Summary
The pH-controlled M2 protein from Influenza is a critical component of the virus, serving as a target for aminoadamantane anti-flu agents that block its H+ channel activity. To better understand its H+-gating mechanism, we investigated M2 in lipid bilayers with a new combination of IR spectroscopies and theory. Linear FTIR spectroscopy was utilized to measure the precise orientation of the backbone carbonyl groups, and 2D-IR spectroscopy was utilized to identify channel-lining residues. At low pH (open-state), our results match previously published ss-NMR and X-ray structures remarkably well. However, at neutral pH (closed-state), our measurements point to a large conformational change, that is consistent with the transmembrane α-helices rotating by one amino acid register: a structural rearrangement not previously observed. The combination of isotope-labelled FTIR and 2D-IR spectroscopies, alongside simulations, provides a non-invasive mean of interrogating structures of membrane proteins in general and ion channels in particular.
Introduction
The M2 protein has become a model system for understanding the H+ gating mechanism of ion channels. M2 assembles into tetramers with a single α-helical transmembrane domain (Duff and Ashley, 1992; Holsinger and Lamb, 1991). Following viral internalization, at the low pH of the endocytic vesicles, histidine residues in the transmembrane domain of M2 are thought to protonate, thereby activating the protein (Pinto et al., 1992; Wang et al., 1995). Upon opening of M2, the viral lumen acidifies, which facilitates the release of the viral contents into the cell. This critical step in the infectious cycle of the virus renders M2 an important target for anti-flu agents, such as amantadine. Finally, M2 is particularly amenable to structural analyses due to the fact that a peptide that encompasses its transmembrane domain, faithfully represents its tetramerization, H+ channel and drug-binding activity (Duff et al., 1992; Hu et al., 2007; Hu et al., 2006; Salom et al., 2000). As a result, it has been possible to structurally characterize the M2 peptide both by solid-state NMR (ss-NMR) (Nishimura et al., 2002) and X-ray crystallography (Stouffer et al., 2008). Therefore the objective of our study was to characterize the structural transition that the M2 channel undergoes upon gating. Towards this end we have employed, and quantitatively assessed a novel combination of 1 and 2D-IR spectroscopies, along with calculations, that is particularly suited for this challenging task: probing the structure and dynamics of membrane proteins in their native environments.
Our approach relies on a combination of linear dichroism measurements and 2D-IR lineshape analysis, which are largely enabled by site-specific isotope labelling. Since isotope labels do not perturb protein structures, they can be placed in sterically confined locations, such as the channel lumen of membrane proteins (like we do herein) that may not tolerate ESR or fluorescence probes. Moreover, 2D-IR spectroscopy holds especially important promise for following the structural kinetics of membrane proteins. Finally, all of these IR measurements are conducted in the protein’s native environment: the lipid bilayer.
The M2 channel is an ideal system to assess the accuracy of our IR methodology due to the fact that it is structurally well-characterized (Nishimura et al., 2002; Stouffer et al., 2008). The utility of our approach is underscored by the results of this study: we discovered that the M2 channel undergoes a large structural rearrangement upon gating that has not been previously detected and which has implications for the drug and gating mechanism of this important drug target.
Results and Discussion
General strategy using isotopic labels
The experimental approach that we have taken relies on linear dichroism measurements and 2D-IR lineshape analyses. To gain site-specific structural information, we used 1-13C=18O labels to spectroscopically isolate individual residues (Torres et al., 2000a; Torres et al., 2001). 13C=18O labelling creates a ca. 60 cm−1 shift in the amide I band, which largely decouples it from the remaining unlabelled amide I modes (Fang and Hochstrasser, 2005; Mukherjee et al., 2006b). As a result, it can be treated as an isolated vibrational mode whose frequency is influenced mostly by its immediate surroundings and has a transition dipole that is independent of the rest of the peptide structure. The unlabeled amide I band that remains after isotope labelling contains significant structural information, and isotope editing can be used to help unravel its structural content. However, in what follows we focus on the labelled modes themselves.
Linear FTIR dichroism yields orientational constraints that are in remarkable agreement with previous experimental data
The dichroisms that we have measured by ATR-FTIR spectroscopy are of amide I vibrational modes of individual carbonyl groups in aligned bilayers. These data are then converted into the angles between the transition dipole moments of the labelled carbonyl groups and the bilayer normal. Since the orientation of the transition dipole moment of the amide I mode relative to the molecular frame is known (Torii and Tasumi, 1992), such dichroism measurements yield the angle between the bilayer normal and the carbonyl group (see Fig. 1d). Thus, henceforth for simplicity we shall refer to our measurements as yielding the angle between the bilayer normal and the C=O bond. The precise procedures for deriving the aforementioned angle is according to protocols that we have established in the past (Manor et al., 2005) and are outlined in the Supplemental Information Methods Section. Briefly, the process consists of: (i) reconstituting the labelled channels in hydrated vesicles, (ii) removal of bulk solvent whilst depositing the membranes on a solid support to create aligned bilayers, (iii) measuring the dichroism using ATR-FTIR spectroscopy, (iv) assessing the bilayer disorder by X-ray scattering, and finally (v) converting the measured dichroisms to the angles of the carbonyl groups with respect to the membrane normal. We followed this procedure for ten residues spread along the length of the peptide (as listed in Fig. 1a) at acidic (pH 4 to 5) or neutral/basic conditions (pH 7 to 9), in which the channel is open or closed, respectively (Pinto et al., 1992).
Figure 1.
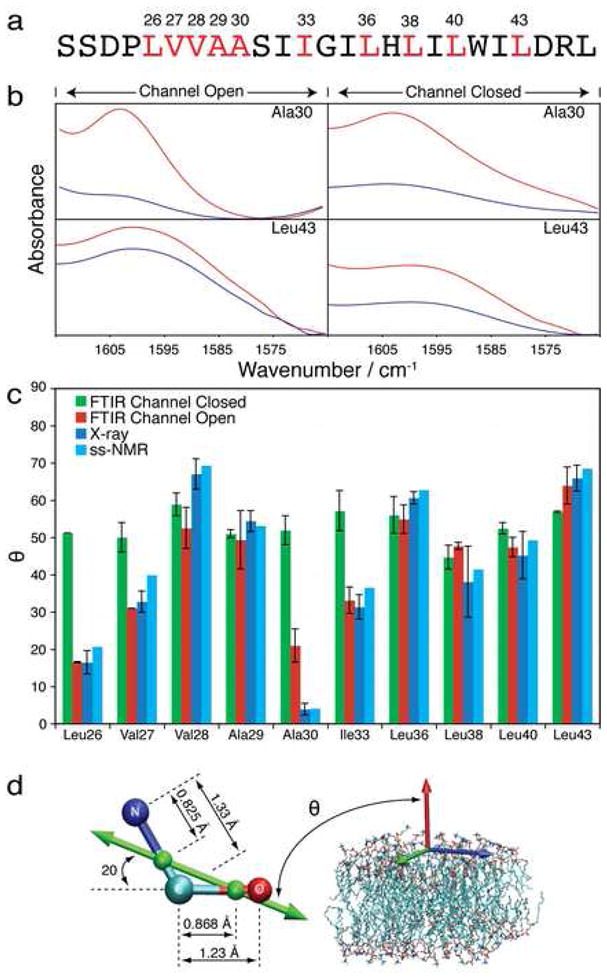
Results of the Linear FTIR spectroscopy measurements. a. Sequence of the M2 transmembrane peptides used in all analyses. The red colouring signifies the 1-13C=18O labelled amino acids. b. Representative ATR-FTIR spectra of the M2 peptide in lipid bilayers focusing on the isotope-edited band of the amide I mode. The spectra were collected with parallel or perpendicular polarized light, red and blue curves, respectively. Spectra of all labels are shown in Supplemental Figures 2. c. A comparison between the tilts of the transition dipole moments of the amide I mode derived from the FTIR studies at acidic (red) or neutral pH (green), and those determined from the X-ray (blue) (Stouffer et al., 2008) and ss-NMR (cyan) (Nishimura et al., 2002) structures. d. Schematic diagram of the orientation of the transition dipole moment of the amide I mode relative to: (i) the amide group’s molecular frame according to (Torii and Tasumi, 1992), and to (ii) the bilayer normal, given by the angle θ.
To demonstrate how sensitive the dichroisms are to the carbonyl orientations in the bilayer, the FTIR spectra of Ala30 and Leu43 are shown in Fig. 1b (FTIR spectra of all labelled peptides are shown in Supplemental Fig. 2). The dichroisms at acidic conditions of Ala30 and Leu43 are 18±8 and 1.4±0.2, respectively. These measurements in turn correspond to dramatically different angles of the carbonyl groups of Ala 30 and Leu43 from the membrane normal of 21±4° and 64±5°, respectively.
The accuracy of our method is assessed by a comparison of the angles we determined using infrared spectroscopy at acidic conditions to the ss-NMR (Nishimura et al., 2002) and X-ray structures (Stouffer et al., 2008). A remarkably good agreement with the previously determined structures is observed (compare the red with the blue and cyan bars in Fig. 1c). The average discrepancy is 6±6° and 7.6±5° between the IR derived angles and the X-ray and ss-NMR structures, respectively. The angular differences between the ss-NMR and X-ray data are 3.2±2°.
Rigid body refinement of an M2 protomer based the experimental constraints
Angular data may be used to construct the backbone structures of proteins by an orientational refinement procedure (Nishimura et al., 2002). Rather than using a flexible peptide model and allowing the dihedral angles to vary for every residue, we have taken a conservative approach and fit our IR bond angles to a rigid helix. The angular refinement consisted of rotating and tilting the helix from the membrane normal and computing the angular deviation from our data, which is plotted in Fig. 2 (top). The minimum deviation occurs for a helix tilted by 27° and rotated by 290°. Despite the fact that the peptide was treated as an ideal helix, this configuration is in close agreement with the structures previously determined by ss-NMR (Nishimura et al., 2002) and X-ray crystallography (Stouffer et al., 2008), with a backbone RMSD between individual helices of 2.4 Å and 2.5 Å, respectively. For comparison, the backbone RMSD between the ss-NMR and X-ray structures is 2.8 Å. Thus, we conclude that the dichroisms provide a quantitatively reliable way of characterizing the backbone structure of membrane proteins.
Figure 2.
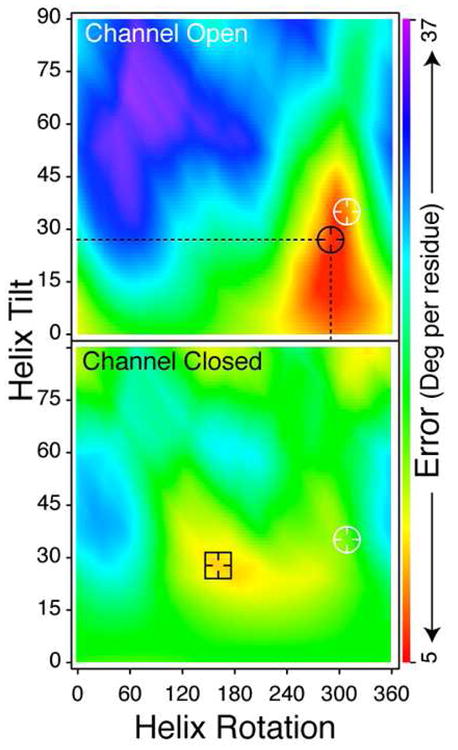
Rigid-body refinement of the M2 protomer according to the FTIR derived angles. Each point in the graph represents a particular combination of tilt and rotation angles for an ideal helix. The colour scale represents the difference per residue (in angles) that the structure has relative to the angles derived from the FTIR study (Fig. 1c). The black circle and square represent the helix tilt and rotation combinations that resulted in the smallest deviation form the experimental results, for the closed and open channel data, respectively. The white circle represents the tilt and rotation of the helices of the X-ray structure (Stouffer et al., 2008).
2D-IR analyses identify the channel’s pore lining residues
We next collected 2D-IR spectra of the same ten isotopically labelled carbonyl groups using a photon echo pulse sequence at acidic conditions. An example 2D-IR spectrum for Ile33 is shown in Fig. 3a (spectra of all labelled peptides are presented in Supplemental Fig. 3). The isotope label appears near 1590 cm−1 and is highlighted with a red arrow in the magnitude spectra. We found in our previous work on the CD3ζ transmembrane peptide (Mukherjee et al., 2006a; Mukherjee et al., 2006b; Mukherjee et al., 2004) that the diagonal linewidths of membrane peptides reflect the disorder of their surrounding electrostatic environments and that one of the biggest factors is the carbonyl’s contact with water, which changes the amount of inhomogeneous broadening (Fang and Hochstrasser, 2005; Fang et al., 2006; Zheng et al., 2007). Since M2 is a transmembrane bundle like CD3ζ, we expect that the diagonal linewidths will scale with the depth of the residue in the bilayer since the lipid tails only weakly interact with the peptide backbone. To extract the linewidths from the data, we followed the same procedure as for CD3ζ (Mukherjee et al., 2006a; Mukherjee et al., 2006b; Mukherjee et al., 2004) and fit the spectra to a sum of 2D lineshapes. The resulting homogeneous and inhomogeneous linewidths are given in Supplemental Tables 1 and 2, as are population relaxation times, which were measured by collecting a series of 2D-IR spectra as a function of the waiting time in the pulse sequence. Each peptide was independently measured and fit at least 3 times in order to ensure the reproducibility of our results and to estimate error bars (60 2D-IR spectra were collected in total for this study). Details of the fitting procedure, fits to the data, and error estimates are presented in the Supplemental Information Section.
Figure 3.
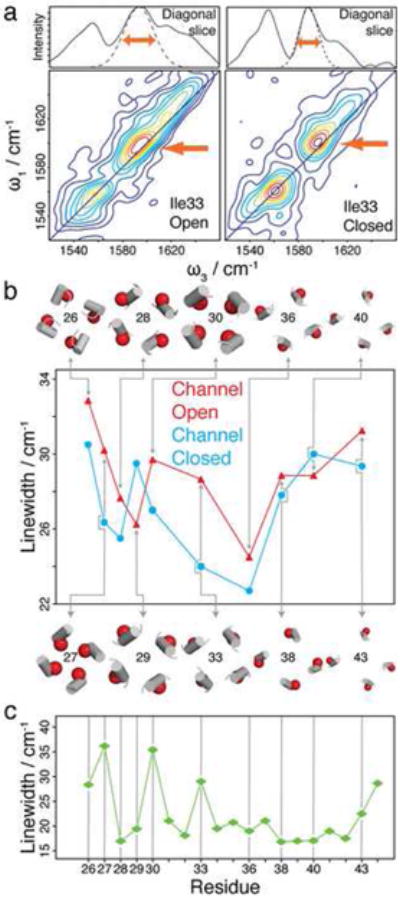
2D-IR spectroscopic measurements and molecular dynamics simulations. a. 2D-IR spectra of Ile33 at acidic (left) and neutral (right) conditions. The isotope labelled peak appears at 1690 cm-1 and is highlighted with a red arrow. (top) Slices along the diagonals of the 2D-IR spectra are shown along with a slice through the fit to the labelled peak that highlights the measured diagonal width (full fitting procedures and data sets are given in the Supplemental Information Section). b. Diagonal linewidths extracted from the 2D-IR spectra of the 10 measured residues at acidic (red triangles) and basic (blue circles) conditions. The positions in the helix bundle of the labelled group (carbonyl oxygen in red), which correlate with the 2D-IR data, were taken from the X-ray structure (Stouffer et al., 2008). The data are presented in a numerical format in Supplemental Tables 1 and 2 in the Supplemental Information Section, which includes error bars. c. Calculated diagonal linewidths from molecular dynamics simulations of the M2 peptide in lipid bilayers. Note the increased linewidths at residues 27, 30, and 33, which are the residues that line the channel pore. Error bars in the calculations are given in the Supplemental Table 3.
The diagonal linewidths of each amide carbonyl are plotted in Fig. 3b (see Fig. 3a, top, for a representative linewidth measurement of Ile33). As expected from our studies on CD3ζ (Mukherjee et al., 2006a; Mukherjee et al., 2006b; Mukherjee et al., 2004), residues at the ends of the peptide near the membrane interface have the largest diagonal linewidths whereas residues in the middle of the membrane have the narrowest linewidth, because the diagonal linewidths scale with the environmental electrostatics. In addition, we also find that the 2D-IR linewidths are sensitive to water inside the channel pore. This effect is best observed at the N-terminal end of the peptide where we were able to label five consecutive residues (Leu26 to Ala30) so that the linewidth of every amino acid was measured for 1.5 turns of the helices. The data in this range reveal an oscillatory trend in linewidths and comparison with the X-ray structure (Stouffer et al., 2008) shows that the linewidths correlate very well to the location of the residue in the helix. This correlation is highlighted in Fig. 3b, where the positions of residues relative to the channel lumen are shown. For example, the inhomogeneous linewidth of Ala30 that is located at the channel lumen is higher than the linewidth of Ala29 that is located in the protein-protein interface. Leu36 has the narrowest linewidth because it is both in the middle of the bilayer and on the outside of the bundle.
Calculation of 2D-IR lineshapes from MD simulation
To further substantiate our observation that the linewidths indicate which residues line the channel pore, we have simulated 2D-IR linewidths from a molecular dynamics trajectory. To accomplish this simulation, we equilibrated the X-ray crystal structure of M2 (Stouffer et al., 2008), assuming that three of the His37 residues are protonated (Chen et al., 2007; Hu et al., 2006), in a hydrated lipid bilayer, followed by a 1 ns molecular dynamics production trajectory. In calculating the spectra, since the amide I modes are isotopically labelled to create a substantial diagonal frequency shift (~60 cm−1) (Torres et al., 2000a; Torres et al., 2001), it is not necessary to include explicit coupling to the unlabeled C=O modes. We calculated the projection of the electric field onto the M2 amide groups at each step in the simulation. The electric fields were then converted into frequencies using a correlation map to give a frequency trajectory, which is used to calculate the 2D-IR spectra (Lin et al., 2008). The details of this procedure are given in the Supplemental Information Section. The simulated linewidths of the 2D-IR magnitude spectra are shown in Fig. 3c. Like the experiment, the calculated linewidths are largest at the terminal ends and narrowest in the middle, which reflect the water content in the membrane. Furthermore, residues 27, 30 and 33, have broad linewidths, consistent with their presence in the pore of the channel, while residues 28, 29, 31 and 32 have narrow linewidths because they are on the outside of the channel or between the helices. This oscillatory trend matches the experimental results for the open channel shown in Fig. 3b. Thus, the simulations support the experimental observation that the linewidths reflect which residues line the channel pore. We know from our previous work that although the correlation between electric field and vibrational frequency is not yet accurate enough to quantitatively simulate IR spectra, (Lin et al., 2008) our method does reproduce the correct trends in the linewidths of membrane proteins caused by water and membrane headgroups. Therefore these simulations support the conclusion that the phase of the oscillatory trend observed in the experiments is a measure of the rotational angle of the helices with regard to the channel pore.
2D-IR reveals a shift in the channel’s pore lining residues
Having established the information content of our 1 and 2D-IR approach using a well-characterized protein structure, we turned to studying the structural changes associated with channel gating by performing the same experiments at neutral/basic conditions when the channel is in its closed state (Pinto et al., 1992). We find that many of the residues change their linewidths as shown for Ile33 in Fig. 3a (all of the 2D-IR data and fits are given in Supplemental Fig. 3, 4 and 5). At neutral conditions, the largest linewidths are still observed at the ends of the helices, and the narrowest in the middle, indicating that the structure of the closed channel lies at about the same depth as when it is open. However, the phase of the oscillation between residues Leu26 and Ala30 is now shifted by about one residue (see Fig. 4 top panel). For instance, Ala30, which had a very large linewidth because it was located inside the pore in the open state now has a linewidth that is comparable to the narrow linewidths of dehydrated residues on the outside of the bundle. In contrast, Ala29 shows the opposite trend, indicating that it is exposed to a much stronger electrostatic environment in the closed state in comparison to the open state, strongly suggesting that it has become hydrated. In comparison, the C-terminal end shows little difference in linewidths between the closed and open states. This may be due to the fact that the protein forms a cone shaped structure in which the N-terminus is well packed and the C-terminal segment forms fewer protein-protein contacts (Stouffer et al., 2008). Thus the accessibility to water of individual residues at the C-terminus is already high and therefore does not depend on their rotational position. We also note that the trend in experimental linewidths of residues 27–30 and 33 in the closed state no longer match the simulations of the open state. Taken together, the most straightforward interpretation of the 2D-IR linewidth changes is that the helices rotate by one amino acid register upon channel activation, such that the channel is lined by different sets of residues in the open and closed states.
Figure 4.
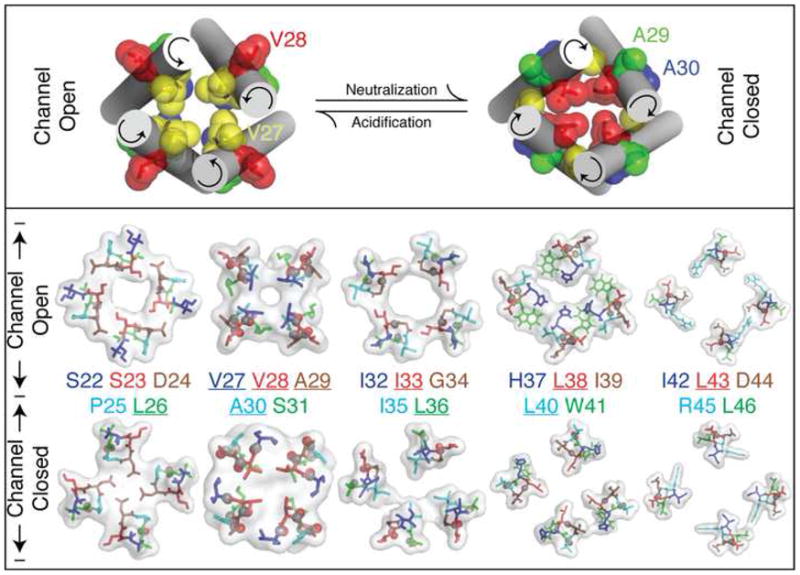
Visualization of the proposed gating mechanism. Top panel: The location of residues Val27 – Ala30 in our models of the closed and open conformations of the M2 channel. Note the different rotational location of the residues that is consistent with the 2D-IR data shown in Fig. 3. Bottom panel: Slices through the M2 tetrameric complexes constructed according to the FTIR derived angles at the two pH conditions. The location of the labelled carbonyl group (underlined residues), are shown in CPK representation.
Large angular changes between the two pH conditions are observed by linear FTIR
To gain a more detailed insight into this structural change, we turned back to our 1D dichroism measurements, which we repeated at neutral conditions. The extracted angles between the labelled C=O groups and the bilayer normal are shown in Fig. 1c (green bars). We find substantial changes in the carbonyl angles from acidic to neutral conditions, especially at residues Leu26, Ala30 and Ala33, where the angles change by >30°, indicating large structural changes in the helices. It is important to realize that an observed change in the angle between a carbonyl bond and the bilayer normal mandates a structural change of the protein. However, a lack of a change in the carbonyl bond angle does not mean that a structural change did not occur, because a single carbonyl group’s projection on the membrane normal does not restrain a structure unambiguously. We also note that it is merely a geometrical coincidence that the angles measured for the closed channel do not exhibit the extent of variation that was observed for the open channel.
Rigid-body refinement suggests a rotation of the helices
To quantify the structural change that accompanies gating, we once again employed rigid body refinement, rotating and tilting the helix to make a new refinement map based on the angles collected at neutral conditions. The new map, shown in Fig. 2 (bottom) has a minimum at the same tilt angle (28º), but with a rotational angle that is 129º different from the configuration obtained at acidic conditions. Thus, according to both the 1 and 2D-IR data sets, the helices conserve their tilt but undergo a large rotation, suggesting that the channel pore is lined by a number of different residues at acidic conditions compared to neutral state. Specifically, if under acidic conditions residue number i lines the pore, at neutral conditions, residue i + 1 faces the pore. Finally, we note that the discrepancy between the experimental results and the refinement model of the closed channel is higher than that of the open channel. In Supplemental Figure 6, we plot the errors for the individual residues and find that the overall error is not larger because of a single residue, but is generally larger for all of the residues. This might indicate that the approximation of an ideal helix is a better assumption in the open state than in the closed state. Regardless, this structure is not in agreement with the ss-NMR or x-ray derived M2 bundles, because the helices are rotated by 100 to 130°.
Fully constructed M2 complex reveals the properties of the open and closed states
To better understand the pore structure and the gating movement that we observed, we constructed the tetrameric complex from the optimized helices at the two pH conditions. Our IR data do not currently yield distance constraints between the helices, which are necessary to reliably construct the ion channel from four individual helices. We therefore constructed the channel complex using the interhelical distances measured in the X-ray structure (Stouffer et al., 2008). Thus, the helices are constrained at the tilt and rotations predicted by our data, translated to proper relative distances, while the side-chains were allowed to relax. Slices through the two complexes, which include a surface representation looking at the channel from the N-terminus are shown in Fig. 4. At neutral conditions, we find that the channel is closed and that at acidic conditions the channel is open, consistent with physiological results (Pinto et al., 1992). Thus, a gating mechanism that is coupled to rotational motion of the helices can explain our results.
The points of contact that seemingly close the channel in neutral conditions are Asp24 and Val28. Asp24 is one of the most conserved residues in the channel, and together with His37 are the only ionizable residues in the membrane vicinity. Therefore it is tempting to speculate that both His37 and Asp24 undergo a change in protonation state upon channel activation. Under acidic conditions His37 would be charged and water accessible, while Asp24 would be neutral and potentially water inaccessible. Under neutral conditions the reverse will transpire, leading to exposure of Asp24 and sequestration of His37. Interestingly that is exactly what we observe in our model shown in Fig. 4. Further experimentation is needed to substantiate the above hypothesis.
Other membrane proteins undergo rotational motion upon gating. For example, the pore lining helices of the acetylcholine receptor are thought to rotate upon binding acetylcholine, although the extent of rotation is still under debate (Law et al., 2005; Miyazawa et al., 2003; Sansom, 1995). Similarly, both the large and small conductance mechanosensitive channels from E. coli contain helices that are thought undergo significant rotation upon activation (Bartlett et al., 2006; Edwards et al., 2005). Finally, thiol-disulfide equilibria mapping (Stouffer et al., 2008) as well as ss-NMR experiments (Bauer et al., 1999; Li et al., 2007; Tian et al., 2003) suggest that the M2 sequence is particularly tuned towards reduced stability that may accompany large motions.
Based on the above we conclude the following: (i) Our low pH results, both in terms of hydration and orientational data, are highly consistent with the previously available structures determined by either ss-NMR (Nishimura et al., 2002) or X-ray crystallography (Stouffer et al., 2008), (ii) Both our 1 and 2D-IR data sets consistently point to a large structural reorganization between acidic and neutral pH, leading to a structure that is significantly different from published ss-NMR (Nishimura et al., 2002) and X-ray (Stouffer et al., 2008) structures.
An important strength of our approach is that we are able to study membrane proteins in the natural environment of a lipid bilayer and, because of the high sensitivity of IR spectroscopy, at near-physiological protein-to-lipid ratios (<1:20 by weight). As a result, we can ascertain the accuracy of structures obtained by X-ray crystallography or ss-NMR, which are performed in detergent micelles or at high protein-to-lipid ratios. The ability to double-check structures in bilayers is important because detergents can alter the energetics and structures of the transmembrane helices, as was documented for human glycophorin A and EmrE (Fisher et al., 1999; Fisher et al., 2003; Soskine et al., 2006). Finally, by altering the association energetics of a transmembrane helical bundle, detergents may shift the equilibrium between two states.
1 and 2D-IR studies can serve to independently assess the validity of a structure
In this study, we find that the structures determined by ss-NMR (Nishimura et al., 2002) and X-ray crystallography (Stouffer et al., 2008) are structurally accurate as compared to our structure, but only if those experiments examined the channel in its open state, not the closed state. High protein-to-lipid ratios, or detergents, as discussed above, could cause this discrepancy but it could also be caused by a difference in pH, since the pH is responsible for gating in M2. pH can be difficult to maintain in samples with reduced hydration, like that used in ss-NMR, and unknown in crystal structures, especially when heavy metals which are used in crystallization may act as acids. Moreover, we note that the X-ray structures obtained at low and high pH (Stouffer et al., 2008) are very similar to one another (transmembrane Ca-RMSD of 1.07Å). This finding may point to the fact that the detergent micelle preferentially stabilizes one state of the protein, irrespective of the pH. The ss-NMR structure was obtained only at a single pH (Nishimura et al., 2002).
Now, with 1 and 2D-IR spectroscopies, it is possible to provide an independent check on the structures of proteins using backbone hydration and conformation data. The ability to probe hydration levels in ion channels without structural perturbation is particularly appealing, especially since 2D-IR lineshapes can be predicted from molecular dynamics simulations (Choi et al., 2005; Jansen et al., 2006; Schmidt et al., 2004; Zhuang et al., 2006), as we demonstrated here.
In summary, we have found that a pH change induces a large-scale conformational change in the channel structure. While our results might be explained by a number of possible structural changes, the 2D-IR linewidths suggest that the channel pore is lined by a different set of residues in the open and closed states. Moreover, this change is highly consistent with our angular constraints derived from linear FTIR spectroscopy.
This change has implications for the binding of amantadine or other drugs to the channel, since a change in residues will change the size and hydrophobicity of the channel. During the course of these experiments, we have also demonstrated the utility of using infrared spectroscopy to quantitatively and qualitatively assess the structure of a membrane peptide channel. These methodologies might find use on other ion channel systems or be applied in a time-resolved manner to study membrane protein dynamics.
Experimental Procedures
Linear FTIR spectra
The synthesis, labelling, purification and lipid reconstitution of the peptides used in this work have been described elsewhere (Torres et al., 2000a; Torres et al., 2000b), and are specified in detail in the Supplemental Information Section. Similarly, the ATR-FTIR spectroscopy and the X-ray Scattering bilayer mosaicity estimation were undertaken as described elsewhere (Manor et al., 2005; Torres et al., 2000b), and specified in detail in the Supplemental Information Section.
2D-IR spectra
The experimental procedure for collecting heterodyned 2D-IR spectra is given in detail elsewhere (Fulmer et al., 2004) and described in the Supplemental Information Section. To extract the 2D lineshape of the labelled amide I mode, we follow our previously established fitting procedure of using the Fourier transform of the 3rd-order response in the limit of Bloch dynamics with a pure dephasing time of T2* and an inhomogeneous distribution of Δ0 (Mukherjee et al., 2004).
pH control
The pH control in our experiments was conducted as follows: In the ATR-FTIR work, HCl, NaOH and phosphate buffer saline (PBS) were used to bring the solution to a final pH of 4, 7 or 9, respectively. The pH was monitored throughout the entire process, including during the removal of the bulk solvent from the sample before measuring the FTIR spectra, and was found to change only marginally in all three preparations. The three different conditions had no visible effect on the FTIR spectra. HCl-treated preparation is addressed as low pH samples throughout this work while PBS or NaOH treated preparations are addressed as neutral/basic pH samples. No differences in the results were obtained between preparations with PBS or NaOH. In the 2D-IR work, the vesicles resided in excess water. For the closed state measurements the pH was checked by blotting a small amount on pH paper. For the open state, the sample was buffered to pH 5.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the NIH (R21AI064797) to MTZ and ITA, a grant form the Israeli science foundation (784/01,1249/05,1581/08) to ITA. JLS thanks NSF (CHE-0750307) for support. Funding was also provided to JLS and MTZ through an NSF CRC grant (CHE-0832584). ITA is the Arthur Lejwa Professor of Structural Biochemistry at the Hebrew University of Jerusalem. The authors wish to thank Prof W.F. DeGrado for thoughtful discussions. Molecular figures were generated by PyMOL (DeLano Scientific LLC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartlett JL, Li Y, Blount P. Mechanosensitive channel gating transitions resolved by functional changes upon pore modification. Biophys J. 2006;91:3684–3691. doi: 10.1529/biophysj.106.088062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer CM, Pinto LH, Cross TA, Lamb RA. The influenza virus M2 ion channel protein: probing the structure of the transmembrane domain in intact cells by using engineered disulfide cross-linking. Virology. 1999;254:196–209. doi: 10.1006/viro.1998.9552. [DOI] [PubMed] [Google Scholar]
- Chen H, Wu Y, Voth GA. Proton transport behavior through the influenza A M2 channel: insights from molecular simulation. Biophys J. 2007;93:3470–3479. doi: 10.1529/biophysj.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JH, Hahn S, Cho M. Amide IIR, VCD, and 2D IR spectra of isotope-labeled alpha-helix in liquid water: Numerical simulation studies. International Journal of Quantum Chemistry. 2005;104:616–634. [Google Scholar]
- Duff KC, Ashley RH. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- Duff KC, Kelly SM, Price NC, Bradshaw JP. The secondary structure of influenza A M2 transmembrane domain. A circular dichroism study. FEBS Lett. 1992;311:256–258. doi: 10.1016/0014-5793(92)81114-2. [DOI] [PubMed] [Google Scholar]
- Edwards MD, Li Y, Kim S, Miller S, Bartlett W, Black S, Dennison S, Iscla I, Blount P, Bowie JU, Booth IR. Pivotal role of the glycine-rich TM3 helix in gating the MscS mechanosensitive channel. Nat Struct Mol Biol. 2005;12:113–119. doi: 10.1038/nsmb895. [DOI] [PubMed] [Google Scholar]
- Fang C, Hochstrasser RM. Two-dimensional infrared spectra of the 13C=18O isotopomers of alanine residues in an alpha-helix. J Phys Chem B. 2005;109:18652–18663. doi: 10.1021/jp052525p. [DOI] [PubMed] [Google Scholar]
- Fang C, Senes A, Cristian L, DeGrado WF, Hochstrasser RM. Amide vibrations are delocalized across the hydrophobic interface of a transmembrane helix dimer. Proc Natl Acad Sci U S A. 2006;103:16740–16745. doi: 10.1073/pnas.0608243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher LE, Engelman DM, Sturgis JN. Detergents modulate dimerization, but not helicity, of the glycophorin A transmembrane domain. J Mol Biol. 1999;293:639–651. doi: 10.1006/jmbi.1999.3126. [DOI] [PubMed] [Google Scholar]
- Fisher LE, Engelman DM, Sturgis JN. Effect of detergents on the association of the glycophorin a transmembrane helix. Biophys J. 2003;85:3097–3105. doi: 10.1016/S0006-3495(03)74728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulmer EC, Mukherjee P, Krummel AT, Zanni MT. A pulse sequence for directly measuring the anharmonicities of coupled vibrations: Two-quantum two-dimensional infrared spectroscopy. J Chem Phys. 2004;120:8067–8078. doi: 10.1063/1.1649725. [DOI] [PubMed] [Google Scholar]
- Holsinger LJ, Lamb RA. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology. 1991;183:32–43. doi: 10.1016/0042-6822(91)90115-r. [DOI] [PubMed] [Google Scholar]
- Hu J, Fu R, Cross TA. The chemical and dynamical influence of the anti-viral drug amantadine on the M2 proton channel transmembrane domain. Biophys J. 2007;93:276–283. doi: 10.1529/biophysj.106.102103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Fu R, Nishimura K, Zhang L, Zhou HX, Busath DD, Vijayvergiya V, Cross TA. Histidines, heart of the hydrogen ion channel from influenza A virus: toward an understanding of conductance and proton selectivity. Proc Natl Acad Sci U S A. 2006;103:6865–6870. doi: 10.1073/pnas.0601944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen TL, Dijkstra AG, Watson TM, Hirst JD, Knoester J. Modeling the amide I bands of small peptides. Journal of Chemical Physics. 2006;125:04431201–04431209. doi: 10.1063/1.2218516. [DOI] [PubMed] [Google Scholar]
- Law RJ, Henchman RH, McCammon JA. A gating mechanism proposed from a simulation of a human alpha7 nicotinic acetylcholine receptor. Proc Natl Acad Sci U S A. 2005;102:6813–6818. doi: 10.1073/pnas.0407739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Qin H, Gao FP, Cross TA. Solid-state NMR characterization of conformational plasticity within the transmembrane domain of the influenza A M2 proton channel. Biochim Biophys Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Shorb JM, Mukherjee P, Zanni MT, Skinner JL. Empirical amide I vibrational frequency map: Application to 2D-IR line shapes for isotope-edited membrane peptide bundles. J Phys Chem B. 2008 doi: 10.1021/jp807528q. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor J, Khattari Z, Salditt T, Arkin IT. Disorder influence on linear dichroism analyses of smectic phases. Biophys J. 2005;89:563–571. doi: 10.1529/biophysj.104.058842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Unwin N. Structure and gating mechanism of the acetylcholine receptor pore. Nature. 2003;423:949–955. doi: 10.1038/nature01748. [DOI] [PubMed] [Google Scholar]
- Mukherjee P, Kass I, Arkin IT, Zanni MT. Picosecond dynamics of a membrane protein revealed by 2D IR. Proc Natl Acad Sci U S A. 2006a;103:3528–3533. doi: 10.1073/pnas.0508833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Kass I, Arkin IT, Zanni MT. Structural disorder of the CD3zeta transmembrane domain studied with 2D IR spectroscopy and molecular dynamics simulations. J Phys Chem B. 2006b;110:24740–24749. doi: 10.1021/jp0640530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee P, Krummel AT, Fulmer EC, Kass I, Arkin IT, Zanni MT. Site-specific vibrational dynamics of the CD3zeta membrane peptide using heterodyned two-dimensional infrared photon echo spectroscopy. J Chem Phys. 2004;120:10215–10224. doi: 10.1063/1.1718332. [DOI] [PubMed] [Google Scholar]
- Nishimura K, Kim S, Zhang L, Cross TA. The closed state of a H+ channel helical bundle combining precise orientational and distance restraints from solid state NMR. Biochemistry. 2002;41:13170–13177. doi: 10.1021/bi0262799. [DOI] [PubMed] [Google Scholar]
- Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- Salom D, Hill BR, Lear JD, DeGrado WF. pH-dependent tetramerization and amantadine binding of the transmembrane helix of M2 from the influenza A virus. Biochemistry. 2000;39:14160–14170. doi: 10.1021/bi001799u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansom MS. Ion-channel gating. Twist to open. Curr Biol. 1995;5:373–375. doi: 10.1016/s0960-9822(95)00076-5. [DOI] [PubMed] [Google Scholar]
- Schmidt JR, Corcelli SA, Skinner JL. Ultrafast vibrational spectroscopy of water and aqueous N-methylacetamide: Comparison of different electronic structure/molecular dynamics approaches. J Chem Phys. 2004;121:8887–8896. doi: 10.1063/1.1791632. [DOI] [PubMed] [Google Scholar]
- Soskine M, Mark S, Tayer N, Mizrachi R, Schuldiner S. On parallel and antiparallel topology of a homodimeric multidrug transporter. J Biol Chem. 2006;281:36205–36212. doi: 10.1074/jbc.M607186200. [DOI] [PubMed] [Google Scholar]
- Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Protein Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii H, Tasumi M. Model-Calculations on the Amide-I Infrared Bands of Globular-Proteins. Journal of Chemical Physics. 1992;96:3379–3387. [Google Scholar]
- Torres J, Adams PD, Arkin IT. Use of a new label, (13)==(18)O, in the determination of a structural model of phospholamban in a lipid bilayer. Spatial restraints resolve the ambiguity arising from interpretations of mutagenesis data. J Mol Biol. 2000a;300:677–685. doi: 10.1006/jmbi.2000.3885. [DOI] [PubMed] [Google Scholar]
- Torres J, Kukol A, Arkin IT. Use of a single glycine residue to determine the tilt and orientation of a transmembrane helix. A new structural label for infrared spectroscopy. Biophys J. 2000b;79:3139–3143. doi: 10.1016/S0006-3495(00)76547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres J, Kukol A, Goodman JM, Arkin IT. Site-specific examination of secondary structure and orientation determination in membrane proteins: the peptidic (13)C=(18)O group as a novel infrared probe. Biopolymers. 2001;59:396–401. doi: 10.1002/1097-0282(200111)59:6<396::AID-BIP1044>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: a role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Kwak K, Fayer MD. Ultrafast 2D IR vibrational echo spectroscopy. Acc Chem Res. 2007;40:75–83. doi: 10.1021/ar068010d. [DOI] [PubMed] [Google Scholar]
- Zhuang W, Abramavicius D, Hayashi T, Mukamel S. Simulation protocols for coherent femtosecond vibrational spectra of peptides. Journal of Physical Chemistry B. 2006;110:3362–3374. doi: 10.1021/jp055813u. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


