Abstract
Previous studies have shown that sucrose promotes changes in the composition of the extracellular matrix (ECM) of plaque-like biofilm (PLB), but its effect on protein expression has not been studied in vivo. Therefore, the protein compositions of ECM of PLB formed with and without sucrose exposure were analyzed by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). For this purpose, a crossover study was conducted during two phases of 14 days each, during which a volunteer wore a palatal appliance containing eight enamel blocks for PLB accumulation. In each phase, a 20% sucrose solution or distilled and deionized water (control) were extraorally dripped onto the blocks 8×/day. On the 14th day, the PLB were collected, the ECM proteins were extracted, separated by two-dimensional gel electrophoresis, digested by in-gel trypsin and MALDI-TOF MS analyzed. In the ECM of PLB formed under sucrose exposure, the following changes compared with the control PLB were observed: (1) the presence of upregulated proteins that may be involved in bacterial response to environmental changes induced by sucrose and (2) the absence of calcium-binding proteins that may partly explain the low inorganic concentration found in ECM of PLB formed under sucrose exposure. The findings showing that sucrose affected the ECM protein composition of PLB in vivo provide further insight into the unique cariogenic properties of this dietary carbohydrate.
Key Words: Dental plaque; Gel electrophoresis, two-dimensional; Mass spectrometry; Sucrose
Sucrose is the most cariogenic carbohydrate because it is acidogenic and serves as substrate for extracellular polysaccharide (EPS) synthesis by cariogenic bacteria. Previous studies have shown that this carbohydrate promotes biochemical (proteins, polysaccharides, ions) changes in the extracellular matrix (ECM) composition of plaque-like biofilm (PLB) [Cury et al., 2000]. Also, these effects are dependent on sucrose concentration [Aires et al., 2006], frequency of exposure [Ccahuana-Vásquez et al., 2007] and time of PLB formation [Vale et al., 2007]. The changes in the ECM composition of PLB are strongly associated with the transition from health to disease, and the carbohydrates are key environmental factors influencing the ECM composition [Cury et al., 2000; Ribeiro et al., 2005]. However, the composition of ECM proteins has not been explored, even though it could potentially play significant roles in the pathogenicity of PLB [as reviewed by Paes Leme et al., 2006].
Indeed, preliminary data have suggested that sucrose and its glucose and fructose moieties induce different changes in the ECM protein composition of PLB formed in situ [Cury et al., 2000], but the proteins were not identified. Other studies have explored in vitro the cellular protein composition of Streptococcus mutans to understand the physiological responses of this cariogenic bacteria under a variety of environmental conditions [Quivey et al., 2000; Svensäter et al., 2000; Wilkins et al., 2002; Welin et al., 2003; Len et al., 2004a, b]. However, little is known about the ECM protein composition of PLBs formed in experimental conditions that mimic saliva properties (salivary flow, buffer capacity, clearance, minerals and protein content), availability of nutrients as well as diversity, selection and competition of microbial species found in the oral cavity, which are relevant to explain the biological properties of PLBs formed in vivo.
Therefore, using proteomic approaches, we evaluated the protein composition of ECM of PLB formed with and without sucrose exposure, to increase our understanding of the cariogenic properties of this most consumed sugar. A preliminary report of major differences in the levels of calcium binding proteins has previously been reported [Paes Leme et al., 2006].
Materials and Methods
Experimental Design
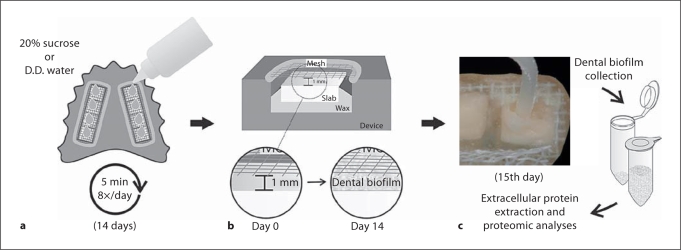
This study was approved by the Research and Ethics Committee of the Piracicaba Dental School (UNICAMP, Piracicaba, SP, Brazil), and involved a crossover in situ design [Cury et al., 1997] conducted in two phases of 14 days each. A healthy 26-year-old volunteer wore an acrylic palatal appliance containing eight human dental enamel blocks [Hara et al., 2003, for details] and 20% sucrose solution or distilled and deionized water (control) was extraorally dripped onto the blocks 8 times a day during each phase of 14 days (fig. 1a, b). The dental PLB formed on the enamel blocks were collected (fig. 1c) 10 h after the last exposure to treatment and subjected to protein extraction and analysis. At least two distinct experiments were performed for each treatment with samples obtained from the same volunteer.
Fig. 1.
Schematic illustration of the experimental design. D.D. = Distilled and deionized.
Extraction of ECM Protein of PLB
Extracellular proteins were extracted according to Cury et al. [2000]. The PLBs were treated with 50 μl of 0.1 N NaOH [Fox and Dawes, 1970] containing 1 mM EDTA [Iacono et al., 1982] for each milligram of PLB wet weight for 1 h at 0°C. The extracts were centrifuged (3,000 g) for 30 min at 4°C and 3 vol of ice-cold acetone were added to the supernatant to precipitate and concentrate the proteins extracted. After standing overnight at −20°C, the pellets were collected by centrifugation (3,000 g) at 4°C for 30 min and after evaporation of residual acetone the pellets were resuspended in 0.125 M Tris-HCl, pH 6.8 plus 0.25 ml of protease inhibitor cocktail (Calbiochem) per gram PLB and stored at −20°C. Contaminants present in the extract, mainly polysaccharide, were eliminated using the 2D clean up kit (GE Healthcare) and protein concentration was determined [Bradford, 1976].
2-Dimensional Electrophoresis and Image Analysis
The first-dimension (isoelectric focusing, IEF) and second-dimension electrophoresis were performed according to Bellato et al. [2004] using 20 μg of proteins for both experimental conditions. IEF was conducted with Immobiline Dry Strip pH 4–7 (18 cm). Strips were rehydrated for 8 h at 20°C with 400 μl IEF solution (8 M urea, 4% CHAPS, 70 mM DTT, 0.8% ampholytes and 0.006% bromophenol blue) in an IPGphor system (GE Healthcare) with current limit 50 μA/strip until focusing reached 70 kVh. After focusing, the proteins were reduced and alkylated by sequential incubation in the following solutions: (1) 20 mg/ml DTT in 0.05 M Tris-HCl, pH 8.4, 2% SDS, 30% glycerol, 6 M urea, 0.006% bromophenol blue and (2) 30 mg/ml iodoacetamide in the same buffer. Then, the strips were directly applied to 8–18% gradient polyacrylamide gels. Molecular-weight markers (Invitrogen) covering the 220- to 10-kDa range were applied at the basic end of the IPG strips. Electrophoresis was carried out for 12 h at 10°C. Following separation in the second dimension, the proteins were visualized with silver staining [Blum et al., 1987]. The experiment was repeated twice for each treatment to check reproducibility and all gels were run in duplicate.
Eight gel (two separate experiments with controls and sucrose treatment in duplicate) images were analyzed (ImageMaster 2D Platinum software, version 5.0; GE Healthcare) as follows. Observed masses for resolved proteins were calculated by comparing their mobility with that of the molecular weight markers, and pI values were calculated according to linearity of the IPG strips using the software. ImageMaster 2D Platinum detection parameters, such as number of smooth, saliency, and minimum area, were adjusted for every selected region of each gel to detect protein spots automatically. Subsequently, each protein spot received an identification number, which was confirmed visually. Spots found along the edges of the gels and streaked spots were not considered for further analysis. For each protein spot, the spot volume was calculated, according to the software manual, as above spot border situated at 75% of the spot height (measured from the peak of the spot), which permitted the automatic subtraction of background. The percentage volume of each spot was determined in relation to the total volume of all the spots in a gel. In order to evaluate the reproducibility between the duplicates and between independent experiments, the correlation coefficient was calculated according to the percent volume of paired spots [Huang, 2004]. Subsequently, a master gel image (average gel) was generated from matched gel sets for each condition by averaging the volume of the spots. Individual spot volumes were expressed as normalized volumes relative to the total detected spot volume. Spots of each master gel of PLB formed in the presence and absence of sucrose were matched and the percent volume of spots was compared. The protein spot levels were considered to have a greater or smaller volume when there was at least a 1.5-fold difference [Wilkins et al., 2002, 2003].
Protein Digestion and Mass Spectrometry Identification
For identification, protein spots were excised, destained [Gharahdaghi et al., 1999] and then digested with trypsin [Wilkins et al., 2001]. Tryptic peptides were analyzed using matrix- assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS; PerSeptive Biosystem Voyager DE-STR) operated in the reflection-delayed extraction mode. Mass lists were used to screen against database including Mascot (www.matrixscience.com) and Protein Prospector (University of California; www.prospector.ucsf.edu/ucsfhtml4.0/msfit.htm) programs using the National Center for Biotechnology Information nonredundant database (NCBI nr 2005.01.06). The parameters for Prospector and Mascot searching were: (1) maximum mass tolerance of 115 ppm, (2) one single miss cleavage, (3) carbamidomethylation of cysteines and (4) at least four peptides required to match. The candidate protein always possessed the highest MOWSE (molecular weight search) score [Pappin et al., 1993]. Since this study evaluated proteins in the PLB originating from human saliva and oral microorganisms, and since the genome of some microorganisms is not currently undergoing sequencing, it was necessary to select some phylogenetically similar species to perform this analysis [Wilkins et al., 2002; Len et al., 2003].
Results
Preliminary analyses using IPG strips between the pH ranges 3–10 and 4.5–5.5 revealed that most of the protein spots were concentrated within the pH range 4–7 (data not shown). Thus, subsequent analyses for the extracellular proteins extracted from the PLBs were evaluated within the pH range 4–7.
Examination of the original two-dimensional gel electrophoresis protein profiles showed 521.5 ± 7.3 and 512.0 ± 37.4 (mean ± SD; n = 2) automatically detected spots for PLBs formed in the absence (control) and presence of sucrose, respectively. The image master gels created by the software showed 445 spots detected in the control and 327 spots in sucrose-treated samples. The reproducibility of image gels was evaluated by comparing the percent volume of spots between gel duplicates and between independent experiments. The correlation coefficient between duplicates in the absence of sucrose was 0.904–0.957 and in the presence of sucrose it was 0.801–0.855. For independent experiments, the correlation coefficient was 0.701–0.964 in the absence of sucrose and 0.810–0.940 in the presence of sucrose. Since the main source of error associated with this form of quantification is biological variations, the variability in the number and percent volume of spots in this study was minimized by repeating the experiment with the same volunteer, and by running duplicate gels for the control and sucrose treatments.
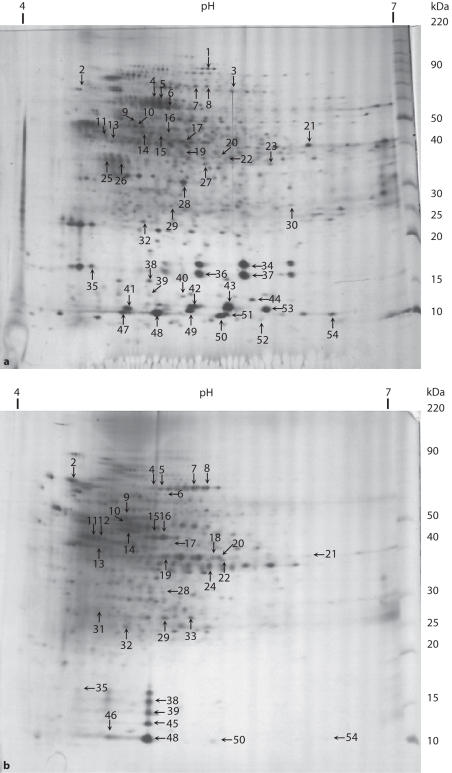
The comparative analysis based on the percent volume of spots of the PLB formed in the absence or presence of sucrose revealed 143 paired spots, of which 78 protein spots with larger volumes and 19 spots with smaller volumes in PLB formed in the presence of sucrose, while 46 protein spots showed a similar abundance in both conditions (fig. 2a, b; tables 1, 2). This comparative analysis also showed 302 and 184 nonpaired spots in the absence and presence of sucrose, respectively. The profiles of salivary and bacterial proteins in the matrix of the PLBs were qualitatively and quantitatively different depending on whether or not sucrose was applied during PLB formation (tables 1, 2).
Fig. 2.
Two-dimensional electrophoresis pattern of ECM proteins of PLB (20 μg) formed in the absence (a) or presence (b) of sucrose. ECM proteins were applied to 4–7 IPG strips followed by electrophoresis on 8–18% polyacrylamide gel. The gels were silver-stained. Spots indicated with numbers were identified by in-gel trypsin digestion and MALDI-TOF MS [reprinted with permission of Paes Leme et al., 2006].
Table 1.
Proteins assigned and their relative abundances in ECM of PLB formed in the presence of sucrose/control (fold change)
| Spot No.a | Protein assigned | Protein information resource database codeb | Sequence coveragec, % | Fold changed |
|---|---|---|---|---|
| 16 | Putative NADP-specific glutamate dehydrogenase | 24379360e/24377287f (B) | 38 | 36.33 |
| 38 | Prolactin-induced proteine | 51094526 (M) | 21 | 6.94 |
| 2 | DnaK proteine | 15900431 (B) | 15 | 6.67 |
| 19 | Putative transposasee | 19746041 (B) | 17 | 6.25 |
| 9 | Translation elongation factor TUe | 15903386 (B) | 31 | 5.40 |
| 15 | Putative NADP-specific glutamate dehydrogenase | 24379360e/24377287f (B) | 29 | 3.94 |
| 20 | Phosphotransferase system, mannose-specific EIIABe | 15902305 (B) | 14 | 3.75 |
| 7 | Pyruvate kinasee | 42519006 (B) | 27 | 3.63 |
| 29 | Amino acid ABC transporter, ATP-binding proteine | 15900712 (B) | 9 | 3.50 |
| 48 | Prolactin-induced proteine | 51094526 (M) | 45 | 3.26 |
| 39 | Prolactin-induced proteine | 51094526 (M) | 32 | 3.03 |
| 8 | Pyruvate kinasee | 42519006 (B) | 20 | 2.67 |
| 17 | Elongation factor Tu | 15599473e/9950496f (B) | 20 | 2.61 |
| 22 | Methionine synthase II (cobalamin-independent)e | 23002637 (B) | 15 | 2.43 |
| 32 | Hypothetical protein SMU.373e | 24378870 (B) | 23 | 1.65 |
| 10 | ATP synthase beta | 15600747e/9951894f (B) | 51 | 1.56 |
| 14 | Tuf | 38606905e/38606877f (B) | 32 | 1.56 |
| 5 | GroEL | 15599581e/576779f (B) | 25 | 1.26 |
| 28 | Translation elongation factor Ts | 46164365e/9949816f (B) | 51 | 1.19 |
| 50 | Prolactin-induced proteine | 51094526 (M) | 36 | 1.19 |
| 4 | GroEL | 15599581e/576779f (B) | 19 | 1.18 |
| 11 | Enolase (isoform 1) | 15900994e/15900994f (B) | 21 | 0.61 |
| 21 | Transporter, putativee | 34398072 (B) | 9 | 0.58 |
| 13 | Putative transposasee | 56707134 (B) | 15 | 0.54 |
| 35 | Putative ribonucleotide reductase (NrdI protein)e | 28896618 (B) | 25 | 0.51 |
| 54 | Prolactin-induced proteine | 51094526 (M) | 45 | 0.38 |
| 6 | GroEL | 15599581e/576779f (B) | 14 | 0.16 |
Refers to the proteins indicated in figure 2.
Proteins of bacterial (B) or mammalian (M) origin.
The percentage of amino acid coverage (peptides observed/theoretical number from sequence data given in database).
Fold change means the ratio of % volume of protein spot of PLB formed in the presence of sucrose/control; the expression is considered enhanced or diminished when the ratio is greater than 1.50- or lower than 0.66-fold, respectively [Wilkins et al., 2002, 2003].
Based on Protein Prospector program, the first candidate using a maximum of 115 ppm of mass tolerance.
Based on Mascot programs, the first candidate using a maximum of 115 ppm of mass tolerance.
Table 2.
Proteins exclusively identified in ECM of PLB formed with and without sucrose exposure
| Spot No.a | Conditions of PLB formation |
Protein information resource database codeb | Sequence coveragec, % | |
|---|---|---|---|---|
| with sucrose | without sucrose | |||
| 12 | Enolase (isoform 2)d | − | 15900994 (B) | 14 |
| 18 | Hypothetical protein Ip_0493d | − | 28377385 (B) | 11 |
| 24 | Enterotoxind | − | 49484068 (B) | 22 |
| 31 | Hypothetical cytosolic proteind | − | 19704518 (B) | 21 |
| 33 | Phosphoglycerate mutase Id | − | 23003585 (B) | 21 |
| 45 | Prolactin-induced proteind | − | 51094526 (M) | 22 |
| 46 | Prolactin-induced proteind | − | 51094526 (M) | 39 |
| 1 | − | Lipase precursord | 49484866 (B) | 6 |
| 3 | − | Putative type I restriction-modification system, specificity determinant; restriction endonucleased | 24379345 (B) | 7 |
| 23 | − | ATP-binding protein, putatived | 33390966 (B) | 12 |
| 25 | − | Hypothetical proteind | 34762127 (B) | 17 |
| 26 | − | Thioesterase domain containing 1d | 8922871 (M) | 19 |
| 27 | − | GTPased | 28378511 (B) | 18 |
| 30 | − | Hypothetical cytosolic proteind | 50914941 (B) | 27 |
| 34 | − | Calcium-binding protein 1 isoform 2d | 13929434 (M) | 18 |
| 36 | − | Calcium-binding protein 1 isoform 2d | 13929434 (M) | 18 |
| 37 | − | Calcium-binding protein 1 isoform 2d | 13929434 (M) | 14 |
| 40 | − | Hypothetical protein XP_498283d | 51467140 (M) | 38 |
| 41 | − | Putative chorismate mutased | 24379707 (B) | 32 |
| 42 | − | S100 calcium-binding protein A9d | 4506773 (M) | 39 |
| 43 | − | S100 calcium-binding protein A9d | 4506773 (M) | 48 |
| 44 | − | Nucleoside diphosphate kinase | 15599002d/9949980e (B) | 56 |
| 47 | − | Hypothetical protein SA V0447d | 15923437 (B) | 30 |
| 49 | − | S100 calcium-binding protein A9d | 4506773 (M) | 47 |
| 51 | − | S100 calcium-binding protein A9d | 4506773 (M) | 42 |
| 53 | − | S100 calcium-binding protein A9d | 4506773 (M) | 31 |
| 52 | − | S100 calcium-binding protein A9d | 4506773 (M) | 31 |
Refers to the proteins indicated in figure 2.
Proteins of bacterial (B) or mammalian (M) origin.
The percentage of amino acid coverage (peptides observed/theoretical number from sequence data given in database).
Based on Protein Prospector program, the first candidate using a maximum of 115 ppm of mass tolerance.
Based on Mascot program, the first candidate using a maximum of 115 ppm of mass tolerance.
Most of the spots were excised from the gels and submitted to in-gel trypsin digestion for identification by mass spectrometry. A total of 54 proteins were identified from the PLB (tables 1, 2). Gel image analysis showed that, in the PLB formed in the presence of sucrose, 17 proteins were more abundant, 6 were less abundant and 4 proteins showed similar abundance to the control group (table 1). Of the proteins identified, 20 were exclusively found in ECM from the control group and 7 when the PLB was formed in the presence of sucrose (table 2).
Database search revealed that the proteins which were identified in the PLB formed in the presence and absence of sucrose were mainly calcium-binding proteins (spots 34, 36, 37, 42, 43, 49, 51, 52 and 53), proteins related to binding properties (spots 23, 38, 39, 45, 46, 48, 50 and 54), proteins associated with stress conditions (spots 2, 4, 5 and 6), proteins related to protein biosynthesis (spots 9, 14, 17 and 28), energy metabolism (spots 7, 8, 10, 11, 12, 20, 29 and 33), amino acid biosynthesis (spots 15 and 16) and other proteins (fig. 2; tables 1, 2).
A number of isoforms were identified in both conditions and these data indicated that the presence of isoforms, in terms of both the number and their abundance, was altered under the two different conditions.
Discussion
The formation of acid end-products during fermentation of dietary sugars by acidogenic bacteria is considered the driving force to generate a cariogenic dental biofilm. However, the effect of this stressful acidic environmental condition on protein expression has been limited to in vitro studies using S. mutans and the analyses have also focused on cellular protein change. In the present study, we investigated in vivo the protein change in the ECM of PLB since sucrose, the most cariogenic dietary carbohydrate, promotes decreases in pH and is also a substrate for EPS synthesis. The ECM has an important role in biofilm structure and function [Sutherland, 2001].
To evaluate the protein composition of the ECM we used short alkali extraction at low temperature to avoid cellular membrane lysis [Fox and Dawes, 1970]. Alkali extraction of ECM components from PLB formed in the presence of sucrose is required to dissolve the high concentration of insoluble EPS usually found in this biofilm [Cury et al., 2000], allowing the extraction of substances that could be trapped in the matrix.
Therefore, the absence of calcium-binding proteins in the ECM of PLB formed under exposure to sucrose (table 2) should not be due to their being trapped in the insoluble EPS. Thus, the presence of calcium-binding proteins, exclusively in ECM of the PLB formed in the absence of sucrose, supports the view that these proteins, in addition to plaque Ca binding [Rose et al., 1993], may act as organic mineral reservoirs in PLB [Paes Leme et al., 2006, for a review]. However, calcium-binding proteins may have other functions which have not yet been reported. On the other hand, prolactin-induced protein was identified in both conditions; however, 2 isoforms were uniquely found and 3 of them were found in greater abundance in the matrix of PLB formed with sucrose (tables 1, 2).
It should be emphasized that several proteins found in the ECM of PLB formed in vivo, such as enolase, translation elongation factors, pyruvate kinase, GroEL, DnaK, and other proteins previously thought to be confined to the cytosol are associated with the cell surface or secreted into the external milieu [Joe et al., 1994; Pancholi and Fischetti, 1998; Len et al., 2003; Wilkins et al., 2003; Len et al., 2004a; Black et al., 2004; Nandakumar et al., 2005; Paddick et al., 2006]. Therefore, the presence of these proteins in the extracellular milieu may not have resulted from cellular lysis during extraction of the extracellular proteins.
Overall, our results showed that specific bacterial stress response proteins are differentially expressed in ECM of PLBs depending on the availability of sucrose (tables 1, 2; fig. 2a, b). These proteins have been found intracellularly in in vitro studies of S. mutans in response to environmental changes [Svensäter et al., 2000; Quivey et al., 2000; Wilkins et al., 2002; Welin et al., 2003; Len et al., 2004a, b], and in the present study they were found in the ECM. The extracellular presence of these proteins may be a consequence of cell death, considering that the PBL collected was 14 days old.
Proteins linked to carbohydrate metabolism, such as pyruvate kinase and mannose-specific EIIAB, were upregulated in PBL formed under sucrose exposure (table 1) in agreement with an in vitro report [Abranches et al., 2006]. Enolase was also identified, one of the isoforms being identified only in the presence of sucrose and the other being downregulated in the presence of sucrose, but the biological functions of the differential expression of the isoforms remain to be clarified. The presence of these proteins might be an indicator of enhanced carbohydrate metabolism in the PLB formed in the presence of sucrose.
The upregulation of ATP synthase beta chain in PLB formed under sucrose exposure (table 1) could reflect bacterial adaptation to acidic stress conditions imposed by eight decreases in pH per day. Even though this enzyme was found in ECM, the result agrees with the predominance of aciduric microorganisms in biofilms formed in situ [Pecharki et al., 2005; Ribeiro et al., 2005; Tenuta et al., 2006]. Also, the stress conditions promoted by low pH induced upregulation of DnaK in the PLB formed under sucrose exposure (table 1) in agreement with Jayaraman et al. [1997], who showed increased levels of dnaK mRNA and intracellular DnaK in S. mutans in a continuous chemostat culture in response to acid shock. Furthermore, most proteins involved in the translation function were also upregulated, such as the translation elongation factors (EF-Tu). Thus, in addition to their regular function in translation elongation, these proteins could behave like chaperones toward protein folding and protection from stress as previously reported in Escherichia coli [Caldas et al., 1998], which could explain their higher levels under the low pH conditions maybe caused by sucrose fermentation (table 1).
However, GroEL, another chaperone, behaved differently from DnaK, since it was downregulated in PLB formed in the presence of sucrose (table 1). This protein is elevated in the intracellular compartment of S. mutans subjected to an acidic environment [Wilkins et al., 2002; Len et al., 2004a]. Apparently, the different isoforms of GroEL found may be regulated by different pathways under different stress conditions as in our study; the control PLB was formed under nutrient-limited conditions, confirming a previous report [Len et al., 2003].
Interestingly, NADP-specific glutamate dehydrogenase was the protein most highly expressed under sucrose exposure (table 1). This enzyme is involved in the intracellular metabolism of amino acids [Wilkins et al., 2001, 2002]. The ammonia produced could be an alternative mechanism used by aciduric bacteria to increase the pH of acidified cytoplasm. The reason for the extracellular occurrence of this protein and its role in this location remain to be investigated, but the fact that it was 30-fold more highly expressed in PLB under sucrose exposure suggests that its presence in the ECM is not an artifact of cell lysis because the control PLB was extracted under the same conditions. It can be speculated whether this protein could act as an adhesin by binding to immobilized host and bacterial proteins through the glutamate-binding domain [Joe et al., 1994].
Many bacterial or salivary protein isoforms were identified in this study. However, neither the role nor the nature of different isoforms, which possibly resulted from processing by bacterial proteases or posttranslational modification, is clear yet. For instance, protein phosphatases, which are important in the phosphorylation process and signal transduction in other organisms [Mukhopadyay et al., 1999; Vijay et al., 2000], were previously found to be more abundant at low pH [Wilkins et al., 2003] and they could have an effect on the PLB formed under sucrose exposure.
It should be emphasized that the proteins found in the PLB could originate from the host and different bacteria; therefore, it was necessary to include several species such as Homo sapiens, Actinomyces, Fusobacterium nucleatum, Lactobacillus, Porphyromonas gingivalis, Prevotella intermedia, Streptococcus anginosus, Streptococcus equi, Streptococcus gordonii, Streptococcus mitis,S. mutans, Streptococcus oralis, Streptococcus pneumoniae, Streptococcus pyogenes, Streptococcus salivarius, Streptococcus sanguinis, Streptococcus sobrinus, Neisseria subflava, Veillonella parvula, and other species, such as Bacillus subtilis, Staphylococcus aureus, and also Pseudomonas aeruginosa to improve the protein identification. However, the fact that the majority of plaque bacteria are not represented in the sequence databases could explain the lack of the identification of many spots.
In summary, these results showed that sucrose induced marked changes in ECM protein composition of PLB formed in vivo. The data provide further insights into how the biochemical and microbiological changes induced by sucrose affect the ECM composition at a proteomic level. The composition of bacteria- and host-derived proteins in the ECM may be modulated by the availability of sucrose, but additional studies should be done using other dietary carbohydrates to check whether this effect is unique to this most cariogenic dietary carbohydrate. Also, in addition to the ECM composition, the intracellular or total protein composition of PLB formed in vivo should be evaluated as a control.
Acknowledgements
This study was supported by FAPESP (99/07185-7; 02/00293-3; 03/01536-0), CNPq (472392/03-4) and Protein Core Facility grant NIH RR14682. We thank Dr. Peter Baker of Protein Prospector and Dr. David Beighton of King's College London for their assistances with the Prospector program to help identify the proteins, Dr. Anderson T. Hara from the Indiana University for designing figure 1, and Dr. Solange M.T. Serrano for critically reviewing the manuscript. The manuscript was based on the thesis submitted by the first author to the Faculty of Dentistry of Piracicaba, UNICAMP, SP, Brazil, as a partial fulfillment of the requirements of the Doctorate Program in Dentistry, concentration in cariology.
References
- Abranches J, Candella MM, Wen ZT, Baker HV, Burne RA. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J Bacteriol. 2006;188:3748–3756. doi: 10.1128/JB.00169-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires CP, Tabchoury CP, Del Bel Cury AA, Koo H, Cury JA. Effect of sucrose concentration on dental biofilm formed in situ and on enamel demineralization. Caries Res. 2006;40:28–32. doi: 10.1159/000088902. [DOI] [PubMed] [Google Scholar]
- Bellato CM, Garcia AKM, Mestrinelli F, Tsai SM, Machado MA, Meinhardt LW. The induction of differentially expressed proteins of Xylella fastidiosa with citrus extract. Braz J Microbiol. 2004;35:235–242. [Google Scholar]
- Black C, Allan I, Ford SK, Wilson M, McNab R. Biofilm-specific surface properties and protein expression in oral Streptococcus sanguis. Arch Oral Biol. 2004;49:295–304. doi: 10.1016/j.archoralbio.2003.12.001. [DOI] [PubMed] [Google Scholar]
- Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Caldas TD, Yaagoubi AEI, Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem. 1998;273:11478–11482. doi: 10.1074/jbc.273.19.11478. [DOI] [PubMed] [Google Scholar]
- Ccahuana-Vásquez RA, Tabchoury CP, Tenuta LM, Del Bel Cury AA, Vale GC, Cury JA. Effect of frequency of sucrose exposure on dental biofilm composition and enamel demineralization in the presence of fluoride. Caries Res. 2007;41:9–15. doi: 10.1159/000096100. [DOI] [PubMed] [Google Scholar]
- Cury JA, Rebello MAB, Del Bel Cury AA. In situ relationship between sucrose exposure and the composition of dental plaque. Caries Res. 1997;31:356–360. doi: 10.1159/000262418. [DOI] [PubMed] [Google Scholar]
- Cury JA, Rebelo MAB, Del Bel Cury AA, Derbyshire MTVC, Tabchoury CPM. Biochemical composition and cariogenicity of dental plaque formed in the presence of sucrose or glucose and fructose. Caries Res. 2000;34:491–497. doi: 10.1159/000016629. [DOI] [PubMed] [Google Scholar]
- Fox DJ, Dawes C. The extraction of protein matrix from human dental plaque. Arch Oral Biol. 1970;15:1069–1077. doi: 10.1016/0003-9969(70)90119-6. [DOI] [PubMed] [Google Scholar]
- Gharahdaghi F, Weinberg CR, Meagher DA, Imai BS, Mische SM. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Hara AT, Queiroz CS, Paes Leme AF, Serra MC, Cury JA. Caries progression and inhibition in human and bovine root dentine in situ. Caries Res. 2003;37:339–344. doi: 10.1159/000072165. [DOI] [PubMed] [Google Scholar]
- Huang CM. Comparative proteomic analysis of human whole saliva. Arch Oral Biol. 2004;49:951–962. doi: 10.1016/j.archoralbio.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Iacono VJ, Mackay BJ, Pollock JJ, Bolot PR, Laqqenhein S, Grossbard BL, Rochon ML. Roles of lysozyme in the host response to periodontopathic organisms. In: Genco RJ, Mergenhagen SE, editors. Host-Bacterial Interactions in Periodontal Diseases. Washington: American Society for Microbiology; 1982. pp. 318–342. [Google Scholar]
- Jayaraman GC, Penders JE, Burne RA. Transcriptional analysis of the Streptococcus mutanshrcA, grpE and dnaK genes and regulation of expression in response to heat shock and environmental acidification. Mol Microbiol. 1997;25:329–341. doi: 10.1046/j.1365-2958.1997.4671835.x. [DOI] [PubMed] [Google Scholar]
- Joe A, Murray CS, McBride BC. Nucleotide sequence of a Porphyromonas gingivalis gene encoding a surface-associated glutamate dehydrogenase and construction of a glutamate dehydrogenase-deficient isogenic mutant. Infect Immun. 1994;62:1358–1368. doi: 10.1128/iai.62.4.1358-1368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Len ACL, Cordwell SJ, Harty DWS, Jacques NA. Cellular and extracellular proteome analysis of Streptococcus mutans grown in a chemostat. Proteomics. 2003;3:627–646. doi: 10.1002/pmic.200300391. [DOI] [PubMed] [Google Scholar]
- Len ACL, Harty DWS, Jacques NA. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology. 2004a;150:1339–1351. doi: 10.1099/mic.0.27008-0. [DOI] [PubMed] [Google Scholar]
- Len ACL, Harty DWS, Jacques NA. Proteome analysis of Streptococcus mutans metabolic phenotype during acid tolerance. Microbiology. 2004b;150:1353–1366. doi: 10.1099/mic.0.26888-0. [DOI] [PubMed] [Google Scholar]
- Mukhopadyay SV, Kapatral V, Xu W, Chakrabarty AM. Characterization of a Hank's type serine/threonine kinases and serine/threonine phosphoprotein phosphatase in Pseudomonas aeruginosa. J Bacteriol. 1999;181:6615–6622. doi: 10.1128/jb.181.21.6615-6622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandakumar R, Nandakumar MP, Marten MR, Ross JM. Proteome analysis of membrane and cell wall associated proteins from Staphylococcus aureus. J Proteom Res. 2005;4:250–257. doi: 10.1021/pr049866k. [DOI] [PubMed] [Google Scholar]
- Paddick JS, Brailsford SR, Rao S, Soares RF, Kidd EA, Beighton D, Homer KA. Effect of biofilm growth on expression of surface proteins of Actinomyces naeslundii genospecies 2. Appl Environ Microbiol. 2006;72:3774–3779. doi: 10.1128/AEM.72.5.3774-3779.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paes Leme AF, Koo H, Bellato CM, Bedi G, Cury JA. The role of sucrose in cariogenic dental biofilm formation – new insight. J Dent Res. 2006;85:878–887. doi: 10.1177/154405910608501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancholi V, Fischetti VA. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273:14503–14515. doi: 10.1074/jbc.273.23.14503. [DOI] [PubMed] [Google Scholar]
- Pappin DJC, Hojrup P, Bleasby AJ. Rapid identification of proteins by peptide-mass fingerprinting. Curr Biol. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- Pecharki GD, Cury JA, Paes Leme AF, Tabchoury CPM, Del Bel Cury AA, Rosalen PL, Bowen WH. Effect of sucrose containing iron (II) on dental biofilm and enamel demineralization in situ. Caries Res. 2005;39:123–129. doi: 10.1159/000083157. [DOI] [PubMed] [Google Scholar]
- Quivey RG, Jr, Kuhnert WL, Hahn K. Adaptation of oral streptococci to low pH. Adv Microb Physiol. 2000;42:239–274. doi: 10.1016/s0065-2911(00)42004-7. [DOI] [PubMed] [Google Scholar]
- Ribeiro CCC, Tabchoury CPM, Del Bel Cury AA, Tenuta LMA, Rosalen PL, Cury JA. Effect of starch on the cariogenic potential of sucrose. Br J Nutr. 2005;94:44–50. doi: 10.1079/bjn20051452. [DOI] [PubMed] [Google Scholar]
- Rose RK, Dibdin GH, Shellis RP. A quantitative study of calcium binding and aggregation in selected oral bacteria. J Dent Res. 1993;72:78–84. doi: 10.1177/00220345930720011201. [DOI] [PubMed] [Google Scholar]
- Sutherland IW. The biofilm matrix – an immobilized but dynamic microbial environment. Trends Microbiol. 2001;9:222–227. doi: 10.1016/s0966-842x(01)02012-1. [DOI] [PubMed] [Google Scholar]
- Svensäter G, Sjögreen B, Hamilton IR. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology. 2000;146:107–117. doi: 10.1099/00221287-146-1-107. [DOI] [PubMed] [Google Scholar]
- Tenuta LM, Ricomini Filho AP, Del Bel Cury AA, Cury JA. Effect of sucrose on the selection of mutans streptococci and lactobacilli in dental biofilm formed in situ. Caries Res. 2006;40:546–549. doi: 10.1159/000095656. [DOI] [PubMed] [Google Scholar]
- Vale GC, Tabchoury CP, Arthur RA, Del Bel Cury AA, Paes Leme AF, Cury JA. Temporal relationship between sucrose-associated changes in dental biofilm composition and enamel demineralization. Caries Res. 2007;41:406–412. doi: 10.1159/000105764. [DOI] [PubMed] [Google Scholar]
- Vijay K, Brody MS, Freudlund E, Price CW. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the sigmaB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- Welin J, Wilkins JC, Beighton D, Wrzesinski K, Fey SJ, Mose-Larsen P, Hamilton IR, Svensäter G. Effect of acid shock on protein expression by biofilm cells of Streptococcus mutans. FEMS Microbiol Lett. 2003;227:287–293. doi: 10.1016/S0378-1097(03)00693-1. [DOI] [PubMed] [Google Scholar]
- Wilkins JC, Beighton D, Homer KA. Effect of acidic pH on expression of surface-associated proteins of Streptococcus oralis. Appl Environ Microbiol. 2003;69:5290–5296. doi: 10.1128/AEM.69.9.5290-5296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JC, Homer KA, Beighton D. Altered protein expression of Streptococcusoralis cultured at low pH revealed by two-dimensional gel electrophoresis. Appl Environ Microbiol. 2001;67:3396–3405. doi: 10.1128/AEM.67.8.3396-3405.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins JC, Homer KA, Beighton D. Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl Environ Microbiol. 2002;68:2382–2390. doi: 10.1128/AEM.68.5.2382-2390.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]