Abstract
Background/Aims
Studies of trace metals in drinking water and tooth enamel have suggested a caries-promoting potential for manganese (Mn). Additionally, Mn has been shown to be essential for the expression of mutans streptococci virulence factors such as the glucan-binding lectin (GBL) of Streptococcus sobrinus. The Streptococcus mutans glucan-binding protein (Gbp) GbpC is the functional analogue of the S. sobrinus GBL. S. mutans Gbps have been shown to contribute to biofilm architecture and virulence. This study was undertaken to examine the effects of Mn on the transcription of genes encoding S. mutans Gbps, including gbpC, along with other critical S. mutans virulence genes.
Methods
Microarray analyses suggested the potential for an Mn effect on Gbp genes. Further investigation of the Mn effects on selected genes was undertaken by performing Northern blots, Western blots, and RT-PCR under conditions of planktonic and biofilm growth in Mn-depleted media or in media containing 50 μM Mn.
Results
Mn resulted in increased expression of gbpC and gtfB, and decreased expression of wapA, in both planktonic and biofilm cultures. The expression levels of gbpA and gbpD were also decreased in the presence of Mn, but only in biofilms. The expression of gtfC was increased in the presence of Mn only in planktonic cultures. The spaP gene was expressed more highly in Mn-supplemented planktonic cultures but less in Mn-supplemented biofilms.
Conclusion
Mn availability affects the expression of multiple S. mutans genes involved in adhesion and biofilm formation. Furthermore, these effects depend on the growth state of the organism.
Key Words: Biofilm, Manganese, Streptococcus mutans virulence
Streptococcus mutans is known to cause dental caries by colonizing in high proportions within localized areas of dental plaque, excreting lactic acid as the main byproduct of fermentation, and being able to efficiently adapt to a lower pH environment [Marsh, 1994]. The adherence process necessary to establish colonization may be by one of two different mechanisms. Sucrose-independent adhesion involves both specific [Lee et al., 1989] and nonspecific [Gibbons and Etherden, 1983] interactions with the complex layer of salivary glycoproteins in the acquired enamel pellicle bound to the tooth surface. SpaP (Ag I/II, AgB, SR, Pac IF, MSL-1) and WapA are S. mutans proteins that have been extensively studied for their roles in sucrose-independent adhesion. Sucrose-dependent adhesion relies on the synthesis of extracellular glucan polymers from sucrose by the action of glucosyltransferase (Gtf) enzymes. S. mutans possesses three different Gtfs, encoded by gtfB, gtfC, and gtfD, each synthesizing unique proportions of water-soluble and -insoluble glucan polymers. The glucans, in concert with glucan-binding proteins (Gbps: GbpA, GbpB, GbpC, and GbpD), promote tenacious adhesion and accumulation on tooth surfaces [Banas and Vickerman, 2003; Burne, 1998; Loesche, 1986].
The attributes of each of the Gbps are still under investigation. It is known that both GbpB and GbpC are part of the cell wall. GbpB is perhaps the most unique among the Gbps. It may function as a peptidoglycan hydrolase and be necessary for cell wall cycling and synthesis [Mattos-Graner et al., 2000, 2001, 2006]. GbpC acts as a surface receptor for glucan and is responsible for dextran-dependent aggregation [Sato et al., 1997]. Both GbpA and GbpD are secreted Gbps that share sequence similarity in their glucan-binding domains with the glucan-binding domains of the Gtfs [Banas et al., 1990; Shah and Russell, 2004]. Both GbpA and GbpC have been reported to influence S. mutans virulence [Hazlett et al., 1998; Matsumura et al., 2003] and Gbps A, C, and D have been shown to contribute to sucrose-dependent biofilm architecture [Hazlett et al., 1999; Lynch et al., 2007; Shah and Russell, 2004]. The GbpD also possesses lipase activity [Shah and Russell, 2004].
The S. mutans GbpC is a functional analogue of the glucan-binding lectin, which is responsible for dextran-dependent aggregation in two other members of the mutans streptococci, Streptococcus criceti and Streptococcus sobrinus [Liang et al., 1989]. Mn was found to be essential for the expression of glucan-binding lectin [Drake et al., 1988]. A role for Mn in bacterial pathogenesis is being recognized for an increasing number of species [Kehres and Maguire, 2003; Papp-Wallace and Maguire, 2006]. This role may take the form of protection from reactive oxygen species via Mn-superoxide dismutase, of a cofactor for metabolic enzymes, or interaction with transcriptional regulators [Jakubovics and Jenkinson, 2001; Martin et al., 1986; Zaharik and Finlay, 2004]. Mutation of Mn transport genes has been linked to a reduced ability of Streptococcus pneumoniae to cause pneumonia in mice [Johnston et al., 2006] and a loss of S. mutans virulence in a rat model of endocarditis [Kitten et al., 2000; Paik et al., 2003]. Multiple studies have suggested a cariogenic potential for Mn [Adkins and Losee, 1970; Beighton 1982, 1983; Glass et al., 1973] but the basis for this association is still uncertain. A recent study [Rolerson et al., 2006] found that the S. mutans SloR metalloregulator for an Fe3+-Mn transport operon could positively regulate the transcription of multiple virulence genes, including gbpB, spaP, and gtfB. Our accompanying study [Arirachakaran et al., 2007] revealed that Mn could affect growth of S. mutans, both at high and low concentrations, and that the presence or absence of Mn affected biofilm formation. Taken together, these observations support the possibility that Mn affects the regulation of S. mutans Gbps, particularly GbpC. This study was undertaken to test this hypothesis.
Materials and Methods
Bacterial Strains and Media
S. mutans serotype c strain UA159 was used throughout the study. The bacteria were stored in brain-heart infusion broth containing 15% glycerol at −80°C. Bacteria were cultured in modified chemically defined medium (SCDM) originallydeveloped by [Terleckyj and Shockman 1975]. Amino acids were replaced by casein hydrolysate (2 g/l). Based on the percentage of individual amino acids in the casein hydrolysate (reference guide of 2001 product catalog for Microbiology of Difco Laboratories, Detroit, Mich., USA), cysteine, glutamic acid, and leucine were added to the medium to a concentration of 200, 30 and 10 mg/1, respectively, to enhance the growth rate. Glucose was added at a concentration of 0.8%. The medium was treated with Chelex 100 (Sigma, USA) to reduce trace metal contaminations and then supplemented with high-purity trace metal salts to provide the optimal concentrations required for maximal growth of the microorganism. Calcium, iron, and magnesium were added to final concentrations of 50, 3.6 and 126 μM, respectively [Aranha et al., 1982, 1986]. When desired, MnSO4 at a final concentration of 50 μM was added. All glassware used was cleaned with 70% nitric acid and rinsed 3 times with distilled water and 3 times with deionized water.
Culture Conditions
Inocula were prepared by serial subculture in Mn-depleted media. Two hundred milliliters of freshly prepared, prewarmed growth media were placed aseptically into Erlenmeyer flasks and then inoculated with 1% of the inocula from cells growing in mid-exponential phase. Cultures were incubated at 37°C in an anaerobic chamber (Forma Scientific, USA) throughout the study unless specified. For isolation of RNA and protein extraction from planktonic cultures, the bacteria were grown to an optical density of 0.2 (early exponential phase) at wavelength 540 nm and then split into two cultures of 100 ml each. Fifty micromolar Mn was added to one of these. The cultures were further incubated for 2 h before collecting. When biofilm bacteria were to be collected, 70 μl of inocula were added to each well of a 24-well polystyrene dish that contained 1.5 ml (4.7% inoculum) 5% sucrose in SCDM with or without Mn. An artificial saliva coating of the wells was prepared as previously described [Landa et al., 1997; Russell and Coulter, 1975]. Briefly, 1 g Lab Lemco (Oxoid), 2 g yeast extract, 2.5 g mucin (Sigma, USA), 0.35 g NaCl, 0.2 g KCl and 0.2 g CaCl2 were mixed well in 1 liter deionized H2O and treated with Chelex 100 at 4°C for 1 h before 0.35 g NaCl, 0.2 g KCl, and 0.2 g CaCl2 were added. This suspension was filter-sterilized and stored at 4°C. When used, the artificial saliva was pipetted into wells, allowed to dry, and UV-sterilized. Prewarmed media, with or without Mn, was then added along with the bacterial inocula. The biofilms were incubated overnight in an anaerobic chamber at 37°C on a slow rotating platform (approximately 5 rpm). After 24 h the spent medium was aspirated and prewarmed new medium was added and incubated for another 1 h. For RNA isolation, the biofilm bacteria were dislodged by sonication (sonic dismembrator 60, Fisher Scientific) and the cells collected by centrifugation at 4,000 rpm for 20 min at 4°C. The bacteria were then washed in phosphate-buffered saline (PBS) and resuspended in RNase-free H2O and stored at −80°C until used.
Protein Extraction
Cell pellets were collected by centrifuging the bacterial cultures at 6,000 rpm at 4°C for 10 min. The supernatant was collected and saved for further protein extraction. The pellets were then resuspended in PBS, transferred to 1.5-ml microtubes, and centrifuged at 14,000 rpm for 5 min at 4°C. The resulting supernatant was discarded, and the pellet resuspended in 75 μl ×4 cracking buffer (1.5 ml 0.5 M Tris HCl, pH 6.8, 1 ml 20% SDS, 0.5 ml β-mercaptoethanol, 3 ml 100% glycerol, 4 ml H2O, bromphenol blue) and 75 μl deionized water. The mixture was incubated at room temperature for 2 h, periodically vortexed, then centrifuged at 10,000 rpm at 4°C for 5 min, and the supernatant transferred to a new 1.5-ml microtube for storage at −20°C until all samples were ready for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblot.
The original culture supernatant was further clarified by centrifugation at 12,000 g for 20 min at 4°C. Protein was precipitated by adding 10 ml 100% trichloroacetic acid, thoroughly mixing with the supernatant, and letting stand for at least 3 h, or as long as overnight, at 4°C. The mixture was centrifuged at 18,000 g for 30 min at 4°C to collect the protein precipitate. The pellets were washed twice in ice-cold acetone and spun down at 14,000 g, dried at 70°C for 5 min, and then resuspended in 250 μl lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.0% Triton X-100, 0.5% Na-deoxycholate). Samples were used immediately or stored at −80°C.
Western Immunoblot
Equal amounts of cell-associated and secreted proteins from planktonic cultures (Quick Start Bradford Protein Assay, Bio-Rad) were resolved by SDS-PAGE. Proteins were transferred onto nitrocellulose membrane (Bio-Rad) using the semidry electroblotter (model HEP-3, Owl Separation Systems) at 80 mA for 1.5 h. The membranes were blocked using 5% skim milk in PBS (pH 6.4) with 2% Tween 20 (PBST) for 1 h at room temperature, then washed twice in PBST for 10 min per wash. Rabbit polyclonal antibody against the glucan-binding domain of GbpA was added at a dilution of 1:5,000 in PBST and incubated overnight. The membranes were then washed 4 × 5 min in PBST. Membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G diluted 1:10,000 in PBST for 30 min at room temperature, and then washed 4 × 5 min in PBST. Signals were developed using Supersignal West Pico Chemiluminescent Substrate (Pierce). Working solutions of the substrates were prepared according to the manufacturer's instructions and added to the membranes for 1 min. Each membrane was exposed to CL-XPosure Film (Pierce) for 30 s.
Extraction of Total RNA
RNA samples were extracted by a hot-phenol method [Sambrook and Russell, 2001] with some modifications. Briefly, frozen bacterial pellets were thawed on ice and centrifuged. Equal amounts (500 μl) of acid phenol-chloroform (pH 4.7, Ambion Inc., USA) and NAES buffer (50 mM sodium acetate, pH 5, 10 mM EDTA, 10% SDS in diethyl pyrocarbonate-treated water) were used to resuspend and lyse the cell pellet. The lysate was transferred into an ice-cold screw-capped cryovial tube stored at −20°C containing 500 μl of 0.1-mm-diameter RNase-free zirconia beads (Biospec Products, Inc., Bartlesville, Okla., USA). Cell disruption was performed by a Mini-Beadbeater-8 cell disruptor (Biospec Products). The lysate was extracted twice with phenol:chloroform (pH 4.7). After centrifugation, the aqueous phase was collected and precipitated with isopropanol and 3 M sodium acetate, pH 5.5 (Ambion). The nucleic acid was collected by centrifugation and washed in 70% ice-cold ethanol, and suspended in diethyl pyrocarbonate (MP Biomedicals, LLC, Ohio, USA) -treated water. The RNA was further treated with RNase-free DNAse I (Ambion); a second treatment was done when necessary as recommended by the manufacturer. RNA was further purified by means of an RNeasy Minelute cleanup column (QIAGEN, Hilden, Germany). Purified RNA was eluted from the column with a final volume of 26 μl of RNase-free water and stored at −80°C until use. RNA quality and quantity were assayed by agarose gel electrophoresis, Bioanalyzer 2100 using RNA LabChips (Agilent Technologies) and by Biophotometer 6131 (Eppendorf) (A260 nm/A280 nm 1.9–2.1). The absence of DNA was verified by polymerase chain reaction (PCR) using primers specific for the S. mutans gene.
Microarrays
A custom Affymetrix array representing antisense oligonucleotides of 17 perfect match and 17 mismatch probes for each gene was manufactured based on the sequence for S. mutans strain UA159 [Ajdić et al., 2002]. The antisense probes were 25mers; the mismatch control probes were identical to the perfect match probes with the exception of a single base difference in the central position. The presence of the mismatched oligonucleotide allowed cross-hybridization and local background to be estimated and subtracted from the perfect match signal. A probe pair was called positive when the intensity of the perfect match probe cell was significantly greater than the corresponding mismatch probe cell. A probe pair was called negative if the situation was reversed. Each probe was tiled in approximately 1 million copies per spot, targeting 1,963 ORFs. To assure minimal cross-hybridization of the probes, genomic repetitive sequences (rRNA, IS elements, transposases and gene duplications) were used for elimination of the nondesired probes. These probes were designed using Affymetrix probe-selection software.
RNA samples from three independent experiments were processed by the array facility at the New York State Department of Health Microarray Core Facility using hybridization, washing, and scanning protocols described by Affymetrix (GeneChip Expression Analysis, 2004). Briefly, biotin-labeled cDNA was prepared from purified RNA samples using GeneChip DNA Labeling Reagent (Affymetrix, 900542) following fragmentation. The nucleic acid was fluorescently labeled by incubating with 10 μg/ml streptavidin-phycoerythrin (Molecular Probes, Eugene, Oreg., USA) and 2 mg/ml bovine serum albumin in 1× MES (100 mM MES, 1 M NaCl, 20 mM EDTA, 0.01% Tween 20, pH 6.6). After the streptavidin solution was removed, an antibody mix was added as the second stain containing 0.1 mg/ml goat-IgG, 5 μg/ml anti-streptavidin antibody and 2 mg/ml bovine serum albumin in ×1 MES.
The arrays were scanned according to manufacturer protocols and analyzed with GeneChip Operating Software (Affymetrix) and GeneSpring GX v7.3 (Agilent Technologies). Data were scaled using the proportional variance RMA method (http://discover.nci.nih.gov/microarrayAnalysis/Affymetrix.Preprocessing.jsp), then normalized to the 50th percentile using GeneSpring per chip normalization, followed by per gene normalization to specific samples. The values for the Mn-depleted samples were set to 1 and the fold change calculated for the Mn-supplemented samples.
Northern Blot Analysis
Glucan-binding protein A (gbpA), glucan-binding protein C (gbpC) and gyrA probes were generated using PCRs obtained with primers shown in table 1. The probes were labeled by digoxigenin-11-uridine-triphosphate (Roche Diagnostics GmbH, Germany). Probe concentrations were determined by immunological detection of dot blotted dilutions of the probe versus control DNA. Northern blot analyses of gbpC and gbpA gene transcription were carried out with 10-μg aliquots of total RNA isolated from S. mutans UA159 strains collected under the conditions described above. Samples were performed in triplicate. The RNA was separated on a 1.2% agarose-formaldehyde denaturing gel (RNA ladder, 0.24–9.5 kb, Invitrogen). The gel was rinsed in ×20 SSC (3 M NaCl, 0.3 M sodium acetate, pH 7.0) for 2 × 15 min to remove formaldehyde from the gel. RNA was transferred to a nylon membrane, positively charged (Roche Applied Science, Germany), using ×20 SSC overnight. After transfer, the membrane was fixed by UV cross-linking using a Spectrolinker XL-1000 (Spectronics Corp.). Hybridization was carried out at 50°C using Dig Easy Hyb (Roche Applied Science, Germany) in a hybridization incubator (Barnstead International, Melrose Park, Ill., USA). Following 18 h of hybridization, the membrane was rinsed twice for 30 min at room temperature with 37°C ×2 SSC (pH 7.0), 0.5% SDS, then twice for 30 min in ×2 SSC (pH 7.0), 0.1% SDS, and for 20 min in ×0.1 SSC, 0.1% SDS. Next the membrane was washed in washing buffer containing maleic acid buffer (0.1 M maleic acid, 0.15 M NaCl, pH 7.5) and 0.3% Tween 20 for 2 min. The membrane was then incubated in blocking solution (10% maleic acid buffer) for 60 min at room temperature. Anti-digoxigenin antibody tagged with alkaline phosphatase was added at a dilution of 1:10,000 and incubated at room temperature for 30 min. The membrane was washed twice in washing buffer, and transferred to equilibration buffer. CSPD (Roche Applied Science) was added as the substrate and the membrane was exposed to X-OMAT film (Eastman Kodak, Rochester, N.Y., USA). The films were analyzed with the Image J image processing program (NIH: rsb.info.nih.gov/ij/).
Table 1.
PCR primers used in this study
| Gene ID | Primer | Sequence | Amplicon, bp |
|---|---|---|---|
| SMU.2112 | gbpA F | 5′-CGCCAATAGTTCTCCAGCCGAT-3′ | 410 |
| gbpA R | 5′-CGAACCAGCGACTGCTGCA-3′ | ||
| SMU.1396 | gbpC F | 5′-GCCATTATGAGTCTCTCATCG-3′ | 478 |
| gbpC R | 5′-GTCACTGGAGGAACTTCCT-3′ | ||
| SMU.772 | gbpD F | 5′-CATGCTGGTGCAATGGTAAC-3′ | 401 |
| gbpD R | 5′-TTCTTCTCACCGCCAATAGC-3′ | ||
| SMU.1004 | gtfB F | 5′-CAGTTGACAAAACTTCTGAAGC-3′ | 347 |
| gtfB R | 5′-TCAACATGCTCAAAGCTCTG-3′ | ||
| SMU.1005 | gtfC F | 5′-GCTTCTGGGTTCCAAGCTAA-3′ | 379 |
| gtfC R | 5′-GGCGCTGTCCATTAACAACT-3′ | ||
| SMU.610 | spaP F | 5′-TCAGGCTGAACTGAAACGTG-3′ | 386 |
| spaP R | 5′-TAGCATTCTCATTGCGTTGC-3′ | ||
| SMU.987 | wapA F | 5′-TCCAGGATCCAGTAACAACG-3′ | 375 |
| wapA R | 5′-GTTGTCGGAACATTCGTTTGA-3′ | ||
| SMU.1509 | rgg F | 5′-TGCTGCCAATGATTTCCAT-3′ | 383 |
| rgg R | 5′-GACGTCGATTTCGAGGTATTTC-3′ | ||
| SMUr04 | 16S rRNA F | 5′-GGGCTTAGTGCCGGAGCTA-3′ | 60 |
| 16S rRNA R | 5′-TTTCAACCTTGCGGTCGTACT-3′ | ||
| SMU.1114 | gyrA F | 5′-CAACCATTAATTCTGTTCGGC-3′ | 455 |
| gyrA R | 5′-CTATTGAGAAGGGTGTCCC-3′ |
Reverse Transcription (RT)-PCR
RNA was isolated and cleaned as described above. Duplicate and triplicate samples were prepared from planktonic and sucrose-independent biofilm growth, respectively. The cDNA was synthesized and amplified according to the manufacturer's protocol (cMaster RTplus PCR system, Eppendorf, USA). Briefly, template RNA was serially diluted from 1 μg to 250 pg. Initial RT was at 50°C for 30 min followed by heating to 94°C for 2 min. Following this step, the subsequent PCR reaction was performed in a total reaction of 20 μl. The amplification program was 25 cycles of denaturation at 94°C for 15 s, annealing (54–58°C; table 1) for 20 s, and extension at 68°C for 1 min. The final elongation was at 68°C for 7 min. The PCR products were run on a 1% agarose gel. Data analysis was performed with the Image J image processing program (NIH: rsb.info.nih.gov/ij/) based on comparison between RNA extracted from the Mn+ and Mn– media, using the 16S rRNA as the control.
Adherence Assay
Planktonic bacteria from early exponential phase (optical density 0.2) were divided equally into Mn+ or Mn– cultures and incubated for 2 h at 37°C. The cells were pelleted, washed, and resuspended in KCl buffer, pH 6.8. The cells were then resuspended to an optical density of 0.8 at 540 nm. One milliliter of each cell suspension was added into saliva-coated wells prepared by modification of the protocol of Vickerman and Jones [1995]. Briefly, freshly collected saliva from a male subject was centrifuged at 15,000 g at 4°C for 15 min. The supernatant was transferred into a new 50-ml tube and incubated at 60°C for 60 min, followed by centrifugation at 15,000 g at 4°C for 15 min [Vickerman and Jones, 1995]. The supernatant was collected and stirred with Chelex 100 at 4°C for 1 h, filter-sterilized and stored at −80°C. When used, the saliva stock was diluted with KCl buffer 1:4, pipetted into 24 microtiter wells, allowed to dry and UV-sterilized. The culture was incubated anaerobically at 37°C for 1 h, then aspirated and washed in KCl buffer solution. After a second aspiration, an equal volume of KCl buffer was added. The adhered bacteria were then removed by sonication. Bacteria were serially diluted and plated on brain-heart infusion agar. Colony-forming units were counted and expressed as the percentage of input cells.
Results
Microarrays were performed in triplicate using RNA isolated from planktonic cultures grown in Mn-depleted or Mn-supplemented media. While our main interest was the effect of Mn on the transcription of genes encoding Gbps, the arrays were performed to place the results in context and also determine if other known virulence genes were susceptible to Mn depletion. The array data indicated that gtfC (+1.4-fold; p = 0.014) and gbpC (+1.7-fold; p = 0.038) were expressed more highly under Mn-supplemented conditions than Mn-depleted conditions (full array results are given in online suppl. table 1, www.karger.com/doi/10.1159/000110883). The expression of wapA declined marginally (–1.3-fold; p = 0.112) and that of gtfB increased marginally (+1.4-fold; p = 0.243), though neither change was statistically significant. The strong induction of the sloABCR Mn transport operon (25-fold induction; p = 0.014) under conditions of Mn deprivation confirmed the utility of the experimental design and function of this operon [Paik et al., 2003; Rolerson et al., 2006]. While the array data provided an estimation of changes in gene transcription, we chose to confirm the influence of Mn on selected virulence genes using both planktonic and biofilm cultures and independent means of verifying transcriptional changes.
Since antisera against GbpA and GbpC were available, Western immunoblots were carried out to determine if differences in protein expression as a function of Mn availability could be detected. GbpA was detected in a concentrated culture supernatant fraction since this protein is secreted extracellularly. GbpC was extracted from the cell pellet since it is anchored to the cell wall. Figure 1 shows that the relative amounts of GbpA were similar whether the bacteria were grown with or without Mn. For GbpC, however, there was more protein associated with the bacteria grown in the presence of Mn.
Fig. 1.
Representative Western immunoblot from three independent experiments. Equal amounts of cell-associated (lanes 1–2) or secreted proteins (lanes 3–4) from planktonic cultures grown in Mn-supplemented or Mn-depleted media were resolved by SDSPAGE. Proteins were transferred onto nitrocellulose membranes and incubated with rabbit polyclonal antibody to GbpC or the glucan-binding domain of GbpA. The secondary antibody was goat anti-rabbit IgG conjugated to horseradish peroxidase. Signals were developed using chemiluminescent substrate and semiquantified by densitometry.
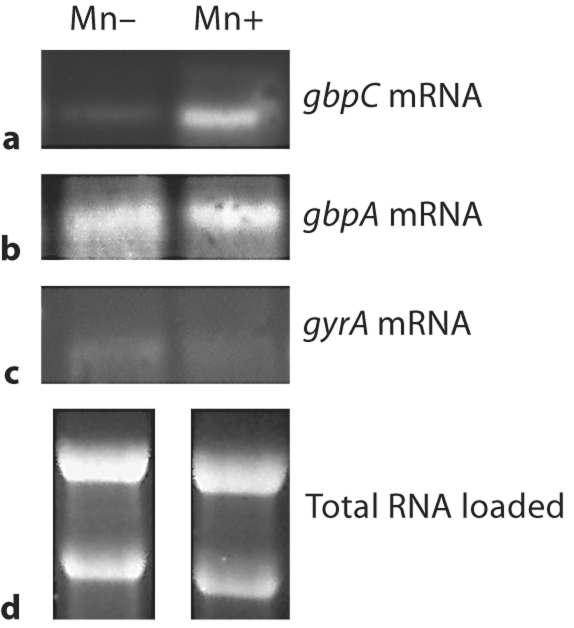
When gbpA and gbpC were further investigated at the transcriptional level by Northern blotting, the gbpC mRNA was significantly increased in bacteria grown in Mn-supplemented media (fig. 2). The gbpA mRNA still demonstrated no difference between the two conditions. The gyrA probe was included with the intention that this gene would serve as the control for normalization. However, it appeared that Mn availability affected gyrA expression. Therefore the amount of loaded RNA was calibrated by running 10 μg of total RNA on a 1.2% denaturing gel (fig. 2).
Fig. 2.
Northern blot of RNA collected from bacteria grown in Mn-supplemented or Mn-depleted media, separated by electrophoresis, blotted onto nitrocellulose and probed for gbpC (a), gbpA (b), or gyrA (c). d Loading control.
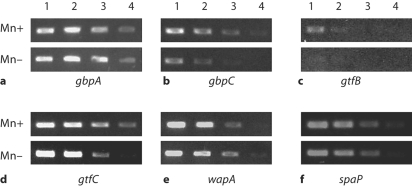
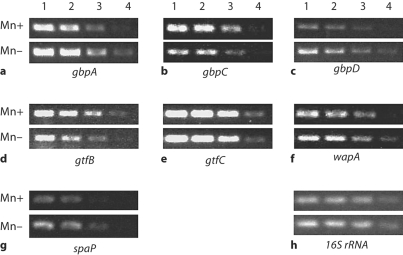
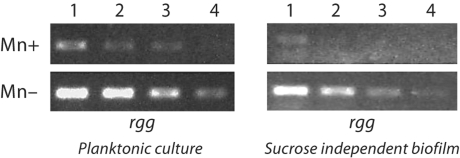
These results confirmed that Mn availability could have differential effects on S. mutans virulence genes. To more broadly examine the effects of Mn on virulence gene expression, a semiquantitative analysis using RT-PCR was performed on S. mutans cultures grown under planktonic or biofilm conditions. The results from the planktonic culture analysis agreed with those from the microarray experiments. The presence of Mn resulted in increased expression of gbpC, gtfB, and gtfC (fig. 3b–d). The expression of the spaP gene was slightly increased. Interestingly, however, the expression of wapA was decreased under Mn-supplemented growth conditions (fig. 3e). When S. mutans was grown in a biofilm culture on saliva-coated polystyrene, the effects of Mn were not always similar to those observed for planktonic cultures. The presence of Mn still resulted in increased expression of gtfB, slight increase in gbpC and decreased expression of wapA (fig. 4). But gtfC expression appeared to be unaffected, and the expression of gbpA, gbpD, and to a lesser extent spaP, all decreased under the Mn-supplemented conditions. Expression of rgg, the S. mutans homologue encoding the regulatory protein Rgg, was also decreased under Mn-supplemented conditions in both planktonic and biofilm cultures (fig. 5).
Fig. 3.
RT-PCR of RNA from planktonic cultures grown in Mn-supplemented or Mn-depleted media. Total RNA from cultures was serially diluted prior to amplification with gene-specific primers (as labeled in a–f) and represented in lanes 1 (most concentrated) to 4 (least concentrated). The PCR products were separated on a 1% agarose gel. The experiments were run in duplicate and, along with the microarray data, confirmed reproducibility. 16S RNA was run as a control.
Fig. 4.
RT-PCR of RNA from biofilm cultures grown in Mn-supplemented or Mndepleted media. Total RNA from biofilm bacteria was serially diluted prior to amplification with gene-specific primers (as labeled in a–h) and represented in lanes 1 (most concentrated) to 4 (least concentrated). The PCR products were separated on a 1% agarose gel. The experiments were run in triplicate to confirm reproducibility. 16S RNA was run as a control.
Fig. 5.
RT-PCR of RNA from either planktonic or biofilm cultures grown in Mn-supplemented or Mn-depleted media. Total RNA from bacteria was serially diluted prior to amplification with rgg-specific primers and represented in lanes 1 (most concentrated) to 4 (least concentrated). The PCR products were separated on a 1% agarose gel. The experiment was performed in triplicate to confirm reproducibility. 16S RNA was run as control.
The decreased expression of wapA when grown in Mn-supplemented media led us to examine whether S. mutans adherence to saliva-coated polystyrene was affected by Mn availability. Data from multiple trials (n = 9) of an adherence assay indicated that the bacteria grown under Mn-supplemented conditions adhered in a significantly lower percentage (3.04 ± 1.14) than organisms grown without Mn (14.54 ± 5.16) when compared by Student's t test (p < 0.001).
Discussion
Studies have suggested that Mn availability can influence caries potential [Adkins and Losee, 1970; Beighton, 1982, 1983; Glass et al., 1973]. Although the effects of Mn likely can be manifested in multiple ways, one plausible manner is to influence the expression of S. mutans genes encoding virulence factors. Previously, [Rolerson et al. 2006] concluded that SloR was a positive regulator of virulence genes such as ropA, spaP, gbpB, and gtfB, among others, and a negative regulator of sloC. This conclusion was based on gene expression levels in a sloR mutant accompanied by gel shift assays that showed binding of SloR to the promoter regions of spaP, sloABC, sloR, and ropA. The absence of negative regulation of the Fe3+-Mn transport operon sloABC should have resulted in a higher intracellular Mn concentration within the sloR mutant thereby creating a second variable in addition to the loss of SloR. In our study, the presence of Mn along with an intact SloR resulted in an increase in the expression of spaP and gtfB, and a decrease in sloABC expression. These results would be expected if Mn were acting as a cofactor for SloR as suggested by [Rolerson et al. 2006]. In contrast, both ropA and gbpB were slightly reduced in expression based on the array results. Therefore, it is possible that an increased Mn concentration was the primary basis for the decreased expression of ropA and gbpB observed by [Rolerson et al. 2006] in the sloR mutant.
The presence or absence of Mn affected the expression of Gbps as well as the virulence genes noted above. The microarray served its role as a screen for changes in transcription. Further analysis using Northern hybridization and RT-PCR confirmed array results, and also documented smaller changes in transcription that were not found to be statistically significant in the array analysis. Sucrose-related virulence genes generally showed higher or steady expression when grown in the presence of Mn either in a biofilm or in a planktonic state. The exceptions were gbpA and gbpD, which showed decreased expression within a biofilm. The GbpA and GbpD have been linked to biofilm elevation [Hazlett et al., 1999; Lynch et al., 2007]. Decreasing levels of these two extracellular Gbps might provide protection against building the height of the biofilm beyond its limits of cohesion. Expression of GbpC did not require Mn which was dissimilar to the functionally analogous glucan-binding lectin of S. sobrinus [Drake et al., 1988]. However, GbpC expression did increase in the presence of Mn compared to expression in the absence of Mn. Whether this increased expression of GbpC is sufficient to lead to increased virulence remains to be determined.
Our accompanying study [Arirachakaran et al., 2007] noted profound differences in mature non-sucrose biofilms grown in the presence or absence of Mn. Gilmore et al. [2003] reported that genes involved in Mn transport were among the most down-regulated in biofilms formed by Streptococcus gordonii thereby linking acquisition of Mn to biofilm maturation. These results may also be explained by an accumulation of Mn within a biofilm. In our accompanying paper [Arirachakaran et al., 2007] we measured dramatically higher concentrations of Mn in dental plaque when compared with salivary concentrations. The presence of Mn influenced the expression of two genes thought to contribute to sucrose-independent adhesion, wapA and spaP. In the planktonic state the spaP gene product was slightly up-regulated and the wapA gene down-regulated in the presence of Mn. These changes accompanied decreased sucrose-independent adherence of bacteria grown under Mn-supplemented conditions. The spaP gene product is often credited with promoting sucrose-independent adhesion and so its increased expression in conjunction with decreased adhesion is contrary to what would be predicted. While it is tempting to speculate that the down-regulation of wapA was responsible for the decreased adhesion, an effect by other genes cannot be ruled out. In particular, the product of the sloC gene is believed to function as an adhesin of the LraI family of lipoproteins in addition to acting as a cell surface ligand for metallic ions [Fenno et al., 1995; Paik et al., 2003]. The increased transcription of sloC in the absence of Mn may be primarily responsible for the clumping phenotype and changes in biofilm formation observed in our accompanying study. These changes mimic those described for a sloR mutant that also had increased expression of sloC [Rolerson et al., 2006]. In biofilm cultures the expression of spaP reversed and showed a slight decrease in the presence of Mn. It can be speculated that down-regulation of sucrose-independent adhesins within the biofilm may indicate they are not needed to maintain biofilm integrity. Alternatively, the relatively higher expression of WapA and SpaP in the absence of Mn may promote the likelihood of adhesion to nutritionally rich sites.
The data clearly suggest that Mn availability has an impact on the expression of S. mutans virulence factors. In some cases the effect may be direct (e.g. cofactoring SloR). But the microarray data revealed a Mn effect on an assortment of genes including the S. mutans homologue encoding the Rgg transcriptional regulator. Rgg is known to positively regulate gtfG expression in S. gordonii [Sulavik et al., 1992]. Therefore, the down-regulation of the S. mutans gtfB under Mn-depleted conditions may indicate that Rgg is necessary for parental levels of gtfB expression. However, the mechanism of regulation likely differs from that in S. gordonii, where parental expression levels of gtfG require the cis presence of rgg [Vickerman and Minick, 2002].
It is conceivable that there are several layers of regulation for S. mutans virulence genes, some of which may be gene-specific and others global. The altered expression of virulence and biofilm-related genes as a function of Mn availability may suggest that S. mutans has evolved means of responding to the nutritional state of its environment. Further investigation will be necessary to determine if host variations in salivary levels of Mn influence the epidemiology and virulence potential of select strains of S. mutans.
Acknowledgments
This research has been supported by grant DE10058 (J.A.B.) from the NIDCR, grant P20RR018741 (D.A.) and the Dental School Research Fund, Chulalongkorn University. We thank Dr. Yutaka Sato, Tokyo Dental College, for kindly providing antisera to GbpC.
References
- Adkins BL, Losee FL. A study of the covariation of dental caries prevalence and multiple trace element content of water supplies. NY State Dent J. 1970;36:618–622. [PubMed] [Google Scholar]
- Ajdić D, McShan WM, McLaughlin RE, Savić G, Chang J, Carson MB, Primeaux C, Tian R, Kenton S, Jia H, Lin S, Qian Y, Li S, Zhu H, Najar F, Lai H, White J, Roe BA, Ferretti JJ. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranha H, Evans SL, Arceneaux JE, Byers BR. Calcium modulation of growth of Streptococcus mutans. J Gen Microbiol. 1986;132:2661–2663. doi: 10.1099/00221287-132-9-2661. [DOI] [PubMed] [Google Scholar]
- Aranha H, Strachan RC, Arceneaux JE, Byers BR. Effect of trace metals on growth of Streptococcus mutans in a Teflon chemostat. Infect Immun. 1982;35:456–460. doi: 10.1128/iai.35.2.456-460.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arirachakaran P, Luengpailin S, Banas JA, Mazurkiewicz JE, Benjavongkulchai E. Effects of manganese on Streptococcus mutans planktonic and biofilm growth. Caries Res. 2007;41:497–502. doi: 10.1159/000110882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA, Russell RR, Ferretti JJ. Sequence analysis of the gene for the glucan-binding protein of Streptococcus mutans Ingbritt. Infect Immun. 1990;58:667–673. doi: 10.1128/iai.58.3.667-673.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas JA, Vickerman MM. Glucan-binding proteins of the oral streptococci. Crit Rev Oral Biol Med. 2003;14:89–99. doi: 10.1177/154411130301400203. [DOI] [PubMed] [Google Scholar]
- Beighton D. The influence of manganese on carbohydrate metabolism and caries induction by Streptococcus mutans strain Ingbritt. Caries Res. 1982;16:189–192. doi: 10.1159/000260596. [DOI] [PubMed] [Google Scholar]
- Beighton D. Manganese. In: Curzon MEJ, Cutress TW, editors. Trace Elements and Dental Disease. Postgraduate Dental Handbook Series. Littleton: Wright; 1983. pp. 237–244. [Google Scholar]
- Burne RA. Oral streptococci… products of their environment. J Dent Res. 1998;77:445–452. doi: 10.1177/00220345980770030301. [DOI] [PubMed] [Google Scholar]
- Drake D, Taylor KG, Doyle RJ. Expression of the glucan-binding lectin of Streptococcus cricetus requires manganous ion. Infect Immun. 1988;56:2205–2207. doi: 10.1128/iai.56.8.2205-2207.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenno JC, Shaikh A, Spatafora G, Fives-Taylor P. The fimA locus of Streptococcus parasanguis encodes an ATP-binding membrane transport system. Mol Microbiol. 1995;15:849–863. doi: 10.1111/j.1365-2958.1995.tb02355.x. [DOI] [PubMed] [Google Scholar]
- Gibbons RJ, Etherden I. Comparative hydrophobicities of oral bacteria and their adherence to salivary pellicles. Infect Immun. 1983;41:1190–1196. doi: 10.1128/iai.41.3.1190-1196.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore KS, Srinivas P, Akins DR, Hatter KL, Gilmore MS. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect Immun. 2003;71:4759–4766. doi: 10.1128/IAI.71.8.4759-4766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass RL, Rothman KJ, Espinal F, Velez H, Smith NJ. The prevalence of human dental caries and water-borne trace metals. Arch Oral Biol. 1973;18:1099–1104. doi: 10.1016/0003-9969(73)90083-6. [DOI] [PubMed] [Google Scholar]
- Hazlett KR, Mazurkiewicz JE, Banas JA. Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect Immun. 1999;67:3909–3914. doi: 10.1128/iai.67.8.3909-3914.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazlett KR, Michalek SM, Banas JA. Inactivation of the gbpA gene of Streptococcus mutans increases virulence and promotes in vivo accumulation of recombinations between the glucosyltransferase B and C genes. Infect Immun. 1998;66:2180–2185. doi: 10.1128/iai.66.5.2180-2185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubovics NS, Jenkinson HF. Out of the iron age: new insights into the critical role of manganese homeostasis in bacteria. Microbiology. 2001;147:1709–1718. doi: 10.1099/00221287-147-7-1709. [DOI] [PubMed] [Google Scholar]
- Johnston JW, Briles DE, Myers LE, Hollingshead SK. Mn2+-dependent regulation of multiple genes in Streptococcus pneumoniae through PsaR and the resultant impact on virulence. Infect Immun. 2006;74:1171–1180. doi: 10.1128/IAI.74.2.1171-1180.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehres DG, Maguire ME. Emerging themes in manganese transport, biochemistry and pathogenesis in bacteria. FEMS Microbiol Rev. 2003;27:263–290. doi: 10.1016/S0168-6445(03)00052-4. [DOI] [PubMed] [Google Scholar]
- Kitten T, Munro CL, Michalek SM, Macrina FL. Genetic characterization of a Streptococcus mutans LraI family operon and role in virulence. Infect Immun. 2000;68:4441–4451. doi: 10.1128/iai.68.8.4441-4451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa AS, van der Mei HC, Busscher HJ. Detachment of linking film bacteria from enamel surfaces by oral rinses and penetration of sodium lauryl sulphate through an artificial oral biofilm. Adv Dent Res. 1997;11:528–538. doi: 10.1177/08959374970110042201. [DOI] [PubMed] [Google Scholar]
- Lee SF, Progulske-Fox A, Erdos GW, Piacentini DA, Ayakawa GY, Crowley PJ, Bleiweis AS. Construction and characterization of isogenic mutants of Streptococcus mutans deficient in major surface protein antigen P1 (I/II) Infect Immun. 1989;57:3306–3313. doi: 10.1128/iai.57.11.3306-3313.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Drake D, Doyle RJ. Stability of the glucan-binding lectin of oral streptococci. J Dent Res. 1989;68:1677. [Google Scholar]
- Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50:353–380. doi: 10.1128/mr.50.4.353-380.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DJ, Fountain TL, Mazurkiewicz JE, Banas JA. Glucan-binding proteins are essential for shaping Streptococcus mutans biofilm architecture. FEMS Microbiol Lett. 2007;268:158–165. doi: 10.1111/j.1574-6968.2006.00576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- Martin ME, Byers BR, Olson MOJ, Salin ML, Arceneaux JEL, Tolbert C. Streptococcus mutans superoxide dismutase that is active with either manganese or iron as a cofactor. J Biol Chem. 1986;261:9361–9367. [PubMed] [Google Scholar]
- Matsumura M, Izumi T, Matsumoto M, Tsuji M, Fujiwara T, Ooshima T. The role of glucan-binding proteins in the cariogenicity of Streptococcus mutans. Microbiol Immunol. 2003;47:213–215. doi: 10.1111/j.1348-0421.2003.tb03389.x. [DOI] [PubMed] [Google Scholar]
- Mattos-Graner RO, Jin S, King WF, Chen T, Smith DJ, Duncan MJ. Cloning of the Streptococcus mutans gene encoding glucan binding protein B and analysis of genetic diversity and protein production in clinical isolates. Infect Immun. 2001;69:6931–6941. doi: 10.1128/IAI.69.11.6931-6941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos-Graner RO, Porter KA, Smith DJ, Hosogi Y, Duncan MJ. Functional analysis of glucan binding protein B from Streptococcus mutans. J Bacteriol. 2006;188:3813–3825. doi: 10.1128/JB.01845-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattos-Graner RO, Smith DJ, King WF, Mayer MP. Water-insoluble glucan synthesis by mutans streptococcal strains correlates with caries incidence in 12- to 30-month-old children. J Dent Res. 2000;79:1371–1377. doi: 10.1177/00220345000790060401. [DOI] [PubMed] [Google Scholar]
- Paik S, Brown A, Munro CL, Cornelissen CN, Kitten T. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport system required for endocarditis virulence and its Mn-dependent repressor. J Bacteriol. 2003;185:5967–5975. doi: 10.1128/JB.185.20.5967-5975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace KM, Maguire ME. Manganese transport and the role of manganese in virulence. Annu Rev Microbiol. 2006;60:187–209. doi: 10.1146/annurev.micro.60.080805.142149. [DOI] [PubMed] [Google Scholar]
- Rolerson E, Swick A, Newlon L, Palmer C, Pan Y, Keeshan B, Spatafora G. The SloR/Dlg metalloregulator modulates Streptococcus mutans virulence gene expression. J Bacteriol. 2006;188:5033–5044. doi: 10.1128/JB.00155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell C, Coulter WA. Continuous monitoring of pH and Eh in bacterial plaque grown on a tooth in an artificial mouth. Appl Microbiol. 1975;29:141–144. doi: 10.1128/am.29.2.141-144.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Sato Y, Yamamoto Y, Kizaki H. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect Immun. 1997;65:668–675. doi: 10.1128/iai.65.2.668-675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah DS, Russell RR. A novel glucan-binding protein with lipase activity from the oral pathogen Streptococcus mutans. Microbiology. 2004;150:1947–1956. doi: 10.1099/mic.0.26955-0. [DOI] [PubMed] [Google Scholar]
- Sulavik MC, Tardif G, Clewell DB. Identification of a gene, rgg, which regulates expression of glucosyltransferase and influences the Spp phenotype of Streptococcus gordonii Challis. J Bacteriol. 1992;174:3577–3586. doi: 10.1128/jb.174.11.3577-3586.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terleckyj B, Shockman GD. Amino acid requirements of Streptococcus mutans and other oral streptococci. Infect Immun. 1975;11:656–664. doi: 10.1128/iai.11.4.656-664.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman MM, Jones GW. Sucrose-dependent accumulation of oral streptococci and their adhesion-defective mutants on saliva-coated hydroxyapatite. Oral Microbiol Immunol. 1995;10:175–182. doi: 10.1111/j.1399-302x.1995.tb00139.x. [DOI] [PubMed] [Google Scholar]
- Vickerman MM, Minick PE. Genetic analysis of rgg-gtfG junctional region and its role in Streptococcus gordonii glucosyltransferase activity. Infect Immun. 2002;70:1703–1714. doi: 10.1128/IAI.70.4.1703-1714.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharik ML, Finlay BB. Mn2+ and bacterial pathogenesis. Frontiers Biosci. 2004;9:1035–1042. doi: 10.2741/1317. [DOI] [PubMed] [Google Scholar]