Synopsis
The past few years have seen the identification of numerous small molecules that selectively inhibit specific Class I isoforms of PI3K yet little has been revealed about the molecular basis for the observed selectivities. Using site directed mutagenesis, we have investigated one of the areas postulated as critical to the observed selectivity. The residues, T886 and K890 of the PI3K γ isoform project towards the ATP binding pocket at the entrance to the catalytic site but are not conserved. We have made reciprocal mutations between those residues in the β isoform (E858 and D862) and those in the α isoform (H855 and Q859) and evaluated the potency of a range of reported PI3K inhibitors. The results show that the potencies of β-selective inhibitors TGX221 and TGX286 are unaffected by this change. In contrast, close analogues of these compounds, in particular the α-isoform selective compound (III), are markedly influenced by the point mutations. The collected data suggests two distinct binding poses for these inhibitor classes, one of which is associated with potent PI3K beta activity and is not associated with the mutated residues, and a second that in accord with earlier hypotheses does involve this pair of non-conserved amino acids at the catalytic site entrance and contributes to the α-isoform selectivity of the compounds studied.
Keywords: PI3kinase, isoforms, selectivity, small molecule inhibitors, site directed mutagenesis, ATP binding pocket
Introduction
Enzymes of the phosphoinositide 3-kinase (PI3K) family are fundamental to numerous intracellular signaling cascades, catalysing phosphorylation at the 3-hydroxyl function of phosphatidylinsoitols. [1, 2] The 3-phosphorylated products - PtdIns(3)P, PtdIns(3,4)P2 and PtdIns(3,4,5)P3 act as second messengers, binding to and activating downstream effector proteins, and in so doing controlling numerous physiological processes such as cell growth, proliferation, adhesion and survival.[2] Given this array of activities, inhibitors of the PI3K activity have been anticipated to have significant therapeutic potential in a range of diseases states, notably cancer,[3-5] thrombosis [6] and immuno-inflammatory diseases.[7, 8]
The PI3K family comprises a variety of sub-types displaying distinct but sometimes overlapping regulation, substrate specificity, tissue expression and kinetic behavior. The class I PI3Ks have been studied in most detail and are characterized by a 110kD catalytic domain, utilization of PtdIns(4,5)P2 as substrate and sensitivity to the pharmacological inhibitors wortmannin and LY294002.[9] The four class I isoforms p110α, p110β, p110γ and p110δ are distinguished by sequence variation that can result in differences in regulatory partners and different pathways to activation. Each individual isoform has been nominated as a target for the treatment of the disease states listed above and for a number of years it has been postulated that for therapeutic benefit without serious side effect potential, inhibitors that could discriminate the individual Class I isoforms of PI3K would be required.[10] From the earliest elucidation of the isoform sequences it was suggested that this would be a difficult task, such was the level of homology around the catalytic site. The release of x-ray co-ordinates for p110γ further reinforced this notion; interactions between the enzyme and the inhibitors LY294002, quercetin, myricetin, wortmannin and staurosporin were exclusively with residues conserved across the class I isoforms.[11] Those reservations though have ultimately proved groundless, as agents showing marked selectivity for one or two isoforms continue to be described. Included amongst those inhibitors are those selective for p110δ described by workers at ICOS,[12] compounds that target p110β/δ as described by Jackson et al, [6] a number of compound classes that selectively inhibit p110γ [8, 13], and most recently agents have been described that selectively inhibit p110α. [14] An armoury of reagents is developing with which to analyse the roles of these enzymes in physiology. As demonstrated recently by Knight et al the combination of these reagents as pharmacological tools provides for compelling cross-referencing in studies of cellular function.[15]
While a great deal of progress has been made towards the development of isoform selective inhibitors of PI3K isoforms, this has been achieved in large part through the application of typical medicinal chemistry hit-to-lead strategies and understanding of the molecular origins of the observed selectivities remains at best rudimentary. Even with a series of inhibitor-enzyme co-crystals (for which there are currently eleven published) the causation of inhibitor selectivity is not obvious. Irrespective of the ligand, the key interactions are with absolutely conserved residues of the catalytic pocket.[11] Moreover the p110γ structure is largely insensitive to the nature of the inhibitor with low RMS values describing the overlap between catalytic site residues in most cases. The only indication that selectivity might be driven by differential dynamic behaviour of the isoforms is provided by significant repositioning of residues in the catalytic site in the co-crystal of p110γ and a p110δ selective compound, PIK39. [15] The lack of crystal data relating to any isoform other than p110γ has been a major stumbling block, and the recent description of p110α hopefully opens the door to new insights relating to inhibitor selectivity. [16]
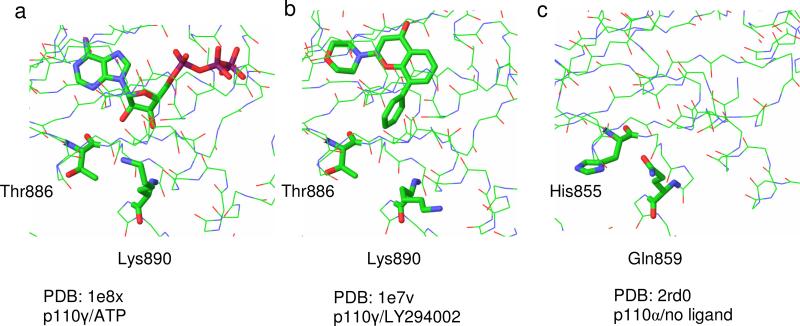
Inspection of the published p110 structures more distant from the direct binding site identifies non-conserved regions that might be expected to play a significant role in conferring some non-uniformity in the catalytic pocket and these have been the subject of previous speculation. Previously a comparison of the γ isoform crystal structure and a 3-dimensional model of the δ isoform was used to identify potential regions of selectivity within the inhibitor binding pocket. Four regions in the γ isoform were identified with the amino acids conferring selectivity proposed as Ala805 and Lys808 (region 1), Ile881 and Ala885 (region 2), Thr886 (region 3) and Lys890 (region 4).[13, 17, 18] We analysed the available x-ray structures of p110γ co-crystallised with ATP and with various inhibitors. In the stretch of non-conserved amino acid residues that sits at the outer edge of the binding pocket from Lys883 to Lys890 of p110γ it was found that Lys890 shows marked variation in side chain placement. As shown in Figure 1, the side chain reaches towards the ribose hydroxyl groups of ATP (PDB code: 1e8x, Figure 1a), but orients away from the aryl ring in LY294002 (PDB code 1e7v, Figure 1b). [11] In sequence alignments and superposition with p110α, the corresponding residue is Gln859 (PDB code 2rd0, Figure 1c).[16] A second residue in this region that orients towards the catalytic site is Thr886, which corresponds to His855 in p110α. These residues appear to exert no conformational influence at the backbone with a close overlay between the isoform structures. In p110β the corresponding residues are Asp862 and Glu858 respectively.
Figure 1. Structural heterogeneity of PI3K catalytic site entrance.
Common views of PI3K crystal structures highlighting structural heterogeneity at binding site entrance are shown.
(a) p110γ/ATP – Lys890 side chain projects towards the ribose moiety of ATP.
(b) p110γ/LY294002 – Lys890 side chain projects away from Ly294002 aryl ring.
(c) p110α/no ligand present – superposition show His and Gln residues replacing Thr and Gln (p110γ) lining the outer edge of the binding pocket.
Our previous studies have focused upon inhibitors elaborated from the non-selective inhibitor LY294002, that in general display a preference for the p110β isoform. The published inhibitors TGX221 and TGX286 [6, 15] have an obvious potential for overlap with the pose adopted by LY294002, such that the elaboration of the structure would be expected to account for the observed increase in potency and selectivity. In seeking to understand this further, our initial models suggested the potential for the binding of these extended structures to be influenced by the residues of the non-conserved region, particularly Thr866 and Lys890 of p110γ or their corresponding residues in other isoforms. In order to test this postulate in the absence of crystallographic data, we chose to make mutations to this region in the hope of observing modified inhibitory potency against the mutant form. In this article we describe the preparation of mutants of p110β and p110α. We have characterized the catalytic activity of these mutants in comparison to the wild type enzymes and measured the varied sensitivity of a panel of inhibitors to these changes.
Materials and Methods
Chemical Synthesis
The compounds of this study were prepared according to reported literature procedures. LY294002, TGX102, TGX132, and compounds (I), (II) and (III) were prepared according to previously described routes.[19] TGX221, TGX286 were generously provided by Kinacia Pty. Ltd. prepared as described in WO 2004016607.[20]
Mutagenesis of PI3K p110α and p110β
Site directed mutagenesis using Pfu Ultra-high fidelity DNA polymerase was carried out using the Quickchange kit (Stratagene) with pBacPAK8/p110α and pBacPAK/p110β as the template DNA. The presence of the correct point mutation was confirmed by DNA sequencing. Recombinant baculoviruses were generated by co-transfection of the appropriate pBacPAK8 transfer plasmid with pBacPAK6 viral DNA (Clontech) into Sf21 insect cells. Recombinant baculovirus plaques were purified and tested for protein expression by Western blot using isoform-specific antibodies. Positive recombinant baculovirus was then amplified to a titre in the range of 107 to 108 pfu/ml.
Protein Expression and Purification
Co-infection of Sf21 cells with p110α baculovirus (wild-type or mutant) and p85 baculovirus was carried out for 48h at 27°C. in serum free insect cell medium Sf900 (Invitrogen). Cells were then pelleted, resusupended in 50mM NaH2PO4, 150mM NaCl pH 7.2 containing 1x complete protease inhibitor cocktail (Roche), sonicated and centrifuged at 17000g for 5 min at 4°C. The resulting supernatant was filtered through a 0.2μm filter and loaded onto a Superose 12 gel filtration column equilibrated in 50mM NaH2PO4, 150mM NaCl pH 7.2 at a flow rate of 0.5ml/min. Fractions were collected and PI3K activity was assayed. Positive fractions were pooled, 30% (v/v) glycerol was added and stored at −80°C.
Co-infection of Sf21 cells with p110β baculovirus (wild-type or mutant) and N-terminal 6-his tagged p85 baculovirus was carried out for 48h at 27°C. in serum free insect cell medium Sf900 (Invitrogen). Cells were then pelleted, resuspended in 50mM NaH2PO4, 300mM NaCl, 10mM imidazole pH 8.0 containing 1x EDTA-free complete protease inhibitor cocktail (Roche), sonicated and centrifuged at 17000g for 5 mins at 4°C. The supernatant was loaded onto a Ni-NTA affinity column, washed with 4 volumes of 50mM NaH2PO4, 300mM NaCl, 20mM imidazole pH 8.0. Bound PI3K was eluted with 50mM NaH2PO4, 300mM NaCl, 2500mM imidazole pH 8.0. Fractions were collected and PI3K activity was assayed. PI3K fractions were pooled, 30% (v/v) glycerol was added and stored at −80°C.
Inhibition Assays
PI3K lipid kinase assays were performed in a total volume of 100 μl in a buffer containing 20 mm HEPES, pH 7.4, 5 mm MgCl2 and 0.25mM EDTA using 170 μg/ml phosphatidylinositol as a substrate. Reactions were started with the addition of 50μm ATP (plus 0.1 μCi of [γ-32P]ATP/assay), incubated at 25 °C for 20 min and terminated by the addition of 100 μl of 0.1 m HCl. Phospholipids were extracted with a mixture of 200μl of chloroform/methanol (1:1) and 500μl of 2M KCl followed by scintillation counting. IC50 was determined using inhibitor concentrations ranging form 3nM to 10μM with one-half log unit between doses. Assays were performed for each compound at n=3. Individual dose response curves were generated, IC50 determined using Prism (GraphPad, USA) and reported as the average of 3 results.
Results and Discussion
1. Design, preparation and catalytic activity of mutants of PI3K p110α and p110β
We set out to examine the participation of the non-conserved residues at the binding pocket entrance in the isoform selectivity of a series of PI3K inhibitors as shown in Figure 2. To do this we prepared mutant forms of p110β in which those residues Glu858 and Asp862 were mutated to the corresponding residues in p110α (His and Gln respectively) and also preparing the reciprocal mutants in p110α. Our hypotheses were that (i) mutation in those residues should have no significant impact on catalytic activity, (ii) that mutation of these residues should have no impact on the inhibitory potency of LY294002, but (iii) that we should witness diminished or enhanced potency of inhibitors against the mutated enzymes, dependent upon the complimentary nature or otherwise of the inhibitor. We hoped that the evaluation of reciprocal pairs of mutants might provide greater insight into the relevance of the specific mutated position than single mutation alone.
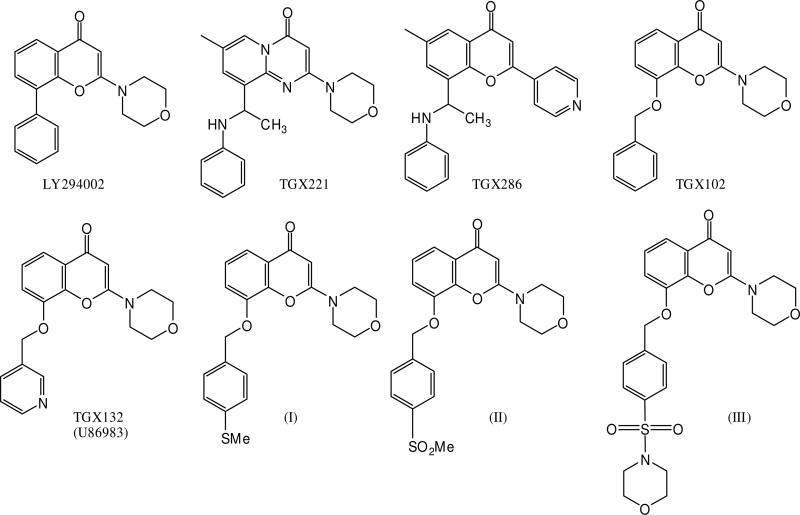
Figure 2. Structure of PI3K inhibitors.
Chemical structures of the PI3K inhibitors used in this study.
Preparation of wild type and mutant enzymes
The p85, p110α and p110β subunits and the site directed mutants of the catalytic domains were successfully expressed using the baculovirus expression system. The wild type p110α and mutants were co-expressed with the native p85 sub-unit, and purified by gel filtration and ion exchange chromatography. For the wild type p110β and mutants, an N-terminal 6-his-tag was incorporated on the p85 sub-unit allowing for purification by immobilized-metal affinity chromatography. The Km for PI has previously been shown to be higher for p110α compared to p110β. Here the Km for wild type p110α and β was 221 and 41 μM respectively (see Table 1). This correlated well with previously reported values for p110α Km of 512μM [21] and for p110β of 130μM [2].
Table 1. Kinetic constants of α and β wild-type and mutant purified PI3K.
Purified PI3K was assayed in the presence of varying amounts of substrate, phosphatidylinositol, in 20 mm HEPES, pH 7.4, 5 mM MgCl2 ,0.25mM EDTA and 50μM ATP (0.1 μCi of [γ-32P]ATP). Assay was carried out according the protocol detailed in the Experimental section. Kinetic constants were estimated using Prism (Graphpad, USA)
| p110α | p110α (Q859D) | p110α (H855E) | p110β | p110β (D862Q) | p110β (E858H) | |
|---|---|---|---|---|---|---|
| Km (μM) | 221.5 | 113.2 | 62.5 | 40.7 | 31.7 | 35.5 |
| Vmax | 6.1 | 7.5 | 6.9 | 10.8 | 13.2 | 13.1 |
The mutant forms of the enzyme fulfilled our key criteria for study. The mutants all exhibit catalytic activity similar to the wild type enzyme as mutagenesis did not affect the Vmax in any mutant. However mutagenesis of p110α resulted in a reduction in the Km value for both the D862Q and E858H mutants, towards that of p110β perhaps indicating that substrate access to the active site is enhanced, while having no effect on catalysis as no change in Vmax was observed. As no increase in Km was observed following p110β mutagenesis (Table 1) other amino acids must also influence substrate access to the active site in p110β.
LY294002 was anticipated to make no distinction between wild type and mutant PI3K enzymes and this was in general found to be the case (Table 2). For the p110α mutants, LY294002 showed identical potency as for the wild type isoforms. This was also true of the E858H-p110β mutant, although inhibitory potency was decreased 2-fold in the case of D862Q suggestive of a subtle change in catalytic site topography.
Table 2. IC50 estimation of effect of inhibitors on purified PI3K isoforms and in vitro mutants.
Purified PI3K (p110β/p85) was assayed in the presence of varying concentrations of the indicated inhibitors 3nM to 10μM with one-half log unit between doses in 20 mM HEPES, pH 7.4, 5mM MgCl2 ,0.25mM EDTA, 170μg/ml phosphatidylinositol and 50μM ATP (0.1 μCi of [γ-32P]ATP). Assay was carried out according the protocol detailed in the Experimental section. IC50 (μM) was estimated using Prism (Graphpad, USA) with each value being the average of three separate IC50 determinations.
| IC50 (μM) | ||||||
|---|---|---|---|---|---|---|
| Inhibitor | p110β WT | p110β (D862Q) | p110β (E858H) | p110α WT | p110α (Q859D) | p110α (H855E) |
| LY294002 | 0.34 | 0.62 | 0.36 | 0.46 | 0.40 | 0.63 |
| TGX221 | 0.029 | 0.032 | 0.0088 | 0.77 | 0.86 | 1.4 |
| TGX286 | 0.0070 | 0.017 | 0.013 | 0.99 | 2.2 | 1.3 |
| TGX102 | 0.18 | 0.20 | 0.13 | 0.77 | 2.5 | 2.3 |
| TGX132 | 2.50 | 1.10 | 1.16 | 0.84 | 5.2 | 4.1 |
| (I) | 0.53 | 0.44 | 0.32 | 0.13 | 0.39 | 0.66 |
| (II) | 0.83 | 0.68 | 0.21 | 0.22 | 0.66 | 0.64 |
| (III) | 4.3 | 0.34 | 0.38 | 0.16 | 0.27 | 0.48 |
2. Assays of inhibitor potency at wild type p110 isoforms
The results of the screen of wild-type enzymes against selected PI3K inhibitors are shown in Table 2. As a priority of this study, the previously described inhibitors TGX221 and TGX286 were confirmed to be potent and selective inhibitors of p110β with IC50's of 29nM and 7.0nM respectively.1 At p110α, they are both much less potent. The benzyloxy-substituted chromone, TGX102, shows moderate p110β potency (IC50 0.18μM) and approximately 4-fold selectivity relative to p110α, which demonstrates that simple homologation of the pendant group is sufficient to gain some selectivity. In contrast, the pyridinyl substituted analogue, TGX132 (U86983)[22] shows a significant drop in p110β potency (IC50 2.5μM) while against p110α potency is unchanged (IC50 0.84μM) relative to TGX102. The inclusion of a p-methylthio, p-methylsulfonyl and p-morpholinosulfonyl substitutent in the pendant benzyloxy ring yields compounds (I), (II) and (III) respectively (Figure 2). They were identified as compounds showing enhanced potency at p110α and modified selectivity profiles relative to their parent compound TGX102 (Table 2 and 3). Compounds (I) and (II) show approximately 4-fold selectivity towards p110α compared to p110β with compound (III) displaying 27 fold selectivity. The observed selectivity for p110α appears to derive from a combination of enhanced potency at p110α and diminished potency at p110β, the latter particularly marked in the case of compound (III) which is 24 fold less potent than TGX102 at p110β.
Table 3. Influence of reciprocal mutation upon p110α/p110β selectivity.
Selectivity ratio considers the mutation of a point residue in p110α for the corresponding residue in p110β and vice versa. The fold change from wild type: values > 1 indicate a preference for the p110α wild type residue; values < 1 indicate a preference for the p110β wild type residue.
| Selectivity ratio | |||||
|---|---|---|---|---|---|
| Compound | α/β (wt) | α/β Gln↔Asp | α/β His↔Glu | Fold change from wt: Gln↔Asp | Fold change from wt: His↔Glu |
| LY294002 | 1.37 | 0.65 | 1.74 | 0.5 | 1.3 |
| TGX221 | 26.3 | 26.7 | 157.1 | 1.0 | 6.0 |
| TGX286 | 142.0 | 126.2 | 106.4 | 0.9 | 0.7 |
| TGX102 | 4.30 | 12.65 | 17.3 | 2.9 | 4.0 |
| TGX132 | 0.34 | 4.59 | 3.56 | 13.7 | 10.6 |
| (I) | 0.25 | 0.88 | 2.05 | 3.6 | 8.3 |
| (II) | 0.27 | 0.98 | 3.11 | 3.6 | 11.6 |
| (III) | 0.036 | 0.80 | 1.25 | 22.2 | 34.8 |
The key outcome of this initial screen with respect to our studies was the apparently incremental gain in p110β potency relative to LY294002 with increased structural elaboration at the pendant ring and the capacity for small structural changes to impact on isoform selectivity. The identification of TGX132, (I), (II) and (III) as having some selectivity for p110α suggests the possibility to manipulate these compounds to yield selective inhibitors at isoforms other than p110β. Also based on our hypothesis, the graded changes in structure and potency might be expected to manifest in the sensitivity of these inhibitors to the site directed mutagens.
3. The influence of p110β mutations on inhibitor potency
As described earlier, we postulated that non-conserved residues at the outer edge of the ATP-binding pocket may be responsible, at least in part, for the observed increase in selectivity of TGX221 and TGX286. The p110β mutants p110β-E858H and p110β-D862Q were designed to test this hypothesis.
In the event, this arm of the hypothesis did not hold. (Table 2) Neither of the mutations gave rise to any diminution of TGX221 potency. In fact, TGX221 was found to inhibit p110β-E858H 3-fold more potently than the wild type form and increasing the selectivity over p110α. The other potent p110β inhibitor from the Kinacia series, TGX286, which showed over 140-fold selectivity, showed approximately 2-fold reduction in potency at p110β-D862Q and p110β-E858H. For TGX102, the inhibition curves derived from the mutated enzymes were indistinguishable from the wild type.
TGX132 from this series, in which inclusion of a pyridinyl methoxy substituent relative to TGX102 resulted in a 12-fold drop in p110β potency (IC50 2.5μM), recovered some of that potency (approximately 2 fold) with mutation of either residue. Similarly, mutation at p110β resulted in modest enhancements in the potency of compounds (I) and (II). However, mutation of p110β had a quite profound effect on the potency of (III) (IC50 4.3μM. v. wild type) shifting the IC50 by over an order of magnitude in both cases, and essentially recovering much of the p110β potency exhibited by TGX102. In other words, the selectivity of compound (III), the most p110α-selective inhibitor of this group appears to largely derive from disfavoured interaction with the acidic Asp862 and Glu858 residues of p110β.
4. The influence of p110α mutations on inhibitor potency
As with the p110β mutants, a general survey of p110α and the mutant isoforms was carried out to determine any influence, positive or negative, that the particular residues have upon activity that might imply a role for those amino acids in determining inhibitor potency (Table 2).
While each of the compounds did show sensitivity to p110α mutation, none of the compounds displayed any preference for the mutated p110α forms over the wild type. Mutations Q859D-p110α and H857E-p110α resulted in TGX132 potency dropping 5-6 fold. Its analogue, TGX102, (IC50 0.77μM v. wild type) which was transparent to p110β mutation, lost 3-fold potency at either mutated p110α isoform. Compounds (I), (II) and (III) were between 2- and 4-fold less potent at mutated forms of p110α showing that potency at p110α was at some level dependent upon the mutated residues.
Incorporation of a residue found in p110β might have been expected to enhance potency of p110β-selective compounds, TGX221 and TGX286, at p110α. In fact, both show some lost potency at p110α upon mutation.
In summary, it would appear that the histidine and glutamine residues are in general positive contributors to the observed p110α potency of this series of compounds. In contrast, the aspartyl and glutamatyl residues found in wild type p110β play no positive role in ligand binding of these compounds.
5. Models for PI3K inhibition based upon reciprocal mutation
In the absence of crystal data relating to either of the isoforms of this study, indirect experimental techniques were utilized to provide support to hypothetical binding models. Here we have tested a hypothesis that a non-conserved region adjacent to the ATP binding site would play a role in the binding affinity of compounds known to have high selectivity for p110β over p110α or vice versa. TGX221 and TGX286 each display strong preference for p110β, and were developed through iterative extension of the pendant aryl group of LY294002. It was anticipated that these compounds would show significant sensitivity to the presence of mutations in the non-conserved region, extrapolating from the available p110γ-LY294002 co-crystal.[11] The assertion that these residues would present in a similar locale to that aligned residues in p110γ has subsequently been confirmed crystallographically.[16] The His855 and Gln859 residues of p110α overlay exquisitely with the Thr886 and Lys890 of p110γ.
The observed potency of the compounds tested in the experiments described in this work is suggestive of a model for binding and selectivity quite different to that envisaged. The resultant hypothesis from this work is in fact the opposite – the selectivity of the potent 110β selective compounds, TGX221 and TGX286, is not determined by the nature of the residues Asp862 and Glu858 of the non-conserved region. In contrast, the remaining data suggests that these residues can and do play a role in the binding of other ligands, inclusive of chromone derivatives of TGX102. As a group of compounds, the substituted analogues of the p110β selective inhibitor, TGX102, all show some level of preference for p110α and, to a greater or lesser extent, the change in p110α/β selectivity appears to be drawn from the residues that have been mutated. The reciprocated nature of the gains and losses of potency observed in these examples forms the basis of the following discussion.
It can be reasonably asserted that potent and selective p110β inhibition is not determined by the non-conserved residues that have been mutated in this study. Indeed, the impression to be gained from the data compiled against the p110 mutants is that this region actually provides some hindrance to p110β selectivity for these potent ligands. On the other hand, it appears that p110 binding of the less potent compounds does have reliance on interactions with the non-conserved region, with altered activity of inhibitors observed for many of the experiments where mutations were introduced in both isoforms, and a clear preference for the residues present in p110α.
Consideration of the effect upon selectivity of reciprocal mutagenesis of the chosen residues provides an interesting view of the results. (Table 3) To do this, we compare the selectivity of the wild type p110α/β IC50 ratios for wild type with the reciprocal mutant pairings, p110α-(Q859D)/p110β-(D862Q) and p110α-(H855E)/p110β-(E858H). For example, TGX221 shows 26-fold selectivity for wild type p110β over wild type p110α. For the reciprocal mutant pairing p110β-(E858H) over p110α-(H857E), TGX221 shows 154 fold selectivity. Introducing reciprocal mutations in that position improved selectivity for p110β by 6-fold.
Any shift from a ratio of greater than one would seem to be indicative of a preference to the residues in this region found in p110α. A shift of less than one suggests a preference for the residues found in p110β. Shifts of an order of magnitude would indicate that the residue at that position plays a significant role in the selectivity profile of the compound. Such a shift was observed for the compounds, TGX132, (I), (II) and (III) in these cases the mutagenesis pairings actually led to a change in the isoform selectivity - switching from p110α preferring as wild type enzymes to p110β preferring as mutant pairings in one or both cases.
This information could be utilized as a significant driver in drug design where the objective may be to achieve enhanced levels of selectivity. For example, the presence of the glutamyl group D862 in p110β prevents even higher TGX221 selectivity from being attained. If increased selectivity is desired, an enhanced compatibility with the glutamyl residue may be the means to achieve it. With TGX286, that is relatively insensitive to these mutations, perhaps such compatibility has been realized, albeit unintentionally.
As shown in the collected data, a specific residue can contribute to enhanced isoform selectivity by either enhancing ligand affinity for the target enzyme or reducing affinity for the “off-target” enzyme. In these results, the greater evidence is that the non-conserved region is primarily able to influence selectivity by inhibiting interactions with an off-target isoform. This is most apparent with compound (III). It is selective for p110α predominantly by virtue of decreases in affinity for the p110β isoform relative to TGX102, rather than through significant gains in p110α potency. The mutagenesis data supports this strongly with mutation of p110β able to recover virtually all the lost potency relative to the analogue, while mutation at p110α gives moderate decreases in potency. In summary, it may be concluded that p110α selectivity over p110β of these compounds is achieved predominantly through the compounds’ hindered interaction with the non-conserved region of the catalytic site of p110β.
An alternate explanation may be derived from the observation that for both p110α mutants the Km for substrate was shifted towards the lower concentration of p110β WT Km indicating that these amino acids at the entrance to the catalytic site of 110α may restrict substrate entry or product release, reflected in the higher p110α Km ie. p110β catalytic site is more accessible than the p110α catalytic site. It may be that the differences between the two isoforms in the structures of the entrance to the catalytic site may contribute to the observed selectivities of inhibitors and their sensitivity to mutation. More detailed kinetic studies may serve to resolve these possibilities.
If we return to the issue of the binding site for the potent p110β inhibitors. That there is an alternate binding pose for these ligands to that hypothesized, is apparent from the high potency at p110β of TGX221, TGX286, and TGX102 relative to LY294002 and the apparent lack of influence exhibited by the non-conserved residues. This study is unable to delineate that binding site but has ruled out a role for non-conserved amino acids at the p110β catalytic site entrance. Interestingly, a model for p110β binding of TGX280 (PIK108) has recently been proposed that explains the observed data quite well.[15] In the x-ray structure of p110γ bound to the ligand PIK39, a p110δ preferring compound, a methionine residue (Met804) undergoes a major, ligand-induced conformational switch creating a binding pocket hidden in othe rcrystal forms of p110γ. In analysing the potential for other ligands to interact with such a conformational variant, PIK-108 (TGX280), a close analogue of TGX221 and TGX286 was successfully modelled into the new binding site, notably accommodating the phenylaminoethyl substituent shared by TGX280, TGX221 and TGX286 [15]. In this model, the mutated residues of the current study might be expected to play no significant role in binding, whereas in models based upon the LY294002 co-crystal structure, these ligands are anticipated to interact with the mutated residues. It should be pointed out that these studies consider only one stereoisomer of these compounds and the active isomer has yet to be identified.
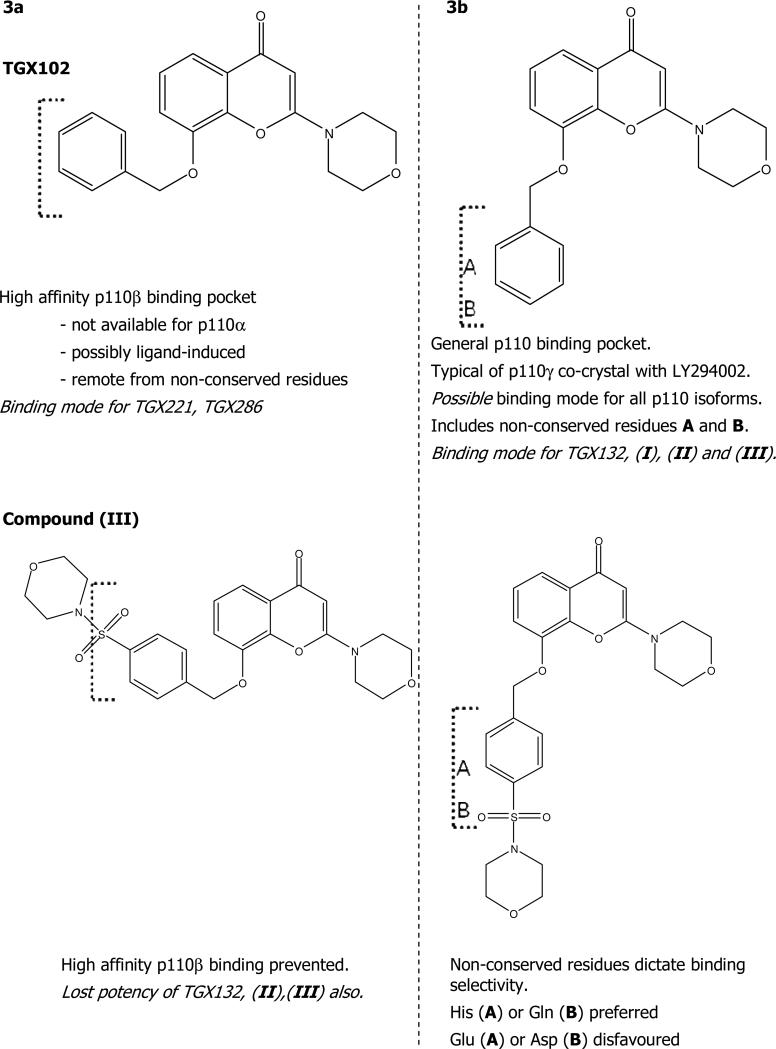
Thus two binding orientations and binding site conformations for these ligand types may be in operation – one associated with potent affinity for p110β that does not depend directly upon the non-conserved amino acids mutated here and another consistent with the LY294002 binding site conformer that is sensitive to the non-conserved residues. This is illustrated in Figure 4, with reference to TGX102 and the substituted analogue (III).
In this figure, TGX102 is able to adopt a suitable conformation to bind and perhaps induce a discrete conformer of p110β not available to p110α, which explains the observed selectivity of that compound as well as TGX221 and TGX286. In the conformational state presented in the LY294002 co-crystal, TGX102 is able to bind p110α with lower affinity and access to the non-conserved residues. It is likely competent to bind in this way to p110β also, but prefers the alternate conformation. Inclusion of the morpholinylsulfonyl substituent as shown in (III) blocks access to the p110β-specific pocket forcing the p110β potency to be determined by interactions at the non-conserved site. In that conformation, the presence of the Glu and Asp residues significantly hinder p110β binding, while the His and Gln sustain p110α binding. The mutagenesis supports this hypothesis by the reversal of selectivity for (III) upon reciprocal mutation of those residues. It can also be envisaged that the presence of the pyridinyl nitrogen of TGX132 or the p-methylthio and p-sulfonyl substituents on compounds (I) and (II) prevent them from binding in the p110β-specific binding site, such that p110β potency has greater dependence upon the non-conserved region.
While the evidence for distinct binding orientations by these ligands is quite strong, it could be reinforced by the rational design of new ligands. To date, only p110β-selective ligands have emerged with high potency from the benzopyranone family, but the altered selectivity displayed by TGX132, (I), (II) and (III) and the ability to manipulate it by mutagenesis suggests that the non-conserved region p110α 852-859 has not been optimally targeted by existing ligands and remains a potential means for engineering enhanced selectivity for other isoforms – a hypothesis recently supported by Ruckle et al.[23] With minimal x-ray data available, the study of inhibitor binding by mutagenesis can yield critical information for drug design and the capacity to use this technique to swap the selectivity profile of a ligand would appear to be an exciting approach also to study selective isoform inhibition.
Supplementary Material
Supplementary Figure 1: Inhibition of PI3K (WT and mutant) with LY294002 Inhibition of purified PI3K enzyme activity by varying concentrations of LY294002
A: Graph shows decrease in activity of PI3Kα WT (■) and PI3Kα mutants H855E (▲) and Q859D (•) upon the addition of increasing concentrations of LY294002
B: Graph shows decrease in activity of PI3Kβ WT (■) and PI3Kβ mutants E858H (▲) and D862Q (•) upon the addition of increasing concentrations of LY294002
Supplementary Table 1: Full IC50 data including error calculations (SD and SEM) for inhibition of WT and mutant PI3K isoforms.
Figure 3. Binding site conformations for morpholinochromone stereoisomers.
Two possible binding orientations and binding site conformations for morpholinochromone stereoisomers may be in operation – (a) one associated with potent affinity for p110β that does not depend directly upon the non-conserved amino acids mutated here and (b) another, consistent with the LY294002 binding site structures, that is sensitive to the non-conserved residues.
Abbreviations
- PI3K
phosphatidylinositol 3-kinase
Footnotes
Inhibitor potency for LY294002 and TGX221 reflect that found in other reports and differences in determined IC50s and those published, are modest and can generally be explained by variation in experimental conditions used for IC50 determination. However, the IC50 against p110β for TGX286 reported by Knight et al. 2006 was 0.12μM, where it was determined to be 7nM in this study, a 17-fold difference. The difference in the concentration of ATP used in this study compared to 10μM used in the literature report cannot account for this variation. However, our experience with this compound suggests that low compound solubility may be an explanation for the discrepancy. The rank order of selectivity is retained, marking this compound as a very potent and selective inhibitor of p110β.
References
- 1.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–675. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 2.Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD. Synthesis and function of 3-phosphorylated inositol lipids. Annual Review of Biochemistry. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 3.Samuels Y, Velculescu VE. Oncogenic Mutations of PIK3CA in Human Cancers. Cell Cycle. 2004;3:1221–1224. doi: 10.4161/cc.3.10.1164. [DOI] [PubMed] [Google Scholar]
- 4.Workman P. Inhibiting the phosphoinositide 3-kinase pathway for cancer treatment. Biochem Soc Trans. 2004;32:393–396. doi: 10.1042/bst0320393. [DOI] [PubMed] [Google Scholar]
- 5.Stephens L, Williams R, Hawkins P. Phosphoinositide 3-kinases as drug targets in cancer. Curr Opin Pharmacol. 2005;5:357–365. doi: 10.1016/j.coph.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CE, Kenche V, Anderson KE, Dopheide SM, Yuan Y, Sturgeon SA, Prabaharan H, Thompson PE, Smith GD, Shepherd PR, Daniele N, Kulkarni S, Abbott B, Saylik D, Jones C, Lu L, Giuliano S, Hughan SC, Angus JA, Robertson AD, Salem HH. PI 3-kinase p110beta: a new target for antithrombotic therapy. Nat Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch E, Katanaev VL, Garlanda C, Azzolino O, Pirola L, Silengo L, Sozzani S, Mantovani A, Altruda F, Wymann MP. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287:1049–1053. doi: 10.1126/science.287.5455.1049. [DOI] [PubMed] [Google Scholar]
- 8.Camps M, Ruckle T, Ji H, Ardissone V, Rintelen F, Shaw J, Ferrandi C, Chabert C, Gillieron C, Francon B, Martin T, Gretener D, Perrin D, Leroy D, Vitte PA, Hirsch E, Wymann MP, Cirillo R, Schwarz MK, Rommel C. Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis. Nat Med. 2005;11:936–943. doi: 10.1038/nm1284. [DOI] [PubMed] [Google Scholar]
- 9.Anderson KE, Jackson SP. Class 1 phosphoinositide 3-kinases. International Journal of Biochemistry & Cell Biology. 2003;35:1028–1033. doi: 10.1016/s1357-2725(02)00270-4. [DOI] [PubMed] [Google Scholar]
- 10.Ward S, Sotsios Y, Dowden J, Bruce I, Finan PM. Therapeutic Potential of Phosphoinositide 3-Kinase Inhibitors. Chemistry & Biology. 2003;11:619–637. doi: 10.1016/s1074-5521(03)00048-6. [DOI] [PubMed] [Google Scholar]
- 11.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural Determinants of Phosphoinositide 3-Kinase Inhibition by Wortmannin, LY294002, Quercetin, Myricetin, and Staurosporine. Molecular Cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 12.Sadhu C, Dick K, Tino WT, Staunton DE. Selective role of PI3K delta in neutrophil inflammatory responses. Biochem Biophys Res Commun. 2003;308:764–769. doi: 10.1016/s0006-291x(03)01480-3. [DOI] [PubMed] [Google Scholar]
- 13.Pomel V, Klicic J, Covini D, Church DD, Shaw JP, Roulin K, Burgat-Charvillon F, Valognes D, Camps M, Chabert C, Gillieron C, Francon B, Perrin D, Leroy D, Gretener D, Nichols A, Vitte PA, Carboni S, Rommel C, Schwarz MK, Ruckle T. Furan-2-ylmethylene Thiazolidinediones as Novel, Potent, and Selective Inhibitors of Phosphoinositide 3-Kinase. J. Med. Chem. 2006;49:3857–3871. doi: 10.1021/jm0601598. [DOI] [PubMed] [Google Scholar]
- 14.Hayakawa M, Kaizawa H, Moritomo H, Koizumi T, Ohishi T, Okada M, Ohta M, Tsukamoto S.-i., Parker P, Workman P, Waterfield M. Synthesis and biological evaluation of 4-morpholino-2-phenylquinazolines and related derivatives as novel PI3 kinase p110[alpha] inhibitors. Bioorganic & Medicinal Chemistry. 2006;14:6847–6848. doi: 10.1016/j.bmc.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 15.Knight ZA, Gonzalez B, Feldman ME, Zunder ER, Goldenberg DD, Williams O, Loewith R, Stokoe D, Balla A, Toth B. A Pharmacological Map of the PI3-K Family Defines a Role for p110[alpha] in Insulin Signaling. Cell. 2006;125:733–747. doi: 10.1016/j.cell.2006.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110alpha/p85alpha complex elucidates the effects of oncogenic PI3Kalpha mutations. Science. 2007;318:1744–1748. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 17.Knight ZA, Chiang GG, Alaimo PJ, Kenski DM, Ho CB, Coan K, Abraham RT, Shokat KM. Isoform-specific phosphoinositide 3-kinase inhibitors from an arylmorpholine scaffold. Bioorganic & Medicinal Chemistry. 2004;12:4749–4759. doi: 10.1016/j.bmc.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 18.Kuang RR, Qian F, Li Z, Wei DZ. Study on improving the selectivity of compounds that inhibit two PI3Ks (gamma and delta). J Mol Model (Online) 2006;12:445–452. doi: 10.1007/s00894-005-0069-8. [DOI] [PubMed] [Google Scholar]
- 19.Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CE, Kenche V, Anderson KE, Dopheide SM, Yuan Y, Sturgeon SA, Prabaharan H, Thompson PE, Smith GD, Shepherd PR, Daniele N, Kulkarni S, Abbott B, Saylik D, Jones C, Lu L, Giuliano S, Hughan SC, Angus JA, Robertson AD, Salem HH. PI 3-kinase p110[beta]: a new target for antithrombotic therapy. Nat Med. 2005;11:507–514. doi: 10.1038/nm1232. [DOI] [PubMed] [Google Scholar]
- 20.Jackson SP, Robertson AD, Kenche V, Thompson P, Prabaharan H, Anderson K, Abbott B, Goncalves I, Nesbitt W, Schoenwaelder S, Saylik D. Inhibition of phosphatidylinositide kinase beta. 2004 International Patent Number WO 2004016607.
- 21.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3'-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol. Cell. Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erickson LA, Bonin PD, Wishka DG, Morris J, Dalga RJ, Williams DJ, Wilson GJ, Hoover JL, Simmons CA, Humphrey SJ, et al. In vitro and in vivo inhibition of rat vascular smooth muscle cell migration and proliferation by a 2-aminochromone U-86983. J Pharmacol Exp Ther. 1994;271:415–421. [PubMed] [Google Scholar]
- 23.Ruckle T, Schwarz MK, Rommel C. PI3Kgamma inhibition: towards an ‘aspirin of the 21st century’? Nat Rev Drug Discov. 2006;5:903–918. doi: 10.1038/nrd2145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Inhibition of PI3K (WT and mutant) with LY294002 Inhibition of purified PI3K enzyme activity by varying concentrations of LY294002
A: Graph shows decrease in activity of PI3Kα WT (■) and PI3Kα mutants H855E (▲) and Q859D (•) upon the addition of increasing concentrations of LY294002
B: Graph shows decrease in activity of PI3Kβ WT (■) and PI3Kβ mutants E858H (▲) and D862Q (•) upon the addition of increasing concentrations of LY294002
Supplementary Table 1: Full IC50 data including error calculations (SD and SEM) for inhibition of WT and mutant PI3K isoforms.