is not essential for meiosis or recombination. In addition, S. pombe does not have crossover interference, the regulation of the number of crossovers and their distribution along chromosomes (Munz 1994). These features allow in S. pombe a study of the essential features of recombination without the complexities of the SC or interference. Comparison of results between different species, such as S. pombe and S. cerevisiae, has revealed both conserved and diverged aspects of meiosis. In this regard, comparison of S. pombe and S. cerevisiae may help deduce the evolution of meiosis.
2 Overview: A Pathway for S. pombe Meiotic Recombination
In our current understanding of S. pombe meiotic recombination there are three stages, the first of which is concurrent with the other two: 1) the overall alignment and then intimate pairing of homologs, 2) the programmed formation of DNA double-strand breaks (DSBs), and 3) the repair of DSBs (Figure 1). Stages 1 and 2 are meiosis-specific, whereas stage 3 shares many functions with mitotic DNA repair. Stage 1, homolog alignment, involves the clustering of telomeres (“bouquet” formation) and the movement of the nucleus back and forth in the cell (“horsetail” formation) (see chapter by Hiraoka). These features are found in most organisms but are exaggerated in S. pombe. Homolog alignment reduces recombination between non-allelic loci with similar sequences (Niwa et al. 2000; Davis and Smith 2006); ectopic recombination between such loci could generate deleterious translocations.
Figure 1.
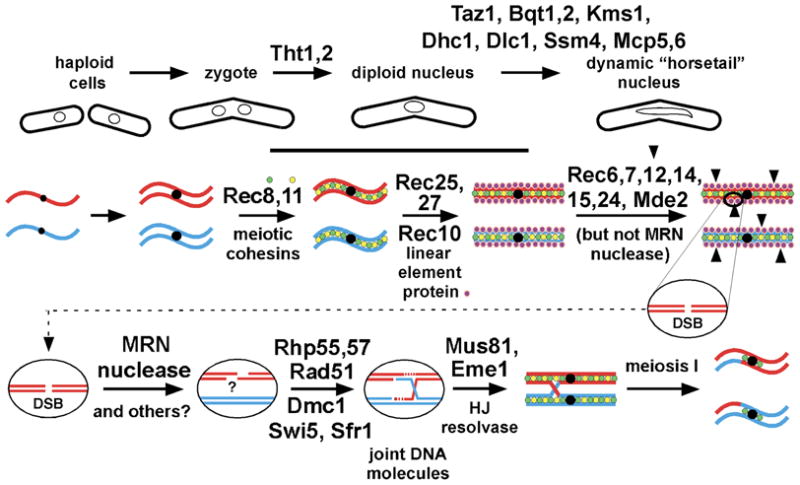
A pathway for meiotic recombination in S. pombe. The upper panels portray the fusion of cells and nuclei and the formation of the “horsetail” nucleus. The middle and lower panels portray the chromosome and DNA events that occur during the horsetail stage, which ceases shortly before meiosis I. Identified gene products required at each stage are indicated above the arrow leading to that stage. Additional proteins required for meiotic recombination, but whose points of action are not clear, include rec13, rec17 – 21, mug1, mug5, pds5, rqh1, mcp7, and meu13 (see sections 2, 3, and 8). MRN, Rad32 (Mre11)-Rad50-Nbs1 complex. Modified from Ellermeier and Smith (2005).
During the horsetail stage chromosomes are replicated and sister chromatid cohesion is modified to allow the unique segregation of homologs at MI (see chapter by Watanabe). The meiosis-specific cohesin subunits Rec8 and Rec11 are important for the formation of linear elements, which are reminiscent of the axial elements of the SC (Lorenz et al. 2004). Linear elements, in turn, appear to be important for the assembly onto the chromosomes (or activation) of the proteins that make DSBs, including the active-site protein Rec12 (Spo11 homolog).
DSBs are made by Rec12 in concert with other proteins (stage 2) and are repaired by interaction with homologous DNA of either the sister chromatid or the homolog (stage 3). Only interhomolog interaction gives rise to the physical connections (chiasmata) that aid homolog segregation at MI, but sister chromatid exchange does occur. The regulation of these two types of repair is an intriguing problem not yet solved. DSB repair occurs in steps. First is the formation of hybrid DNA, which has one strand from each parental DNA, and one or two Holliday junctions, an intermediate with two crossed, single strands connecting the parental duplexes. Second is the resolution of the Holliday junction(s) into linear duplexes, which may occur in either the crossover or non-crossover configuration. Regulation of this outcome is also an intriguing, largely unsolved problem.
In the following sections we discuss each of these stages of meiotic recombination. Tables 1 – 3 list the known S. pombe gene products required for wild-type levels of meiotic recombination. (Most of the primary references are given in these tables rather than in the text.) These gene products are listed according to the stage at which they play the most prominent role; however, some may act at more than one stage. Mutations in additional genes (rec13 and rec17 – rec21) reduce meiotic recombination frequencies (DeVeaux et al. 1992), but these mutations have not been placed in genes identified otherwise.
Table 1.
Genes required for recombination and nuclear fusion, nuclear movement, bouquet formation, or chromosome alignment
| Gene | Protein size (kDa) | Approx. extent of reduction by mutation | Putative S. cerevisiae ortholog (~% identity) | Inferred primary activity | References |
|---|---|---|---|---|---|
| tht1 | 63 | —a | Kar5 (19) | Nuclear fusion | Tange et al. 1998 |
| tht2 | 23 | 1000a | —b | Nuclear fusion | Martin-Castellanos et al. 2005 |
| taz1 | 75 | 5 | Tbf1 (30) | Binds telomere repeats | Cooper et al. 1998; Niwa et al. 2000 |
| rap1 | 80 | —c | Rap1 (22) | Binds Taz1 | Chikashige and Hiraoka 2001; Kanoh and Ishikawa 2001 |
| bqt1 | 12 | 5 | — | Connects telomeres and SPB | Martin-Castellanos et al. 2005; Chikashige et al. 2006 |
| bqt2 | 14 | 5 | — | Connects telomeres and SPB | Martin-Castellanos et al. 2005; Chikashige et al. 2006; Davis and Smith 2006 |
| sad1 | 58 | —c | Mps3 (17) | SPB component | Hagan and Yanagida 1995 |
| kms1 | 69 | 5 | — | SPB component | Shimanuki et al. 1997; Miki et al. 2002 |
| mcp6 | 38 | 5 | — | SPB component | Saito et al. 2005 |
| dhc1 | 484 | 5 | Dyn1 (25) | Dynein motor protein | Yamamoto et al. 1999 |
| dlc1 | 12 | 10 | YER071C (21) | Dynein accessory factor | Miki et al. 2002 |
| ssm4 | 77 | 10 | Nip100 (24) | Binds Dhc1 | Niccoli et al. 2004 |
| mcp5 (num1) | 111 | 5 | Num1 (24) | Binds dynein to microtubules | Saito et al. 2006; Yamashita and Yamamoto 2006; C. Ellermeier, pers. comm. |
| pds5d | 139 | 5 | Pds5 (24) | Sister chromatid cohesin partner | Tanaka et al. 2001; Wang et al. 2002; Ding et al. 2006; unpublished data |
| rqh1 (rec9)d | 150 | 5 | Sgs1 (37) | RecQ family helicase | Stewart et al. 1997; J. Young, pers. comm. |
| mug1d | 41 | 5 | Uso1 (23) Sla2 (23) | Martin-Castellanos et al. 2005; C. Ellermeier, pers. comm. | |
| mug5d | 21 | 5 | — | Martin-Castellanos et al. 2005; C. Ellermeier, pers. comm. |
tht1 is recombination-proficient in azygotic meiosis (that of an established diploid); tht2 was not tested. tht1 is assumed to be as deficient in zygotic meiosis (that immediately following mating) as tht2.
No ortholog is obvious.
Not determined. Requirement for meiotic recombination is assumed, based on the protein being required for telomere clustering during meiosis.
Role in recombination is uncertain. See section 3.
Table 3.
Genes required for recombination and DSB or mismatch repair
| Gene | Protein size (kDa) | Approx. extent of reduction by mutation | Putative S. cerevisiae ortholog (~% identity) | Inferred primary activity | References |
|---|---|---|---|---|---|
| rad32 | 74 | 15 | Mre11 (42) | MRN component; nuclease | Tavassoli et al. 1995 |
| rad50 | 150 | —a | Rad50 (34) | MRN component; ATPase | — |
| nbs1 | 69 | — | Xrs2 (limited)b | MRN component | — |
| rad51c | 40 | 15 | Rad51 (71) | Strand exchange protein | Grishchuk and Kohli 2003 |
| dmc1 | 36 | 3 | Dmc1 (63) | Strand exchange protein | Grishchuk and Kohli 2003 |
| rad55 | 39 | 5 | Rad55 (32) | Rad51 accessory protein | Grishchuk and Kohli 2003 |
| rad57 | 40 | 3 | Rad57 (37) | Rad51 accessory protein | Grishchuk and Kohli 2003 |
| swi5 | 10 | 5 | Sae3 (24) | Rad51 and Dmc1 accessory protein | Ellermeier et al. 2004 |
| sfr1 | 34 | 10 | Mei5 (limited)d | Rad51 and Dmc1 accessory protein | Unpublished data |
| rlp1 | 42 | 2 (xo only)e | hXRCC2 (31) | Rad51 accessory protein | Grishchuk and Kohli 2003 |
| mcp7 | 24 | 10 | Mnd1 (26) | Acts with Dmc1 | Saito et al. 2004 |
| meu13 | 25 | 10 | Hop2 (27) | Acts with Dmc1 | Nabeshima et al. 2001; Saito et al. 2004 |
| rad54 | 97 | 1 f | Rad54 (54) | Chromatin remodeling | Catlett and Forsburg 2003 |
| rdh54 | 93 | 3 f | Rdh54 (33) | Chromatin remodeling | Catlett and Forsburg 2003 |
| mus81 | 69 | 50 (xo only)g | Mus81 (28) | Holliday junction resolvase | Osman et al. 2003; Smith et al. 2003 |
| eme1 | 83 | — | Mms4 (10) | Holliday junction resolvase | Boddy et al. 2001 |
| rad22 | 52 | 3 h | Rad52 (32) | Rad51 accessory protein? | van den Bosch et al. 2002 |
| rti1 | 42 | 0.8 h | Rad52 (32) | Rad51 accessory protein? | van den Bosch et al. 2002 |
| swi4 | 115 | 2 – 3 | Msh3 (33) | Tornier et al. 2001 | |
| pms1 | 88 | gc effectsi | Pms1 (33) | Mismatch repair (MMR)j | Fleck et al. 1999 |
| msh2 | 110 | gc effectsi | Msh2 (42) | Mismatch repair (MMR) | Fleck et al. 1999 |
| msh6 | 141 | gc effectsi | Msh6 (39) | Mismatch repair (MMR) | Tornier et al. 2001 |
| swi10 | 29 | gc effectsi | Rad10 (37) | Mismatch repair (NER)k | Fleck et al. 1999 |
| rad13 | 126 | gc effectsi | Rad2 (35) | Mismatch repair (NER) | Kunz and Fleck 2001 |
| exo1 | 64 | gc effectsi | Exo1 (29) | 5′→ 3′ ds DNA exonuclease | Szankasi and Smith 1995 |
Not determined. Requirement for meiotic recombination is assumed, based on other phenotypes similar in rad32 mutants (for rad50 and nbs1) or in mus81 mutants (for eme1).
Regions of 79 and 27 amino acids show ~22% and 37% identity, respectively.
Also called rhp51.
A region of 160 amino acids shows ~21% identity.
Reduction affects crossovers (xo) only; gene conversions are modestly increased in frequency.
The rad54 rdh54 double mutant has greatly reduced spore viability and is severely defective in DSB repair.
Crossovers are reduced by factors of 10 – 90; gene conversions are reduced by a factor of <2.
The rad22 rti1 double mutant has greatly reduced spore viability (van den Bosch et al. 2002) and is defective in recombinational repair of meiotic DSBs (Young et al. 2004)
Frequency of intragenic recombinants (gene convertants) is decreased or increased by factors of up to 35, but generally less, depending on the markers tested for recombination.
Mismatch repair.
Nucleotide excision repair.
S. pombe typically grows as haploid cells. When cells of opposite mating type meet under starvation conditions, the cells and then their nuclei fuse to form a diploid. Unless nutrients are supplied, the diploid immediately undergoes meiosis (see chapter by Moreno and Martin-Castellanos). Two mutants, tht1 and tht2, are deficient in nuclear fusion and produce essentially no interhomolog recombinants among spores. Although not reported, intersister recombination in these mutants is expected to be high, since DSBs are formed and repaired as in wild-type. Each nucleus undergoes an aberrant meiosis, sometimes with only one nuclear division.
3 Nuclear Movement Promotes Chromosome Alignment: “Bouquet” and “Horsetail” Formation
Before the outset of meiosis the centromeres are clustered at the spindle-pole body (SPB), the fungal equivalent of the centrosome. As meiosis proceeds, the telomeres cluster and replace the centromeres at the SPB, to form the meiotic “bouquet” arrangement of chromosomes (Chikashige et al. 1994). The SPB leads the nucleus back and forth in movement across the cell, and the nucleus becomes elongated and curved, like a horse’s tail. Meiotic recombination is reduced by a factor of ~5 in all of the tested mutants deficient in bouquet or horsetail formation (Table 1). In these mutants DSBs are formed and, where tested, repaired with nearly wild-type frequency and kinetics. Presumably, repair occurs most frequently by interaction with the sister chromatid, with some residual interhomolog interaction accounting for the observed recombinants.
Bouquet formation requires two meiosis-specific gene products, Bqt1 and Bqt2. These small proteins appear to act as a complex to glue the telomeres to the SPB. Throughout the life cycle, Taz1 binds to telomeres and to Rap1. The Bqt1-Bqt2 complex forms a meiosis-specific bridge between Rap1 and Sad1, a component of the SPB, thereby joining the telomeres to the SPB. In the absence of Taz1, Rap1, Bqt1, or Bqt2 the nucleus moves but, since the telomeres are not attached to the SPB, the nucleus does not assume the characteristic horsetail shape, and chromosomes are not properly aligned.
The bouquet restricts ectopic recombination, which can cause deleterious genome rearrangements. In S. pombe, ectopic recombination occurs 10 – 1000 times less frequently than allelic recombination (Virgin and Bailey 1998). Mutations in kms1 and bqt2 affect bouquet formation and increase up to 20-fold the frequency of meiotic ectopic recombination. Attachment of telomeres to the SPB during bouquet formation may restrict recombination to sequences equivalent distances from the anchored telomeres. This spatial constraint would favor allelic over ectopic recombination.
Horsetail movement requires the dynein components Dhc1 (heavy chain) and Dlc1 (light chain), the dynactin component Ssm4, and the SPB components Mcp6 (meiotic coiled-coil protein) and Kms1. During meiosis the SPB is linked to the dynein motor complex via Kms1 and perhaps the dynactin complex. Dynein is the motor that moves the nucleus, led by the SPB, along the microtubule arrays in the cell. In dhc1, dlc1, and ssm4 mutants, the nucleus does not move and homologs do not align, although the telomeres become attached to the SPB. Attachment of dynein to microtubules at the cell cortex, which generates the force for horsetail movement, requires Mcp5 (Num1).
Additional genes are placed at the bottom of Table 1 because the corresponding mutants produce recombinant frequencies ~5 times lower than that of wild type and, where tested, make and repair DSBs with nearly wild-type kinetics and frequencies, as is the case for the bouquet- and horsetail-defective mutants discussed above. Pds5 aids loading of the Rec8 cohesin subunit (see sections 4 and 5.2); in pds5 mutants the chromosomes are hypercompacted, and horsetail shape is aberrant. Rqh1 is a homolog of the E. coli RecQ and S. cerevisiae Sgs1 helicases; rqh1 was identified (as rec9) in the initial screen for S. pombe meiotic Rec− mutants (Ponticelli and Smith 1989). mug1 and mug5 are meiotic up-regulated genes; the mutants make aberrant asci indicative of chromosome missegregation. Further analyses are required to determine the stage at which these proteins promote recombination.
4 Meiosis-specific Sister Chromatid Cohesins: Behavior Change
During or shortly after meiotic replication, the meiosis-specific cohesin subunits Rec8 and Rec11 are recruited to the chromosomes, where they largely replace the mitotic cohesin subunits Rad21 and Psc3 (see chapter by Watanabe). During mitotic division Rad21 is cleaved by separase (Cut1) to allow sister chromatid segregation. During the first meiotic division, Rec8 located in the chromosomes arms is, like Rad21, cleaved by separase. However, unlike Rad21, Rec8 at the centromeres is protected from separase by Sgo1 (Kitajima et al. 2004). This differential cleavage allows sisters to separate distal to the crossovers that hold homologs together but maintains cohesion between sisters at the centromeres (Figure 1). Thus, the change in cohesins permits segregation of homologs, rather than sisters, at MI. The role of Rec11 is less clear; its location primarily in the arms suggests involvement in arm cohesion, but as noted in section 5.3 Rec11 and Rec8 are also required for recombination.
5 DSB Formation by Rec12: Preparation and Partnership
5.1 S. pombe: A Second Eukaryote with Directly Observed Meiotic DSBs
DSBs were postulated to initiate meiotic recombination (Resnick 1976; Szostak et al. 1983) many years before their demonstration in S. cerevisiae at hotspots of recombination (Sun et al. 1989; Cao et al. 1990). Searches for DSBs in S. pombe were first successful when whole chromosomes and large restriction fragments were examined (Cervantes et al. 2000). Aided by a mutant, rad50S (see section 7.1), DSBs were later found at the genetically well-characterized hotspot M26 (Steiner et al. 2002; see section 6.1). Meiotic DSBs have not, to our knowledge, been directly observed in other organisms. They have been inferred, however, from the requirement for Spo11 homologs for successful meiosis or recombination, from the Spo11-dependent fragmentation of chromosomes in DSB repair-deficient mutants (Pasierbek et al. 2001; Puizina et al. 2004), or from the appearance on meiotic chromosomes of foci of a particular form of histone H2 that is thought to be a signal of DSBs (see chapter by Lichten). The direct detection of DSBs in S. pombe opened the way for the discovery of natural S. pombe hotspots, discussed below, and the study of other intermediates of recombination.
5.2 Modification of Sister Chromatid Cohesion: A Foundation for Meiosis-specific DSB Formation
As noted above, the substitution of the Rec8 and Rec11 cohesin subunits for their Rad21 and Psc3 mitotic counterparts dramatically modifies the segregation behavior of chromosomes during meiosis. Rec8 and Rec11 also initiate a series of events that lead to meiotic DSBs. Current evidence indicates that, after Rec8 and Rec11 are placed on chromosomes, the Rec25 and Rec27 proteins, perhaps as a complex, form foci on the chromosomes. These proteins in turn allow the loading of Rec10, a major component of linear elements (Lorenz et al. 2004; see section 5.3). Finally, Rec7 and presumably the other proteins required for DSB formation, including Rec12, are recruited to the chromosomes (Lorenz et al. 2006). Thus, the modification of chromosomes both for their unique segregation and for high-level recombination appears to be initiated at the time of meiotic replication (see section 4).
5.3 Formation of Linear Elements: Structures Reminiscent of the Synaptonemal Complex
Electron microscopy of thin sections of S. pombe meiotic cells or of spreads of their nuclear contents fail to reveal the synaptonemal complexes (SC) common to most organisms (Olson et al. 1978; Bähler et al. 1993; Loidl 2006). Structures similar to one part of the SC, however, are observed and are designated linear elements (LinEs; Bähler et al. 1993; Lorenz et al. 2004). The classical SC is composed of a central element between two parallel lateral elements connected by transverse filaments when homologs are fully aligned and intimately paired. Before this pairing, the lateral elements are called chromatid cores or axial elements; they encase the bases of the chromatin loops of each sister chromatid pair (see chapters by Suja and by McKim). The LinEs of S. pombe appear similar to axial elements, but LinEs do not show the parallel alignment of lateral elements in paired chromosomes and do not extend the full length of the chromosomes as do the axial elements of the SC (Bähler et al. 1993). The role of LinEs, like that of the SC, is not clear, but rec8 and rec10 mutants have aberrant or no LinEs, respectively, and are recombination-deficient (Molnar et al. 1995, 2003; Lorenz et al. 2004; Loidl 2006).
Fluorescence microscopy of meiotic cells or nuclear spreads reveals structures likely identical to the LinEs, and this analysis confirms the close connection between LinEs and recombination. During meiosis the LinE component Rec10 first forms discrete nuclear foci and then filaments (Figure 2), whose numbers and morphological classes approximate those of the LinEs seen by electron microscopy. Formation of Rec10 filaments requires Rec8, Rec11, Rec25, and Rec27 (Lorenz et al. 2004; C. Martin-Castellanos, pers. comm.). All of these proteins are required for full levels of recombination, presumably by allowing the loading or activity of the DSB-forming complex.
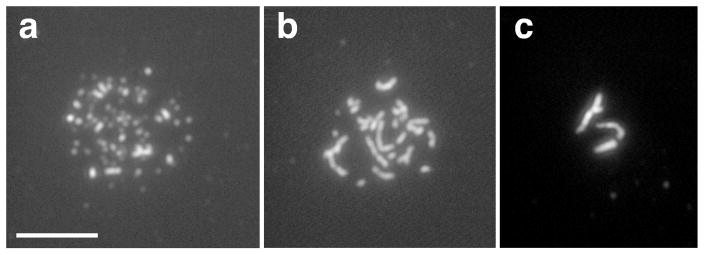
Figure 2.
Linear elements containing Rec10 protein. Spreads of meiotic nuclei were stained using an antibody to Rec10 protein. Dots (a), filaments (b), and bundles (c) appear to occur in that order during meiosis. The filaments and bundles appear to reflect the linear elements seen by electron microscopy (see text). Bar indicates 5 μm. Figure supplied by J. Loidl.
5.4 Rec12: The Active Site Protein for DSB Formation
Meiotic DSBs are formed by Rec12, the S. pombe homolog of Spo11, in conjunction with other proteins. S. cerevisiae Spo11 becomes covalently linked to the 5′ ends of the DNA at a DSB, presumably via a tyrosine residue that is essential for DSB formation and recombination (Keeney 2001). The corresponding tyrosine in Rec12 is also required for DSB formation and recombination (Cervantes et al. 2000), and covalent linkage of Rec12 to DNA has been inferred from chromatin-immunoprecipitation studies using an epitope-tagged version of Rec12 (R. Hyppa, pers. comm.). Spo11 homologs from a wide variety of organisms, including S. pombe Rec12, have amino acid sequence similarity to that of an archeal DNA topoisomerase, whose crystal structure reveals a dimer with the two active-site tyrosine residues pointed into a cleft plausibly holding DNA during catalysis of DSB formation (Nichols et al. 1999). Thus, the mechanism of meiotic DSB formation appears to be highly conserved and closely related to that of type II topoisomerases (see chapter by Keeney).
5.5 Other Proteins Essential for DSB Formation: Potential Rec12 Partners and Regulators
Rec12 does not make DSBs on its own but requires numerous other proteins. The cascade of proteins noted in sections 5.2 and 5.3 appear to be needed for the proper localization of Rec12, and other proteins are needed for Rec12 activity. rec12 mutants have no detectable meiotic recombination above the level in mitotic cells and no detectable DSBs (Young et al. 2002). This is also true for rec6, rec7, rec10, rec14, rec15, rec24, and mde2 mutants (Table 2), indicating that their gene products are essential for Rec12 action. The corresponding proteins may be partners for Rec12, perhaps in a complex with it, much as several proteins activate S. cerevisiae Spo11 by forming a complex with it (see chapter by Keeney). Loading of Rec7 requires Rec10 (Lorenz et al. 2006) and presumably also the proteins needed for Rec10 loading (Rec8, Rec11, Rec25, and Rec27; see above). It is noteworthy that the MRN complex (see section 7.1) is not required for DSB formation in S. pombe, although its homolog MRX in S. cerevisiae is required (Cao et al. 1990; Young et al. 2004); in both organisms the complex is required for DSB repair. Other differences in the control of DSB formation and repair are discussed in section 13.
Table 2.
Genes required for recombination and meiosis-specific sister chromatid cohesion, linear element formation, or DSB formation
| Gene | Protein size (kDa) | Approx. extent of reduction by mutation | Putative S. cerevisiae ortholog (~% identity) | Inferred primary activity | References |
|---|---|---|---|---|---|
| rec8 | 64 | 5 – 500a | Rec8 (20) | Sister chromatid cohesion | Ponticelli and Smith 1989; Parisi et al. 1999; Ellermeier and Smith 2005 |
| rec11 | 107 | 5 – 500 | Irr1(Scc3) (20) | Sister chromatid cohesion | Ponticelli and Smith 1989; Ellermeier and Smith 2005 |
| rec25 | 17 | 15 | —b | Aids linear element formation | Martin-Castellanos et al. 2005 |
| rec27 | 16 | 15 | — | Aids linear element formation | Martin-Castellanos et al. 2005 |
| rec10 | 90 | 1000 | Red1 (v. limited)c | Linear element component | Ponticelli and Smith 1989; Lorenz et al. 2004; Ellermeier and Smith 2005 |
| rec12 | 39 | 1000 | Spo11 (30) | Makes DSBs | DeVeaux et al. 1992; Cervantes et al. 2000 |
| rec6 | 21 | 1000 | — | Putative Rec12 partner | Ponticelli and Smith 1989; Cervantes et al. 2000 |
| rec7 | 38 | 1000 | Rec114 (9) | Putative Rec12 partner | Ponticelli and Smith 1989; Cervantes et al. 2000; Molnar et al. 2001 |
| rec14 | 33 | 1000 | Ski8 (25) | Putative Rec12 partner | DeVeaux et al. 1992; Cervantes et al. 2000 |
| rec15 | 21 | 1000 | — | Putative Rec12 partner | DeVeaux et al. 1992; Cervantes et al. 2000 |
| rec24 | 40 | 1000 | — | Putative Rec12 partner | Martin-Castellanos et al. 2005 |
| mde2 | 23 | 300 | — | Putative Rec12 partner | Gregan et al. 2005 |
| hsk1 | 58 | 10 | Cdc7 (37) | Protein kinase; substrate unknown | Ogino et al. 2006 |
| atf1 | 60 | 15d | Sko1 (25) | Stress response transcription factor | Kon et al. 1997; Fox et al. 2000 |
| pcr1 | 19 | 15d | — | Stress response transcription factor | Kon et al. 1997; Fox et al. 2000 |
The extent of reduction depends on the interval measured. See section 6.3.
No ortholog is obvious.
A region of 64 amino acids has ~27 % identity.
Reduction only at M26 and related hotspots.
In S. cerevisiae meiotic replication is essential for DSB formation (Borde et al. 2000; Smith et al. 2001), but in S. pombe the situation is less clear. As in S. cerevisiae, DSBs appear after replication (Cervantes et al. 2000), but replication can be severely inhibited by mutations or by hydroxyurea with only slight diminution of DSB formation, provided the replication checkpoint is inactivated (Tonami et al. 2005). Hydroxyurea blocks transcription of several meiotic genes required for DSB formation, including mde2, and replication checkpoint mutations relieve this block (Ogino and Masai 2006). Conversely, a particular hsk1 mutant does not form detectable DSBs under conditions in which meiotic replication appears normal (Ogino et al. 2006). Hsk1 protein kinase is required for mitotic replication, via phosphorylation of the MCM (mini-chromosome maintenance) complex; the hsk1 mutant tested may lack a second function needed for DSB formation, such as phosphorylation of Rec12 or one or more of its putative partners. Meiotic replication and DSB formation in S. pombe may be normally coupled by a checkpoint mechanism but not obligatorily coupled as appears to be the case in S. cerevisiae.
6 DSB Hotspots and Coldspots: Regulating Where Recombination Occurs
Rec12 does not make DSBs uniformly across chromosomes; rather, there are sites or regions with DSBs at above-average frequency (hotspots) and below-average frequency (coldspots). Hotspots and coldspots were first identified genetically as chromosomal intervals with higher or lower than average intensity of recombination (Gutz 1971; see chapter by Jeffreys). Wild-type chromosomes in all organisms tested have such hot and cold intervals, but the S. pombe mutation ade6-M26, which creates a hotspot, has been especially informative.
6.1 M26 : A Eukaryotic Sequence-specific Hotspot
The ade6-M26 mutation recombines with other ade6 mutations ~10 times more frequently than does the closely linked M375 mutation (Gutz 1971). By tetrad analysis M26 also converts~10 times more frequently than does M375, and it converts preferentially to ade6+. M26 is a single bp mutation G → T that creates the sequence 5′ ATGACGT 3′, each nucleotide of which is important for hotspot activity (Ponticelli et al. 1988; Szankasi et al. 1988; Schuchert et al. 1991). This sequence is bound by the Atf1-Pcr1 “stress response” transcription factor, which is essential for M26 hotspot activity (Wahls and Smith 1994; Kon et al. 1997). An iterative binding and PCR-amplification scheme identified 5′ GNVTATGACGTCATNBNC 3′ as a consensus sequence for Atf1-Pcr1 binding to DNA, and mutations creating this sequence in ade6 have hotspot activity greater than that of M26 itself (Steiner and Smith 2005b). Sequences closely related to this consensus occur in the wild-type S. pombe genome, and the majority of 15 such loci tested are hotspots of DSB formation (Steiner and Smith 2005a). In the one case tested, in the cds1 gene, this sequence is also a hotspot of recombination. This appears to be the first case of meiotic recombination hotspots being successfully predicted from a genome’s sequence. [In S. cerevisiae, sites bound by the Bas1 transcription factor are hot- or coldspots (Mieczkowski et al. 2006).] Collectively, the M26-like sequences may account for a few percent of all of the meiotic DSBs and recombination. Other transcription factors may account for additional DSBs and recombination.
The molecular basis of the M26 hotspot is partially understood. During meiosis the chromatin in and near ade6 becomes more sensitive (“open”) to exogenous micrococcal nuclease in an M26 sequence-and Atf1-Pcr1 factor-dependent manner (Mizuno et al. 1997, 2001; Yamada et al. 2004). DSB formation at and around M26 depends on Rec12 and Pcr1 (Steiner et al. 2002). Among M26-like sequences, there is a strong correlation between DSB frequency and hotspot activity, leaving no doubt that these DSBs are causally related to recombination. The M26 sequence is not sufficient, however, for hotspot activity. Most transplacements of the ade6-M26 gene, with >1 kb of DNA to each side of M26, to a distant site do not manifest hotspot activity (Ponticelli and Smith 1992). Presumably, the chromatin structure is influenced by nucleotide sequences >1 kb away from M26 and is more “open” at the endogenous ade6 locus. The features of “open” chromatin that permit DSB-formation are currently unknown but may involve binding of Rec12 or its putative partners to proteins that “open” the chromatin (see chapter by Lichten).
6.2 Hotspots in Large Intergenic Regions: Another Role for “Junk” DNA?
Surveys for DSBs across large regions of wild-type S. pombe chromosomes reveal prominent DSB hotspots roughly 50 – 100 kb apart separated by regions with few if any DSBs (Young et al. 2002). Each of these hotspots appears to be a cluster of DSB sites spread over ~1 – 3 kb. Among 24 such prominent DSB hotspots examined, 21 fall in intergenic intervals markedly larger than the mode of 0.4 kb (Wood et al. 2002): 15 of these 21 DSB hotspots are in intergenic intervals >4 kb, and the smallest of these 21 intervals is 1.9 kb (unpublished data). The nucleotide sequences responsible for these prominent hotspots have not been determined. They may be collections of transcription factor binding sites exemplified by M26, just discussed. Alternatively, the primary role of this apparently “junk” intergenic DNA may be to promote meiotic recombination.
6.3 Region-specific Activation by Cohesins: Megabase-scale Control of DSB Formation
Early studies showed that rec8, rec10, and rec11 mutants are far more deficient for recombination at the ade6 locus, the basis for their isolation (Ponticelli and Smith 1989), than in several other intervals tested (DeVeaux and Smith 1994). For example, in rec8Δ and rec11Δ mutants ade6 recombination is reduced by a factor of ~500, whereas recombination in many other intervals is reduced by a factor of 10 or less. In rec10Δ mutants recombination is strongly reduced throughout the genome, although the initial mutant rec10-109, a double missense, behaves much like the rec8Δ and rec11Δ mutants (Ellermeier and Smith 2005). The intervals with the least reduction in rec8 and rec11 mutants appear to be toward the ends of the chromosomes, although no clear pattern has been established (Parisi et al. 1999). Nevertheless, the strongly affected intervals are large – up to a few Mb.
The basis for this remarkable regional specificity is not entirely clear. It is noteworthy that in a rec8Δ mutant Rec10 forms short patches that may correspond to short LinEs seen by electron microscopy (Molnar et al. 1995; Lorenz et al. 2004). Rec8 and Rec11 meiosis-specific cohesin subunits are required for the cascade resulting in the loading of Rec12 (see section 5). These cohesin subunits do not entirely replace the mitotic cohesin subunit Rad21, residual levels of which remain along meiotic chromosomes (Yokobayashi et al. 2003). In the absence of Rec8 or Rec11, Rec10 may be able to load onto Rad21-bound intervals and lead to DSBs in those intervals; the Rec10-109 mutant protein may be active with Rad21 but not with Rec8 or Rec11.
6.4 Recombination in DSB-poor Intervals: Action at a Distance or Novel Lesions?
Between the prominent hotspots noted in section 6.2 are regions of 50 – 100 kb with few if any DSBs. Nevertheless, in the intervals tested crossovers occur at an intensity (cM per kb) close to that in intervals with prominent DSB hotspots (Young et al. 2002). The origin of these crossovers is currently unclear. The hypothesis that crossovers are generated by distant DSBs (Smith 2001; Young et al. 2002) was not supported by direct and indirect tests (Cromie et al. 2005). Perhaps there are DNA lesions other than DSBs that occur in the DSB-poor regions and lead to crossovers. If so, these lesions must depend on Rec12 and the tyrosine at its putative active site, for a mutant lacking this tyrosine is completely deficient for meiotic recombination (Cervantes et al. 2000). Rec12 may generate recombinogenic single-strand lesions, such as nicks and gaps, in some intervals and DSBs in others. Alternatively, some Rec12-dependent DSBs may not be detectable by the methods used.
6.5 Coldspots: Forbidden Regions for Recombination
Reciprocal recombination (crossing-over) occurs throughout most of the genome with nearly uniform intensity of 0.16 cM/kb (Young et al. 2002). Two regions, however, appear to have essentially no recombination. The first recognized is the 15 kb “K region” between the two silent mating-type loci, mat2 and mat3, which are separated by <0.002 cM, indicating a recombination intensity <0.1% of the genome average (Egel 1984). Recombination also appears to be rare within centromeres: loci flanking cenII and cenIII recombine with an intensity (cM/kb) <3% of the genome average (Nakaseko et al. 1986; C. Ellermeier, V. Tseng, and G. Smith, unpublished data).
The reduced recombination appears to be due to heterochromatin at these loci. There are multiple copies of several different repeats in the centromeres (Wood et al. 2002), and ~4.3 kb of the mat K region shares 96% identity with parts of two of these repeats (Grewal and Klar 1996). In the K region and at centromeres, heterochromatin blocks expression of inserted genes, and mutations that alter chromatin structure, such as swi6 (HP-1 homolog) or rik1 (putative partner for the Clr4 histone methyltransferase), allow both gene expression and recombination in these intervals (Egel et al. 1989; Klar and Bonaduce 1991; C. Ellermeier and G. Smith, unpublished data). Presumably, wild-type “closed” chromatin in these intervals prevents DSB formation by Rec12. The biological advantage of such cold regions is not clear, but recombination in these intervals may interfere with normal mating-type switching (K region) or chromosome segregation (centromeres) (see chapters by Petersen and by Watanabe).
7 Processing of Rec12-generated DSBs: Converting a Lesion into a Recombinogenic DNA-Protein Complex
Rec12-generated DSBs must be processed into a DNA-protein complex capable of initiating strand exchange with a homologous duplex. The major steps are thought to be removal of bound Rec12 from the 5′ strand termini of the DSB; resection of the 5′ strands to give long 3′ single-strand (ss) DNA overhangs; and loading of strand exchange proteins onto the single-stranded (ss) DNA (Table 3).
7.1 The MRN complex Is Needed for Removing Rec12 from DSBs But Not for DSB Formation
The Mre11-Rad50-Nbs1 complex (MRN) is a widely conserved eukaryotic protein complex with both exo- and endonuclease activities and is essential for meiotic recombination in all tested organisms. Consistent with this conserved phenotype, S. pombe rad32 (MRE11 ortholog) and rad50 mutants have strongly reduced spore viability and meiotic recombination and fail to repair meiotic DSBs (Tavassoli et al. 1995; Young et al. 2004). In contrast, S. cerevisiae MRN null mutants fail to make breaks (see sections 5.5 and 13).
The three components of the MRN complex have distinct roles. Rad50, an ATPase, is a member of the structural-maintenance-of-chromosomes (SMC) family of proteins, which have a long coiled-coil hairpin-like structure. This structure may allow Rad50 to co-ordinate events at the two sides of a meiotic DSB. The nuclease domain of Rad32 is required for processing DSBs; Rad50 appears to regulate this nuclease, as the rad50S (K81I) mutant accumulates Rec12-DNA complexes (Young et al. 2002; R. Hyppa, pers. comm.). The Nbs1 subunit is also believed to be regulatory.
S. pombe MRN nuclease-deficient mutants, like MRN null mutants, accumulate meiotic DSBs (Young et al. 2002, 2004; J. Farah, pers. comm.). Despite the 3′ to 5′ exonuclease polarity of the human and S. cerevisiae enzymes (Paull and Gellert 1998; Usui et al. 1998), it has been suggested that the MRN nuclease is required for 5′ end resection. However, S. cerevisiae MRN appears to cleave DNA ~10 – 40 nucleotides from the covalently linked Spo11 (Neale et al. 2005). S. pombe MRN nuclease mutants are also unable to remove Rec12 from the sites of DSBs (E. Hartsuiker, pers. comm.), and they can carry out meiotic recombination initiated by the I-SceI endonuclease (J. Farah, pers. comm.), which, unlike Rec12, does not remain covalently linked to the DNA. These data suggest that the sole nucleolytic role of MRN is to remove Rec12 from the ends of meiotic DSBs, with 5′ resection being carried out by another enzyme. The responsible exonuclease is unknown. Exonuclease I has the appropriate specificity but seems to play little role other than mismatch correction in S. pombe meiotic recombination (Szankasi and Smith 1995; see section 10).
7.2 Loading Strand-Exchange Proteins: Many Actors with Overlapping Roles
Strand-exchange proteins, bound to ss DNA, generate the joint molecule intermediates of recombination. However, these proteins alone are unable to compete with ss DNA binding (SSB) proteins for DNA. Consequently, from bacteriophages to humans, accessory proteins are needed to assist in their loading. S. pombe possesses several such accessory proteins: Rad22, Rti1, the Rhp55-Rhp57 and Swi5-Sfr1 complexes, and possibly the Rlp1 protein. Rad22 and Rti1 (also called Rad22B) appear to have redundant functions, but a suppressor mutation commonly found in rad22 strains has complicated their analysis (Doe et al. 2004). The S. pombe Rhp55, Rhp57, and Rlp1 proteins are paralogs of the strand exchange protein Rad51 (also called Rhp51; Grishchuk and Kohli 2003). The Rhp55-Rhp57 and Swi5-Sfr1 complexes promote Rad51 and Dmc1 loading onto ss DNA (Haruta et al. 2006). Mutations in rlp1, rhp55, rhp57 or the double rhp55-rhp57 mutation have relatively mild effects on meiotic recombination frequencies and spore viability (Grishchuk and Kohli 2003). Mutations in swi5 or sfr1 also show a moderate reduction in meiotic recombination and slightly lowered spore viability (Young et al. 2004; unpublished data). In contrast to the single mutants, double mutants affecting both the Rhp55-Rhp57 and the Swi5-Sfr1 complexes have severe defects in spore viability and recombination, similar in magnitude to a dmc1 rad51 double mutant (Ellermeier et al. 2004), suggesting that the two complexes possess redundant functions. However, the exact roles of these proteins, and the distinctions between them, remain unclear.
8 Strand Invasion and Partner Choice
By analogy with recombination in other organisms, the action of Rad22, Rti1, Rlp1, Rhp55-57 and Swi5-Sfr1 is believed to produce DNA ends with 3′ overhangs coated in strand exchange proteins. Through the process of strand invasion, this nucleoprotein complex generates joint molecules between homologous DNA duplexes that can then be processed to give recombinants.
8.1 The Dmc1 and Rad51 Strand Exchange Proteins: Finding a Homologous Partner for Recombination
The archetypal DNA strand exchange protein is Escherichia coli RecA, which is loaded onto ss DNA by the RecBCD or the RecFOR proteins to form a nucleoprotein filament capable of strand exchange (see chapter by Prevost). The filament pairs with the complementary strand of a homologous duplex, displacing the other strand to form a displacement loop (D-loop). This joint molecule is held together by a region of hybrid DNA, having one strand from each parent. Finally, RecA cycles off the DNA after hydrolysis of ATP.
S. pombe possesses two RecA structural and functional homologs – Rad51, expressed in all cells of all studied eukaryotes, and Dmc1, meiosis-specific but absent from some species. Mutations in rad51 and dmc1 have quite distinct effects on meiotic recombination. Although both mutations show an approximately 5-fold reduction in crossing-over, the effect of the rad51 mutation on gene conversion (non-reciprocal recombination) is much stronger. The spore viability of a rad51 mutant is very low, while that of a dmc1 mutant is close to the wild-type level (Grishchuk and Kohli 2003). The relative spore viabilities can be explained by the observation that in dmc1 mutants meiotic DSBs are repaired, but in rad51 mutants they are not (Young et al. 2004; see section 13). The recombination and spore viability phenotypes of the double mutant are much more severe than those of the single mutants, suggesting some redundancy in the function of the two proteins (Grishchuk and Kohli 2003). Double mutant analyses suggest that Dmc1, Swi5, and Sfr1 function in one branch of a pathway and Rhp55 and Rhp57 in another (Ellermeier et al. 2004; Figure 1). In addition, the Mcp7-Meu13 complex, which is homologous to S. cerevisiae Mnd1-Hop2, also appears to act specifically with Dmc1. Mutations in either mcp7 or meu13 have mild spore-viability defects and substantial recombination defects (Saito et al. 2004), but the exact function of this complex is unclear. Further studies are needed to elucidate these aspects of S. pombe meiotic recombination.
8.2 The Rhp54 and Rdh54 Proteins: Enabling Strand Exchange in a Chromatin Context?
S. pombe Rhp54 and its paralog Rdh54 are members of the Swi2 (Snf2) family of proteins, many of which remodel chromatin. Rhp54 is expressed both mitotically and meiotically, while Rdh54 is meiosis-specific (Catlett and Forsburg 2003). rhp54 and rdh54 mutants show mild defects in recombination, spore viability, and meiotic DSB repair, but the double mutant is severely defective. The S. cerevisiae orthologs of rhp54 and rdh54 appear to alter chromatin structure to facilitate Rad51 and Dmc1 strand exchange (Heyer et al. 2006). The S. pombe proteins may act similarly, since Rhp54 interacts with Rad51, and Rdh54 interacts with both Rad51 and Dmc1 (Catlett and Forsburg 2003). Mutations in S. pombe rdh54 have meiotic phenotypes similar to those of dmc1 mutations (high spore viability and successful repair of DSBs but reduced recombination), suggesting that these two meiosis-specific proteins may act in the same pathway.
8.3 Intersister vs. Interhomolog Recombination: Any Partner Will Do?
In meiosis there are almost always three homologous DNA targets with which a recombinogenic DNA end can interact – the sister chromatid or either of the two chromatids of the homolog. Interhomolog recombination might be expected to be favored over intersister recombination because meiotic recombination must produce crossovers between homologs for their proper segregation (see chapter by McKim). Results from S. cerevisiae support this idea (see section 12). However, genetic studies and physical analysis of joint molecules in S. pombe have demonstrated that intersister recombination does occur and is actually more frequent than interhomolog recombination (Cromie et al. 2006). How this is reconciled with the necessity of producing interhomolog crossovers is discussed in section 12. Nevertheless, S. pombe does appear to have mechanisms that specifically promote interhomolog recombination (see section 3). As discussed above, dmc1 mutants have reduced recombination frequencies, but they repair meiotic DSBs and have high spore viability. This suggests that Dmc1 promotes the repair of DSBs using homologs, but, if Dmc1 is absent, breaks are repaired using sister chromatids. The distinct phenotypes of S. cerevisiae vs. S. pombe dmc1 mutants may result from a mechanism, present in S. cerevisiae but not S. pombe, that inhibits intersister recombination (see sections 12 and 13).
9 Joint Molecule Resolution
In the current canonical model of meiotic crossing over (Szostak et al. 1983; Sun et al. 1989), after strand invasion and D-loop formation the second end of the initiating DSB anneals to the D-loop (Figure 3). Branch migration at each side of the D-loop then generates two Holliday junctions (double HJs, Figure 3 right) connecting the homologous duplexes. Cleavage and rejoining of appropriate pairs of strands in the two HJs can then generate a crossover.
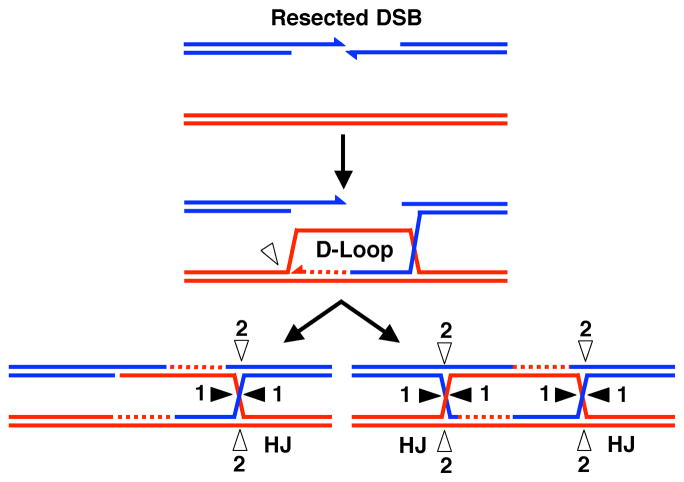
Figure 3.
A scheme that produces either crossover or non-crossover recombinants from single or double Holliday junctions. Resection of a DSB produces two 3′ single strand overhangs, one of which invades a homologous duplex, producing a D-loop. A single HJ results if this D-loop is cut before second end capture (left). A double HJ results if the D-loop remains uncut before second end capture (right). In both cases resolution of the HJ(s) results in crossover or non-crossover products, depending on the strands cleaved.
9.1 Single Holliday Junctions: An Unexpected Recombination Intermediate
Electron microscope studies of meiotic joint molecules in S. pombe reveal DNA structures different from those predicted by the current canonical model and previously observed in S. cerevisiae (Cromie et al. 2006). Instead of double HJs, the majority of molecules contain single HJs (Figure 4). Two dimensional gel electrophoretic analyses are also consistent with a majority of single HJs. The single HJ and double HJ mechanisms may differ only by the timing of cleavage of the strand forming the D-loop: cleavage before second end capture produces a single HJ (Figure 3, left), whereas cleavage after second end capture allows formation of a double HJ (Figure 3, right).
Figure 4.
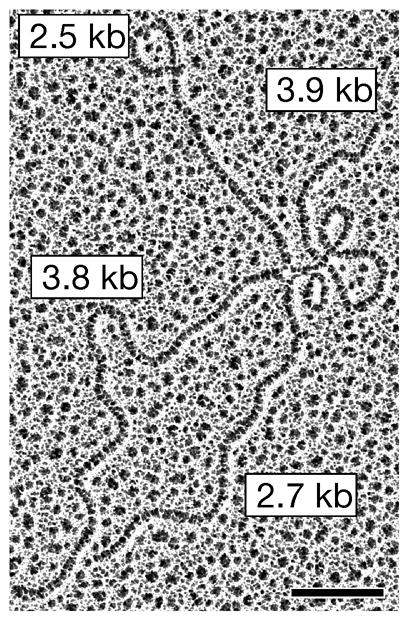
Single Holliday junction intermediates in S. pombe meiotic recombination. DNA from meiotic mus81 mutant cells was separated by two-dimensional gel electrophoresis, and DNA in the joint molecule region was visualized with an electron microscope. The junction of the four DNA duplex segments is splayed out in a “traffic circle” due to formamide in the spreading mixture. Bar indicates 0.2 μm. From Cromie et al. (2006).
9.2 Mus81-Eme1: The Meiotic Holliday Junction Resolvase of S. pombe
To generate crossovers from joint molecules, the HJs must be cleaved and the broken strands ligated. The S. pombe Mus81-Eme1 endonuclease can resolve HJs and closely related structures (Boddy et al. 2001). mus81 and eme1 mutants have, as far as tested, indistinguishable phenotypes which are those expected of nuclear HJ resolvase mutants. Most notably, HJs accumulate in mus81 mutant meiosis (Cromie et al. 2006). Physical and genetic assays show that meiotic crossovers, which are expected to depend on an HJ resolvase, are greatly reduced in mus81 mutants, whereas genetic assays show that gene conversions without crossovers, which are not necessarily resolvase-dependent, are essentially unaffected (Osman et al. 2003; Smith et al. 2003). In mus81 mutants the asci are aberrantly shaped, spore viability is very low, and there is a single mass of apparently entangled DNA. These phenotypes are indicative of chromosome segregation failure, as expected from chromosomes remaining held together by unresolved HJs. The phenotypes of mus81 mutants can be relieved by expression of a bacterial HJ resolvase. As expected, in meiosis rec12Δ is epistatic to mus81Δ and eme1Δ, indicating that Mus81-Eme1 is required for meiotic DSB repair. Amino acid substitutions affecting the Mus81 nuclease active site have a phenotype indistinguishable from that of a complete deletion. Collectively, these observations indicate that the complex’s role in crossing-over is endonucleolytic resolution (Boddy et al. 2001; Osman et al. 2003; Smith et al. 2003).
The archetypal HJ resolvase, E. coli RuvC, cleaves intact HJs by symmetrical cleavages of two strands; the cleaved product is a good substrate for DNA ligase. Mus81-Eme1 purified from S. pombe and human cells cleaves intact HJs, although not symmetrically. All reported preparations of Mus81-Eme1 show higher activity on three-strand junctions than on intact HJs, but nicked HJs are preferred over the 3-strand junctions, with cleavage occurring at the site on the HJ strand opposite the nick (Doe et al. 2002; Gaillard et al. 2003; Osman et al. 2003). Mus81-Eme1 may readily cleave 3-strand junctions because they resemble nicked HJs. Osman et al. (2003) have proposed a model of recombination involving Mus81-Eme1 cleavage of nicked HJs. However, Mus81-Eme1 purified from S. pombe and human cells slowly cleaves one strand of an intact HJ and then rapidly cleaves the nicked product (Gaillard et al. 2003); this may be the relevant in vivo activity, perhaps stimulated by other factors. Asymmetric HJ cleavage would prevent immediate ligation of the cut strands, but repair DNA synthesis and single-strand flap-processing would allow the required ligation.
10 Mismatch Correction
Gene conversion is a form of recombination involving the non-reciprocal transfer of sequence information between homologous DNA sequences. It is observed as heterozygous marker segregation other than the normal Mendelian 2:2 among the four spores in an ascus and is often produced as an outcome of mismatch correction. Hybrid DNA is generated between homologous loci undergoing strand exchange, and, if the loci are non-identical in sequence, the hybrid DNA will contain mismatches. Repair of such mismatches can lead to gene conversion, seen as 3:1 segregation among the four spores. Unrepaired mismatches segregate at the first round of DNA replication after meiosis and so can be identified as post-meiotic segregation (PMS) events, detectable as a sectored colony arising from one spore.
In S. pombe both mismatch repair (MMR) and nucleotide excision repair (NER) function in the repair of mismatches arising during meiotic recombination. Presumably, after mismatch recognition by MMR proteins and strand incision by unknown factors, exonuclease I removes part of a strand with one of the mismatched nucleotides, and repair DNA synthesis restores complementarity. MMR appears to repair efficiently small insertion or deletion loops and all base mismatches other than C/C. The C/C mismatch is repaired, but inefficiently, by the NER system, which can also repair other base mismatches in the absence of the MMR pathway (Fleck et al. 1999). The MMR and NER gene products demonstrably required for these processes are shown in Table 3. In meiotic crosses involving the ade6-M26 hotspot essentially all mismatch correction (identified as 3:1 marker segregation) is abolished in the absence of both the MMR and NER pathways, and only PMS events are seen, at elevated frequency. One class of PMS event has hybrid DNA in one spore (~70% of all asci with non-2:2 segregation), and another class has hybrid DNA in two spores (~30%). The first class may reflect a mismatch in the region resected adjacent to a meiotic DSB (asymmetric heteroduplex), and the second class a mismatch in a more distant region where branch migration formed an HJ (symmetric heteroduplex). Thus, both symmetric and asymmetric hybrid DNA forms appear to be frequent in S. pombe meiosis.
The fission yeast swi4 gene encodes a homolog of the budding yeast MMR protein Msh3. However, rather than a meiotic MMR defect, mutations in swi4 exhibit a mild deficiency in both intragenic and intergenic recombination. The function of Swi4 in meiotic recombination is unclear.
11 Relation of Gene Conversion and Crossing-over
Gene conversion indicates hybrid DNA at a marked locus and thus strand exchange. Markers flanking this locus may or may not undergo crossing-over. Until recently, it was generally believed that there was a single pathway of homologous recombination with an HJ intermediate containing hybrid DNA. At the HJ resolution stage, if the pairs of complementary strands cleaved in the HJs are chosen at random, crossovers and non-crossovers would be equally frequent (Figure 3). Recent work in both prokaryotes and eukaryotes has thrown doubt on this model and has suggested that crossovers and non-crossovers are generated by different pathways (Cromie and Leach 2000; Allers and Lichten 2001). The mechanism that generates non-crossovers is unclear but may involve sequential DNA synthesis, DNA unwinding, and annealing, termed “synthesis-dependent strand annealing” (SDSA; see chapter by Haber). Gene conversion could occur in the hybrid DNA envisaged as a part of the SDSA model.
In S. pombe crossovers accompany gene conversions ~75% of the time rather than 50%, as predicted by the random HJ resolution model, or ~40%, as observed in S. cerevisiae (Grimm et al. 1994; Cromie et al. 2005). In addition, as noted in section 9.2, mus81 resolvase mutations have very little effect on the frequency of gene conversions that lack an associated crossover (Osman et al. 2003; Smith et al. 2003). This suggests that these non-crossovers do not result from HJ resolution. It therefore appears that in S. pombe crossovers result from HJ resolution and non-crossovers from a second mechanism, such as SDSA, although HJ resolution may contribute to some non-crossover events.
12 Species-specific Strategies for Ensuring, With or Without Interference, the Crossovers Required for Segregation
Meiotic crossovers generate new combinations of alleles that increase genetic diversity in the population, and in most organisms crossovers also aid the correct segregation of homologs at the first meiotic division. Intersister recombination achieves neither of these aims. Why then does S. pombe show a bias towards intersister events, while S. cerevisiae shows a bias towards interhomolog events (Cromie et al. 2006)? Similarly, why is crossover interference, which reduces the probability of crossovers occurring close together, present in S. cerevisiae and not in S. pombe (Munz 1994)?
The bias to interhomolog recombination seen in S. cerevisiae appears to result from a barrier to intersister recombination events (Niu et al. 2005), i.e., it is a form of regulation of recombination, as is crossover interference. Some of the 16 chromosomes of S. cerevisiae are very small, as short as 230 kb, and all are smaller than the smallest of S. pombe (3500 kb). If the total number of crossovers in S. cerevisiae were distributed randomly across the DNA (i.e., if there were no interference), then these small chromosomes would receive no crossover ~10% of the time and would frequently missegregate. Interference may ensure that crossovers are distributed so that small chromosomes always receive at least one. Interhomolog bias may be a further adaptation to ensure enough interhomolog crossovers on small chromosomes without increasing the number of DSBs.
In contrast to S. cerevisiae, S. pombe has only three, large chromosomes. Random (Poisson) distribution of crossovers suffices to ensure that each chromosome receives at least one interhomolog crossover, since there is an average of 10, 15, and 20 interhomolog crossovers per meiotic cell (Munz et al. 1989). The average number of crossovers on the smallest chromosome is high enough to prevent missegregation, without needing to regulate recombination through crossover interference (Munz et al. 1989) or a barrier to intersister recombination. In this view S. pombe uses a meiotic recombination system “pared down” to its fundamentals. The obvious success of this scheme raises the question of why other organisms regulate crossovers rather than alter the size and number of chromosomes or increase the number of DSBs so that unregulated recombination would allow successful chromosome segregation.
13 Differences Between S. pombe and S. cerevisiae Meiotic Recombination: A Reprise
At all steps of meiotic recombination significant differences are seen between S. pombe and S. cerevisiae – in the production and processing of DSBs, in the loading of strand exchange proteins, in choice of partner for the strand exchange reaction, in the structure of joint molecules, and in the processing of joint molecules into recombinant products. These differences may account for the differences, noted in sections 1, 3, and 11 and below, in the occurrence of interference, the frequency of ectopic recombination, and the frequency of crossovers associated with gene conversion.
Several components for the formation and processing of meiotic DSBs differ markedly in S. pombe and S. cerevisiae. Each species has proteins essential for DSB formation that have no obvious ortholog in the other species (Table 2). Furthermore, the Rec8 cohesin subunit is required for DSB formation in S. pombe but not in S. cerevisiae; it is required for DSB repair in S. cerevisiae but perhaps not in S. pombe (Klein et al. 1999; Ellermeier and Smith 2005). Conversely, the MRN nuclease complex is required for DSB formation in S. cerevisiae but not in S. pombe; it is required for DSB repair in both species (Cao et al. 1990; Young et al. 2004). Presumably, Rec8 and MRN are required indirectly, and differentially in the two species, for the assembly of the Rec12 (Spo11) complex at sites of DSB formation. DSB repair in both species may require the MRN nuclease to remove Rec12 (Spo11) linked to the DNA (see section 7.1). The role of Rec8 in DSB repair in S. cerevisiae is unclear.
There are several differences between S. cerevisiae and S. pombe relating to Rad51 accessory proteins. First, the function of the S. cerevisiae Rad52 protein appears to be carried out by two partially redundant Rad52 homologs in S. pombe. Interestingly, rad52−/− knockout mice are viable and fertile (Rijkers et al. 1998), suggesting that mammals may be more similar in this regard to S. pombe than to S. cerevisiae. Second, mutations in the rad55, rad57 and rdh54 genes have very severe recombination defects in S. cerevisiae, but the S. pombe ortholog mutants have only mild defects (Petes et al. 1991; Catlett and Forsburg 2003; Grishchuk and Kohli 2003). Third, the S. pombe Swi5-Sfr1 complex appears to function in both mitosis and meiosis, unlike the S. cerevisiae meiosis-specific Sae3-Mei5 ortholog. This is consistent with the ability of Swi5-Sfr1 to promote both Rad51 and Dmc1 activity (Haruta et al. 2006), whereas Sae3-Mei5 is believed to interact only with Dmc1 (Hayase et al. 2004). Fourth, there appears to be no Rlp1 ortholog in S. cerevisiae, but mammals have an ortholog, Xrcc2 (Khasanov et al. 2004).
The mechanics of strand invasion are not known to differ between S. pombe and S. cerevisiae. However, significant differences are seen in the choice of DNA used as the target of strand invasion; i.e., in S. pombe intersister events are preferred, while in S. cerevisiae interhomolog events predominate (see sections 8.3 and 12). These differences appear to be related to differences in the phenotypes of dmc1 mutants. S. pombe dmc1 mutants successfully repair meiotic DSBs, presumably using sister chromatids. In contrast, in dmc1 mutants of S. cerevisiae strain SK1 DSBs remain unrepaired (Bishop et al. 1992), apparently due to a barrier to intersister recombination that involves the Hop1, Mek1 and Red1 proteins (Niu et al. 2005). The apparent absence of this barrier in S. pombe may explain why intersister events are more frequent in S. pombe than in S. cerevisiae.
The structures of recombination joint molecules in S. pombe and S. cerevisiae are distinctly different. As discussed in section 9.1 most recombination joint molecules in S. pombe contain single HJs rather than the double HJs seen in most S. cerevisiae joint molecules and predicted by a current recombination model (Szostak et al. 1983). A variation on this model can account for either a single HJ or a double HJ arising from a D-loop and producing recombinants (Figure 3).
In S. pombe the Mus81-Eme1 complex appears to be the only meiotic nuclear HJ resolvase. However, despite the wide conservation of these two proteins among eukaryotes and the similar mitotic phenotypes of the mutants, the importance of these proteins in meiosis appears to vary between organisms. In S. cerevisiae crossovers are only mildly reduced by mus81 or mms4 (eme1) mutations, and in mice mus81 mutants are viable and fertile (de los Santos et al. 2003; McPherson et al. 2004). S. cerevisiae and mice may have a second crossover pathway that requires the Msh4 and Msh5 proteins, which are apparently absent from S. pombe. Interestingly, mus81-dependent crossovers in S. cerevisiae, like those of S. pombe, are not subject to crossover interference (de los Santos et al. 2003).
The similarities of meiotic recombination in S. pombe and S. cerevisiae indicate that the basic process of DSB formation and repair is widely conserved and perhaps universal. But the multiple differences suggest that the regulation of meiotic recombination is variable between species and may have arisen independently during evolution. Further understanding of these similarities and differences may provide insight into how meiotic recombination arose, presumably from a mitotic precursor.
Acknowledgments
We are grateful to Chad Ellermeier, Joe Farah, Edgar Hartsuiker, Randy Hyppa, Cristina Martin-Castellanos, and Jennifer Young for unpublished data; Josef Loidl for Figure 2; and Luther Davis, Joe Farah, Yasushi Hiraoka, Josef Loidl, Yoshinori Watanabe, and an anonymous reviewer for helpful comments on the manuscript. Our research is supported by research grant R01 GM031693 from the National Institutes of Health of the United States of America.
Abbreviations
- DSB
double-strand break
- MI
first meiotic division
- SC
synaptonemal complex
- LinE
linear element
- SPB
spindle-pole body
- MRN
Mre11-Rad50-Nbs1 complex
- MCM
mini-chromosome maintenance
- SDSA
synthesis-dependent strand annealing
- ss
single-stranded
- SSB
ss DNA binding protein
- HJ
Holliday junction
- MMR
mismatch repair
- NER
nucleotide excision repair
Footnotes
Prepared for Recombination and Meiosis, edited by Richard Egel, in the series “Genome Dynamics and Stability” to be published by Springer-Verlag
References
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Bähler J, Wyler T, Loidl J, Kohli J. Unusual nuclear structures in meiotic prophase of fission yeast: a cytological analysis. J Cell Biol. 1993;121:241–256. doi: 10.1083/jcb.121.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Borde V, Goldman ASH, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Catlett MG, Forsburg SL. Schizosaccharomyces pombe Rdh54 (TID1) acts with Rhp54 (RAD54) to repair meiotic double-strand breaks. Mol Biol Cell. 2003;14:4707–4720. doi: 10.1091/mbc.E03-05-0288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Farah JA, Smith GR. Meiotic DNA breaks associated with recombination in S. pombe. Mol Cell. 2000;5:883–888. doi: 10.1016/s1097-2765(00)80328-7. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Hiraoka Y. Telomere binding of the Rap1 protein is required for meiosis in fission yeast. Curr Biol. 2001;11:1618–1623. doi: 10.1016/s0960-9822(01)00457-2. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Ding DQ, Funabiki H, Haraguchi T, Mashiko S, Yanagida M, Hiraoka Y. Telomere-led premeiotic chromosome movement in fission yeast. Science. 1994;264:270–273. doi: 10.1126/science.8146661. [DOI] [PubMed] [Google Scholar]
- Chikashige Y, Tsutsumi C, Yamane M, Okamasa K, Haraguchi T, Hiraoka Y. Meiotic proteins Bqt1 and Bqt2 tether telomeres to form the bouquet arrangement of chromosomes. Cell. 2006;125:59–69. doi: 10.1016/j.cell.2006.01.048. [DOI] [PubMed] [Google Scholar]
- Cooper JP, Watanabe Y, Nurse P. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature. 1998;392:828–831. doi: 10.1038/33947. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Leach DR. Control of crossing over. Mol Cell. 2000;6:815–826. doi: 10.1016/s1097-2765(05)00095-x. [DOI] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Rubio CA, Hyppa RW, Smith GR. A natural meiotic DNA break site in Schizosaccharomyces pombe is a hotspot of gene conversion, highly associated with crossing over. Genetics. 2005;169:595–605. doi: 10.1534/genetics.104.037176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Smith GR. Dynein promotes achiasmate segregation in Schizosaccharomyces pombe. Genetics. 2005;170:581–90. doi: 10.1534/genetics.104.040253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L, Smith GR. The meiotic bouquet promotes homolog interactions and restricts ectopic recombination in Schizosaccharomyces pombe. Genetics. 2006;174:167–177. doi: 10.1534/genetics.106.059733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVeaux LC, Smith GR. Region-specific activators of meiotic recombination in Schizosaccharomyces pombe. Genes Dev. 1994;8:203–210. doi: 10.1101/gad.8.2.203. [DOI] [PubMed] [Google Scholar]
- DeVeaux LC, Hoagland NA, Smith GR. Seventeen complementation groups of mutations decreasing meiotic recombination in Schizosaccharomyces pombe. Genetics. 1992;130:251–262. doi: 10.1093/genetics/130.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding DQ, Sakurai N, Katou Y, Itoh T, Shirahige K, Haraguchi T, Hiraoka Y. Meiotic cohesins modulate chromosome compaction during meiotic prophase in fission yeast. J Cell Biol. 2006;174:499–508. doi: 10.1083/jcb.200605074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe CL, Ahn JS, Dixon J, Whitby MC. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J Biol Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
- Doe CL, Osman F, Dixon J, Whitby MC. DNA repair by a Rad22-Mus81-dependent pathway that is independent of Rhp51. Nucleic Acids Res. 2005;32:5570–5581. doi: 10.1093/nar/gkh853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R. Two tightly linked silent cassettes in the mating-type region of Schizosaccharomyces pombe. Curr Genet. 1984;8:199–203. doi: 10.1007/BF00417816. [DOI] [PubMed] [Google Scholar]
- Egel R, Willer M, Nielsen O. Unblocking of meiotic crossing-over between the silent mating-type cassettes of fission yeast, conditioned by the recessive, pleiotropic mutant rik1. Curr Genet. 1989;15:407–410. [Google Scholar]
- Ellermeier C, Smith GR. Cohesins are required for meiotic DNA breakage and recombination in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 2005;102:10952–10957. doi: 10.1073/pnas.0504805102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier C, Schmidt H, Smith GR. Swi5 acts in meiotic DNA joint molecule formation in Schizosaccharomyces pombe. Genetics. 2004;168:1891–1898. doi: 10.1534/genetics.104.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleck O, Lehmann E, Schär P, Kohli J. Involvement of nucleotide-excision repair in msh2 pms1-independent mismatch repair. Nature Genet. 1999;21:314–317. doi: 10.1038/6838. [DOI] [PubMed] [Google Scholar]
- Fox ME, Yamada T, Ohta K, Smith GR. A family of CRE-related DNA sequences with meiotic recombination hotspot activity in Schizosaccharomyces pombe. Genetics. 2000;156:59–68. doi: 10.1093/genetics/156.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard P-HL, Noguchi E, Shanahan P, Russell P. The endogenous Mus81-Eme1 complex resolves Holliday junctions by a nick and counternick mechanism. Mol Cell. 2003;12:747–759. doi: 10.1016/s1097-2765(03)00342-3. [DOI] [PubMed] [Google Scholar]
- Gregan J, Rabitsch PK, Sakem B, Csutak O, Latypov V, Lehmann E, Kohli J, Nasmyth K. Novel genes required for meiotic chromosome segregation are identified by a high-throughput knockout screen in fission yeast. Curr Biol. 2005;15:1663–1669. doi: 10.1016/j.cub.2005.07.059. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Klar AJ. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics. 1997;146:1221–38. doi: 10.1093/genetics/146.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Bahler J, Kohli J. M26 recombinational hotspot and physical conversion tract analysis in the ade6 gene of Schizosaccharomyces pombe. Genetics. 1994;135:41–51. doi: 10.1093/genetics/136.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk AL, Kohli J. Five RecA-like proteins of Schizosaccharomyces pombe are involved in meiotic recombination. Genetics. 2003;165:1031–1043. doi: 10.1093/genetics/165.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutz H. Site specific induction of gene conversion in Schizosaccharomyces pombe. Genetics. 1971;69:317–337. doi: 10.1093/genetics/69.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan I, Yanagida M. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J Cell Biol. 1995;129:1033–1047. doi: 10.1083/jcb.129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta N, Kurokawa Y, Murayama Y, Akamatsu Y, Unzai S, Tsutsui Y, Iwasaki H. The Swi5-Sfr1 complex stimulates Rhp51/Rad51 and Dmc1-mediated DNA strand exchange in vitro. Nat Struct Mol Biol. 2006;13:823–830. doi: 10.1038/nsmb1136. [DOI] [PubMed] [Google Scholar]
- Hayase A, Takagi M, Miyazaki T, Oshiumi H, Shinohara M, Shinohara A. A protein complex containing Mei5 and Sae3 promotes the assembly of the meiosis-specific RecA homolog Dmc1. Cell. 2004;119:927–940. doi: 10.1016/j.cell.2004.10.031. [DOI] [PubMed] [Google Scholar]
- Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanoh J, Ishikawa F. spRap1 and spRif1, recruited to telomeres by Taz1, are essential for telomere function in fission yeast. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- Keeney S. Mechanism and control of meiotic recombination initiation. Curr Topics Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- Khasanov FK, Salakhova AF, Chepurnaja OV, Korolev VG, Bashkirov VI. Identification and characterization of the rlp1+, the novel Rad51 paralog in the fission yeast Schizosaccharomyces pombe. DNA Repair (Amst) 2004;3:1363–1374. doi: 10.1016/j.dnarep.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Klar AJS, Bonaduce MJ. swi6, a gene required for mating-type switching, prohibits meiotic recombination in the mat2-mat3 “cold spot” of fission yeast. Genetics. 1991;129:1033–1042. doi: 10.1093/genetics/129.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell. 1999;98:91–103. doi: 10.1016/S0092-8674(00)80609-1. [DOI] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic recombination hotspot in Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1997;94:13756–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz C, Fleck O. Role of the DNA repair nucleases Rad13, Rad2 and Uve1 of Schizosaccharomyces pombe in mismatch correction. J Mol Biol. 2001;313:241–53. doi: 10.1006/jmbi.2001.5054. [DOI] [PubMed] [Google Scholar]
- Loidl J. S. pombe linear elements: the modest cousins of synaptonemal complexes. Chromosoma. 2006;115:260–271. doi: 10.1007/s00412-006-0047-7. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Wells JL, Pryce DW, Novatchkova FE, Eisenhaber F, McFarlane RJ, Loidl J. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 2004;117:3343–3351. doi: 10.1242/jcs.01203. [DOI] [PubMed] [Google Scholar]
- Lorenz A, Estreicher A, Kohli J, Loidl J. Meiotic recombination proteins localize to linear elements in Schizosaccharomyces pombe. Chromosoma. 2006;115:330–340. doi: 10.1007/s00412-006-0053-9. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Blanco M, Rozalen AE, Perez-Hidalgo L, Garcia AI, Conde F, Mata J, Ellermeier C, Davis L, San-Segundo P, Smith GR, Moreno S. A large-scale screen in S. pombe identifies seven novel genes required for critical meiotic events. Curr Biol. 2005;22:2056–2062. doi: 10.1016/j.cub.2005.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherson JP, Lemmers B, Chahwan R, Pamidi A, Migon E, Matysiak-Zablocki E, Moynahan ME, Essers J, Hanada K, Poonepalli A, Sanchez-Sweatman O, Khokha R, Kanaar R, Jasin M, Hande MP, Hakem R. Involvement of mammalian Mus81 in genome integrity and tumor suppression. Science. 2004;304:1822–1826. doi: 10.1126/science.1094557. [DOI] [PubMed] [Google Scholar]
- Mieczkowski PA, Dominska M, Buck MJ, Gerton JL, Lieb JD, Petes TD. Global analysis of the relationship between the binding of the Bas1p transcription factor and meiosis-specific double-strand DNA breaks in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:1014–1027. doi: 10.1128/MCB.26.3.1014-1027.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F, Okazaki K, Shimanuki M, Yamamoto A, Hiraoka Y, Niwa O. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol Biol Cell. 2002;3:930–946. doi: 10.1091/mbc.01-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K-i, Emura Y, Baur M, Kohli J, Ohta K, Shibata T. Remodeling of chromatin structure around a single nucleotide mutation in ade6-M26 that creates a meiotic recombination hotspot in fission yeast. Genes Dev. 1997;11:876–886. doi: 10.1101/gad.11.7.876. [DOI] [PubMed] [Google Scholar]
- Mizuno K-i, Hasemi T, Ubukata T, Yamada T, Lehmann E, Kohli J, Watanabe Y, Iino Y, Yamamoto M, Fox ME, Smith GR, Murofushi H, Shibata T, Ohta K. Counteracting regulation of chromatin remodeling at a fission yeast cAMP responsive element-related recombination hotspot by stress-activated protein kinase, cAMP-dependent kinase and meiosis regulators. Genetics. 2001;159:1467–147. doi: 10.1093/genetics/159.4.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Bahler J, Sipiczki M, Kohli J. The rec8 gene of Schizosaccaromyces pombe is involved in linear element formation, chromosome pairing and sister-chromatid cohesion during meiosis. Genetics. 1995;141:61–73. doi: 10.1093/genetics/141.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Parisi S, Kakihara Y, Nojima H, Yamamoto A, Hiraoka Y, Bozsik A, Sipiczki M, Kohli J. Characterization of rec7, an early meiotic recombination gene in Schizosaccharomyces pombe. Genetics. 2001;157:519–532. doi: 10.1093/genetics/157.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar M, Doll E, Yamamoto A, Hiraoka Y, Kohli J. Linear element formation and their role in meiotic sister chromatid cohesion and chromosome pairing. J Cell Sci. 2003;114:1719–1731. doi: 10.1242/jcs.00387. [DOI] [PubMed] [Google Scholar]
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics. 1994;137:701–707. doi: 10.1093/genetics/137.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munz P, Wolf K, Kohli J, Leupold U. Molecular Biology of the Fission Yeast. Academic Press; San Diego: 1989. Genetics overview; pp. 1–30. [Google Scholar]
- Nabeshima K, Kakihara Y, Hiraoka Y, Nojima H. A novel meiosis-specific protein of fission yeast, Meu13p, promotes homologous pairing independently of homologous recombination. EMBO J. 2001;20:3871–3881. doi: 10.1093/emboj/20.14.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaseko Y, Adachi Y, Funahashi S, Niwa O, Yanagida M. Chromosome walking shows a highly homologous repetitive sequence present in all the centromere regions of fission yeast. EMBO J. 1986;5:1011–21. doi: 10.1002/j.1460-2075.1986.tb04316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niccoli T, Yamashita A, Nurse P, Yamamoto M. The p150-Glued Ssm4p regulates microtubular dynamics and nuclear movement in fission yeast. J Cell Sci. 2004;117:5543–5556. doi: 10.1242/jcs.01475. [DOI] [PubMed] [Google Scholar]
- Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–6188. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, Hollingsworth NM. Partner choice during meiosis is regulated by Hop1-prompted dimerization of Mek1. Mol Biol Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Shimanuki M, Miki F. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interaction in fission yeast meiosis. EMBO J. 2000;19:3831–3840. doi: 10.1093/emboj/19.14.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Hirota K, Matsumoto S, Takeda T, Ohta K, Arai K, Masai H. Hsk1 kinase is required for induction of meiotic dsDNA breaks without involving checkpoint kinases in fission yeast. Proc Natl Acad Sci USA. 2006;23:8131–8136. doi: 10.1073/pnas.0602498103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino K, Masai H. Rad3-Cds1 mediates coupling of initiation of meiotic recombination with DNA replication: Mei4-dependent transcription as a potential target of meiotic checkpoint. J Biol Chem. 2006;281:1338–1344. doi: 10.1074/jbc.M505767200. [DOI] [PubMed] [Google Scholar]
- Olson LW, Eden U, Egel-Mitani M, Egel R. Asynaptic meiosis in fission yeast? Heriditas. 1978;89:189–199. [Google Scholar]
- Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: A role for Mus81-Eme1 in meiosis. Mol Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- Parisi S, McKay MJ, Molnar M, Thompson MA, van der Speck PJ, van Drunen-Schoenmaker E, Kanaar R, Lehmann E, Hoeijmakers JHJ, Kohli J. Rec8p, a meiotic recombination and sister chromatid cohesion phosphoprotein of the Rad21p family conserved from fission yeast to humans. Mol Cell Biol. 1999;19:3515–3528. doi: 10.1128/mcb.19.5.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasierbek P, Jantsch M, Melcher M, Schleiffer A, Schweizer D, Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Petes TD, Malone RE, Symington LS. Recombination in yeast. In: Broach J, Jones E, Pringle J, editors. The Molecular and Cellular Biology of the Yeast Saccharomyces. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1991. pp. 407–521. [Google Scholar]
- Ponticelli AS, Smith GR. Meiotic recombination-deficient mutants of Schizosaccharomyces pombe. Genetics. 1989;123:45–54. doi: 10.1093/genetics/123.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli AS, Smith GR. Chromosomal context dependence of a eukaryotic recombinational hot spot. Proc Natl Acad Sci USA. 1992;89:227–231. doi: 10.1073/pnas.89.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponticelli AS, Sena EP, Smith GR. Genetic and physical analysis of the M26 recombination hotspot of Schizosaccharomyces pombe. Genetics. 1988;119:491–497. doi: 10.1093/genetics/119.3.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell. 2004;16:1968–1978. doi: 10.1105/tpc.104.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick MA. The repair of double-strand breaks in DNA: a model involving recombination. J Theo Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rijkers T, Van Den Ouweland J, Morolli B, Rolink AG, Baarends WM, Van Sloun PP, Lohman PH, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Tougan T, Kasama T, Okuzaki D, Nojima H. Mcp7, a meiosis-specific coiled-coil protein of fission yeast, associates with Meu13 and is required for meiotic recombination. Nucleic Acids Res. 2004;32:3325–3339. doi: 10.1093/nar/gkh654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito TT, Tougan T, Okuzaki D, Kasama T, Nojima H. Mcp6, a meiosis-specific coiled-coil protein of Schizosaccharomyces pombe, localizes to the spindle pole body and is required for horsetail movement and recombination. J Cell Sci. 2005;118:447–459. doi: 10.1242/jcs.01629. [DOI] [PubMed] [Google Scholar]
- Saito TT, Okuzaki D, Nojima H. Mcp5, a meiotic cell cortex protein, is required for nuclear movement mediated by dynein and microtubules in fission yeast. J Cell Biol. 2006;173:27–33. doi: 10.1083/jcb.200512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuchert P, Langsford M, Käslin E, Kohli J. A specific DNA sequence is required for high frequency of recombination in the ade6 gene of fission yeast. EMBO J. 1991;10:2157–2163. doi: 10.1002/j.1460-2075.1991.tb07750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimanuki M, Miki F, Ding D-Q, Chikashige Y, Hiraoka Y, Horio T, Niwa O. A novel fission yeast gene, kms1+, is required for the formation of meiotic prophase-specific nuclear architecture. Mol Gen Genet. 1997;254:238–249. doi: 10.1007/s004380050412. [DOI] [PubMed] [Google Scholar]
- Smith GR. Homologous recombination near and far from DNA breaks: Alternative roles and contrasting views. Annu Rev Genet. 2001;35:243–274. doi: 10.1146/annurev.genet.35.102401.090509. [DOI] [PubMed] [Google Scholar]
- Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81 Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KN, Penkner A, Ohta K, Klein F, Nicolas A. B-type cyclins CLB5 and CLB6 control the initiation of recombination and synaptonemal complex formation in yeast meiosis. Curr Biol. 2001;11:88–97. doi: 10.1016/s0960-9822(01)00026-4. [DOI] [PubMed] [Google Scholar]
- Steiner WW, Smith GR. Natural meiotic recombination hot spots in the Schizosaccharomyces pombe genome successfully predicted from the simple sequence motif M26. Mol Cell Biol. 2005a;25:9054–9062. doi: 10.1128/MCB.25.20.9054-9062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Smith GR. Optimizing the nucleotide sequence of a meiotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2005b;169:1973–1983. doi: 10.1534/genetics.104.039230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner WW, Schreckhise RW, Smith GR. Meiotic DNA breaks at the S. pombe recombination hotspot M26. Mol Cell. 2002;9:847–855. doi: 10.1016/s1097-2765(02)00489-6. [DOI] [PubMed] [Google Scholar]
- Stewart E, Chapman CR, Al-Khodairy F, Carr AM, Enoch T. rqh1+, a fission yeast gene related to the Bloom’s and Werner’s syndrome genes, is required for reversible S phase arrest. EMBO J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Szankasi P, Smith GR. A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- Szankasi P, Heyer WD, Schuchert P, Kohli J. DNA sequence analysis of the ade6 gene of Schizosaccharomyces pombe: Wild-type and mutant alleles including the recombination hotspot allele ade6-M26. J Mol Biol. 1988;204:917–925. doi: 10.1016/0022-2836(88)90051-4. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange Y, Horio T, Shimanuki M, Ding DQ, Hiraoka Y, Niwa O. A novel fission yeast gene, tht1+, is required for the fusion of nuclear envelopes during karyogamy. J Cell Biol. 1998;140:247–258. doi: 10.1083/jcb.140.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavassoli M, Shayeghi M, Nasim A, Watts FZ. Cloning and characterization of the Schizosaccharomyces pombe rad32 gene: a gene required for repair of double strand breaks and recombination. Nucleic Acids Res. 1995;23:383–388. doi: 10.1093/nar/23.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonami Y, Murakami H, Shirahige K, Nakanishi M. A checkpoint control linking meiotic S phase and recombination initiation in fission yeast. Proc Natl Acad Sci USA. 2005;102:5797–5801. doi: 10.1073/pnas.0407236102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tornier C, Bessone S, Varlet I, Rudolph C, Darmon M, Fleck O. Requirement for Msh6, but not for Swi4 (Msh3), in Msh2-dependent repair of base-base mismatches and mononucleotide loops in Schizosaccharomyces pombe. Genetics. 2001;158:65–75. doi: 10.1093/genetics/158.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- van den Bosch M, Zonneveld JB, Vreeken K, de Vries FA, Lohman PH, Pastink A. Differential expression and requirements for Schizosaccharomyces pombe RAD52 homologs in DNA repair and recombination. Nucleic Acids Res. 2002;30:1316–1324. doi: 10.1093/nar/30.6.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin JB, Bailey JP. The M26 hotspot of Schizosaccharomyces pombe stimulates meiotic ectopic recombination and chromosomal rearrangements. Genetics. 1998;149:1191–1204. doi: 10.1093/genetics/149.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls WP, Smith GR. A heteromeric protein that binds to a meiotic homologous recombination hotspot: correlation of binding and hotspot activity. Genes Dev. 1994;8:1693–1702. doi: 10.1101/gad.8.14.1693. [DOI] [PubMed] [Google Scholar]
- Wang SW, Read RL, Norbury CJ. Fission yeast Pds5 is required for accurate chromosome segregation and for survival after DNA damage or metaphase arrest. J Cell Sci. 2002;115:587–598. doi: 10.1242/jcs.115.3.587. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yamada T, Mizuno K, Hirota K, Kon N, Wahls WP, Hartsuiker E, Murofushi H, Shibata T, Ohta K. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, West RR, McIntosh JR, Hiraoka Y. A cytoplasmic dynein heavy chain is required for oscillatory nuclear movement of meiotic prophase and efficient meiotic recombination in fission yeast. J Cell Biol. 1999;145:1233–1249. doi: 10.1083/jcb.145.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A, Yamamoto M. Fission yeast Num1p is a cortical factor anchoring dynein and is essential for the horse-tail nuclear movement during meiotic prophase. Genetics. 2006;173:1187–1196. doi: 10.1534/genetics.105.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobayashi S, Yamamoto M, Watanabe Y. Cohesins determine the attachment manner of kinetochores to spindle microtubules at meiosis I in fission yeast. Mol Cell Biol. 2003;23:3965–3973. doi: 10.1128/MCB.23.11.3965-3973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JA, Schreckhise RW, Steiner WW, Smith GR. Meiotic recombination remote from prominent DNA break sites in S. pombe. Mol Cell. 2002;9:253–263. doi: 10.1016/s1097-2765(02)00452-5. [DOI] [PubMed] [Google Scholar]
- Young JA, Hyppa RW, Smith GR. Conserved and nonconserved proteins for meiotic DNA breakage and repair in yeasts. Genetics. 2004;167:593–605. doi: 10.1534/genetics.103.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]