Abstract
Since the work of Golgi and Cajal, light microscopy has remained a key tool for neuroscientists to observe cellular properties. Ongoing advances have enabled new experimental capabilities using light to inspect the nervous system across multiple spatial scales, including ultrastructural scales finer than the optical diffraction limit. Other progress permits functional imaging at faster speeds, at greater depths in brain tissue, and over larger tissue volumes than previously possible. Portable, miniaturized fluorescence microscopes now allow brain imaging in freely behaving mice. Complementary progress on animal preparations has enabled imaging in head-restrained behaving animals, as well as time-lapse microscopy studies in the brains of live subjects. Mouse genetic approaches permit mosaic and inducible fluorescence-labeling strategies, whereas intrinsic contrast mechanisms allow in vivo imaging of animals and humans without use of exogenous markers. This review surveys such advances and highlights emerging capabilities of particular interest to neuroscientists.
Keywords: two-photon fluorescence, super-resolution, fiber optics, laser-scanning, fluorescence labeling, transgenic mice
THE CHANGING ROLE OF MICROSCOPY IN NEUROSCIENCE
The light microscope has long been one of neuroscientists’ cardinal tools. When used together with Golgi’s technique for staining a sparse population of cells, the light microscope provided the data that drove the famous debate between Golgi and Cajal about whether the nervous system was composed of cells or a syncytium (Cajal 1906, Golgi 1906). Although neuroscientists historically used light microscopy mainly to inspect histological specimens for studies of cellular morphology and the brain’s cyto-architecture, optical microscopy has progressed to where it now routinely provides information about cellular and circuit dynamics, on timescales ranging from milliseconds to months. In addition to this considerable expansion in usage, the basic character of the data provided by the light microscope has also evolved.
Early studies in light microscopy treated images as data represented in pictorial form. These images were either observed directly by eye or captured by photography, but in both cases the data were inspected visually. Over the past few decades, digital image acquisition and laser-scanning microscopy have transformed the data microscopes typically provide into a numerical format, with a specified number of bits per image pixel. This transition has in turn facilitated computational approaches to image data analysis. Today, the ready availability of fast computers is fueling another generation of microscopy techniques that reaches an even higher level of abstraction. Several methods we discuss involve the acquisition of raw images that often lack a straightforward relationship to the structures present in the sample, at least in a way readily discernible by eye (Figures 3, 4, 10, 11). Instead, representations of the sample are reconstructed computationally. The acquired data with these methods are sufficiently divorced from the final reconstructions that the latter images should perhaps be understood as computed hypotheses about the underlying biological reality. These hypotheses may come with statistical likelihoods weighing the evidence and expressing the degree of confidence in the image representation. This reconceptualization of images as hypotheses can have practical consequences by forcing scientists to focus on the ideas being tested and image interpretations needing scrutiny, and to design experiments and analyses accordingly, rather than on optimizing images visual qualities. Optimal configurations for hypothesis testing might even produce results surprisingly poor to the eye. This gradual shift by microscopists toward logical abstraction mimics what has already occurred in other fields, such as astronomy, radiology, and particle physics, in which imaging has long been pivotal to scientific discovery (Galison 1997).
Figure 3.
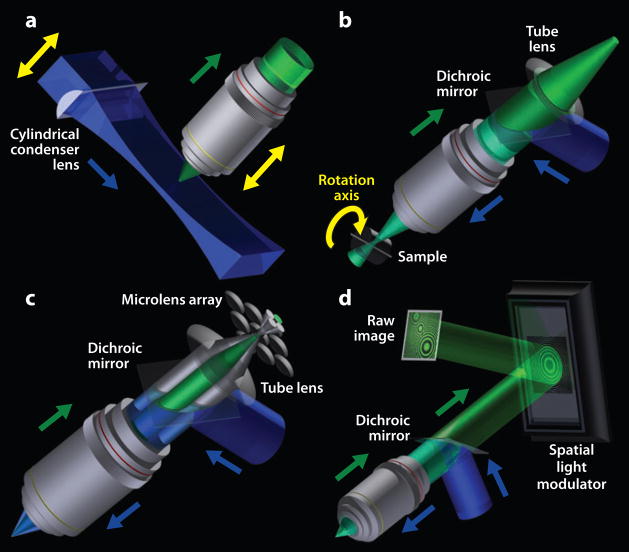
Three-dimensional imaging using linear contrast mechanisms. Fluorescence is depicted here as the contrast mechanism, but alternative contrast mechanisms are available for these techniques. (a) Objective-coupled planar illumination microscopy (OCPI). A cylindrical condenser lens focuses illumination to a sheet of light 4–10 μm thick, exciting fluorescence within an optically sectioned plane. The cylindrical lens and the objective lens are mechanically coupled and move together, enabling fast volume scanning. (b) Optical projection tomography (OPT). OPT reconstructs a 3D sample volume by acquiring a large set of 2D projection images from a rotating sample in a manner analogous to X-ray computed tomography (CT). Each pixel within an individual 2D projection contains a mixture of fluorescence signals originating from many sample voxels. By combining information from projections acquired over multiple angular orientations of the specimen, one can unmix or back compute these signals by using a Radon transform to determine the fluorescence intensities of the individual voxels. (c) Light field microscopy. An array of microlenses situated in an image plane creates a corresponding array of microimages. Light rays originating from different locations in the sample are focused by different microlenses to distinct microimages; rays from nearby locations are focused to the same microimage. Light rays coming from the same neighborhood in the specimen but emerging at different angles reach different pixels within the same microimage. (Light propagation is shown here only for a single neighborhood on the optical axis, with only four cones of light emerging at four distinct angles). The combination of both position and angular information in the set of raw 2D images provides a description of a light field. This in turn allows the sample to be viewed from different perspectives (Figure 4b) or to be synthetically focused at different depths (not shown). (d) Nonscanning holographic fluorescence microscopy. A spatial light modulator (SLM) projects a hologram-generating pattern that reflects two spherical waves of distinct curvature for each emitter in the sample. These two spherical waves interfere at the detector, creating a hologram of the sample (raw image) in which concentric circles encode the emitters’ lateral positions and depths in the circles’ locations and densities, respectively. Three holograms are sequentially recorded per image frame, each using a distinct phase offset for the reflection pattern on the SLM. The holograms are superposed and convolved with a holographic response function to reconstruct the spatial distribution of emitters.
Figure 4.
Image volumes acquired using linear contrast mechanisms. (a) One slice from an image volume obtained by objective-coupled planar illumination microscopy, revealing neuronal somata labeled with the Ca2+-sensitive indicator Oregon Green BAPTA-1 am in the mouse vomeronasal sensory epithelium (Holekamp et al. 2008). (b) Light field microscopy images of Golgi-stained rat neurons. The four images shown were reconstructed from a single raw image, a portion of which is shown (yellow inset). Each image was constructed as the array of intensities from all pixels located at a specific position within the individual microimages, as schematized (orange inset: left pixels, blue; right pixels, red; center pixels, white; top pixels, green). The resulting images from the four distinct angular perspectives are shown within the color corresponding boxes. The degree of accessible parallax is most evident in the looping processes in the lower left of each image (Levoy 2009). (c) Three-dimensional (3D) gene expression maps in Drosophila melanogaster generated using optical projection tomography (OPT). Signals from autofluorescence (green) and bright field readouts of both β-galactosidase activity (blue) and exoskeleton (red) are shown (McGurk et al. 2007). (d) 3D reconstruction of an E12.5 mouse embryo using OPT imaging of fluorescence signals after immunostaining with a monoclonal anti-neurofilament 160 conjugated to Alexa Fluor 488 (green). The fluorescence image is superimposed on a bright field anatomical OPT image (gray) (image courtesy of J. Sharpe). (e) Time evolution of gene expression patterns (Scx-GFP) in mouse limb buds monitored by OPT. Two views of the 3D gene expression patterns are shown for 0, 13, and 19 h (Boot et al. 2008).
Figure 10.
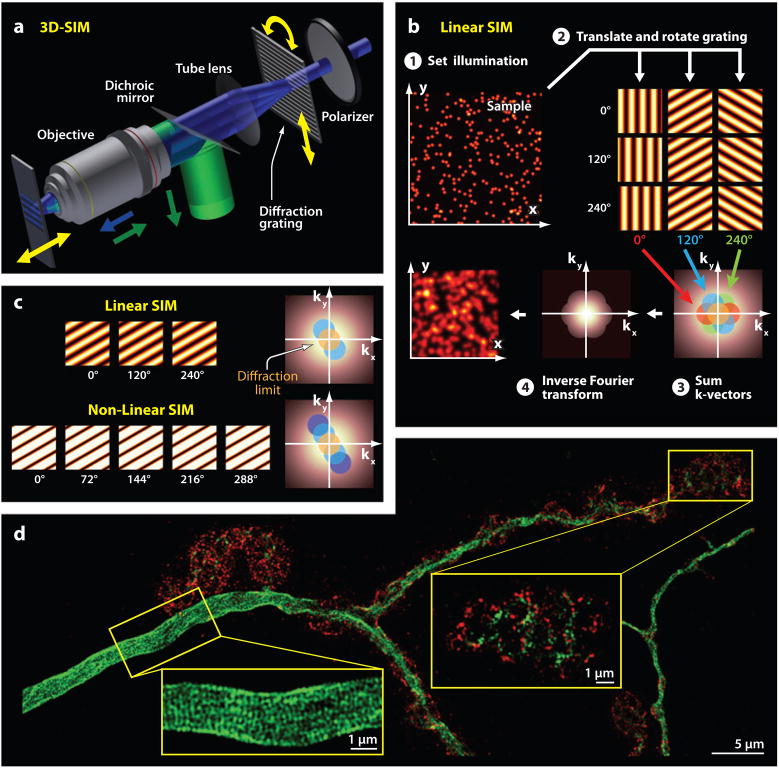
Structured illumination microscopy (SIM). (a) A 3D SIM microscope employs periodic illumination patterns generated by the interference of multiple beams at the sample plane. To permit structured illumination imaging of the finest resolution, the spatial frequencies of the illumination patterns must closely approach the highest values the microscope can transmit. (b) The sequence of steps in 2D linear SIM. The sample is illuminated with a sequence of sine wave spatial patterns that are generated by the interference of two mutually coherent illumination beams. A set of images is acquired using at least three different orientations of the sine pattern, produced by using different in-plane rotations of the grating (three columns marked by angular orientation labels in color). Each pattern orientation is imaged using at least three different phase shifts, produced by using different in-plane translations of the grating (three rows marked by phase shift labels in white). These periodic illumination patterns generate moiré-like interference patterns when mixed (convolved) with the set of spatial frequencies present in the sample structure (i.e., in the spatial frequency transform of the distribution of fluorophores). Thus, although a microscope cannot directly transmit high spatial frequency information beyond the diffraction limit, deliberately inducing aliasing encodes information in the resulting beat pattern between the spatial frequencies present in the illumination and in the sample. Computationally extracting this encoded information and moving it back to its original position in frequency space, and then performing an inverse Fourier transform, yields a super-resolution image with about a twofold improvement over the resolution of conventional microscopy. In 3D SIM, three spatially coherent beams interfere at the sample, creating an illumination pattern with added structure in the axial direction. The axial component of the illumination structure encodes high-axial-resolution sample information in the image data, in a fashion similar to what happens in the lateral direction. The axial information does not require phase-shifting to extract but can be accessed simply by acquiring a focal series with the focal planes spaced twice as densely as normal. (c) Nonlinear SIM, for example using saturating illumination, provides even higher resolution than does linear SIM by applying sine wave illumination patterns that, at their peaks, produce saturating fluorescence excitation. This saturation adds a nonlinearity to the dependence of excitation on illumination intensity and thereby creates higher-order harmonics in the spatial modulation of the fluorescence excitation efficiency. To reconstruct images containing these higher-order spatial frequencies, a greater number of raw images must be acquired with different translations and rotations of the grating than that in linear SIM. Nonlinear SIM allows images to be constructed with about a fourfold improvement in resolution as compared with conventional microscopy but puts greater demands on the phototolerance of the sample. (d) 3D linear SIM reveals axonal and synaptic organization in wild-type Drosophila muscle 4 neuromuscular junction stained for Ank2-L (green) and colabeled with cell adhesion molecule Fas II (red). Insets show greater detail in densely labeled regions (Pielage et al. 2008).
Figure 11.
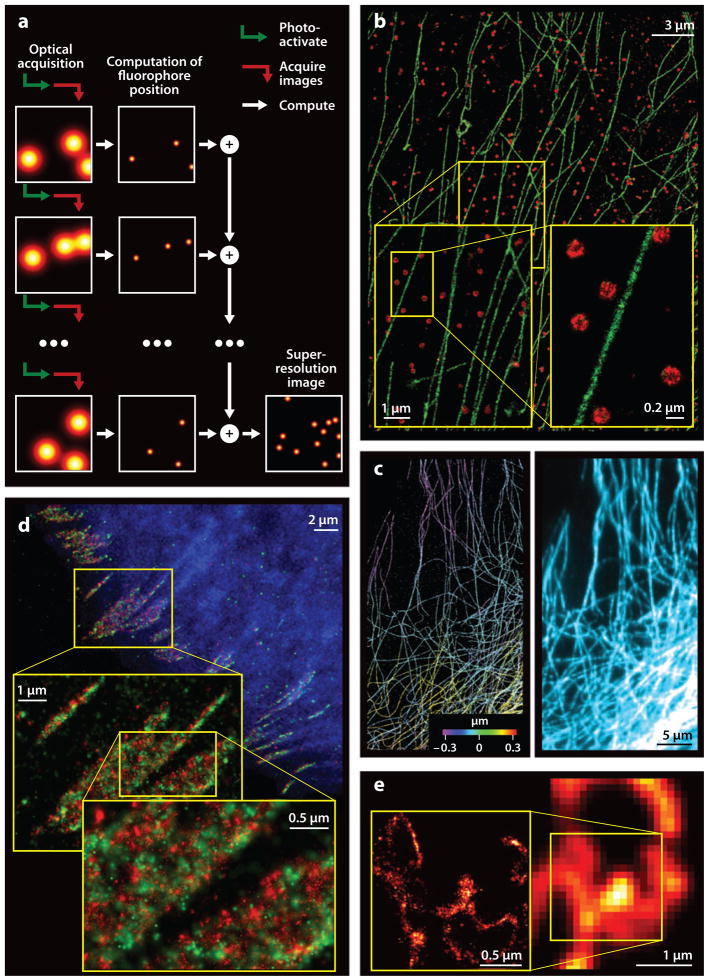
Super-resolution microscopy based on localization of single molecules. (a) To create super-resolution images based on fluorescence signals from single molecules, a sequence of raw images is acquired in which only a sparse subset of the available fluorophores has been activated in each image. This procedure enables each fluorophore to be localized to within sub-diffraction-limited bounds. A super-resolution image results from the sum of the localized fluorophores. (b) Multicolor stochastic optical reconstruction microscopy (STORM) images of clathrin-coated pits labeled with Cy3-Alexa 647 (red) and microtubules labeled with Cy2-Alexa 647 (green) in a BS-C-1 cell shown at three different magnifications (Bates et al. 2007). (c) 3D STORM (left) and corresponding diffraction-limited (right) images of microtubule networks in kidney epithelial (BS-C-1) cells (Huang et al. 2008b). By introducing astigmatism into the optical pathway, the axial position of each fluorophore is encoded in the ellipticity of the spatial distribution of photons emitted from that fluorophore and captured on the camera. Axial position is indicated in color in the 3D-STORM image. (d) Multicolor photoactivated localization microscopy (PALM) image showing the distribution of adhesion complex proteins in a fibroblast (HFF-1) cell. Epi-fluorescence image of mCerulean-tagged actin (blue) overlaid with a PALM image of Dronpa-labeled paxillin ( green) and tdEos-labeled vinculin (red). Images are shown at three different magnifications (Shroff et al. 2007). (e) An image acquired by total internal reflection microscopy (TIRF) of a COS-7 cell expressing the lysosomal transmembrane protein CD63 fused to the photoactivatable fluorescent protein Kaede and the corresponding PALM image (left inset) (Betzig et al. 2006).
Owing to advances on multiple technological fronts, the present generation of light microscopes can provide data about spatial scales and experimental situations far beyond what was feasible even a decade ago. The past few years have produced remarkable progress in microscopy, including several new optical methods available to help neuroscientists probe ultrastructural issues, as well as other methods for visualizing cellular dynamics in behaving animals. Advances in automation and image analysis are propelling capabilities for rapid screening and large-scale anatomical reconstruction. Gains in optics and computational techniques, as well as an expanding set of contrast markers and functional indicators, underlie much of the recent advancement. However, improvements in complementary areas including tissue processing, animal preparations, and genetic strategies for fluorescence labeling have been equally important.
In this review, we survey progress in light microscopy, mainly over the past three years, with an eye to those advances poised to impact the practice of neuroscience. Many of the areas we discuss are experiencing rapid growth or already have a substantial research literature. Thus, our goal in writing this review was not to convey all relevant technical details. Rather, we sought to make neuroscientists aware of the growing capabilities they have at their disposal, to convey basic strengths and limitations of each approach, and to help readers decide which techniques might be most appropriate for their own research. In choosing topics, we deliberately omitted several exciting areas, except in passing, owing to the prior existence of excellent reviews covering these areas. These omissions include photostimulation, optogenetics, and fluorescent optical indicators. Recent progress on these fronts has helped energize the field and complements the topics we present (Giepmans et al. 2006, Gorostiza & Isacoff 2008, Miyawaki 2005, Shaner et al. 2005, Zhang et al. 2007).
ADVANCES IN OPTICAL INSTRUMENTATION
Two-Photon Microscopy: Improvements in Penetration Depth
An ongoing challenge in light microscopy concerns how to image deep within dense tissues. Over the range of near ultraviolet (UV), visible, and near infrared (NIR) wavelengths commonly used for biological light microscopy, it is light scattering and not absorption that generally dominates the attenuation of ballistic light propagation. The scattering length expresses the distance over which scattering will reduce light intensities by a factor of 1/e, and typical scattering lengths in the brain for visible and NIR light are in the ~25–100 μm (Yaroslavsky et al. 2002) and ~100–200 μm (Kleinfeld et al. 1998, Oheim et al. 2001) ranges, respectively. By comparison, the corresponding absorption lengths are in the millimeter range. Thus, imaging ~500 μm deep into brain tissue poses the challenge that both illumination and returning signals suffer from multiple e-fold attenuations by scattering of ballistic light propagation. For comparison, the adult mouse neocortex can be >1000 μm thick. Given the desire to look as deeply into the brain as possible, it is crucial to develop imaging modalities that are robust to light scattering.
An established approach for imaging deep into tissue involves two-photon fluorescence excitation (Denk et al. 1990) or other nonlinear optical processes that convert two or more incoming photons into at least one outgoing photon of distinct color. With such processes, the rate of signal generation rises nonlinearly as a function of illumination intensity. To achieve the high instantaneous intensities needed at the focus while keeping the time-averaged intensity below the limit of what the specimen can tolerate, it is common to use ultrashort-pulsed lasers emitting brief (~80–250 fs) but intense pulses. The quadratic dependence of two-photon excitation on illumination intensity leads to a spatial confinement of the excitation volume because of the steep falloff in excitation rate outside the focal plane. This confinement in excitation provides inherent optical sectioning, which can be used to mitigate the effects of emission scattering. Because fluorescence emissions originate from a single, confined focal volume, the emission photons convey useful information about the fluorescence intensity at that focal volume regardless of whether they scatter en route to the detector. Thus, the emission photons need only be collected in as great a number as possible, rather than imaged in a manner that preserves information about their apparent point of origination.
The ability to use scattered emissions as useful signals significantly increases imaging depths into tissue. The NIR wavelengths commonly used for two-photon excitation also reduce scattering of the illumination. Together, these two effects have led to penetration depths of 500–750 μm into the brain, depending on the tissue (Helmchen & Denk 2005). Two-photon imaging also reduces photodamage by using NIR excitation photons of energies lower than those of UV or visible wavelengths and by spatially confining fluorescence excitation, which is associated with photo-induced toxicity. Note that the approach to optical sectioning and signal collection in two-photon microscopy differs substantially from that in confocal microscopy, which uses a pinhole to restrict signals to those photons originating from the laser focus. Confocal microscopy is far less robust to scattering, permitting only ~25–50-μm imaging depths in optically dense tissue, because the pinhole blocks photons that originated from the laser focus but that have scattered (Sabharwal et al. 1999). Confocal imaging is further hampered in dense tissue by its typical use of visible excitation, which scatters more than the NIR illumination used in two-photon microscopy. The reliance on one-photon fluorescence excitation also increases phototoxicity outside the focal plane owing to the lack of excitation confinement. Overall, the advantages of two-photon microscopy have made it the leading microscopy technique for imaging deep within the intact brain or thick brain slices. Still, methods that could extend imaging depths into the brain, even modestly, would be welcomed, in part because such methods would provide opportunities to examine cells in the deeper neocortical layers in live animals.
Five main approaches are currently being explored to extend the penetration of two-photon microscopy. Three of these approaches aim to improve the generation of fluorescence signals, the fourth improves signal collection, and the last uses a thin probe composed of microlenses, termed a “microendoscope,” to guide light to and from deep tissue. The first of these five methods takes inspiration from observational astronomy and involves the use of adaptive optics to correct for deformations in the excitation wavefront that degrade the quality of the focal spot.
Adaptive optical techniques seek to correct both spherical aberrations, which grow progressively worse the deeper one focuses light with a microscope objective into even a uniform medium, as well as lensing effects, which can arise within tissue owing to refractive index inhomogeneities (Albert et al. 2000, Booth et al. 2002, Neil et al. 2000, Rueckel et al. 2006, Sherman et al. 2002, Zhu et al. 1999). A main challenge concerns how one deduces the wavefront deformations occurring within an individual brain. Strategies involve optical assessment of the wavefront (Booth et al. 2002, Neil et al. 2000, Rueckel et al. 2006, Zhu et al. 1999) as well as computational search techniques that seek to optimize signal generation (Albert et al. 2000, Sherman et al. 2002, Wright et al. 2007). Neither approach has been sufficiently successful to date to merit widespread adoption for brain imaging. Nonetheless, this is an area to watch for potentially exciting future developments.
Another approach to improving signal generation in two-photon microscopy employs ultrashort-pulsed regenerative laser amplifiers, which produce pulses of higher energies but usually at lower repetition rates as compared with the ultrashort-pulsed lasers commonly used for two-photon imaging. The amplified pulses retain efficacy to excite fluorescence at greater depths into tissue, which has permitted demonstrations of imaging up to ~850–1000 μm deep into the intact mouse brain (Beaurepaire et al. 2001, Theer et al. 2003). Drawbacks include the considerable cost and size of regenerative amplifiers and their near lack of wavelength tunability. The reduction in pulse repetition rates to ~1–1000 kHz also limits the speed of image-acquisition by raster scanning because each image pixel must receive illumination from at least one laser pulse. More economical, compact amplifier sources with higher repetition rates would facilitate progress, and so the ongoing improvements to ultrashort-pulsed fiber laser amplifiers are of considerable interest.
Although most researchers performing two-photon imaging have used Ti:sapphire lasers, which can have wavelength tuning ranges as broad as ~690–1080 nm, some investigators have pursued the use of alternative, fixed wavelength sources such as Nd:YLF (1.047 μm) (Squirrell et al. 1999), Yb:KYW (~1.033 μm) (Vučinić & Sejnowski 2007), or Cr:forsterite (~1.23–1.27 μm) (Chu et al. 2001, Liu et al. 2001) ultrashort-pulsed lasers. The use of longer wavelengths for excitation can improve depth penetration by diminishing scattering of both the illumination and the often-attendant long wavelength fluorescence emissions. Wavelengths of lasers with Yb-ion-doped gain media overlap those of Ti:sapphire lasers, but the overlap occurs toward the end of Ti:sapphire’s usable range, where the powers produced by Yb-ion-based lasers and amplifiers can be considerably greater. Cr:fosterite lasers operate in a transparent spectral window in which light absorption by water remains modest and the reduction in light scattering within tissue is even greater owing to the longer wavelength (Chu et al. 2001, Liu et al. 2001, Squirrell et al. 1999, Vučinić & Sejnowski 2007). Two main factors in the past that limited the utility of these alternative sources for two-photon imaging was the relative dearth of NIR and red fluorescent markers, especially functional indicators, and a lack of commercial availability. As molecular probes become increasingly available for use with longer wavelengths, these and other alternative ultrashort-pulsed lasers may find more applications.
A complementary approach to extending the penetration of two-photon imaging into tissue involves increasing the collection of fluorescence emissions. Because emissions must only be routed to a photodetector, not projected in an image-preserving manner, it appears possible to raise the numerical aperture (NA) of the collection pathway above that of the microscope objective used for laser beam delivery. One proposal is to equip the objective with an auxiliary nonimaging parabolic or ellipsoidal mirror surrounding the lens body to increase the collection aperture (Vučinić et al. 2006); simulations indicated this might increase collection efficiencies by up to a factor of four as benchmarked against a normal 40 × 0.8 NA water immersion microscope objective. This or similar approaches to collecting more photons seem viable, but such augmented objective lenses are not commercially available.
Finally, two-photon imaging deep within tissue can be accomplished by microendoscopy (Jung et al. 2004, Jung & Schnitzer 2003, Levene et al. 2004). The microendoscope is a thin but rigid optical probe, typically 350–1000 μm in diameter (Figure 1a), which inserts into tissue and conducts light to and from deep tissue locations ( Jung & Schnitzer 2003, Levene et al. 2004). Thus, microendoscopy increases the reach of laser-scanning microscopy into tissue up to the centimeter range (Llewellyn et al. 2008). The microendoscope probe typically comprises 1–3 gradient refractive index (GRIN) microlenses, which use internal variations in the refractive index, rather than the curved refractive surfaces employed by conventional lenses, to guide light (Flusberg et al. 2005a, Göbel et al. 2004, Jung et al. 2004, Jung & Schnitzer 2003, Levene et al. 2004, Monfared et al. 2006, Piyawattanametha et al. 2006) (Figure 1a).
Figure 1.
Fluorescence microendoscopy for imaging deep within tissue. (a) Photograph of compound doublet microendoscope probes 350 μrn, 500 μm, and 1000 μrn in diameter. Each doublet probe is composed of two gradient refractive index (GRIN) microlenses, an endoscopic objective lens (shorter lens elements at the bottom of the photograph) and a relay lens (longer elements with dark coating). The endoscopic objective lens has a higher numerical aperture (NA) value and provides micron-scale resolution. The relay lens has a lower NA and provides the length needed to reach deep tissue (Jung et al. 2004). Each minor tick on the scale bar is 1 mm. (b) Optical schematic for two-photon microendoscopy. A GRIN microendoscope is inserted into tissue to image cells ~1 mm to > 1 cm beneath the external surface. A microscope objective lens focuses ultrashort-pulsed laser illumination (red beam) to just above the top face of the microendoscope probe. The laser focal spot is scanned laterally in this plane (scanning indicated by yellow arrows), typically in a raster pattern. The microendoscope probe projects and demagnifies the laser-scanning pattern to the focal plane within tissue (scanning indicated by yellow arrows), generally at a working distance ~150–800 μm from the tip of the endoscope probe. Within the GRIN microendoscope, the beam undergoes a gradual refocusing owing to the smoothly varying radial refractive index profile. A portion of the two-photon excited fluorescence generated at the focal volume in tissue returns back through the microendoscope probe and microscope objective and is captured by a photodetector. A computer controls the laser-scanning process and reconstructs the resulting image. Three-dimensional image stacks are acquired by combining 2D lateral scanning of the laser focal spot with axial adjustments to the position of the microscope objective, which changes the plane of laser-scanning both above the microendoscope and within tissue. (c) Average intensity projection of a 3D image stack acquired by two-photon microendoscopy, showing a CA1 hippocampal neuron expressing YFP under the control of the Thy1 promoter. The microendoscope probe extended through neocortical tissue but did not enter hippocampus and was positioned just dorsal to CA1 tissue layer stratum oriens. Images within the 3D stack (540 μm in total axial extent) were collected at working distances of 160–700 μm from the tip of the microendoscope, across a 185-μm-diameter field of view. The axial range of this stack covers the tissue layers stratum oriens and stratum pyramidale and part of stratum radiatum; thus, the basal dendrites, cell body, and proximal apical dendrites of the pyramidal neuron were sampled during image acquisition. The microendoscope used has lateral and axial optical resolution limits of ~0.9 ~m and ~10–12 μm, respectively (image courtesy of R. Barretto).
In most designs, the microendoscope acts as an optical relay. If the laser focal spot is scanned just above the top face of the microendoscope probe that lies outside tissue, the probe projects and demagnifies the scanning pattern to a focal plane inside tissue (Figure 1b). Because the microendoscope probe is composed of lenses and is not a pixilated fiber bundle, adjustment of the axial position of the laser focus just above the probe leads to corresponding focal adjustments within tissue. Thus, with the probe held at a fixed location in tissue, two-photon microendoscopy permits the acquisition of 3D image stacks (Piyawattanametha et al. 2006), which can extend up to ~500–650 μm in depth measured from the tip of the microendoscope probe (Figure 1c).
There is considerable flexibility in the choice of microendoscope probes’ specifications: Physical lengths of ~0.5–3 cm, optical working distances of ~150–800 μm, NAs of ~0.4–0.75, and fields of view of ~100–1000 μm are the approximate ranges of typical values. There are, however, important tradeoffs within these ranges between the different parameters. For example, longer working distances generally imply reduced NA values, though to a lesser degree for the larger diameter probes. The moderate costs of microendoscope probes relative to those of microscope objectives facilitate the acquisition of a large set of complementary designs for use in different situations. Resolution values achieved to date by microendoscopy (~0.9–1.2 μm lateral, ~10–12 μm axial) (Flusberg et al. 2005b, Jung et al. 2004, Levene et al. 2004) have not been limited by diffraction, but rather by optical aberrations within the endoscope probes. We expect that further optical engineering will yield next-generation microendoscopes capable of diffraction-limited performance. The small size of GRIN microlenses also permits their incorporation into miniaturized, fiber-optic two-photon microscopes (Engelbrecht et al. 2008, Flusberg et al. 2005b, Göbel et al. 2004, Hoy et al. 2008, Jung et al. 2008, Le Harzic et al. 2008) (see section below on Fiber-Optic Microscopy). However, microendoscopes have neither the tapered shapes nor the small diameters of electrode tips, so neuroscientists need to plan surgical strategies and routes of insertion carefully when placing microendoscopes into the brain to minimize disruption to tissue. Locating the tip of the microendoscope just outside, and not within, the brain structure of interest can help lessen effects in the area being imaged. Nonetheless, it is best to perform control studies to check for any notable effects on tissue in each new experimental configuration.
In addition to two-photon fluorescence, the imaging modalities demonstrated to be compatible with microendoscopy include epifluorescence (Flusberg et al. 2008; Jung et al.2004; Murayama et al. 2007, 2009), confocal (Knittel et al. 2001), and second-harmonic generation imaging (Fu & Gu 2007, Llewellyn et al. 2008). Microendoscopy applications in neuroscience have included in vivo imaging of cochlear microanatomy and circulation (Monfared et al. 2006), CA1 hippocampal neurons (Figure 15d) (Deisseroth et al. 2006, Jung et al. 2004), layer V pyramidal neurons (Levene et al. 2004; Murayama et al. 2007, 2009), and the contractile dynamics of striated muscle sarcomeres in both mice and humans (Figure 13a–f) (Llewellyn et al. 2008). The use of microendoscopy for long-term imaging of cells deep in the mammalian brain is also emerging (Deisseroth et al. 2006), which should facilitate longitudinal studies of how cellular properties might change over the course of learning or aging, brain disease, or in response to new therapeutics (see section below on Long-Term Imaging).
Figure 15.
Time-lapse fluorescence microscopy in live mammals. (a) In vivo two-photon microscopy images of the growing dendrite of a layer 2/3 pyramidal neuron that expresses GFP under the control of the Thy1 promoter in mouse somatosensory cortex (GFP-M line). The first three panels show the dendrite following cortical deletion of pten (phosphatase and tensin homolog on chromosome 10). CaMKII-Cre was used to excise floxed pten. Beginning in postnatal week 11, the mouse received daily doses (intraperitoneal injection) of rapamycin, which inhibits the same pathway as does pten. The last three panels show the same dendrite exhibiting a loss of filopodia-like spines near the tip (week 11–12) and stopped growth without retraction (week 12–15) (based on Chow et al. 2009). (b) Long-term in vivo two-photon imaging of layer 2/3 axon dynamics in adult mouse neocortex. Layer 2/3 neuron precursors were transfected with GFP by in utero electroporation at E16. Chronic glass-covered cranial windows were implanted in the hemisphere contralateral to the site of electroporation in adult mice (>3 months of age). At least four weeks after window implantation, callosal projections of layer 2/3 neurons in barrel cortex were imaged by in vivo two-photon microscopy at four-day intervals. Day 0 is the first day of imaging. Examples of axonal bouton dynamics are indicated by arrows (yellow arrows, stable boutons; green arrows, new boutons; red arrows, disappearing boutons). Note the extensive growth and retraction of the tip of the axon between days 12 and 40 (blue dashed lines). Images are projections of 8–16 slices, 1.5 μm apart, in which some distracting processes have been digitally removed for display purposes. Abbreviations: EPB, en passant boutons; TB, terminaux boutons (images courtesy of C. Portera-Cailliau). (c) In vivo time-lapse imaging of dendritic spine formation and elimination in the mouse cortex, as imaged at two time points through a thinned skull. Repeated two-photon imaging of two dendritic branches (top and bottom) at four (left panels) and six (right panels) weeks of age reveals spine elimination (arrowheads) and formation (large arrows) as well as filopodium turnover (small arrows) in control (top) and sensory-deprived (bottom) mouse barrel cortices (based on Zuo et al. 2005b). (d) Images of CA1 hippocampal pyramidal cell bodies expressing YFP under the control of the Thy1 promoter, as imaged by two-photon microendoscopy in a live mouse. On day 0, a guide tube was surgically implanted so that the microendoscope probe could be inserted repeatedly to the same tissue location just dorsal to CA1. The image acquired on day 239 after the initial surgery (green pseudocolor, left) closely resembles that acquired on day 241 (red pseudocolor, middle), as shown by the yellow portions in the merge of the two (right) (based on Deisseroth et al. 2006). (e) Spine imaging after stroke: long-term in vivo two-photon imaging of dendritic spines in layer 5 pyramidal neurons in the sparsely labeled primary somatosensory cortex of Thy1 -GFP-M mice. Imaging through a chronically implanted glass-covered cranial window began eight days before and ended three months after unilateral permanent middle cerebral artery (MCA) occlusion. Only dendrites in peri-infarct cortex, 1–3 mm away from the edge of infarct, were imaged. Panels show high-resolution in vivo imaging of dendritic spines before and after stroke at representative time points. Stable spines are indicated by yellow arrows. Thin blue arrows at day +4 after stroke indicate regions of local beading/swelling after stroke. Images are projections of 8–15 slices 1.5 μm apart (images courtesy of R. Mostany and C. Portera-Cailliau). (f) In vivo two-photon imaging of neocortical dendrites and axons (green) near an amyloid deposit (red) in cortical layer 1 of a mouse model of Alzheimer disease at six months of age. Although most spines (arrowheads) and varicosities (asterisks) were stable over two days, some structural changes [e.g., spine loss (arrows) and varicosity formation (double arrows)] occurred near the deposits (based on Tsai et al. 2004). (g) A dorsal root ganglion sensory axon of a Thy1 -GFP-S mouse in vivo before, immediately, and 48 h after transection with a small pin. The mouse was perfused after the 48-h time point, and the same axon (boxed area in +48 h panel) was re-identified in cut sections using confocal microscopy (fourth panel). Two bulbs formed on the transected axon (yellow insets; high power views below); one formed near the lesion site (left bulb), whereas the other died back from the lesion through acute axonal degeneration (Kerschensteiner et al. 2005). Degenerative fragments can be seen between the bulb and the lesion site along the original path of the axon. (For comparison to the in vitro images, the red insets show +48 h views of the bulb in vivo at higher magnification.) (Images are courtesy of T. Misgeld and M. Kerschensteiner.)
Figure 13.
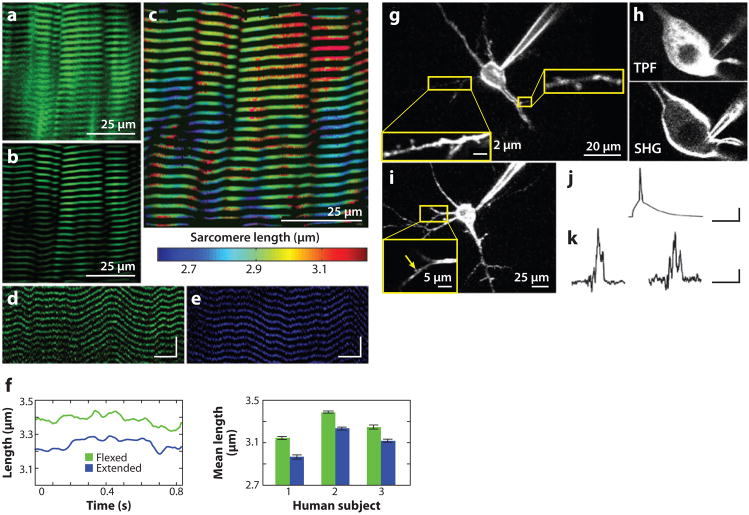
Second-harmonic generation (SHG) imaging reveals the dynamics of striated muscle sarcomeres and neuronal membrane voltage. (a–f) In vivo imaging of striated muscle sarcomeres by SHG microendoscopy. (a) Raw and (b) band-pass-filtered SHG images acquired using a 350-μm-diameter microendoscope inserted into the lateral gastrocnemius of an anesthetized mouse. (c) Map of sarcomere length variations within the images of (a) and (b). (d, e) Line-scan images acquired at 488 Hz in the extensor digitorum muscle of a human subject using a 350-μm-diameter microendoscope with digits of the hand held flexed (d) or extended (e). Scale bars: 100 ms (horizontal), 10 μm (vertical). (f, left) Time variations of mean sarcomere lengths from the same data sets as in (d) and (e). (Right) Average sarcomere lengths as determined from multiple images in each of three human subjects with the digits of the hand held either flexed (green) (n = 12, 17, and 11 trials, respectively) or extended (blue) (n = 10, 10, and 7 trials, respectively). Error bars are standard errors of the mean (a–f based on Llewellyn et al. 2008). (g–k) Imaging neuronal membrane potential by SHG microscopy. (g) SHG imaging of a layer 5 pyramidal neuron from a brain slice of mouse primary visual cortex stained with FM 4-64. Spines on a basal dendrite and an oblique dendrite are readily visible (insets). (h) Comparison between membrane contrast of the same neuron imaged with two-photon fluorescence (TPF) (top) and SHG (bottom). The strong fluorescence signal from the cytoplasm in the TPF image is not present in the SHG image. (i) SHG measurements of spines during action potential invasion. Representative layer 5 pyramidal neuron used for action potential measurements by FM 4-64 SHG imaging. A lower-resolution projection from an image stack reveals the neuron. A high-resolution image (inset), in which the dendritic spine is indicated by the yellow arrow. (j) A single action potential was initiated by current injection at the soma [scale bars: 50 ms (horizontal), 30 mV (vertical)], and (k) the resulting SHG signal changes were measured in the soma (average trace of four recordings, left) and individual dendritic spines (average trace of seven recordings, right). Each recording is a result of averaging ~40 action potentials. Scale bars: 50 ms (horizontal), 2.5% ΔSHG/SHG (vertical). (Images g–k are based on Nuriya et al. 2006.)
Two-Photon Microscopy: Improvements in Speed and Laser-Scanning Mechanisms
Harmonizing with the goal of imaging deep into tissue are the aims of sampling large tissue volumes and doing so at fast data-acquisition rates. The latter two aims are crucial for neuroscientists wishing to monitor the activities of large populations of individual cells with sufficient time resolution to follow fast biological processes such as cellular Ca2+ or voltage dynamics. Two-photon microscopy usually employs one laser beam scanned in a raster pattern over the sample. This configuration leads to basic trade-offs among the frame-acquisition rate, field of view, and signal-to-noise ratio. Parallel streams of complementary research on molecular probes and optical hardware seek to improve the speed and dynamic range of fluorescent functional indicators, as well as the speed and three-dimensional range of laser-scanning mechanisms. Two-photon microscopy conventionally employs a pair of nonresonant galvanometric scanning mirrors, which are individually limited to line scans of ~1–2 kHz. With typical images of 128 × 128 pixels or more, this restricts frame acquisition rates to about <16 Hz. However, alternative scanning strategies for attaining higher speeds are emerging. Work in recent years has yielded particular progress in methods for 3D laser-scanning.
One approach to faster scanning splits the excitation beam into a set of multiple “beamlets”, which are focused to distinct spots and scanned together across the sample to permit parallel data acquisition from multiple locations (Figure 2a). Such techniques come in different varieties but are often collectively termed multifocal multiphoton microscopy (MMM) (Bewersdorf etal. 1998, Buist etal. 1998). These methods require means for creating the beamlets and recording multiple channels of spatially distinct signals. Typically, either a microlens array (Bahlmann et al. 2007, Bewersdorf et al. 1998, Buist et al. 1998, Kim et al. 2007) or a series of beam splitters (Fittinghoff et al. 2000, Fricke & Nielsen 2005, Lévêque-Fort et al. 2004, Nielsen et al. 2001) divides the laser beam, and a fast camera (Bahlmann et al. 2007, Bewersdorf et al. 1998, Buist et al. 1998, Nielsen et al. 2001) detects the emissions. High-power laser sources can allow up to ~64 beamlets of sufficient intensity to excite two-photon fluorescence (Kim et al. 2007, Niesner et al. 2007). This permits increases in frame rates (e.g., ~640 Hz over a 64 × 64 μm2 total imaging area with a 6 × 6 beam array) (Bahlmann et al. 2007) because each beamlet must be scanned across only a portion of the image. However, having a dense set of focal points can diminish the efficacy of optical sectioning owing to interference between mutually coherent beamlets. [For related reasons, approaches to fast two-photon imaging that involve scanning a focused line of laser excitation also significantly compromise optical sectioning (Brakenhoff et al. 1996).] Time-multiplexing the pulses at the picoseconds scale can alleviate this problem by having the pulses reach the focal plane at different times (Andresen et al. 2001, Egner & Hell 2000, Fittinghoff & Squier 2000).
Figure 2.
Laser-scanning techniques for two-photon fluorescence microscopy. (a) Multifocal two-photon microscopy. A microlens array creates a grid of laser beamlets. A 2D (x-y) scanning mirror sweeps the resulting grid of focal spots across an intermediate focal plane conjugate to the specimen plane. Unlike in conventional two-photon microscopy, the fluorescence signals from the array of focal spots in the specimen are imaged onto a camera. (b) Three-dimensional (3D) laser-scanning using a piezoelectric objective mount. Axial vibration of the mount together with the use of conventional scanning mirrors permits fast 3D scanning. (c) Data acquired by 3D scanning using a piezoelectric focusing mount. (Top) A reference image stack from mouse neocortical layer 2, in which cells are stained with the Ca2+ indicator Oregon Green BAPTA-1 am (green) and the astrocyte-specific marker sulforhodamine 101 (SR101) (red). Dual-stained astrocytes appear yellow. (bottom) The spiral 3D scan trajectory (red curve) cuts across three ellipsoids (green, blue, and yellow) that represent the volumes of interest (VOIs) of three example cell bodies selected from the reference stack. Scale bars: 25 μm. (d) Ca2+ activity traces. Line-scan segments within the VOIs selected from panel c were assigned to the three color corresponding cells. For each cell, the fluorescence signals from all sections were averaged, revealing the time courses of the Ca2+ signals (green, blue, and yellow traces at right). Stimulation via an extracellular micropipette consisted of several series of 5 bursts of 10 pulses at 100 Hz (1-s burst interval). Scale bars: 10 s (horizontal), 30% ΔF/F (vertical) (right). (Panels c and d are from Göbel et al. 2007.) (e) 3D laser-scanning using acousto-optic deflectors (AODs). Two successive pairs of orthogonally oriented AODs permit both lateral (x and y) and axial (z) steering of the focal spot by adjustment of the center acoustic frequencies and the net acoustic frequency chirp, respectively. Beam propagation is depicted for two different focal locations. (f) Two-photon fluorescence image of a rat CA1 pyramidal neuron obtained by 3D laser-scanning with AODs. The neuron was fluorescently labeled with 50 μM Alexa 594 and 200 μM of the Ca2+ indicator Oregon Green BAPTA-1 am. The image is a composite of maximum intensity projection images from eight image stacks that together span the entire cell. The overlaid yellow circles represent the magnitudes of Ca2+ transients (ΔF/F amplitudes) measured at a total of 146 recording sites during individual back-propagating action potentials. (Image courtesy of K. Kelleher, Y. Liang, and P. Saggau.) (g) Fast scanning of line illumination with auxiliary optical sectioning provided by temporal focusing. A cylindrical lens focuses a laser beam to a line incident on a diffraction grating, which spatially disperses the light’s wavelength components. A tube lens focuses each wavelength component to a line on the back aperture of the microscope objective, creating a beam with a smoothly varying wavelength gradient in cross section. The objective focuses the beam to a line of illumination on the specimen, such that all wavelength components recombine at the focal plane owing to temporal focusing effects (Tal et al. 2005). Thus, the laser pulses are of shortest duration at the specimen plane, which promotes optical sectioning. A galvanometric (galvo) mirror scans the line of illumination across the sample.
The Achilles’ heel of multifocal imaging is its vulnerability to emission scattering, which induces cross talk between the different signal channels. Particularly at greater imaging depths for which scattered emissions are the norm, it becomes increasingly challenging to prevent mutual contamination of the signals originating from adjacent excitation volumes (Kim et al. 2007, Niesner et al. 2007). A detection setup with a multianode photomultiplier tube instead of a camera can be somewhat less sensitive to emission scattering by facilitating computational strategies for reducing cross talk, but cross talk is not abolished and still increases with imaging depth (Kim et al. 2007). The division of laser power into multiple channels also reduces the capability for imaging in deep tissue by limiting the power that can be used in each channel to compensate for the attenuation of the illumination by scattering. As higher power lasers emerge, this latter drawback to multifocal imaging will be ameliorated, but emission cross talk would be exacerbated by a use of higher powers to create denser arrays of focal spots.
A few studies have utilized the higher frame rates available in multifocal two-photon microscopy for studies of cellular dynamics. Two studies examined the Ca2+ dynamics of mouse hippocampal CA1 neurons in brain slices (Crépel et al. 2007, Goldin et al. 2007), and another used 64 laser spots for fast in vivo monitoring of Ca2+ dynamics in neurons of the Calliphora vicina (blowfly) visual system (Kurtz et al. 2006). Multifocal two-photon imaging has not yet been demonstrated in live mammals, nor has fast 3D-scanning multifocal imaging emerged.
However, there are several laser-scanning strategies arising for fast, continuous sampling of 3D tissue volumes. One approach builds on the established means for creating 3D image stacks using mechanical movements of either the specimen or the microscope objective. Instead of using successive step-wise axial adjustments, however, the objective lens is set in continuous motion using a piezoelectric actuator, and 3D scan trajectories are created through the combined use of a pair of galvanometric mirrors for lateral scanning (Figure 2b) (Göbel et al. 2007). This approach has allowed 3D data to be acquired at volume acquisition rates of ~10 Hz and has enabled continuous monitoring of neuronal and astrocytic Ca2+ dynamics in up to several hundreds of cells across a ~250 × 250 × 250-μm3 volume in the intact rat neocortex (Göbel & Helmchen 2007, Göbel et al. 2007) (Figure 2c,d). Typical microscope objectives have masses of ~150–250 g that limit the speed of mechanical scanning. One can compensate computationally for temporal lags between the drive signals and the actual movement of the objective (Göbel et al. 2007). Advantages of objective scanning include the substantial tissue volumes that can be sampled, the relative ease of implementation, and the feasibility of using spiral and even user-defined scanning trajectories to sample a large number of cell bodies. Limitations include the speed restrictions and potential susceptibilities to motion artifacts when imaging in awake animals, since with complex 3D scanning trajectories it may be harder to apply image registration algorithms that correct for motion artifacts (see Imaging in Awake Behaving Animals below). Furthermore, because each cell is only sampled at a few image pixels, signal-to-noise ratios are generally lower than in 2D laser-scanning imaging performed at comparable frame rates. Overall, 3D laser-scanning by piezoelectric actuation of the objective is poised to enable unprecedented dynamical studies of hundreds to even thousands of cells in vivo, albeit for now with ~100-ms time resolution.
To achieve faster imaging of functional signals, investigators can use noninertial laser-scanning strategies that permit higher speeds. Acousto-optic deflectors (AODs) steer laser beams by using the diffraction of light by sound waves in a crystal. Changing the frequency of the sound waves alters the propagation angle of the diffracted beam. This method enables the use of a one-dimensional scanning mechanism whose speed limit is mainly set by the acoustic delays needed to modify the sound frequencies in the crystal (~10–30 μ is for typical apertures). Thus, a pair of orthogonally oriented AODs permits beam steering in the two lateral dimensions (Bullen et al. 1997). Moreover, frequency chirping of the acoustic waves permits fast axial scanning (Reddy et al. 2008, Reddy & Saggau 2005, Vučinić & Sejnowski 2007). The degree of frequency chirp affects the collimation of the laser beam—because the acoustic frequency and thus the diffraction angle varies across the beam’s cross-sectional profile—resulting in converging or diverging beams that can be used to modify the axial focal position in the sample. Two basic strategies for 3D scanning exist, involving either two (Vučinić & Sejnowski 2007) or four (Reddy et al. 2008, Reddy & Saggau 2005) AODs.
The former approach to 3D scanning offers higher optical throughput, a smaller footprint, and reduced group velocity dispersion, but at the cost of some distortions in the shape of the focal spot and the inability to dwell with the laser spot stably at axially displaced locations (Vučinić & Sejnowski 2007). The latter approach involves somewhat greater group velocity dispersion but offers more versatility in the choice of scanning trajectory (Figure 2e) (Reddy et al. 2008, Reddy & Saggau 2005). This approach enables fully random-access scanning patterns across the 3D field of view, for high-speed monitoring of a predefined sequence of cells or sites. For example, four AODs have been used to monitor the 3D dynamics of this method, a diffraction grating spectrally back-propagating action potentials within neuronal dendritic trees (Figure 2f) over fields of view as large as ~200 × 200 × 50 μm3 (Reddy et al. 2008). Acquisition cycle times are fast, often in the tens of kilohertz, as determined by the number of sampled points, the re-positioning time (~15 μs) per point, and the dwell time (~5 μs) per point (Reddy et al. 2008). Both two and four AOD scanning strategies benefit from corrective methods for compensating the spatial and temporal dispersion that arise within the AODs (Zeng et al. 2006). To date, the use of AODs has been limited to in vitro studies, but we expect experiments that use fast AOD-based laser-scanning in the intact brain will emerge soon.
Temporal focusing is a means of providing auxiliary optical sectioning that can aid rapid scanning strategies employing ultrashort-pulsed laser illumination (Durst et al. 2006, 2008; Oron et al. 2005; Papagiakoumou et al. 2008; Tal et al. 2005; Zhu et al. 2005). In this method, a diffraction grating spectrally disperses an incident laser beam, which is then collimated and focused onto the sample by the microscope objective (Figure 2g). Because the different wavelengths of light are spatiotemporally separated until they reach the common focal plane, outside the focal plane the spectral bandwidth is effectively reduced at each location and hence the pulse duration is broadened. The pulse recompression and recombination of the different wavelengths that occur at the focal plane provide the “temporal focusing” and help confine the generation of two-photon excitation. Thus, the temporal two-photon excitation. Thus, the temporal two-photon excitation. Thus, the temporal restoring some of the optical sectioning capabilities of two-photon imaging configurations, such as those involving fast scanning with a line of illumination (Figure 2g), in which optical sectioning is compromised as compared with the usual raster-scanning mode (Tal et al. 2005) (see multifocal imaging, above). As compared with raster-scanning a focal spot, the division of laser power across a swept line of illumination does reduce two-photon excitation. Nevertheless, as ultrashort-pulsed laser amplifiers become more widely used for imaging, the combination of temporal focusing and line-illumination-scanning may gain traction for high-speed two-photon microscopy.
Two-Photon Microscopy: Reducing Photobleaching and Photodamage
In addition to achieving greater penetration and faster scanning, recent work has broadened appreciation for how the repetition rate of pulsed illumination can affect fluorophores’ total time-integrated signal emission. Increasing the time between illumination pulses to the microseconds timescale or longer allows excited fluorophores to relax electronically from dark, meta-stable spin triplet states, which are usually on the dominant pathways to photobleaching and phototoxicity, instead of absorbing another photon that would increase the propensity for photoinduced toxicity (Donnert et al. 2007a). Of course, a reduction in pulse repetition rate also implies a reduction in the fluorescence emission rate, even if each fluorophore lasts for a greater number of excitation cycles.
By comparison, the photophysics underlying an alternative strategy for reducing photodamage in two-photon microscopy by raising pulse repetition rates remains less well understood (Ji et al. 2008). This strategy relies on the observation that in two-photon microscopy both photobleaching (Chen et al. 2002, Patterson & Piston 2000) and photodamage (Hopt & Neher 2001) rise with a dependence on the instantaneous illumination intensity that is steeper than quadratic. Thus, in the regimes in which pulse energies are limited by phototoxicity, a net increase in signal should be obtainable by raising the pulse rate while keeping the energy per pulse approximately constant. Alternatively, the rate of photodamage might be reduced while keeping signal rates constant by increasing pulse rates but reducing pulse energies. Empirically, by increasing the usual ~80 MHz rate of pulse delivery in two-photon imaging up to ~128-fold via the use of a “pulse splitter,” it was possible to either increase signals or reduce photobleaching in fixed and live tissues (Ji et al. 2008). Strikingly, this result implies that a major route for photobleaching depends on photophysical effects at the picoseconds timescale, much briefer than typical ~1–10-ns fluorescence lifetimes or the microseconds lifetimes of dark triplet states.
Volumetric Imaging Using One-Photon Fluorescence and Other Linear Contrast Mechanisms
In parallel with the ongoing progress in two-photon microscopy, alternative approaches to imaging in 3D using one-photon fluorescence excitation are arising, often for applications complementary to those suited for two-photon imaging. Other traditional linear contrast mechanisms, such as light absorption, can often be used as well. As compared with two-photon imaging, all the volumetric imaging methods discussed in this section suffer from relatively poor depth penetration in scattering tissue. However, these methods generally perform well in small, optically transparent samples such as embryos or small organisms and so are often used to study developmental processes. Optical clearing of histological specimens can further permit the study of large tissue volumes. Moreover, some linear contrast imaging methods do permit fast, 3D functional imaging in relatively transparent organisms such as zebrafish embryos (T. Anderson, L. Grosenick, and S. Smith, unpublished data) or in tissues with reduced scattering such as the vomeronasal epithelium (Holekamp et al. 2008). The use of conventional light sources, as opposed to the ultrashort-pulsed lasers used for two-photon imaging, helps to reduce costs.
Light-sheet microscopy (Fuchs et al. 2002, Voie et al. 1993) resembles a less commonly used form of confocal microscopy, the theta (Lindek et al. 1997, Lindek et al. 1999, Stelzer & Lindek 1994) or dual-axis confocal (Wang et al. 2003, Webb & Rogomentich 1999) microscope, but with planar illumination (Figure 3a). Variants of light-sheet microscopy are known as selective plane illumination microscopy (Huisken et al. 2004) or objective-coupled planar illumination (OCPI) (Holekamp et al. 2008). The optical axes for light delivery and collection are oriented at an angle to one another, usually 90 degrees. Cylindrical lenses of modest NA and long working distances are generally used for creating a sheet of illumination. A high NA microscope objective is typically used in the light collection pathway to maintain high resolution and signal-gathering power. A key advantage of light-sheet illumination is the restriction of fluorescence generation to a single plane, which is not the case in conventional epi-fluorescence microscopy. The use of sheet illumination leads to a large reduction in background fluorescence, which improves the attainable imaging depth by partially mitigating the loss of contrast with depth. Sheet illumination also reduces photobleaching occurring at locations not being imaged, which in turn improves sample viability, such as for time-lapse imaging (Holekamp et al. 2008, Huisken et al. 2004). For example, light sheet microscopy was recently used to track the live development of an entire zebrafish embryo, over multiple hours and at single-cell resolution, in an embryo expressing a green fluorescent protein (GFP) fusion that localizes to chromatin (Keller et al. 2008).
If the plane of illumination is rapidly scanned in tandem with the focal plane of the imaging lens, then light sheet fluorescence microscopy can enable fast functional volumetric imaging (Figure 4a) (Holekamp et al. 2008). Although scattering still limits the depths in tissue at which cellular dynamics can be tracked, in tissues with cell bodies within ~150 μm of the tissue surface, such as the vomeronasal epithelium, investigators have tracked Ca2+ dynamics in up to ~700 neuronal somata with one volume acquisition occurring every 2 s over a 150 μm × 425 μm × 250 μm region (Holekamp et al. 2008). Alternatively, this technique has been used to monitor Ca2+ dynamics in 88 cells within a single plane at 200 Hz (Holekamp et al. 2008).
Another set of applications for light-sheet fluorescence microscopy involves efficient examination of histological specimens (Dodt et al. 2007). Rather than cut tissue samples into a large number of slices that must be separately imaged and then realigned computationally, light-sheet microscopy permits high-resolution data to be acquired from an entire intact tissue specimen. A key element of this approach is the process of “optically clearing” the sample to minimize the internal variations in refractive index. This method vastly reduces light scattering and permits large, millimeter-sized fixed tissue samples, such as embryos, or even an entire mouse brain, to be imaged without slicing (Dodt et al. 2007). The attainable axial resolution varies in part with the specimen’s lateral extent since this will dictate the minimum thickness of the illumination sheet that can be used without significant diffraction occurring over the breadth of the sample.
An alternative volumetric technique for studying cleared histological specimens is optical projection tomography (OPT) (Sharpe 2004, Sharpe et al. 2002), which is in several respects an optical analog of X-ray computed tomography (CT). To date, OPT has used either fluorescence or light absorption for contrast. A key similarity with X-ray CT is that 3D images are reconstructed via a “back computation” process that starts with a series of raw 2D projection images. To collect this data, the sample is rotated while an ensemble of 2D projection images are acquired across a wide set of different angular perspectives. Contrast intensities from each voxel in the 3D reconstruction can then be deduced by a back computation algorithm involving the same type of Radon transform used in X-ray CT. OPT permits reconstruction at resolutions as fine as ~5–10 μm of fixed 3D specimens that are ~1–10 mm across (Figure 3b). Unlike with other one-photon volumetric techniques, the resolution of OPT can be readily made isotropic by using fine rotations of the sample. High-resolution reconstructions require a large number of projection images, particularly with large sample volumes (Sharpe 2004). Thus, most applications of OPT have involved fixed specimens, such as of small organisms (McGurk et al. 2007) (Figure 4c) or embryos (Sharpe et al. 2002) (Figure 4d). Recently, OPT permitted the visualization of limb development in a mouse limb bud culture over multiple time points and without optical clearing, which led to a modest loss of resolution (Figure 4e) (Boot et al. 2008).
Like OPT, light field microscopy involves the computational reconstruction of 3D information encoded within a set of raw 2D data sets (Levoy 2009, Levoy et al. 2006, 2009). However, in light-field microscopy the raw data are not even projection images, but rather abstract representations of 3D information. Thus, this technique well illustrates the ongoing rise in abstraction in light microscopy, a trend that will surely increase with time. Light field microscopy employs a microlens array, placed close to the image plane in front of the camera, to segregate information about signal photons’ originating locations and directions of propagation (Figures 3c, 4b). The microlens array achieves this segregation by transforming the type of raw data gathered by the camera from an ensemble of pixel intensity values into an ensemble of micro-images (e.g. 16 × 16 to 20 × 20 pixels in size) (Figure 4b yellow and orange insets). Each micro-image contains the signals originating from a fixed location within the sample; each pixel within a micro-image represents the intensity of light from that location reaching the camera by a specific propagation route. The maintenance of information about signal origin and direction of propagation implies that 2D images can be reconstructed at different viewing angles by assembling only those pixels with the same coordinate sets across all the micro-images. By applying a “synthetic aperture,” one can also computationally adjust the depth of field in the 2D reconstructions by choosing the maximum propagation angle of the returning rays that contribute to the image. This manipulation is analogous to adjusting the diameter of an iris in the back aperture of a microscope objective.
Computational refocusing at different planes is also possible in light field microscopy. Thus, by assembling a stack of 2D images computationally focused to a sequence of planes, and then applying a deconvolution, 3D reconstructions are also achievable. Because no scanning is needed for this form of volumetric imaging, 3D volume-acquisition rates are limited strictly by photon collection and camera speed. To date, light field microscopy has allowed functional imaging of Ca2+ signals in zebrafish across ~250 × 250 × 70 μm3 3D volumes at 4 Hz (T. Anderson, L. Grosenick, S. Smith, unpublished data). However, researchers face basic trade-offs between the achievable lateral resolution and the number of perspectives or axial planes sampled because with all else held constant, a microlens array that yields an increase in the number of camera pixels per micro-image also yields coarser spatial sampling in the lateral dimensions. Because the relative size of the spacings between the individual microlenses and camera pixels is important for setting the boundaries of this trade-off, progress in light field microscopy will be stimulated by advances in both camera and microlens array technology.
By comparison, progress in holographic microscopy hinges in part on advances in the designs of programmable spatial light modulators (SLM), which can digitally synthesize 2D hologram-generating patterns. Holographic microscopy encodes 3D information by interfering a reference wave, often a plane wave, with spherical waves radiating either toward the sample as illumination or from the sample’s emitters. This interferometric approach to encoding 3D data is basic to the holographic microscopy methods, which have existed for some time (Gabor 1948) and in multiple forms (Poon et al. 1995). Holographic techniques fall into two categories: scanning and nonscanning.
In recent versions of scanning holographic microscopy, illumination reflects off a programmable phase SLM that induces distinct phase shifts in the light reflecting off each of its pixels. This allows the synthesis of an arbitrary, 3D pattern of illumination that can be used to excite fluorescence or other optical processes concurrently at a chosen set of multiple locations in the sample (Lutz et al. 2008, Nikolenko et al. 2008, Papagiakoumou et al. 2008). This illumination strategy also works with two-photon fluorescence excitation (Nikolenko et al. 2008, Papagiakoumou et al. 2008). Alternatively, by scanning the synthesized illumination patterns one can in principle reconstruct an entire volumetric image with reduced background excitation owing to the 3D control of illumination. Current SLM technology enables the illumination patterns to be updated at frame rates up to 60 Hz. The freedom to synthesize arbitrary illumination patterns is also restricted by the number of pixels on the SLM (to date up to ~1920 × 1200 pixels) available for encoding the Fourier transform of the desired pattern. To date, SLMs have permitted concurrent illumination of tens of sample locations and arbitrarily shaped spatial regions, such as the areas occupied by cells (Lutz et al. 2008, Nikolenko et al. 2008, Papagiakoumou et al. 2008).
Holographic imaging has also recently been implemented in a nonscanning manner by reflecting the collected light signals off a single, centered holographic pattern projected onto the SLM (Rosen & Brooker 2008). In this version of holographic microscopy, each emitter in the sample provides two interfering spherical waves with different curvatures reflecting off the SLM. This embeds a separate interferometric hologram for each emitter onto a camera. Three holograms are sequentially recorded per image frame, each using a distinct phase-offset for the reflection pattern on the SLM (Figure 3d) (Rosen & Brooker 2008). The holograms are superposed and convolved with a holographic response function to reconstruct the 3D spatial distribution of emitters.
An alternative use for holographic techniques in microscopy involves the creation of arbitrary illumination patterns for photouncaging or photostimulation (Lutz et al. 2008), including with two-photon excitation for studies in thick tissue (Nikolenko et al. 2008, Papagiakoumou et al. 2008). Thus, holographic illumination methods may provide complementary alternatives to AOD-based approaches for random access imaging and uncaging, albeit with more modest update rates for changing the patterns of illumination but capable of illuminating many points in the sample simultaneously. Similarly, the concepts of light field microscopy can also be used to synthesize arbitrary 3D patterns of illumination by reversing the light path and replacing the camera with a spatially modulated light source (Levoy et al. 2009). Thus, both light field and holographic methods can be used to stimulate not only a set of arbitrarily chosen points, but also a set of arbitrarily chosen regions, such as a collection of cell bodies. Each of these techniques offers unique advantages. Viable spatial light-intensity modulators for light field illumination include digital micromirror devices (DMDs), which can update much faster (up to ~ 1–10 kHz binary image refresh rates) than the programmable phase SLMs used for holographic setups (~60 Hz). However, with light-field illumination a substantial fraction of the light source power would in general be lost because many pixels of the DMD might be dark. In practice, phase SLMs preserve ~50% of the incoming light to synthesize holograms (Nikolenko et al. 2008, Papagiakoumou et al. 2008). SLMs can also synthesize high-resolution patterns in 3D (Lutz et al. 2008, Nikolenko et al. 2008, Papagiakoumou et al. 2008), whereas light field illumination sacrifices lateral resolution as axial resolution improves. Thus, light field strategies for photostimulation permit faster update rates, but holographic strategies can currently generate finer spatial patterns. We expect both illumination techniques will serve key applications in neuroscience.
AUTOMATED AND HIGH-THROUGHPUT MICROSCOPY
Addressing several classes of important issues in neuroscience will require the acquisition and analysis of massive data sets. For example, huge amounts of data will ultimately be needed to provide detailed knowledge about the development and higher-order connectivity patterns of distributed neural circuits, to characterize statistically the variations between nervous systems across and within species, and to conduct high-throughput imaging-based screens for mutations or drugs that affect nervous system function or disease progression. In many such cases, it has long been possible to acquire the type and quality of optical data needed but not the full data sets required to address the core scientific issues. Often lacking are the tools for rapid image collection, automated image analysis, and efficient management of vast image data repositories. In the 1990s, the fields of bioinformatics and drug discovery realized sweeping improvements in the speeds and costs of data acquisition, in large part owing to increased parallelization and automation. These high-throughput approaches have inspired and paved the way for emerging approaches to automation in neuroscience using light microscopy. As in bioinformatics, although the raw data will be worth inspecting manually or at least spot sampling, the crux of automated microscopy often lies in the ability to perform rapid sample manipulations or to distill higher-order statistical conclusions that need not be apparent by eye.
Microfluidic Methods for High Sample Throughput Under the Microscope
One growing approach to the acquisition of large data sets borrows directly from bioinformatics by using microfluidic chips for rapid manipulation of specimens and solution conditions under the microscope, as well as for high sample throughput. Microfluidic “lab-on-a-chip” devices have the virtue of optical transparency, a prerequisite for high-resolution microscopy, and allow high-throughput exchange of samples or sample conditions. In neuroscience, most microfluidic applications for microscopy have involved the model species Caenorhabditis elegans, owing to the ease of manipulating the positions and orientations of these round worms by fluidic control (Figure 5), the battery of available genetic techniques, and the ease of creating mutant libraries (Allen et al. 2008, Chalasani et al. 2007, Chronis et al. 2007, Chung et al. 2008, Cui et al. 2008, Guo et al. 2008, Heng et al. 2006, Hulme et al. 2007, Rohde et al. 2007, Zeng et al. 2008). By allowing worms to be controlled while spatially confined, microfluidic chips present alternatives to traditional means of immobilizing worms for study, such as by anesthesia or cooling (Zeng et al. 2008). Furthermore, Ti:sapphire lasers and amplifiers, familiar to neuroscientists from two-photon imaging, can perform precise micro-surgical procedures in C. elegans, such as cutting a single axonal projection (Allen et al. 2008, Guo et al. 2008, Zeng et al. 2008). 3D two-photon images of subcellular processes can be acquired in awake animals by using highly stable immobilization techniques (Zeng et al. 2008). Microfluidic chips can even be equipped with postsurgical feeding chambers to facilitate survivability for long-term studies (Guo et al. 2008). C. elegans also has the distinction of being the only animal whose entire neuronal wiring diagram, or “connectome,” has been fully characterized (White et al. 1986). Thus, the combination of high-throughput microfluidics with automated image acquisition and analysis should facilitate image-based decision-making algorithms that incorporate prior knowledge about neuronal and synaptic identities.
Figure 5.
Microfluidic devices for automated manipulation of C. elegans under the microscope. (a) Microfluidic worm sorter consisting of control channels and valves that direct flow of worms. (b) A worm immobilized for imaging by suction against microposts. Fluorescent mec-4::GFP-expressing touch neurons are visible. (c) A microchamber chip for large-scale worm screening. The chip has been loaded with dye to facilitate visualization. Each chamber can be independently addressed and loaded. (d) A worm immobilized against circularly oriented microposts in preparation for imaging. (Inset) High-resolution fluorescence image acquired through a microfluidic chip’s glass substrate; a single touch neuron is indicated by a white arrow. (Panels a–d are from Rohde et al. 2007.) (e) Chip for determining how channel dimensions affect the dynamics and efficiency of worm larvae movement. The worms’ ages and velocities are (top to bottom) for each image: L1, υ < 0.1 mm/s; L2/3, υ < 1.4 mm/s; L4, υ = 1.0 mm/s; young adult, υ = 0.3 mm/s; and L4, υ < 0.1 mm/s (images courtesy of D. Albrecht and C. Bargmann). (f) Microfluidic worm sorter loaded with dye for visualization. This chip is equipped with additional temperature control and positioning mechanisms. (g) punc-25-YFP-RAB-5 expression in (top) wild-type and (bottom) unc-16−/− mutants. An online machine vision algorithm automatically sorted ~400 worms/h on the basis of cell and puncta organization with 96.5% sorting accuracy after one round on the chip shown in f. White arrows indicate cell bodies, whereas the yellow arrow indicates a punctum (Chung et al. 2008). (h) Microfluidic chip for rapid fluorescence imaging of worm neural responses to olfactory stimuli (Chalasani et al. 2007). (i) Microfluidic worm corral loaded with dye for visualization. (j) Bright field image of immobilized worm in channel. (k) Fluorescence image of GFP-labeled synaptic puncta within a worm imaged in a microchannel (images i–k courtesy of D. Chiu). (l) Time-lapse sequence showing the development of an axon of the HSNL motor neuron, including the transport of GFP-labeled synapse-specific proteins down the axon (Allen et al. 2008).
Because of how they facilitate sample manipulation, throughput, and imaging, microfluidic devices are especially well suited for sorting and screening large populations of worms. Chips can automatically align a worm in a predefined orientation, in which it can be manually or automatically categorized and sorted for later study (Chung et al. 2008, Rohde et al. 2007). Research groups that historically spent tedious hours at the microscope screening worms for mutations will increasingly adopt automated approaches combining microfluidic manipulations with automated image acquisition and machine vision algorithms for image analysis. For example, one system has demonstrated the ability to automatically detect and sort rare mutations at a rate of ~400 worms inspected per hour (Chung et al. 2008). After training the computer under human supervision, worms were screened for abnormal patterns of YFP expression within synaptic endosomes along the nerve cord (Figure 5f,g) (Chung et al. 2008). Other processes that might be tracked by automated means include neurogenesis, neurite extension, and cell migration. Microfluidics have also proven useful in characterizing functional Ca2+ responses of olfactory neurons to many odorants screened in rapid succession (Figure 5h) (Chalasani et al. 2007, Chronis et al. 2007).
Such methods synergize with existing approaches used in massive screens, such as genetic mutagenesis, RNAi-based gene knockdowns, and the use of drug libraries. However, automated microscopy techniques will be able to gather statistics well beyond what is typically possible by labor-intensive manual studies, enabling researchers to identify both faintly discernable and rare phenotypes. The microfluidic techniques being developed currently for use with worms should also find applications with other model organisms, such as Drosophila embryos (Chen et al. 2004, Dagani et al. 2007, Lucchetta et al. 2005) or zebrafish. Moreover, the techniques of automated image acquisition and analysis are also applicable well beyond the realm of microfluidics applications. For example, computer controlled time-lapse microscopy has been used to monitor thousands of cultured neurons in vitro in a study of a Huntington disease model, automatically identifying neurons with inclusion bodies of aggregated huntingtin protein and repeatedly revisiting the same cells to track their death and survival (Figure 6a) (Arrasate et al. 2004).
Figure 6.
Techniques for automated and multiscale microscopy. (a) Automated microscopy. Neurons were cotransfected with a morphological marker, mRFP (red), and a mutant huntingtin protein fused to GFP, Httex1-Q47-GFP (green). The expression levels of mutant huntingtin and cell fates were tracked automatically in thousands of neurons over multiple days (Arrasate et al. 2004). (b) Multiscale microscopy as illustrated in a series of increasingly magnified images of a medium spiny neuron from mouse nucleus accumbens. (Left) Mosaic of low-magnification, transmitted light images of mouse striatum. (Left inset) Contrast-enhanced, close-up of an individual neuron. (Middle) Two-photon fluorescence image of the same neuron labeled with Lucifer Yellow and Alexa Fluor 568. (Right) Tomographic reconstruction of a dendrite by electron microscopy and the corresponding segmentation of dendritic spines from the same neuron (images courtesy of E. Bushong, M. Martone, and M. Ellisman, available online at the Cell Centered Database, http://ccdb.ucsd.edu). (c) Array tomography. Volumetric image of mouse vibrissa somato-sensory cortex. The image reveals synapses labeled by rhodamine-conjugated antisynapsin I (small red objects), cellular nuclei (DAPI-DNA, blue), autofluorescence from blood cells (large red objects), and a subset of pyramidal neurons labeled by FITC-conjugated anti-GFP (green) and was rendered from 134 sections, each 200-nm thick (image courtesy of K. Micheva and S. Smith; Micheva & Smith 2007). (d) All-optical histology. Maximum intensity projections of fluorescently labeled vasculature in the mouse vibrissa somato-sensory cortex. A penetrating arteriole is highlighted in yellow (P.S. Tsai, P. Blinder, J.A. Kaufhold, B. Friedman, and D. Kleinfeld, unpublished data).
Advances in Tissue Processing for Automated Microscopy
Although applications involving rapid exchange of samples or solutions can be well suited for microfluidic approaches, another set of applications for high-throughput imaging involves automated inspection of histological brain tissue specimens. The latter applications are progressing on the basis of recent advances in tissue processing and preparation. A key goal is the reconstruction of 3D-image data sets across large blocks of brain tissue, ideally with the possibility of using multiple fluorescent labels concurrently. A number of approaches are arising.
Several techniques draw on established methods for tissue processing in electron microscopy. One longstanding technique known as correlative microscopy provides complementary types of information by inspecting the sample using both optical and electron microscopy (Gaietta et al. 2002, 2006; Giepmans et al. 2005; Larson et al. 2005; Sun et al. 2007; Toni et al. 2007; van Rijnsoever et al. 2008). The optical images can facilitate the identification of specific cell types or molecules tagged with fluorescent markers, ideally while the cells are alive (Gaietta et al. 2002, 2006; Larson et al. 2005; Toni et al. 2007; van Rijnsoever et al. 2008). The optical data also provide anatomical coordinates to guide subsequent electron microscopy studies of nanometer-scale ultrastructure, such as synapses (Figure 6b). Dual labels for light and electron microscopy facilitate this process (Sosinsky et al. 2007). The bleaching of fluorescent dyes (Deerinck et al. 1994, Maranto 1982) can drive the oxidation of a secondary compound to form an electron-dense precipitate, which can subsequently be visualized in electron microscopy. When proteins have been tagged with a tetracysteine motif, the ReAsH biarsenical label can drive very specific photo-oxidation (Gaietta et al. 2002). Some fluorescent quantum dots can also readily be imaged by electron microscopy (Giepmans et al. 2005). Electron microscopy can access ~1 nm ultrastructural information in biological samples (Sosinsky et al. 2007), making it a suitable complement to light microscopy. However, when ultrastructural resolutions of >10 nm will suffice, the techniques of super-resolution fluorescence microscopy (see below) should emerge as powerful alternatives for acquiring nanoscale information directly from optical images, often while the sample is still alive.
Another technique that builds on tissue preparation approaches for electron microscopy is array tomography (Figure 6c) (Micheva & Smith 2007). This method for obtaining 3D image reconstructions involves fixing a tissue specimen in acrylic resin; cutting sections as thin as ~50 nm from the specimen using an ultramicrotome; mounting these sections on glass slides; fluorescence immuno-labeling the tissue; imaging all the fluorescently labeled specimens (to date generally by conventional epi-fluorescence microscopy); and computing a 3D reconstruction of the original tissue block. Array tomography has several virtues. First, the cut sections can be far thinner than the axial resolutions of conventional epi-fluorescence or laser-scanning microscopes. Thus, the axial resolution of the 3D reconstruction is not limited by microscope optics, only by the thickness of the ~50–100-nm tissue slices. Second, blocks of tissue millimeters thick can be sectioned in this way. This method enables, for example, 3D reconstructions over the entire depth of a mouse’s neocortex at ~200 nm lateral and ~50 nm axial resolution. Third, unlike with some approaches in super-resolution microscopy (see section below), array tomography places no inherent restrictions on the type of fluorophores used. Together, these benefits have enabled volumetric reconstructions far superior to those attainable by confocal microscopy, opening a broad set of new neuroscience applications.
For example, using array tomography it should be possible to categorize and assess the spatially varying densities of specific types of synapses across the entire depth of neocortex or other millimeter-sized specimens. Other applications might involve analyzing human brain tissue toward statistically characterizing both normal and diseased brains in terms of circuit, cellular, and molecular parameters. Aiding such characterizations, the sections in array tomography can undergo multiple rounds of immuno-labeling, stripping, and imaging, which allows images to be taken with multiple sets of fluorescent tags and then coregistered. Alternatively, the tissue strips can also be examined by correlative approaches employing scanning electron microscopy if still higher resolution information is desired. Finally, the compatibility of array tomography with nearly all forms of fluorescence microscopy, including the super-resolution techniques, makes this method particularly versatile. To date, the tissue slicing in array tomography is a bottleneck step requiring long hours by a skilled operator. By comparison, the image acquisition using a computer-controlled microscope and specimen stage has been automated. In the future, a greater degree of mechanical automation during tissue slicing will improve array tomography’s ease of use and throughput.
A demonstration of how automated slicing might work directly under the microscope is knife-edge scanning microscopy, in which individual sections down to ~0.5–1.0-μm thick are sliced during simultaneous imaging (Mayerich et al. 2008). A diamond knife is mechanically coupled to a microscope such that the knife is oriented perpendicular to the optical axis and lying within the focal plane. The tissue block is oriented at 45° angles to both the optical axis and the knife blade, allowing the cut sections to be imaged as they are sliced and pass over the blade. 3D reconstructions are computed from the set of raw 2D images. This approach has enabled rapid (~70 mm3 per hour at ~0.6 μm lateral resolution) optical imaging of large tissue volumes (~1 cm3) (D. Mayerich, personal communication; Mayerich et al. 2008). Initial studies with knife-edge microscopy used Golgi and Nissl stains for contrast, but genetically encoded labels could also be envisaged.
As an alternative to mechanical slicing of tissue, a tissue block-face imaging technique termed all-optical histology builds on laser machining and plasma-mediated ablation methods using ultrashort-pulsed regenerative laser amplifiers (Figure 6d) (Tsai et al. 2003). Alternating rounds of two-photon fluorescence imaging and tissue ablation are used to acquire 3D image stacks ~100–150 μm in thickness, followed by laser ablation of the volume just imaged to reveal the tissue below. The depth of ablation is typically chosen to be slightly less than the imaged depth to ensure overlap of images between successive cycles of imaging and ablation. The advantages of this approach include the inherent registration of the images, the method’s speed (~20 mm3 of tissue examined per hour appears feasible with an optimized system), and thus the ability to obtain 3D renderings of substantial blocks of tissue. Unlike in array tomography, the axial resolution is limited to that of conventional two-photon microscopy. Because imaging occurs immediately following optical ablation, as in knife-edge microscopy, fluorescent markers should ideally be impregnated within the tissue block beforehand or expressed genetically, as opposed to being introduced by immunolabeling methods after slicing. However, after optical ablation the tissue surface appears to retain normal immunoreactivity (Tsai et al. 2003), so it should be possible to immunostain after each round of tissue ablation, albeit with a considerable reduction in overall processing speed.
Another means of automated slicing is the automatic tape-collecting lathe ultramicrotome (ATLUM), a tour de force of mechanical engineering in which a block of tissue is progressively shaved while rotating about an axis (Hayworth et al. 2007, Lichtman & Hayworth 2009). The shaved tissue sections can be very thin (<30 nm) and are captured automatically on a reel of carbon-coated Mylar tape. This approach requires careful feedback stabilization of the thickness of tissue being shaved but provides a compact, long-term means of storing the sliced tissue in an addressable manner. This collection technique implies that desired locations in the preserved brain could be accessed again by stereotactic coordinates. To date, ATLUM has primarily been used as a means of preparing large tissue blocks for scanning electron microscopy, but optical imaging using either transmitted light or multicolor fluorescence has been demonstrated following tissue slicing by ATLUM (Lichtman & Hayworth 2009).
In the past few years, there has been considerable discussion within the neuroscience community as to whether it might be feasible by current techniques to generate high-resolution wiring diagrams of neural circuits in an automated or semi-automated manner. The ATLUM is one means of tissue preparation that might facilitate acquiring the requisite large data sets. Fully automated versions of the tissue slicing in array tomography might provide competitive alternatives. Although some groups are pursuing circuit reconstructions by electron microscopy, others are considering the use of automated optical microscopy, either alone or in conjunction with correlative electron microscopy approaches. The use of new multicolor fluorescence labeling strategies, such as the Brainbow approaches discussed below (Lichtman et al. 2008, Livet et al. 2007), may help by labeling dense sets of individual neurons, each of which might be traceable using color as a distinguishing marker. Plainly, the reconstruction of large circuits involving densely packed neurons is a challenge that will hinge on progress in image segmentation and tracing algorithms as much as on labeling and imaging techniques. Computational image analysis, not image acquisition, often remains the bottleneck in volume reconstruction (Zhang et al. 2008). However, in part because the optical, labeling, and tissue preparation techniques for neural circuit reconstruction are still evolving, the corresponding set of computational tools for tracing neurites and creating 3D circuit reconstructions is also still germinating. Two recent reviews on circuit reconstruction (Smith 2007) and bioimaging informatics (Peng 2008) discuss some of the hurdles involved.
IMAGING IN AWAKE BEHAVING ANIMALS
A longstanding goal for the use of microscopy in neuroscience has been to develop techniques that permit cellular-level brain imaging in actively moving mammals, toward uncovering and understanding the cellular dynamics underlying specific behaviors. Although in many respects microscopy might appear ill suited for studies in behaving animals, the need for such studies is considerable since anesthesia precludes behavior and substantially alters neural and astrocytic activity (Greenberg et al. 2008, Movshon et al. 2003, Nimmerjahn et al. 2007, Pack et al. 2001, Rinberg et al. 2006). The goal of visualizing individual cells’ activity in actively behaving mammals long remained elusive, but in the past two years two complementary approaches were successfully demonstrated. One of these approaches relies on alert, head-restrained animal preparations that can be studied with conventional microscopy instrumentation. The other approach involves the use of optical fiber to guide light to and from freely behaving animal subjects. Both approaches are compatible with the use of mice, which is important because of the wide availability of genetically modified mouse lines, including fluorescently labeled tool mice and mouse models of brain disease (see sections on fluorescent tool mice and chronic imaging).
Imaging in Awake, Head-Restrained Animal Subjects
In primates, awake, head-restrained preparations have allowed functional brain mapping using intrinsic optical signatures of brain activity (Grinvald et al. 1991) and studies of spatiotemporal responses from neuronal aggregates using voltage-sensitive dyes (Chen et al. 2006, 2008b; Quraishi et al. 2007; Raffi & Siegel 2007; Seidemann et al. 2002; Slovin et al. 2002; Yang et al. 2007). Intrinsic signal imaging in awake, body-restrained rats has facilitated examinations of how cerebral hemodynamics differs between anesthetized and awake subjects (Martin et al. 2002, 2006). Voltage-sensitive dye imaging in head-restrained but behaving mice has enabled the spatiotemporal dynamics of sensory and motor processing to be visualized in the neocortical whisker system with sub-columnar spatial resolution (Ferezou et al. 2007, Petersen 2009, Petersen et al. 2003).
However, cellular-level imaging in awake, head-restrained animals has only recently emerged (Dombeck et al. 2007, Nimmerjahn et al. 2007). By habituating a mouse to periods of head restraint under a conventional upright two-photon microscope, it was possible to visualize the simultaneous Ca2+ dynamics of large numbers of individual cells while the mouse was free to walk or run on a stationary exercise ball (Figure 7a). This assay builds on the longstanding use of head restraint in neuroscience to study locomotor responses in flies walking on a trackball (Götz & Wenking 1973). The more recent two-photon microscopy studies in behaving mice examined Purkinje neuron and Bergmann glial Ca2+ dynamics within the cerebellar vermis (Nimmerjahn et al. 2007) (Figure 7b–e) as well as Ca2+ dynamics of layer 2/3 neurons, astrocytes, and neuropil in the hind-limb sensory neocortex (Dombeck et al. 2007) (Figure 7f–h). In both of these brain areas, there were neurons and astrocytes whose Ca2+ activity correlated with locomotion, opening the door to further studies of the potential roles these cells might play in motor behavior. The use of head-restrained rodent preparations should also generalize to studies of other, nonmotor behaviors. Head-restrained preparations will further be valuable for studies in quietly resting animals, such as for comparing cellular activity patterns that arise in the awake and anesthetized brain. Initial two-photon microscopy studies of visual processing in awake rats have already revealed substantial differences between the two states (Greenberg et al. 2008, Greenberg & Kerr 2009).
Figure 7.
Microscopy in awake, head-restrained animals. (a) Apparatus for microscopy in an awake, head-restrained mouse. Cellular dynamics are imaged while simultaneously monitoring the movement of the mouse’s exercise ball. (b) Images of cerebellar cortex labeled by the Ca2+ indicator Oregon Green BAPTA-1 am (green) and the astrocyte marker sulforhodamine 101 (SR101) (red). SR101 co-staining allows online distinction between neurons (green) and astrocytic cells (yellow) in the molecular (left) and Purkinje cell (right) layers. (c) Example traces of displacements in x and z during mouse motion, showing the motion artifacts associated with running speed. Scale bars: speed, 20 cm/s; displacements, 5 μm; time, 10 s (panels a–c based on Nimmerjahn et al. 2007, 2009; Nimmerjahn 2009). (d) Contours of 48 Purkinje cell dendritic tree segments identified within two-photon Ca2+ imaging data acquired in an awake head-restrained mouse, superimposed on an image of the average fluorescence. (e) Ca2+ activity traces from four of the numbered dendritic tree segments shown in (d). Vertical ticks indicate Ca2+ spikes, as determined from threshold crossing events of the Ca2+ signals, after deconvolution to correct for the dye’s Ca2+ binding kinetics. Scale bars: 3 std. dev. (vertical) and 2 s (horizontal) (panels d and e based on Mukamel et al. 2007). (f, left) A false color time-projection image, acquired by two-photon microscopy over a 5-min period, 150 μm deep within the hind limb sensory cortex (S1HL) of an awake behaving mouse. The tissue was labeled by the Ca2+ indicator Calcium Green-1-am (green) and the astrocyte marker SR101 (red), such that neurons and astrocytes appear green and yellow, respectively. (Right) Regions of interest in which Ca2+ activity was monitored, example traces of which are shown in panel (h). (g) Simultaneous traces of the mouse’s locomotor speed and estimated brain displacements. Scale bars: speed, 20 cm/s; displacements, 5 μm; time, 10 s. (h) Ca2+ activity traces from four neurons outlined in (f), as obtained after correction of the image data for brain motion. Ticks indicate positive-going transients, with an estimated <5% motion-induced false positive error rate. Scale bars: 20% ΔF/F and 10 s (panels f–h based on Dombeck et al. 2007).
Key technical issues for imaging studies in behaving animals concern the motion artifacts that can arise owing to brain displacements during active behavior. Given that typical frame rates for in vivo two-photon microscopy are only ~2–20 Hz, brain displacements can differentially affect the individual line scans within a single image frame (Dombeck et al. 2007). However, even during active locomotion, brain displacements can be kept at modest levels (~1–5 μm) by firm mechanical restraint of the cranium and surgical steps that aid stabilization of the brain (Figure 7c, g). Moreover, many lateral image displacements are correctable offline by image registration algorithms (Dombeck et al. 2007) (see below). It is more difficult to correct axial motion artifacts in software strictly using 2D data sets. Nonetheless, axial motions might be mitigated either by image registration approaches relying on volumetric data or by active stabilization of the focal plane using online measurements of brain displacements (Fee 2000). Even without such corrective measures, the present data sets are of sufficiently high quality so as to provide insights about both neuronal and astrocytic dynamics within densely sampled local networks during active behavior. By comparison, multielectrode electrophysiological recordings generally sample neurons spaced much farther apart (usually ≥100 μm), owing to constraints on electrode density, and inherently miss astrocytes.
One limitation of imaging in head-restrained animals is the intrinsic restriction on animal behavior. Although subjects have the freedom to ambulate in place or even perform tongue or forepaw manipulations, the range of motions and thus the set of behaviors that can be explored remain constrained. Imaging sessions are also limited in duration by the periods of restraint animals can tolerate. On the other hand, in many paradigms the imposed limits on animal behavior are likely to prove advantageous. Virtual reality approaches, in which the animal views a computer screen, navigates simulated environments, and responds to stimuli, may also extend the range of behaviors that can be studied with head fixation. A caveat is that, even for behaviors amenable to head restraint, behavioral performance may be substantially altered. This includes but is surely not limited to the changes in locomotor gait that occur during head fixation. Thus, it will be valuable whenever possible to compare results obtained in head-restrained subjects to those acquired by fiber-optic microscopy in freely behaving animals.
Fiber-Optic Microscopy in Freely Behaving Animals
Just as the use of head–restrained alert preparations in imaging started with studies of intrinsic signals, so too the use of fiber optics in freely behaving subjects initially involved intrinsic optical signatures of brain activity. In the 1990s, a few studies explored the use of optical fiber bundles to image intrinsic functional signals in awake behaving cats (Rector et al. 1993, Rector & Harper 1991). The optical fibers both delivered illumination to the animal and returned signals to a remote camera. However, with just a bare fiber bundle and no lenses, the images produced were of poor quality and low resolution. Nevertheless, the idea of using fiber optics to study freely behaving animals continued to develop and was later applied to behaving mice (Adelsberger et al. 2005, Ferezou et al. 2006, Yamaguchi et al. 2001). In these later studies, optical fibers either were used as single point probes that monitored but did not image the tissue’s characteristics (Adelsberger et al. 2005, Yamaguchi et al. 2001) or were used in devices that did not permit individual cells to be distinguished (Ferezou et al. 2006, Murayama et al. 2007, Poe et al. 1996). Voltage-sensitive dye imaging in active mice permitted elegant examination of aggregate neuronal signals associated with sensory processing in the whisker system (Ferezou et al. 2006). Moreover, the first fiberoptic two-photon microscope was introduced in a pioneering study that used a ~25-g microscope to study freely moving rats (Helmchen et al. 2001).
In this portable two-photon microscope, a piezoelectric actuator accomplished laser scanning by vibrating the illumination fiber at mechanically resonant frequencies in the two lateral dimensions (Helmchen et al. 2001). Fiber delivery of femtosecond pulses for two-photon excitation was achieved with the aid of a pair of diffraction gratings to compensate for group velocity dispersion arising in the optical fiber. The microscope allowed imaging of erythrocyte flow and neuronal Ca2+ transients when used in a line-scanning mode. However, motion artifacts that arose when the rats were moving made functional imaging challenging during periods of active behavior. The microscope’s size also precluded its use in mice or lighter weight rats. Nonetheless, this seminal study not only inspired additional technological improvements, including further miniaturization of both one- and two-photon fluorescence microscopes, but also effectively conveyed that the imaging of cells in freely behaving animals was a feasible pursuit.
More recent portable two-photon microscopes for brain imaging have had masses as small as ~1–4 g, in part by employing GRIN microlenses similar to those used for microendoscopy (Engelbrecht et al. 2008, Flusberg et al. 2005b, Göbel et al. 2004, Piyawattanametha et al. 2006). Miniaturized laser-scanning mechanisms under exploration have included microelectromechanical systems (MEMS) scanning mirrors (Piyawattanametha et al. 2007, Piyawattanametha et al. 2006) as well as non-resonant (Sawinski & Denk 2007) or resonant vibration of the illumination optical fiber (Engelbrecht et al. 2008, Flusberg et al. 2005b). MEMS scanning mirrors (e.g., 1 × 1 mm2 or 0.75 × 0.75 mm2 in size) that are microfabricated in silicon allow line-scanning rates of up to ~ 3.5 kHz, which is sufficient to permit frame acquisition rates of ~30 Hz (Piyawattanametha et al. 2006) or fast tracking of erythrocyte flow by line-scanning (Figure 8a–c). An alternative approach to laser-scanning drives the illumination fiber in a resonant, spiral pattern at frame rates up to 25 Hz (Figure 8d,e) (Engelbrecht et al. 2008). This latter approach has allowed investigators to visualize cerebellar Purkinje cell Ca2+ activity in anesthetized rats (Figure 8f,g). To date no two-photon microscope based on micro-optics has provided data from freely behaving animals, but the steady technological progress suggests such an advance might arrive soon. In comparison, a miniaturized high-speed epi-fluorescence microscope based on micro-optics has already achieved cellular-level Ca2+ imaging in freely moving mice (Flusberg et al. 2008) (Figure 8h,i,j).
Figure 8.
Portable microscopy using miniaturized optics and scanners. (a–c) Portable two-photon microscope based on a microelectromechanical systems (MEMS) scanning mirror. (a) Cutaway view of a computer-aided design (CAD) model of the microscope. NIR ultrashort laser pulses are delivered to the device via the photonic bandgap fiber. Illumination exiting this fiber passes through a collimating lens and reflects off the MEMS scanning mirror. A micro-optic assembly focuses the laser light onto the specimen and projects fluorescence signals into a multimode collection fiber. The collimating lens is mounted within a mechanical shuttle to allow focal adjustments in the plane of excitation within the sample. (b) Two-photon fluorescence image of neocortical capillaries in an anesthetized mouse, obtained using the microscope after labeling the blood plasma with the dye fluorescein-dextran. The frame acquisition rate was 4 Hz, and 8 frames were averaged. (c) Line-scanning data acquired over the region indicated in (b) and acquired at 560 Hz. As erythrocytes passed through the vessels, the fluorescence signal transiently diminished, creating the appearance of dark diagonal streaks. Velocities of individual erythrocytes were found by computing the slopes of individual streaks. Scale bars: 0.5 s (horizontal), 20 μm (vertical) (panels a–c based on Deisseroth et al. 2006; Piyawattanametha et al. 2007). (d) Portable fiber-optic spiral-scanning two-photon microscope. A photonic bandgap fiber guides ultrashort-pulsed laser illumination (red) to the microscope. The overhanging fiber cantilever is driven in a spiral pattern at mechanical resonance by the piezoelectric scanner. The micro-optic assembly images the light emitted from the fiber tip onto the specimen plane. Fluorescence emissions (green) are collected by a large-core multimode fiber and detected remotely by a photomultiplier tube (not shown). (e) Photograph of the microscope schematized in (d). (f) Regions marked in color exhibited Ca2+ activity within cerebellar Purkinje cell dendrites of anesthetized rats as recorded using the microscope of (d) and (e). Cells were stained with the fluorescent Ca2+ indicator Oregon Green BAPTA-1 am. (g) Normalized fluorescence traces (ΔF/F) reveal Ca2+ transients from the color corresponding regions in (f). Scale bars: 5 s (horizontal), 10% ΔF/F (vertical) (panels d–g based on Engelbrecht et al. 2008). (h–j) A high-speed 1.1-g miniaturized epi-fluorescence microscope for use in freely moving mice. (h) Cutaway view of a CAD model of the microscope. The fiber bundle delivers illumination from a mercury arc lamp and returns the fluorescence image for projection onto a high-speed camera. The microscope, which is mounted on the mouse’s head, contains three microlenses and a miniaturized gear assembly for focusing. (i, left) Colored lines mark the perimeters of 17 individual Purkinje cell dendritic tree segments that were identified in a freely behaving mouse using the microscope in (h). Purkinje cells were labeled with the Ca2+ indicator Oregon Green BAPTA-1 am. (Right) Average of four frames in which the blue-filled cell (left) exhibited a Ca2+ spike. (j) Mean waveforms for dendritic Ca2+ spikes are averages of 37 (red) and 23 (blue) Ca2+ spikes from the color corresponding filled cells in (i). Scale bars: 0.1 s (horizontal), 0.5% ΔF/F (vertical) (panels h–j based on Flusberg et al. 2008).
This portable, 1.1-gram epi-fluorescence microscope permitted frame acquisition rates up to 100 Hz, enabling fast imaging of Ca2+ dynamics in cerebellar Purkinje neurons and of erythrocyte circulation in hippocampus and neocortex (Flusberg et al. 2008). The device used an image-transmitting bundle of optical fibers, a tiny planetary gear system for focusing, and three microlenses including interchangeable objectives of varying lengths for imaging deep or surface tissues (Figure 8h). When used to study Purkinje neurons of the cerebellar vermis, this portable microscope revealed the dendritic Ca2+ spikes of these cells (Figure 8i,j), which are known to closely reflect spike inputs from climbing fiber axons originating in the inferior olive. When the mice were actively moving, the Purkinje neurons exhibited higher rates and pairwise synchrony levels of Ca2+ spiking, as compared with periods of rest. These findings highlight the applicability of high-speed fiber-optic epi-fluorescence microscopy to studies of how neuronal activity patterns may vary across behavioral and physiological states.
Compared with fiber-optic two-photon microscopy (Engelbrecht et al. 2008, Flusberg et al. 2005b, Helmchen et al. 2001), distinct advantages of fiber-optic epi-fluorescence microscopy are its faster acquisition rates, lack of mechanical scanning, broader fields of view, and greater depth of field. These properties all help to confer greater robustness to motion artifacts. Disadvantages of epi-fluorescence microscopy include the more limited optical penetration depth, the increased background fluorescence, and the attendant photobleaching outside the focal plane. However, for cells in accessible tissues such as the olfactory bulb or cerebellar cortex, deeper-lying cells possessing superficial dendrites, or cells reachable by microendoscopy lenses (Murayama et al. 2007, 2009), high-speed fiber-optic epi-fluorescence microscopy should permit a range of studies in behaving mice, including mouse models of brain disease. We expect both two-photon and epi-fluorescence fiber-optic microscopes, as well as head-restrained approaches to imaging in behaving animals, will all find complementary usages in neuroscience.
Analysis of Functional Ca2+ Signals from Large-Scale In Vivo Imaging Data
Methods for loading synthetic fluorescent Ca2+ indicators into large populations of cells have enabled recent imaging studies of the dynamics of up to hundreds of individual neurons and astrocytes in live animals (Greenberg et al. 2008, Kerr et al. 2005, Mrsic-Flogel et al. 2007, Nimmerjahn et al. 2004, Ohki et al. 2006, Orger et al. 2008, Stosiek et al. 2003, Sullivan et al. 2005, Sumbre et al. 2008). Data analysis challenges arising from these studies concern the need for reliable, efficient extraction of Ca2+ activity traces from large sets of individual cells, in both awake and anesthetized animals. Hurdles include the correction of motion artifacts, identification of cellular signal sources, and in many cases the detection of individual action potentials. In principle, these challenges should also pertain to imaging studies of membrane voltage dynamics, but there have not yet been comparably large optical recordings of voltage dynamics across many individual cells in live mammals, owing to the current limitations of voltage-sensitive indicators.
In live animals, brain displacements correlated with physiological rhythms can create image motion artifacts. In awake or behaving animals, brain displacements associated with voluntary animal movements can compound the challenge. With images collected by highspeed fiber-optic epi-fluorescence microscopy in freely behaving mice, lateral motion artifacts were correctible by registering images to a common reference frame, leaving residual jitter at the sub-pixel level (Flusberg et al. 2008). However, if the camera’s acquisition rate was reduced below 75 Hz, some of the individual images appeared blurry, indicating there were noticeable levels of brain displacement occurring during the acquisition of single frames. Thus, with laser-scanning in vivo two-photon Ca2+-imaging, for which frame rates can be as low as ~2–4 Hz (Dombeck et al. 2007, Helmchen et al. 2001), rigid coregistration of entire frames will in general be insufficient to remove fast artifacts. However, many faster artifacts are correctible by laterally registering an image’s individual line-scans to one another, since typical line-scanning rates are ~100–2000 Hz. One method for performing this alignment relied on a probabilistic, hidden Markov model of brain displacement (Dombeck et al. 2007). Alternatively, there are alignment methods that permit deformations of the entire image frame to minimize errors due to brain displacements (Greenberg et al. 2008, Greenberg & Kerr 2009).
In addition to algorithms that correct for brain motion, there is also a need for automated methods for identifying the individual cellular signal sources within large-scale Ca2+-imaging data. Many recent studies have involved manual identification of cell bodies, performed by outlining regions of interest (ROI) by eye within static images (Dombeck et al. 2007, Flusberg et al. 2008, Greenberg et al. 2008, Kerr et al. 2005, Niell & Smith 2005). However, as the number of cells in a typical data set grows, manual approaches to identifying cells will become tedious and unwieldy. Semi-automated approaches have grouped pixels together into ROIs on the basis of their signal correlations (Ozden et al. 2008), but since there are still manual steps involved such approaches cannot be easily scaled to handle the largest available data sets without undue human labor.
Moreover, ROI analyses have mainly been based on heuristic, preconceived notions of what defines the geometric and other characteristics of specific cell types (Göbel et al. 2007, Ohki et al. 2006, Ozden et al. 2008). When these characteristics can be well defined, morphological filters can help find cells of the desired type in an automated or semiautomated fashion (Ohki et al. 2006). However, with the most commonly used synthetic Ca2+ indicators, morphological filters will have considerable difficulty in identifying neuronal dendrites or fine glial processes because these structures do not stand out with high contrast and often blend into the image background except during periods of Ca2+ activation. There are also emerging approaches for decomposing an imaging data set into its constituent signal sources on the basis of general statistical principles (Mukamel et al. 2007). This form of automated approach to cell sorting, which has been based on independent components analysis (Brown et al. 2001, Mukamel et al. 2007, Reidl et al. 2007), does not make assumptions about cells’ locations, shapes, or dye labeling patterns. Rather, cell sorting by independent components analysis relies on the statistics of dynamical signals, implying that even multiple, spatially overlapping cells occupying some of the same pixels can often be separated without significant cross talk. In such cases, ROI analysis tends to suffer badly from cross talk. However, owing to the reliance on the statistics of cells’ dynamics, independent components analysis can overlook cells with very low rates of activity, whereas an ROI analysis might find these cells on the basis of their appearance.
After identification of individual cells within Ca2+-imaging data there is often the question of whether either individual neuronal action potentials or rates of neuronal spiking can be found from the dynamical traces of Ca2+ activity. Nearly all neuron types have voltage-sensitive Ca2+ channels, so action potentials often have stereotyped signatures within Ca2+ activity traces. However, the underlying spike trains are often obscured by noise, the kinetics of Ca2+ binding to the indicator, and the cell’s own Ca2+ buffering mechanisms. Recent studies have applied a variety of approaches for estimation of spike rates or extraction of neuronal spike trains in a digital form. Given knowledge of the Ca2+ signal for a single action potential, generally a fast rise in fluorescence followed by a slower, often exponential decline (Helmchen et al. 1996), template matching (Greenberg et al. 2008, Kerr et al. 2005), or temporal deconvolution (Yaksi & Friedrich 2006) approaches can yield spike trains or rate estimates, depending in part on sampling rates and the intervals between spikes relative to the decay time of the Ca2+ signal for one spike. A simple deconvolution can be improved by using additional information regarding the amplitude and spectrum of noise in the data (Holekamp et al. 2008). More generally, one can apply machine learning techniques to paired imaging and electrophysiological data sets toward creating algorithms for spike recognition (Sasaki et al. 2008). Just as with the development of spike sorting methods in electrophysiology, significant effort will be required to compare the strengths and weaknesses of different approaches to cell sorting and spike extraction across a wide variety of cell types and experimental parameters.
SUPER-RESOLUTION OPTICAL MICROSCOPY
Many fundamental neural phenomena arise over sufficiently small length scales to have eluded visualization by diffraction-limited optical techniques. The sizes of synaptic structures are often just within the resolution limits of conventional light microscopy, and many features of dendritic spines and axonal boutons have remained indiscernible. Molecular processes of biochemical signaling and macromolecular interactions also involve spatial scales smaller than those traditionally accessible by light microscopy. Fine processes densely packed within neuropil resist anatomical tracing by light microscopy if adjacent fibers cannot be adequately distinguished. However, advances in “super-resolution” microscopy have made optical examinations of nanometer-scale phenomena tractable. Although diffraction limits how tightly light can be focused in traditional microscopy, by the judicious manipulation of fluorophores and photoswitches it is possible to obtain optical images of structures at sub-diffraction length scales (Figures 9–11). With such super-resolution tools, neuroscientists can now use light to interrogate “nanoscale” structures once accessible only by electron microscopy. Although correlative light and electron microscopy techniques can still provide images of finer (~1 nm) resolutions than those (~20 nm) regularly produced today by super-resolution fluorescence microscopy, for many applications in neuroscience the latter will suffice.
Figure 9.
Stimulated emission depletion microscopy. (a) Jablonski energy diagram illustrating the competing processes of stimulated emission (red) and spontaneous fluorescence emission (green) following electronic excitation. Colored arrows indicate absorptive and radiative transitions. Black arrows indicate nonradiative vibrational relaxations. (b) In STED microscopy, two concentric laser beams are scanned together across the sample. An inner excitation beam excites fluorophores in the sample within a diffraction-limited focal spot (blue). An outer doughnut-shaped beam depletes the population of excited fluorophores at the periphery of the excited region (red) via stimulated emission. Thus, only the excited fluorophores within the central hole of the doughnut emit fluorescence. These remaining excited fluorophores can occupy an area of diameter well below the diffractive limit, even though both laser beams are subject to diffractive limitations. By tuning the power of the depletion laser beam, one can adjust the size of the region over which the population of excited fluorophores is fully depleted. This in turn modulates the diameter of the central fluorescence emission region and thereby the point-spread function. (c) Optical schematic of the propagation of the two laser beams used for STED microscopy. (d) STED image of vimentin (immunolabeled with the dye Atto 590) in a neurite of a retinoic acid differentiated human neuroblastoma (SH-SY5Y). (e) Confocal image of the same sample (images in d and e courtesy of S. Hell, acquired by supercontinuum STED microscopy as described in Wildanger et al. 2008). (f) Single section image of dendritic spines of a YFP-positive CA1 pyramidal neuron in an organotypic hippocampal slice, acquired by live-cell STED microscopy. (g) Volume-rendered reconstruction of a stack of STED images (Δz = 0.5 μm). Panels (f) and (g) are based on Nägerl et al. 2008.
Early improvements to the resolution of light microscopy were attained by interferometric manipulations that shrunk the size of the point-spread function by raising the effective numerical aperture used to excite and/or gather optical signals. Interferometric approaches include 4Pi (Hell et al. 1997, Hell & Stelzer 1992) and I5M microscopy (Gustafsson 1999, Gustafsson et al. 1999, Shao et al. 2008), two methods that use an opposed pair of high-numerical aperture objective lenses to deliver and gather light at a common focal plane. Overall, these early methods were especially successful in improving axial resolution by up to ~sevenfold and to as good as ~80–150 nm in the case of 4Pi microscopy (Gugel et al. 2004). However, the interferometric methods did not free the resolution limit from the constraints of diffraction. Nor did these methods achieve widespread adoption by biologists, largely because of the technical challenges involved in implementation.
Once microscopists began to use fluorophores’ photophysical and photochemical switching effects, diffraction no longer had a constraining role in setting resolution limits. Demonstrated resolution limits have fallen to scales as fine as ~10 nm (Betzig et al. 2006), and ease of use has increased. Today, we stand at the brink of a widespread adoption of super-resolution techniques. In this section, we explore the properties and implications of the three main forms of optical microscopy that provide resolution limits unconstrained by diffraction. All three categories of super-resolution microscopy discussed below are now either commercially available or are soon to become so.
Photophysical Means of Sharpening the Point-Spread Function in Laser-Scanning Microscopy
The first proposed means of breaking the constraints of diffraction is an approach that relies on the photophysics of stimulated emission to narrow the point-spread function, the image of an ideal point emitter, in laser-scanning fluorescence microscopy (Hell & Wichmann 1994). Stimulated emission depletion (STED) microscopy achieves sub-diffraction-limited resolution by both exciting and depleting the available population of fluorophores using a pair of overlapping, concentric laser beams scanned together (Figure 9a–c) (Klar & Hell 1999, Klar et al. 2000). The first beam excites fluorophores lying within a diffraction-limited spot to an excited electronic state. However, the second “STED” beam, which is shaped in cross section like a doughnut, prevents fluorescence at the periphery of the excited area by using stimulated emission, in which the fluorophores return to the ground state by emitting light of the same color as the STED beam rather than emitting fluorescence photons (Figure 9a,b). Thus, only fluorophores at the center of the first beam’s focus are able to fluoresce, which sharpens the point-spread function to the “hole of the doughnut” left unaffected by the STED beam. The ratio of the STED beam’s intensity to the value of intensity needed just to overcome the spontaneous rate of fluorescence emission from the excited state dictates the size of the zone in which fluorescence is still possible. Thus, a more intense STED beam leads to finer resolution (Harke et al. 2008). Using this approach to spatially isolate the set of available fluorophores, which allows sequential examination of neighboring fluorophores by scanning, STED microscopy has achieved imaging resolutions as fine as 16 nm (Donnert et al. 2006, Westphal & Hell 2005), and even ~6 nm in a solid state diamond sample (Rittweger et al. 2009). In theory, the resolution can be made infinitely fine using STED, but in practice the power of the depletion beam cannot be raised beyond what competing processes of photo-damage, photobleaching, and background fluorescence excitation will permit.
As in other forms of laser-scanning imaging, there are important trade-offs in STED microscopy among resolution, signal-to-noise ratio, imaging speed, and deleterious processes such as photobleaching. Because the dual-beam STED strategy produces fluorescence signals from a region of tunable size, and not from individual emitters as do super-resolution approaches based on single-molecule detection, STED microscopy offers considerable control of these trade-offs and so allows versatile optimization under different conditions. STED microscopy has reached frame rates up to 28 Hz at 62-nm resolution in a 2.5 × 1.8 μm2 field of view, albeit with only a few collected signal photons per pixel (Westphal et al. 2008).
Notable results in neuroscience obtained by STED microscopy include several pertaining to the organization of synapses, such as the finding that synaptotagmin I molecules residing in synaptic vesicle membranes remain clustered together in patches on the cell membrane following vesicle release (Willig et al. 2006). STED microscopy has also allowed visualization of the ring-like structures of the protein bruchpilot at the Drosophila neuromuscular junction (Kittel et al. 2006), the SNARE protein syntaxin (Sieber et al. 2006) and its dynamics (Sieber et al. 2007), and the organization of nicotinic acetylcholine receptors (Kellner et al. 2007). Beyond the synapse, STED microscopy has revealed the clustering of the amyloid precursor protein (Schneider et al. 2008) and patterns of channel localization in olfactory neurons (Lin et al. 2007). STED microscopy is readily applicable to live cells and has been used to image the endoplasmic reticulum (Hein et al. 2008), track synaptic vesicle movement in axonal boutons (Westphal et al. 2008), monitor membrane lipid dynamics (Eggeling et al. 2008), and interrogate structural changes of dendritic spines during chemically induced LTP in hippocampal neurons (Figure 9f,g) (Nägerl et al. 2008).
Many technological enhancements to STED microscopy are already underway. Imaging speeds will be raised by multifocal scanning strategies (Hofmann et al. 2005). Additional sculpting of the point-spread function along the axial dimension permits 3D super-resolution imaging (Klar et al. 2000, Schmidt et al. 2008). Novel fluorophores that can better tolerate STED beams of greater intensities without bleaching would enable higher resolutions (Donnert et al. 2006). Dual color STED imaging has already emerged (Donnert et al. 2007b, Meyer et al. 2008). For examination of diffusive effects and reaction kinetics over nanometer-sized length scales, STED techniques can be married with the longstanding approaches of fluorescence recovery after photobleaching (FRAP) (Sieber et al. 2007) and fluorescence correlation spectroscopy (FCS) (Eggeling et al. 2008). Although STED microscopy typically employs pulsed lasers, it was recently implemented using continuous wave lasers, improving ease of use for many users and broadening the set of suitable lasers and fluorescent probes, albeit at the cost of increased photobleaching (Hein et al. 2008, Willig et al. 2007).
STED microscopy is the founding member of a broader family of methods, known as reversible saturable optical linear fluorescence transitions (RESOLFT) imaging, which also includes imaging based on ground-state depletion (GSD) (Bretschneider et al. 2007, Hell & Kroug 1995) and photoactivatable fluorophores (PA-FP) (Dedecker et al. 2007, Hofmann et al. 2005, Schwentker et al. 2007). In all these methods, the core idea remains to temporarily switch off the fluorescence of the indicator to squeeze the effective point-spread function of a scanning microscope. In GSD, instead of depleting fluorophores at the periphery of the excitation spot by stimulated emission, the fluorophores are transiently shelved in a dark triplet state of a long lifetime (Bretschneider et al. 2007). However, the probability of transitioning into a permanently dark, photo-bleached state is raised when the fluorophore is in a triplet state, which complicates the implementation of GSD. PA-FP RESOLFT microscopy relies on fluorophores that can be photoactivated by light of a different color than that used for exciting fluorescence. In STED microscopy, the depletion beam must compete with the nanosecond timescales of fluorescence emission and so requires substantial intensities (~ 10–100 MW/cm2). By comparison, photoactivatable fluorophores can be switched off using much lower beam intensities (~ 10–1000 W/cm2). However, with the photoactivatable fluorophores explored so far, the switching kinetics limit acquisition speeds to ~ 1–50 ms/pixel (Hell 2007, Hofmann et al. 2005). Cross talk between activation and excitation of the fluorophores, as well as the restriction to photoactivatable or photoswitchable fluorophore species, pose other limits to this technique (Hofmann et al. 2005). Overall, STED and other members of the RESOLFT family will allow neuroscientists to address many questions about nanoscopic features of live cells and synapses, with sufficient speed to follow a large set of protein and structural re-arrangements in real time.
Super-Resolution Imaging by Using Patterned Illumination to Encode Fine Spatial Details
The term structured illumination microscopy (SIM) refers to a set of techniques that can provide resolution at length scales finer than the normal limits by illuminating the sample with a sequence of periodic patterns of high spatial frequencies, close to the limit of what the microscope can physically transmit (Gustafsson 1999, 2000; Lukosz & Marchand 1963). The set of raw images is then used to extract information computationally about still higher spatial frequencies and thus provides fine spatial details (Figure 10).
In the simplest such approach, known as linear SIM (Figure 10a–c) (Gustafsson 1999, 2000), each raw image is a product of the illumination pattern with the spatial distribution of fluorophores in the sample. When considered in the Fourier or spatial frequency domain, the spatial frequencies contained in one of the raw images are those obtained mathematically through a convolution of the illumination’s spatial frequency transform with the spatial frequency transform of the distribution of fluorophores. Thus, information about spatial frequencies that cannot normally be observed is encoded within the moiré-like beat patterns that arise between the two frequency sets in the Fourier domain (Figure 10b). The highest spatial frequency so encoded is the sum of the conventional resolution limit and the illumination spatial frequency, or typically about twice the conventional limit. By acquiring a set of several images using different angular orientations and phase shifts for the illumination pattern, returning the spatial information obtained from this image set it to its original position in frequency space, and then applying an inverse Fourier transform, images in linear SIM can be reconstructed at a resolution twice as fine as that of conventional microscopy (Gustafsson 1999, 2000). Similar ideas can be applied to the axial illumination to encode more information about structure along that axis (Bailey et al. 1993) or to allow 3D SIM (Figure 10a) (Gustafsson et al. 2008, Pielage et al. 2008, Schermelleh et al. 2008). However, as with the 4Pi and I5M approaches to improving resolution, the resolution limit of linear SIM is still constrained by the physics of diffraction. This is not so in nonlinear SIM, in which diffraction is no longer the constraining factor (Gustafsson 2005, Heintzmann et al. 2002).
Nonlinear SIM provides even higher resolution by exploiting a nonlinear relationship between the intensity of fluorescence excitation and the rate of fluorescence emission. This nonlinearity effectively induces higher-order spatial harmonics in the pattern of fluorescence excitation, increasing the amount of information that can be encoded in the beat patterns (Figure 10c) (Gustafsson 2005, Heintzmann et al. 2002). One straightforward means for inducing such a nonlinear relationship with sine wave illumination patterns is to have the peaks of the sine waves reach saturating excitation intensities. This approach enables saturating structured illumination microscopy (SSIM), the simplest form of nonlinear SIM. In SSIM, the resolution is set by the number of spatial frequency harmonics that can be extracted while staying above background noise levels. For example, if one harmonic is used, then the resolution limit is approximately threefold smaller than the conventional limit. To date, the most harmonics used in addition to the fundamental frequency has been three, which enabled lateral resolutions of ~50 nm (Gustafsson 2005).
Strengths of SIM are that it records images in a wide field mode without scanning and thus, in principle, could examine very large fields of view, given camera chips of sufficient numbers of pixels. Another virtue is that neither linear nor nonlinear SIM requires fluorophores to be photoswitched or photoactivated, so both approaches work well with conventional fluorophores. This advantage allows the brightest markers and those least susceptible to bleaching to be chosen, making SIM well suited for multicolor imaging (Schermelleh et al. 2008). One disadvantage is that the saturating intensities used in SSIM can promote photobleaching or photodamage. In the future, photoactivated fluorophores might enable nonlinear forms of SIM that employ substantially reduced illumination intensities (Gustafsson 2000, 2005). Although SSIM breaks the diffraction limit, the accurate extraction of information about fine spatial scales requires high signal-to-noise ratios and minimal specimen drift over the time needed to acquire a raw image set. Frame rates are mainly limited by the need to record multiple raw images per reconstruction, which can take many seconds to acquire (Gustafsson 2005).
Neuroscience studies employing linear SIM include an investigation of structural abnormalities in Drosophila synapse organization, with 100-nm lateral and 250-nm axial resolution (Figure 10d) (Pielage et al. 2008). 3D SIM has recently revealed actin microtubule cytoskeletons and synaptonemal complexes (Gustafsson et al. 2008) and permitted imaging of single nuclear pore complexes in mammalian cells (Schermelleh et al. 2008). These studies represent just the beginning, and a key development to watch for is the application of SIM to study live cells, which will necessitate reductions in the required illumination powers.
Super-Resolution Imaging Based on Localization of Single Fluorophores
Several super-resolution techniques rely on the detection and localization of single fluorophores to reconstruct images at resolutions unconstrained by diffraction. This approach builds on a large body of research in single-molecule biophysics that probed the properties of single fluorescence emitters and used these to study macromolecules (Betzig & Chichester 1993, Moerner 2002, Moerner & Kador 1989, Weiss 1999). Single molecule–based approaches to super-resolution imaging localize fluorophores to within tight bounds limited not by diffraction but by photon-counting statistics (Moerner 2007).
Building on work showing that single molecules could be imaged (Ambrose & Moerner 1991) and spatially localized to within uncertainties below the far-field diffraction limit (van Oijen et al. 1998), fluorescence imaging with one-nanometer accuracy (FIONA) enabled researchers to track the individual, 74-nm steps of the molecular motor myosin V, settling a debate about the motility mechanism (Yildiz et al. 2003). FIONA accurately localizes a spatially sparse set of single fluorophores by calculating the centroid position of each fluorophore’s emitted photons. The accuracy of localization is limited not by the diffraction-limited size of an emitter’s image on the camera, but rather by the number of detected photons available for the centroid calculation (Michalet & Weiss 2006, Thompson et al. 2002). [This is akin to the statement in probability theory that a standard error of the mean can be much smaller than the standard deviation, given a large set of observations drawn from the same distribution (Bobroff 1986).] Note that this approach works well only if the spatial distributions of photons from different emitters do not overlap on the camera. Once this condition is violated, the centroids of individual emitters can no longer be calculated accurately.
Recent techniques known as stochastic optical reconstruction microscopy (STORM) (Figure 11a–c) (Bates et al. 2007; Huang et al. 2008a,b; Rust et al. 2006) and photoactivated localization microscopy (PALM) (Figure 11a,d,e) (Betzig et al. 2006; Hess et al. 2006; Juette et al. 2008; Shroff et al. 2007, 2008) build on the basic ability to localize single fluorophores by dividing a denser set of fluorophores into a sequence of sparse fluorophore distributions, each of which has minimal overlap in the images of the individual emitters (Figure 11). Thus, if one finds the individual fluorophores’ locations for each of these sparse subsets, an entire image can be reconstructed by summing the locations of all fluorophores found within all the subsets. The key trick is the means of imaging a sparse subset of the fluorophores at any one time.
This trick is provided by the use of photoswitchable fluorophores, which can be switched on from a dark state using light of a different color than that used for fluorescence excitation. PALM and STORM typically rely on photoactivatable, genetically encoded fluorescent proteins (Betzig et al. 2006, McKinney et al. 2009, Shroff et al. 2007, 2008, Testa et al. 2008, Vaziri et al. 2008) or photoswitchable dye molecules, such as pairs of a cyanine dye and another shorter-wavelength chromophore that together serve as individual photo-switchable fluorophores (Bates et al. 2005, 2007; Huang et al. 2008a,b; Rust et al. 2006) and that can be covalently conjugated (Conley et al. 2008). The details of how a cyanine dye molecule can be photo-switched remain poorly understood, but the shorter-wavelength “activator” dye in the pair is typically used to photo-activate the “reporter” cyanine fluorophore. After being activated, the reporter emits fluorescence when excited by light of the same color that drives the inactivation transition, until inactivation actually occurs. In addition to cyanine dyes or photo-activatable fluorescent proteins, other photo-switchable or photo-uncaged fluorophores can also be used for PALM and STORM (Betzig et al. 2006, Fölling et al. 2007, Lord et al. 2008, Rust et al. 2006).
To activate a sparse set of fluorophores starting from a denser and dark population of emitters, a brief pulse of activation light is applied. The resulting stochastic set of activated fluorophores is then imaged, with sufficient mean numbers of photons collected to permit the desired accuracy of localization. Before activating the next set of emitters, the current set that has already been localized must be inactivated. Inactivation of fluorescent proteins and caged dyes is typically accomplished by photobleaching, which permanently darkens the fluorophores once inspected; the cyanine dyes are inactivated into a dark state that permits subsequent reactivation. Armed with these approaches one can assemble a super-resolution image using repeating sequences of activation, imaging, and either bleaching or inactivation (Figure 11a)
Note that the notion of resolution in PALM and STORM differs somewhat from that in conventional imaging modalities (Ober et al. 2004, Ram et al. 2006, Watkins & Yang 2004). PALM and STORM accurately localize each emitter, but to achieve a high-resolution image, the set of sampled emitters must also densely cover the specimen (Shroff et al. 2008). Meeting the latter proviso typically requires both high labeling densities and many rounds of fluorophore activation. Although the first PALM and STORM images required minutes to hours to localize a sufficient number of emitters (Betzig et al. 2006, Rust et al. 2006), subsequent optimization of excitation and the number of raw frames acquired per image have reduced acquisition times to the ~30-s range (Shroff et al. 2008). It appears further optimization of imaging speed will hinge on the development of brighter probes and faster cameras. Faster acquisition speeds are important for accurate fluorophore localization because drift and sample motion on ~10-nm scales are presently the limiting factors (Betzig et al. 2006, Rust et al. 2006). In longer recordings, drift can be corrected to the precision with which bright, fluorescent beads can be tracked across each frame (Betzig et al. 2006, Rust et al. 2006). To date, researchers have used stochastic localization techniques to image cellular adhesion complexes (Shroff et al. 2007, 2008), mitochondrial networks (Bock et al. 2007, Huang et al. 2008a, van de Linde et al. 2008, Vaziri et al. 2008), endocytotic machinery (Bates et al. 2007, Huang et al. 2008b), microtubule systems (Bates et al. 2007; Egner et al. 2007; Heilemann et al. 2008; Huang et al. 2008a,b), membrane proteins (Betzig et al. 2006, Manley et al. 2008), and localization patterns of regulatory proteins in bacteria (Biteen et al. 2008).
There are several approaches for extending PALM and STORM to 3D imaging. Initial efforts permitted <100-nm axial localization. In one approach, an astigmatism was deliberately introduced into the optical pathway, allowing each fluorophore’s axial position to be encoded at ~50-nm resolution by the shape of its photon distribution on the camera (Huang et al. 2008a,b; Kao & Verkman 1994). In another approach, a 3D point spread function was fit to a pair of images acquired at two distinct focal planes, providing axial information (Juette et al. 2008, Prabhat et al. 2004). Other approaches bring ~20-nm axial localization to super-resolution microscopy. Enhanced axial resolution can be achieved via an interferometric detection scheme that employs two objectives and three or more cameras (Shtengel et al. 2009, von Middendorff et al. 2008). Similar resolution gains can be realized using a single camera and a microscope in which the point-spread function has been shaped like a double-helix, thereby encoding axial information in the angular orientation of the emitted photon distribution (Pavani et al. 2009, Pavani & Piestun 2008). All these methods for 3D super-resolution imaging have to date been limited to specimens a few microns or so in thickness. However, to inspect specimens of greater thickness, optical sectioning techniques such as two-photon excitation might be combined with the above approaches for obtaining nanoscale resolution in 3D (Fölling et al. 2008a, Vaziri et al. 2008).
Although PALM and STORM rest on similar ideas, different laboratories have implemented these methods using different types of photoactivatable fluorophores with distinct strengths and limitations. Genetically expressed photoactivatable fluorescent proteins are compatible with live cell imaging, demonstrated at ~0.03 Hz (Shroff et al. 2008), and high-resolution single molecule tracking (Manley et al. 2008, Ram et al. 2008). However, after multiple rounds of activation, imaging, and bleaching, there are progressively fewer unbleached fluorescent proteins remaining, implying a declining density of fluorophores available for activation. Presently there is also a limited set of photoactivatable fluorescent proteins, but more are emerging including some reversibly switchable varieties (Andresen et al. 2008, Biteen et al. 2008, McKinney et al. 2009, Stiel et al. 2008, Subach et al. 2009). Although the brighter photoactivatable fluorescent proteins such as Kaede and EosFP have yielded hundreds of detected photons before bleaching (Betzig et al. 2006, McKinney et al. 2009), photoswitchable cyanine dyes have provided up to ~6000 detected photons per switching cycle (Huang et al. 2008a), facilitating accurate localizations. The use of cyanine dye pairs also permits a substantial number of different color labels for multicolor imaging (Bates et al. 2007). However, cyanine dye photoswitching requires at least a low concentration of thiol-based reducing agents (Bates et al. 2005). Labeling intracellular proteins with dyes also poses some generic challenges. Immunolabeling is a common approach for dye-labeling proteins of interest, but the physical size of antibodies limits the density of fluorophore labels and thus imaging resolution. Direct conjugation of photoswitchable dyes to primary rather than secondary antibodies would improve this limit (Huang et al. 2008b). Direct coupling of dye molecules to specific proteins through hybrid chemical genetic approaches (Fernández-Suárez et al. 2007, Griffin et al. 1998, Popp et al. 2007) should facilitate super-resolution imaging by capitalizing on the specificity of genetically encoded tags in living cells and the brightness of synthetic fluorophores. Single molecule–based imaging has also been implemented with conventional fluorophores using ground-state depletion (Fölling et al. 2008b) by optically shelving most of the fluorophores into long-lived dark triplet states and imaging the remaining fluorophores. This approach opens up a much wider catalog of usable fluorophores for single molecule imaging but involves higher levels of unintended photoactivation during fluorescence excitation. The stochastic localization methods have also been extended to encode polarization anisotropies, in addition to emitter positions (Gould et al. 2008, Testa et al. 2008). In the future, stochastic localization techniques should be applicable to large fields of view without compromising speed or resolution. However, this will require cameras with large numbers of pixels and fast acquisition speeds, as well as wide-field, high-NA microscope objective lenses.
Outlook for Super-Resolution Imaging
All the main super-resolution microscopy approaches are likely to find their own niches within neuroscience. Thus, super-resolution imaging should permit studies of synaptic and macromolecular rearrangements in response to biochemical, synaptic, or electrical stimuli and will likely also be useful for tracing axons and deducing patterns of neuronal connectivity. In many cases there may be a choice between multiple super-resolution techniques adequate to address neuroscientists’ questions about ultra-structure. With the ability to localize single dye molecules, PALM and STORM may often be especially well suited for probing macromolecular interactions. STED may be particularly suited for studies of diffusion and rapid reaction kinetics, given this method’s ability to park the laser beams at a single spot for fast, continual data acquisition. Although not yet demonstrated, SIM will likely soon be extended to live specimens, which would be a boon, given the method’s ability to use conventional fluorophores. The reconstruction of neural wiring diagrams may also be greatly aided by the combination of automated tissue processing techniques and super-resolution microscopy. The quality of the images provided will dictate the degree of difficulty in the computational challenges of image segmentation and axon tracing. Animals with patterns of fluorescence labeling carefully designed to facilitate the identification of neural circuitry and synaptic connections may be particularly important (see mouse genetic strategies below).
COHERENT OPTICAL CONTRAST MECHANISMS
Three types of laser-scanning microscopy based on coherent forms of optical contrast have found a growing set of applications in neuroscience research. As in two-photon microscopy, each imaging approach described here requires scanning one or more laser focal spots within the sample. However, unlike the majority of fluorescence techniques, these coherent techniques are often (but not always) used without exogenous labels, thereby exploiting the intrinsic optical contrast mechanisms of tissue itself. Avoidance of fluorescence as the contrast mechanism helps to reduce phototoxicity, which can occur via photophysical side pathways after fluorescence excitation. The ability to inspect unstained tissues facilitates potential clinical applications.
Optical Coherence Tomography
Optical coherence tomography (OCT) relies on backscattering of light as the contrast mechanism and provides optical sectioning in a way analogous to how forms of ultrasound imaging use round-trip time-of-flight measurements to determine the depths of signal sources (Huang et al. 1991). Because light travels too fast for the travel delays to be measured directly, OCT uses a low-coherence interferometer to compare the optical path lengths of light backscattered from objects in the sample with the path length of the interferometer’s reference arm. The low-coherence interferometer in OCT is often implemented in optical fiber, owing to the resulting portability and ease of alignment, the availability of economical but excellent fiber optic components for telecom wavelengths, and the benefits for making miniaturized imaging probes. The accuracy to which the optical path length comparisons can be performed, and thus OCT’s axial resolution, is inversely proportional to the spectral bandwidth of the illumination source. Thus, investigators have used broad bandwidth but spatially coherent light sources (~50–400 nm) to attain axial resolutions from tens of microns down to the submicron range (Aguirre et al. 2006b, Cense et al. 2004, Drexler et al. 1999, Leitgeb et al. 2004, Potsaid et al. 2008, Považay et al. 2002, Srinivasan et al. 2006, Wojtkowski et al. 2004). A weakly focused illumination beam is usually scanned laterally across the sample, either in 1D for the acquisition of a 2D cross-sectional image or in 2D for the acquisition of an image volume. Diffractive considerations govern the width of the beam waist and thus OCT’s lateral resolution.
The best-developed application for OCT is the inspection of the live human retina (Drexler & Fujimoto 2008). The resolution, sensitivity, and acquisition speed of OCT have all improved to where, in combination with adaptive optical techniques for correcting the optical wavefront aberrations that arise in the eye, ultrahigh-resolution OCT systems can image across the retinal laminae and even resolve individual human photoreceptors (Figure 12a) (Zawadzki et al. 2008). OCT’s speed of acquisition (e.g. ~36 frames/s with 50-μs line exposure time; Zawadzki et al. 2008) is important for overcoming potential motion artifacts from saccadic eye movements. Careful comparisons to histological tissue specimens have been important for proper image interpretation because of confounds that can arise between absorption and scattering and from uncertainty in the biological features that scatter light (Anger et al. 2004, Gloesmann et al. 2003). However, the capability for simultaneously visualizing human photoreceptors and other aspects of retinal microanatomy will not only permit improved diagnostics for clinical ophthalmology, but also might allow comprehensive studies of how human visual perception correlates with detailed aspects of the eye’s construction and vascular responses. Forms of OCT that detect Doppler shifts incurred in the sample (Chen et al. 1997, Izatt et al. 1997) are capable of measuring blood flow speeds in the human retina (Leitgeb et al. 2003, White et al. 2003) and have also permitted 3D reconstructions of cerebral microcirculatory speeds in mice (Wang et al. 2007b, Wang & Hurst 2007).
Figure 12.
Emerging methods for coherent generation of intrinsic optical contrast: optical coherence tomography and coherent anti-Stokes Raman scattering microscopy (CARS). (a) Image acquired by adaptive optical, ultrahigh-resolution-optical coherence tomography (AO-UHR-OCT) of the retina of a live 35-year-old volunteer. The image was acquired at 4.5° nasal-retinal location by an AO-UHR-OCT system with 3.5-μm axial resolution. The focus of the adaptive optical system was set at the photoreceptor layers, and 500 axial depth scans were performed along a 0.5-mm line to generate the image. Fine details of the photoreceptor layer reveal individual human photoreceptors (bottom). Scale bars: 100 μm (top), 50 μm (bottom). Abbreviations for anatomical assignments: Ch, choriocapillaris and choroid; GCL, ganglion cell layer; I/OS, inner/outer segment junction; INL, inner nuclear layer; IPL, inner plexiform layer; OLM, outer limiting membrane; ONL, fibers of Henle with outer nuclear layer; OPL, outer plexiform layer; RNFL, retinal nerve fiber layer; RPE, retinal pigment epithelium; and VM, Verhoeff’s membrane created by end tips of cone photoreceptor outer segments (based on Zawadzki et al. 2008). (b, c) CARS microscopy images of lysophosphatidylcholine-induced swelling of myelin sheaths that envelope axons in excised guinea pig spinal tissue. The pump and Stokes laser beams for CARS microscopy were colinearly focused in the imaging plane of the neuronal axons. (b) CARS images of normal myelin sheathes obtained using excitation light (pump and Stokes beams) polarized parallel (left) or perpendicular (right) to the axons’ orientation. (c) CARS images of swollen myelin sheaths using excitation light that is polarized parallel (left) or perpendicular (right) to the axons’ orientation (panels b and c are based on Fu et al. 2007).
Although OCT has, to date, mainly enabled imaging of anatomical structures or blood flow speeds, possibilities for functional measurements of brain activity by OCT are under exploration. Several OCT studies have examined intrinsic changes in light reflectance that occur in response to visual stimulation in isolated vertebrate (Yao et al. 2005) and mammalian (Bizheva et al. 2006) retinae, as well as in the retinae oflive rats (Srinivasan et al. 2006) and human subjects (Srinivasan et al. 2009). The use of functional OCT for 3D intrinsic signal mapping in the mammalian brain is also being explored (Aguirre et al. 2006a, Chen et al. 2008a, Maheswari et al. 2002, Maheswari et al. 2003, Rajagopalan & Tanifuji 2007). This might provide a 3D alternative to the conventional use of intrinsic optical reflectance signatures of brain activity for functional brain mapping. However, more work remains to be done toward understanding the depth dependence of activity-related reflectance changes. A few in vitro studies have also used OCT to detect action potentials in excised invertebrate nerves, relying on small changes in refractive index or nanometer-scale changes in axonal volume or geometry that modulate a nerve’s light scattering properties (Akkin et al. 2004, 2007; Fang-Yen et al. 2004; Lazebnik et al. 2003). Further development of this approach would be of interest because it might conceivably lead to functional measurements in the intact nervous system, such as in the optic nerve in the context of clinical ophthalmology.
Coherent Anti-Stokes Raman Scattering Microscopy
Coherent anti-Stokes Raman scattering (CARS) microscopy creates images by using intrinsic signatures of molecular vibrations within the sample for contrast generation. A molecular vibration of interest is probed using nonlinear interactions in the sample between three photons from two laser beams, known as the pump beam and the Stokes beam, whose frequency difference is tuned to match the vibrational resonance. The CARS signal, which has a frequency equal to the sum of the pump laser beam frequency and that of the resonance, rises linearly with the intensity of the Stokes beam and quadratically with the intensity of the pump beam. This enables 3D optical sectioning, as in other forms of nonlinear optical microscopy. However, a nonresonant background signal also arises from the specimen even when the frequency difference of the two lasers is detuned from a molecular resonance. This nonresonant background can overwhelm fainter CARS signals from vibrational resonances. Thus, unlike with two-photon imaging for which femtosecond laser pulses are generally used, in CARS microscopy it is common to use pulse durations of picoseconds, which permit superior signal-to-background ratios because the narrower spectral bandwidths of these pulses are better matched to those of vibrational Raman bands (Cheng et al. 2001).
The integration of NIR lasers, interferometric detection strategies, epi-detection approaches for the study of nontransmitting specimens, and polarization-sensitive techniques (Cheng 2007, Volkmer et al. 2001, Zumbusch et al. 1999) has greatly improved early forms of CARS microscopy (Duncan et al. 1982). The strongest CARS signals tend to come from vibrations of C-H bonds, which are rich within lipids. Thus, for neuroscientists, CARS offers an intrinsic signal for inspecting nerve myelination and its degradation in vitro (Figure 12b,c), in animal disease models, and potentially in clinical contexts (Fu et al. 2007, Wang et al. 2005). CARS microscopy has allowed imaging of single myelinated axons in the live mouse brain (Fu et al. 2008). Additional in vivo studies have involved imaging of mouse brain tumors (Evans et al. 2007) and rat spinal cord (Wang et al. 2007a).
Just recently, a technique that may supplant CARS microscopy in many applications has emerged. Stimulated Raman scattering (SRS) microscopy is another form of coherent Raman imaging that provides information about molecular vibrations, but SRS microscopy does not suffer from a nonresonant background signal, greatly aiding detection sensitivity and interpretation of signals (Freudiger et al. 2008). Like CARS microscopy, SRS imaging appears well suited for examination of myelin sheaths and other tissues rich in lipids.
Second-Harmonic Generation Microscopy
Second-harmonic generation (SHG) microscopy relies on coherent frequency doubling of the illumination for generating optical contrast. The nonlinear SHG process converts two incoming photons into one outgoing photon of twice the frequency and permits inherent optical sectioning. Unlike with two-photon excited fluorescence, SHG does not involve light absorption and then reemission after a ~1–10-ns delay. Rather, frequency doubling is essentially instantaneous, and the outgoing light is coherent with the illumination. Biological specimens that consist of highly ordered but directionally asymmetric molecular assemblies, such as collagen or striated muscle fibers, tend to produce strong SHG signals owing to the dependence of frequency doubling on a broken spatial inversion symmetry within the sample material structure (Campagnola & Loew 2003). Because SHG does not require absorption, it is often well generated over a broader range of illumination frequencies than is typical for fluorescence excitation.
A recent application of SHG of pertinence to the study of motor control and neuromuscular diseases involves in vivo imaging of sarcomeres, the basic contractile units of striated muscle (Llewellyn et al. 2008). Sarcomere force-extension relationships are a crucial determinant of a muscle’s force production, but previous methods for examining sarcomeres and their extension lengths generally required excision of muscle fibers out of their native physiological and biomechanical contexts and thereby removed them from the nervous system’s control signals. By inserting microendoscopes (350–1000 μm in diameter) (Figure 1a) into live muscles (Llewellyn et al. 2008), investigators have imaged the extension lengths of individual sarcomeres by using the strong SHG signals that are thought to arise in sarcomeres’ myosin motor tails (Campagnola & Loew 2003, Campagnola et al. 2002, Plotnikov et al. 2006) (Figure 13a–f). In this way, they could visualize the 3D structures of muscle fibers, characterize sarcomeres’ micron-scale extension lengths in vivo, and monitor sarcomeres’ millisecond-scale contractile dynamics (Figure 13a–f).
The application of minimally invasive SHG microendoscopy to animal models and human subjects (Llewellyn et al. 2008) opens the door to novel studies of how signals from the nervous system control body movements, to in vivo imaging studies of animal models of neuromuscular disorders, and potentially to new clinical diagnostics, surgical strategies, and means of monitoring disease progression. Many neurological diseases of motor control, such as muscular dystrophy, as well as disorders such as muscle contractures caused by stroke or cerebral palsy, likely involve disruptions to sarcomere lengths and force-extension relationships (Plotnikov et al. 2008, Pontén et al. 2007). Further, multiple genetic diseases of motor control have recently been traced to mutations in sarcomeric proteins (Laing & Nowak 2005) and may cause disruptions in sarcomere structures and lengths (Plotnikov et al. 2008). Because the microendoscopes used for sarcomere imaging can be inserted through hypodermics of sizes comparable to those for conventional electromyography (EMG), future approaches may combine SHG microendoscopy with EMG to allow simultaneous monitoring of the electrical signals driving muscle contraction and the resulting mechanical responses.
A key application of SHG microscopy that does not rely on intrinsic signals involves the insertion into neuronal membranes of a chromophore sensitive to electric fields, thereby allowing imaging of membrane voltage transients. This combines the advantages inherent to nonlinear optical microscopy, including optical sectioning and significant penetration into thick tissue, with those of SHG, including lack of light absorption and relative enhancement of signals from ordered cell membranes as opposed to cytoplasmic regions, for the study of neuronal voltage dynamics. Upon incorporation into neuronal membranes, certain molecules with asymmetric structure can respond rapidly to electric fields with changes in their electro-optic or electro-chromic properties, changes in mean angular orientation relative to the membrane surface, or a combination of these effects (Jiang et al. 2007, Pons et al. 2003, Pons & Mertz 2006). Such effects can yield sizable voltage-dependent SHG signals in neurons [e.g. ~7.5% (Dombeck et al. 2005) to ~10%–12% (Nuriya et al. 2006) changes per 100 mV in brain slices, and up to ~25% per 100 mV in cultured neurons (Nemet et al. 2004)].
Investigators have demonstrated the use of SHG microscopy to measure neuronal action potentials in cultured Aplysia neurons using a membrane-bound styryl dye (Dombeck et al. 2004), in cultured hipppocampal pyramidal neurons using a retinal photopigment (Nemet et al. 2004), and in mammalian brain slices using FM 4–64 (Araya et al. 2006, Dombeck et al. 2005, Nuriya et al. 2006). Typically, voltage responses are measured by fast line-scanning of the laser focal spot over the sample and then averaging signals from multiple stimulation trials. The modest dynamic range ofSHG signals has generally made the detection of voltage responses at the level of single trials challenging, albeit not impossible (Sacconi et al. 2006, 2008).
SHG membrane-potential imaging with the dye FM 4–64 has permitted direct measurements of membrane potential in intracellularly filled mouse pyramidal neurons, enabling the first studies of membrane potential dynamics in dendritic spines (Figure 13g–k) (Nuriya et al. 2006). With this dye, the mechanisms of voltage sensitivity appear to be purely electro-optic (Jiang et al. 2007). SHG is sensitive to voltage changes on fast (<1 ms) timescales, and the changes in membrane potential in spines during action potential invasion were detectable (Figure 13i–k) (Nuriya et al. 2006). A later study using similar methods of monitoring membrane potential in layer 5 mouse neocortical pyramidal neurons reported a negative correlation between the lengths of dendritic spine necks and the transmission of voltage changes between the soma and the spine heads (Araya et al. 2006).
MOUSE GENETIC MOSAIC STRATEGIES FOR FLUORESCENCE LABELING
The cloning of GFP opened the door to a wide set of possibilities for expressing fluorescent labels under genetic control (Chalfie et al. 1994). Today, fluorescent proteins of many color variations (so-called XFPs) have been created, permitting concurrent visualization of multiple genetically expressed fluorescent tags. Because absorption cross-sections for two-photon fluorescence excitation tend to be spectrally broad, a single wavelength setting for the illumination often suffices in two-photon microscopy to excite more than one fluorophore species. For neuroscientists, the introduction of mice that brightly expressed XFPs in neuronal subsets under the control of the Thy1 promoter furthered imaging opportunities in many directions (Feng et al. 2000). These mice facilitated live animal imaging studies, including studies of motor axon dynamics and neuromuscular junction formation (Keller-Peck et al. 2001, Nguyen et al. 2002, Walsh & Lichtman 2003), as well as of dendritic spine turnover (see next section). Other notable genetic techniques for creating fluorescently labeled “tool mice” have since arisen, some of which build on the proven success of using the Thy1 promoter to drive high expression levels of fluorescent protein. This section discusses genetic mosaic strategies for fluorescence labeling, in which a single adult animal has different sets of somatic cells with distinct genotypes.
All the mosaic strategies described here rely on Cre recombinase mediated DNA recombination. The enzyme Cre recombinase (Cre) recognizes specific directional DNA sequences, the canonical loxP site and its variants, which can be inserted to flank DNA segments of interest (sometimes called “floxed” segments). Cre recombinase can bind to two loxP sites and promote recombination of the DNA between the two sites. If both loxP sites have the same directional orientation, recombination excises the floxed segment. If the loxP sites have the opposite directional orientation, recombination inverts but does not excise the floxed segment. Translocation recombination is also possible between separate strands of DNA, leading to an exchange of DNA sequences starting from the loxP sites. As elaborated below, this set of possibilities permits sophisticated manipulations of gene expression. The wide availability of Cre driver mouse lines, in which Cre recombinase is expressed within specific populations of cells, as well as driver lines permitting expression of ligand-inducible forms of Cre facilitate the spatiotemporal control of DNA recombination and thus of XFP expression.
Brainbow Tool Mice
As useful as the original Thy1-XFP mice are, recently created mosaic mice with neurons labeled in a rainbow of colors should further facilitate experimentation by allowing dense sets of individual cells and their overlapping processes to be distinguishable by color. Livet et al. (2007) have described two transgenic strategies for creating such multicolor “Brainbow” mice (Figure 14a–c). The ability to trace neuronal processes for mammalian neural circuit reconstructions may be greatly aided in Brainbow mice by the pantheon of spectrally separable tags (see Automated Microscopy section). Brainbow mice may also serve neuronal lineage analyses, allowing individual neuronal progenitor cells and their progeny to be labeled in a common color from among a large palette of possibilities. For experiments that were previously feasible only with the sparsely expressing lines of the original Thy1-XFP mice, Thy1-Brainbow mice may enable larger data sets to be acquired from each animal because the capabilities for spectral separation may allow use of lines with denser neuronal labeling patterns.
Figure 14.
Fluorescent tool mice for microscopy studies. (a) Two strategies for creating fluorescently labeled Brainbow mice. (Top) In the Brainbow-1 construct, three sets of incompatible lox sites (shaded gray triangles) enable three mutually exclusive Cre-mediated recombinations. Excisions (dash-dot lines) of XFP-coding segments of DNA are possible. (Bottom) In the Brainbow-2 construct, two sequential invertible XFP-coding segments of DNA between pairs of loxP sites (blue triangles) undergo Cre-mediated recombinations. Both inversions (dashed lines) and excisions (dash-dot lines) are possible. In both strategies, only the first XFP following the promoter is expressed. (b) Fluorescence images of an axon tract in the brain stem and (c) the hippocampal dentate gyrus of Brainbow mice. The Brainbow mice from which these images were taken exhibited an estimated 90–160 distinct colors, as a result of the co-integration of several tandem copies of the transgene into the mouse genome and the independent recombination of each by Cre recombinase. The images were obtained by the superposition of separate red, green, and blue imaging channels (images a–c are based on Lichtman et al. 2008). (d) Fluorescent labeling using mosaic analysis with double markers (MADM). Chimeric genes containing loxP sites (blue triangles) between split exons of GFP and RFP, or vice versa, are positioned on homologous chromosomes. (XFP-coding regions are shown as halves of filled colored pentagons.) The chimeric genes on each chromosome are abbreviated as GR and RG, respectively. After DNA replication (G2 phase, left), Cre-mediated interchromosomal recombination of the reciprocally chimeric gene pair GR/RG (middle), followed by one of two types of chromosomal recombination during mitosis (G2-X or G2-Z segregation), generates a daughter-cell pair with different fluorescence qualities (right). Outcomes for conditional gene knock-out in daughter cells after mitosis are shown using symbols to signify the placement of wild-type (+) or mutant knocked-out (−) genes distal to the XFP-coding regions. Whereas unlabeled and double-labeled daughter cells are heterozygous for the gene mutation, singly labeled daughter cells are homozygous for either the knock-out mutation (GFP-labeled) or the wild-type gene (RFP-labeled). (e) Hippocampal dentate gyrus of a GR/RG; Nestin-Cre/+ mouse (P15) showing distinct labeling of neuronal cell types in the hillus, granule cell layer, and molecular layer. Blue structures indicate (4′-6-Diamidino-2-phenylindole) (DAPI) nuclear stain (images d and e are based on Zong et al. 2005). (f) Cerebral cortex of a GR/RG; Nestin-Cre/+ mouse (P2) showing green, red, and double-labeled neurons (image courtesy of J.S. Espinosa and L. Luo). (g) Single-neuron labeling with inducible Cre-mediated knockout (SLICK). (top) In the SLICK construct, YFP and CreERT2 are coexpressed under the control of back-to-back copies of the Thy1 promoter. (bottom) In progeny of a cross between SLICK mice and transgenic mice created with the construct shown, administration of tamoxifen induces temporally controlled Cre-mediated conditional transgene expression. Use of a different construct enables conditional gene knock-out. (h) Conditional transgene expression using SLICK. Progeny of a cross between SLICK (line V) and R26R mice were treated for five days with tamoxifen. In the R26R Cre reporter strain, a transcriptional stop sequence is floxed upstream of a lacZ reporter gene coding for β-galactosidase at the ROSA26 locus. Two weeks following tamoxifen treatment, YFP-labeled pyramidal neurons in the CA1 hippocampal region exhibited robust lacZ expression revealed by immunofluorescent staining for β-galactosidase (images g and h are based on Young et al. 2008). Cyan, green, orange, red and yellow fluorescent proteins are abbreviated as CFP, GFP, OFP, RFP, and YFP, respectively.
The DNA constructs used to generate Brainbow mice exploit Cre-mediated recombination. In the Brainbow-1 strategy (Figure 14a, top), distinct XFP exons are floxed by three mutually exclusive types of lox sites, the canonical loxP site and two incompatible lox site variants (Livet et al. 2007). Upon exposure to Cre recombinase, one of three mutually exclusive gene excisions can occur, resulting in the expression of one of three distinct XFPs following recombination. A fourth color can be used to indicate the lack of recombination.
The Brainbow-2 strategy (Figure 14a, bottom) exploits inversion of floxed XFP-coding segments. Cre recombinase can invert each segment multiple times, so the segments randomly settle in either a forward or backward orientation, yielding two different XFP-coding possibilities. Additionally, excisions can occur in Brainbow-2 (Figure 14a, bottom). The total set of inversion and excision possibilities yields a random expression of one of four XFPs.
For both Brainbow strategies, the random integration of multiple Thy1-Brainbow constructs into the mouse genome led to an even wider set of color labels owing to the large set of combinatorial possibilities for XFP expression from each incorporation site. Crossing Thy1-Brainbow mouse lines with Cre driver transgenic mouse lines led to ~100 different color hues arising from the combinatorial expression of three or more XFPs, which can be expressed in a wide variety of glial and neuronal labeling patterns. For example, Brainbow mice have permitted multicolor imaging of densely packed neurons and their processes, such as in peripheral motor nerves; cerebellar granule cells and mossy fibers; brain stem axonal tracts (Figure 14b); and the hippocampal dentate gyrus (Figure 14c). In one of the Brainbow-1 mouse lines astrocytes were also labeled, an unexpected effect since Thy1 usually drives expression in neuronal subsets.
The Brainbow strategy will likely undergo refinements that will continue to increase its utility. The current use of the Thy1 promoter limits expression of the XFPs to late embryonic and postnatal stages and hinders use of the strategy for early developmental studies. For cases in which immuno-staining is required, it is unfortunate that antibody labeling cannot differentiate between some of the fluorescent proteins because of their structural similarity (for example, GFP, YFP, and CFP). GFP also has close spectral similarity to YFP and CFP, so in Brainbow-2 lines a nuclear localization signal was tagged to GFP to aid disambiguation of the fluorescence signals (Livet et al. 2007). As the number of available XFPs grows, the combinatorial possibilities for Brainbow color labeling will expand even further. Moreover, as the super-resolution microscopy techniques continue to develop, the ability to trace individual axons within densely labeled axonal bundles in Brainbow mice should also improve.
Combining Mosaic Fluorescence Labeling and Conditional Gene Knock-out
In addition to permitting expression of multiple color markers as in Brainbow, mosaic genetic techniques are well suited for studying sparse sets of genetically modified neurons among a wild-type background population of cells, and often for examining spatiotemporal patterns of circuit formation in the developing brain. For such studies in mice, an approach called mosaic analysis with double markers (MADM) yields mosaic mice expressing two fluorescent markers and has the option of performing a conditional gene knock-out (Zong et al. 2005). The MADM strategy was inspired in part by a technique called mosaic analysis with a repressible cell marker (MARCM) for use in Drosophila (Lee & Luo 1999) and uses Cre-loxP mediated interchromosomal recombination in somatic cells to create mosaic animals (Figure 14d–f). MADM mice start life with homologous chromosomes containing reciprocally chimeric genes knocked in at identical chromosomal loci. If GFP and a red fluorescent protein (RFP) are used as the two markers, the chimeric genes contain split exons of either GFP/RFP or RFP/GFP (written here as N-terminus/C-terminus), with a loxP site lying within an intron between the two exons (Figure 14d). Because the intron interrupts the coding sequences of the XFPs in different reading frames, functional fluorescent proteins are not produced from the original chimeric genes. Fluorescent protein expression is enabled only when MADM mice are crossed with a mouse Cre driver line to introduce Cre-recombinase in designated cell populations.
In the progeny of this cross, after DNA replication in G2 phase (Figure 14d, left), Cre-mediated interchromosomal recombination events can occur across arms of the two chromosomes prior to mitosis (Figure 14d, middle). These comparatively rare interchromosomal recombination events yield two chromosomes that each have a reconstituted XFP-encoding gene in one of the chromatids. There are then two possibilities for how mitosis can proceed (Figure 14d, right). In an X segregation, in which the recombinant chromatids from the two chromosomes are distributed to different daughter cells, one daughter cell will be heterozygous for a functional GFP and the other heterozygous for a functional RFP (Figure 14d, right top). If a Z segregation occurs, with the recombinant chromatids from the two chromosomes reaching the same daughter cell, one daughter cell will be double labeled with both GFP and RFP and the other will be unlabeled (Figure 14d, right bottom). Note that if recombination occurs in a G1 phase or postmitotic cell, this cell will be double labeled.
To accomplish a mosaic pattern of conditional gene knock-out, a mutation of interest is placed distal to the chimeric gene on one arm of the chromosome, such as the GFP/RFP arm. If X segregation occurs following G2 recombination, the GFP-labeled cell will be homozygous for the mutation, whereas the RFP-labeled cell will be homozygous for the wild-type gene. If Z segregation occurs, both the unlabeled and the double-labeled daughter cells will be heterozygous for the mutation. Thus, with MADM, the genotype of any labeled cell can be discerned by its color. This labeling strategy facilitates tightly controlled experiments because in individual mice there will be fluorescently labeled mutant and nonmutant cells available for inspection.
Cell labeling using MADM has been demonstrated across a variety of areas in the mouse brain, including hippocampus, dentate gyrus (Figure 14e), neocortex (Figure 14f), cerebellum, and retina. MADM has also permitted clonal analyses of neurogenesis, differentiation, and axonal projection patterns of cerebellar granule cells (Espinosa & Luo 2008, Zong et al. 2005). To date, only the well-characterized ROSA26 chromosomal locus that lies on the long arm of chromosome 6 has been employed as the knock-in site, presently limiting the conditional knock-out to those genes lying distal to ROSA26. Another limitation of the existing MADM mice that will improve in the future concerns the insufficient brightness of the RFP for in vivo imaging. To date, visualization of the RFP marker has been demonstrated only by immunolabeling. As these aspects of MADM are refined, the approach will facilitate in vivo imaging studies of neuronal circuit establishment, with direct comparisons between mutant and wild-type cells in the same brain area.
Another mosaic technique for fluorescence labeling of single cells is single-neuron labeling with inducible Cre-mediated knock-out (SLICK) (Figure 14g–h) (Young et al. 2008). To generate SLICK mice, separate back-to-back copies of the Thy1 promoter drive the two genes coding for YFP and for CreERT2, a ligand-inducible form of Cre-recombinase that is induced by injection of the drug tamoxifen (Figure 14g, top). To create a mosaic conditional genetic knockout or a conditional transgenic (Figure 14g, bottom), SLICK mice are crossed with another mouse line in which a gene of interest is either floxed or preceded by a floxed transcriptional stop sequence, respectively. Systemic administration of tamoxifen at chosen time points permits temporal control over Cre recombinase activity in the progeny of the cross.
To demonstrate the strategy, multiple lines of SLICK mice have been created and characterized. In one example, SLICK mice were crossed with a Cre reporter strain in which a floxed STOP precedes a lacZ sequence coding for β-galactosidase. Upon tamoxifen administration, lacZ expression as assessed by immunofluorescent staining for β-galactosidase and YFP fluorescence were jointly visible in brain slices, illustrating dual expression of YFP and CreERT2 within single cells (Figure 14h). SLICK mice have also been used to study the elimination of synaptic transmission at the adult mouse neuromuscular junction by a mosaic conditional knock-out of the choline acetyltransferase (Chat) gene, which encodes an enzyme involved in acetylcholine synthesis. In YFP-labeled motor axons, neuromuscular junctions stably persisted in adult animals even four to eight weeks after depletion of the ChAT protein (Young et al. 2008). The coupling of fluorescence labeling and conditional knock-out, which should ideally be checked for each floxed gene, was found to be >95% for the Chat knock-out. A key feature of SLICK is that in an in vivo imaging study there is the possibility of inspecting the same individual neurons before and after the induction of gene knock-out.
As with MADM mice, SLICK mice are well suited for examination of the effects of conditional gene knock-out on neurons’ morphology and integration into functional circuits. Both MADM and SLICK enable mosaic genetic manipulation of cells, but the two approaches offer distinct advantages. SLICK offers temporal control via inducible genetic manipulation of neuron types in which the Thy1 promoter is active, which often starts in late embryonic development and continues into adulthood. MADM is used to study mitotic cells, which for neuronal precursors generally implies during early brain development. A strength of MADM is the simultaneous generation of two colors of fluorescently labeled cells that allow internal controls (RFP- and double-labeled cells) for conditional gene knock-out studies. Following tamoxifen administration, the YFP-labeled cells generated in SLICK are very likely, but not absolutely guaranteed, to have the conditional gene knock-out. MADM can also be used to study mutations of any kind lying distal to a MADM chromosomal knock-in site, whereas SLICK requires floxed conditional alleles.
CHRONIC MOUSE PREPARATIONS FOR LONG-TERM IMAGING STUDIES
An ability to monitor individual neurons repeatedly by long-term time-lapse imaging is crucial for studies of how the nervous system evolves during development, aging, learning, or other life experiences, or over disease progression. Chronic mouse preparations permitting time-lapse imaging are well established for studies of the peripheral nervous system (Lichtman et al. 1987, Purves et al. 1986, Purves & Hadley 1985) and cranially implanted tumors (Melder et al. 1995, Yuan et al. 1994). However, XFP-labeled transgenic mice have recently facilitated studies of neurons in the periphery (Bishop et al. 2004, Gan et al. 2003, Walsh & Lichtman 2003) and, in combination with two-photon microscopy, have enabled chronic imaging studies within the mammalian brain.
A main advantage of chronic imaging preparations concerns the experimental designs that can generally be achieved by longitudinal studies, in which one follows individual animals over time, as compared to population studies, in which different sets of animals are inspected at distinct time points. Information about dynamics in individual subjects is inherently absent when each subject is examined only once, e.g., at postmortem. For disease studies, analysis of within-subject dynamics is often crucial because it can reveal how early cellular-level symptoms might predict disease outcomes or how initial cellular signatures of disease might be reversed by intervention. Longitudinal imaging can also permit a substantial reduction in the number of animals needed because each animal ideally provides data at all time points, not just one.
In 2002, two different chronic mouse preparations emerged for in vivo time-lapse two-photon imaging of neuronal dendrites and spines. The two groups responsible both examined layer 5 neocortical neurons in Thy1-XFP mice over periods of weeks to months (Grutzendler et al. 2002, Trachtenberg et al. 2002). One of the chronic preparations drew on earlier usages of implanted glass “cranial windows” that had enabled seminal intravital fluorescence microscopy studies of brain tumor angiogenesis (Melder et al. 1995, Yuan et al. 1994). By implanting glass windows into the cranium it was possible to monitor spines on large portions of the dendritic trees of XFP-labeled neocortical pyramidal neurons, repeatedly for up to ~30 days after surgery and ~100 μm below the pial surface of the brain (Trachtenberg et al. 2002). The other chronic preparation involved shaving the mouse’s cranium to be sufficiently thin (~30–50 μm) such that two-photon imaging could be performed through the skull to inspect neocortical neuronal dendrites up to ~ 100–200 μm below the pia (Grutzendler et al. 2002). The skull regrows and repeated thinning renders it opaque, so the thinned skull preparation generally provided data only at a few time points (e.g., ~4) but up to ~18 months apart (Zuo et al. 2005a). The two methods yielded different results regarding the long-term stabilities of dendritic spines, which generated debate in part about methodologies (see below) (Xu et al. 2007), but results obtained with both approaches did suggest the majority of layer 5 pyramidal cell spines in the adult brain are stable over months (Holtmaat et al. 2005, Zuo et al. 2005a).
Multiple laboratories have since used chronic imaging techniques to study the stability and turnover of dendritic spines from layer 2/3 and layer 5 neocortical pyramidal neurons under various experimental manipulations in adult mice (Chow et al. 2009; Holtmaat et al. 2005, 2006; Knott et al. 2006; Majewska et al. 2006; Trachtenberg et al. 2002; Zuo et al. 2005a,b) (Figure 15a,c,e). Other studies have examined axonal dynamics (Figure 15b), (De Paola et al. 2006, Nishiyama et al. 2007, Portera-Cailliau et al. 2005), dynamic remodeling in layer 2/3 interneurons (Lee et al. 2006, 2008), redistribution of PSD-95 in dendrites of layer 2/3 pyramidal neurons (Gray et al. 2006), as well as the dynamics of neurogenesis and integration of new dendrites in the olfactory bulb (Mizrahi 2007, Mizrahi et al. 2006). Long-term imaging is also being established for use in deeper brain tissues using methods that allow the repeated insertion of microendoscope probes to the same tissue site (Figure 1c), for example to visualize CA1 hippocampal pyramidal neurons by two-photon microendoscopy (Figure 15d) (Deisseroth et al. 2006).
Another important application of time-lapse microscopy in XFP-expressing mice concerns the study of animal models of brain disease or injury because repeated imaging allows the temporal dynamics of disease and recovery to unfold within individual subjects (Misgeld & Kerschensteiner 2006, Pan & Gan 2008). Several studies have investigated the dynamics of dendritic spines proximal to an ischemic stroke (Figure 15e) (R. Mostany & C. Portera-Cailliau, unpublished observations; Winship & Murphy 2008, Zhang et al. 2005), and another has examined spine dynamics in a mouse model of prion disease (Fuhrmann et al. 2007). By crossing mouse models of Alzheimer disease with XFP-expressing lines, other time-lapse studies have monitored neuronal (Figure 15f) (Brendza et al. 2005, Meyer-Luehmann et al. 2008, Spires et al. 2005, Tsai et al. 2004) and microglial (Koenigsknecht-Talboo et al. 2008, Meyer-Luehmann et al. 2008) changes at the sites of amyloid plaques. Not all time-lapse imaging studies of disease have required two-photon microscopy. XFP-expressing mice have also facilitated studies of spinal cord injury by confocal microscopy (Figure 15g) (Davalos et al. 2008; T. Misgeld, unpublished data). Furthermore, time-lapse SHG microscopy has permitted imaging of fibrillar collagen during the growth of brain tumors (Brown et al. 2003, Perentes et al. 2009).
Among the groups using time-lapse imaging in the brain to study dendritic spines, discussion has arisen over whether the different surgical preparations can affect measurements of spine stability. With the cranial window preparation, layer 5 pyramidal neurons in mouse whisker barrel cortex had spine turnover rates of ~50% per month in 6–10-week old mice (Holtmaat et al. 2005, Trachtenberg et al. 2002). Results from the thinned skull preparation suggested this turnover rate was ~10% per month in the visual cortex (Grutzendler et al. 2002). These seemingly conflicting results stimulated discussion about the factors that might influence both the real and the measured stability of dendritic spines (Knott & Holtmaat 2008, Majewska et al. 2006, Xu et al. 2007). Biological factors that might influence spine turnover rates in long-term imaging studies include the animals’ ages (Holtmaat et al. 2005, Zuo et al. 2005a), experimental manipulations such as whisker trimming (Holtmaat et al. 2006, Zuo et al. 2005b) or visual deprivation (Hofer et al. 2009), and differences between brain areas (Holtmaat et al. 2005, Majewska et al. 2006, Zuo et al. 2005a). Glial activation beneath an implanted cranial window was proposed as a factor, or as a symptom of factors, that could promote spine turnover (Xu et al. 2007). However, other studies that used both thinned skull and cranial window assays for imaging experiments did not report any notable differences between the two in spine turnover rates (Chow et al. 2009, Majewska et al. 2006; see also Holtmaat et al. 2009). Still other factors may influence the ability to make accurate determinations of spine turnover. These include user-dependent variability in spine-counting methods, the brightness and patterns of fluorescence labeling, and any potential differences that might exist between the two surgical preparations in the capacity to resolve the smallest and likely the most labile spines (Holtmaat et al. 2005, 2006).
Given these sources of potential variability, it will be important for scientists to benchmark results across laboratories and to demonstrate equal capabilities for resolving spines and comparable criteria for scoring spine stability. It seems equally critical to have experimental designs that, whenever possible, permit quantitative comparisons across control and experimental sets of animals to isolate the factors that may influence neuronal dynamics. As these issues become better explored and controls become standardized, chronic in vivo imaging will become an even more powerful tool for neuroscientists to study gradual changes in the intact brain.
OUTLOOK
The past few years have been exceptionally productive for the development of new microscopy techniques. Major breakthroughs have occurred on multiple technological fronts, impacting neuroscientists’ abilities to examine the nanoscale, create large-scale tissue reconstructions, and image cellular properties in live animals. These newfound capabilities will further expand the role of microscopy in neuroscience research. Many of the methods discussed here remain in early stages of development, but as these approaches progress we will start to see sophisticated combinations of techniques, such as combinations of complex fluorescence-labeling strategies with in vivo or super-resolution microscopy. Another subset of techniques is intended for the acquisition of massive data sets, which pose unprecedented challenges for neuroscientists regarding data management, analysis, and mining.
Overall, the technological complexity of image-based experimentation in neuroscience continues to grow, and in the long run, the highly collaborative approaches that arose in other imaging disciplines such as medical imaging or astronomy might become common in certain specialties within neuroscience, requiring large-scale automated imaging and massive data sets. In addition to the potential alterations in collaborative norms, the basic conceptual understanding of an image will also evolve. As explored in this review, a growing number of techniques do not directly capture the final images of the specimen, but rather create them computationally on the basis of optical data of various forms. Image data sets of many types now require extensive, often model-based, computational analysis just to be interpreted. As microscopy grows in logical abstraction, our images— increasingly processed, reconstructed, filtered, deconvolved, and distilled—will assume some of the attributes of computational hypotheses. This view of images as mere hypotheses, rather than as direct depictions of reality, should not deter us from image-based experimentation. On the contrary, the notion of images as hypotheses should push us toward new logical and statistical strategies for hypothesis testing.
SUMMARY POINTS
Techniques for data acquisition by light microscopy have grown increasingly abstract and reliant on computational approaches. Many techniques now involve computational reconstructions of the sample based on a set of raw images that may lack an obvious visual relationship to the sample’s structure. Computational approaches can also help to correct motion artifacts, identify cells automatically, and extract traces of cellular activity.
Progress in laser-scanning two-photon fluorescence microscopy permits fast 3D data acquisition. This has allowed functional Ca2+ imaging across the volumetric extent of large dendritic tree segments in vitro, as well from hundreds of individual neurons in live animals.
Several forms of volumetric microscopy relying on one-photon fluorescence are emerging for use in relatively transparent specimens, enabling fast 3D functional imaging and time-lapse studies of development.
Strategies for high-throughput, automated forms of microscopy are becoming potent for screening small animal species and creating large-scale reconstructions of histological tissue specimens. Computational machine vision and image segmentation techniques are growing increasingly crucial for online control of automated experiments and reaching conclusions based on vast amounts of imaging data.
Miniaturized fluorescence microscopes based on fiber optics permit cellular-level brain imaging in freely behaving animals. A complementary approach is to perform imaging in alert, head-restrained animals, which permits the use of conventional microscopes but constrains animal behavior.
Three main categories of super-resolution light microscopy afford new opportunities to examine the structures and functions of cells, synapses, and macromolecular interactions by using light to probe length scales finer than the optical diffraction limit.
Microscopy techniques that rely on intrinsic optical contrast mechanisms are enabling clinical neuroscientists to examine cells in human subjects, without use of exogenous contrast agents. Intrinsic signals permit in vivo imaging of retinal photoreceptors, nerve myelination, and striated muscle sarcomeres with micron-scale resolution.
The combination of genetically engineered mice expressing targeted fluorescent markers and chronic mouse imaging preparations has facilitated longitudinal in vivo imaging studies of cellular properties, in both healthy animals and mouse models of brain disease. Mosaic and inducible labeling strategies, some combined with conditional gene knockout, permit novel analyses of neural lineages and connectivity patterns.
FUTURE CHALLENGES
We have chosen these challenges as examples of technological achievements that would address key needs of neuroscience research across the different areas covered by this review:
Construct forms of fluorescence microscopy that allow functional imaging at ~1–10-ms temporal resolution across labeled populations of thousands of individual cells.
Identify an intrinsic optical contrast parameter that permits functional imaging of membrane voltage in live animals, with cellular-level and millisecond-scale resolution.
Develop techniques that permit live-cell, multicolor super-resolution imaging across 3D fields of view sufficient in size to cover entire dendritic trees, thereby facilitating studies with nanoscale resolution of how individual neurons globally regulate macromolecular interactions.
Create high-speed illumination techniques for random access control of neural activity patterns by photostimulation, with millisecond- and micron-scale precision across volumes of brain tissue >1 mm3.
Develop mouse genetic techniques that spectrally encode synaptic connectivity patterns in dense sets of targeted neurons and axons using multicolored fluorescent labels.
Generate automated forms of microscopy that permit functional brain imaging across large numbers of individual animals in a massively parallel fashion.
Develop computational methods of image analysis that will permit, in conjunction with automated approaches to tissue preservation and imaging, large-scale reconstructions of neural circuits in a high-throughput manner.
Create microscopy methods for use in freely behaving animals that can be used continually over multiple weeks, permitting studies of how patterns of neural activity, gene expression, and macromolecular interactions can evolve over long timescales pertinent to development and many learned behaviors.
Acknowledgments
This review greatly benefited from critical feedback on portions of the text and contributions of images generously provided by many neuroscientists and microscopists. For this substantial help we gratefully thank D. Albrecht, T. Anderson, C. Bargmann, R. Barretto, E. Betzig, E. Bushong, P. Carlton, J.-X. Cheng, D. Chiu, G. Davis, D. Dombeck, M. Ellisman, C. Engel-brecht, J.S. Espinosa, G. Feng, S. Finkbeiner, W. Gan, M. Gustaffson, S.W. Hell, F. Helmchen, T. Holy, D. Kleinfeld, M. Levoy, J. Lichtman, H. Lu, L. Luo, M. Martone, D. Mayerich, T. Misgeld, W.E. Moerner, U.V Nägerl, A. Nimmerjahn, C. Portera-Cailliau, M. O’Connell, J. Rosen, P. Saggau, T. Sasaki, J. Sharpe, K. Shen, H. Shroff, S. Smith, D. Tank, P. Tsai, J. Trachtenberg, D. Vučinić, J. Werner, C. Xu, M.F. Yanik, R. Yuste, R. Zawadzki, X. Zhuang, and Y. Ziv. We particularly thank P. Saggau, C. Xu, and X. Zhuang for extended discussions.
Our laboratory’s work on microscopy is supported by a Stanford Bio-X fellowship (B.A.W.), a Stanford Graduate Student fellowship (L.D.B.), NSF Graduate Fellowships (L.D.B. and E.A.M.), and grants from NINDS, NIBIB, NIDA, ONR, NSF, and the Packard, Coulter, and Beckman Foundations (M.J.S.).
- Scattering length
the characteristic distance in tissue or other material over which the intensity of ballistically propagated light attenuates by a factor of 1/e owing to light scattering
- Two-photon fluorescence excitation
widely used nonlinear process in which two photons electronically excite the fluorophore, which can then exit a fluorescence photon
- Nonlinear optical imaging
imaging techniques based on light-matter interactions requiring near simultaneous interactions of the molecule with two or more photons to generate contrast
- Optical sectioning
Restriction of signals to those from sources lying within a thin plane in the specimen. Signals from other planes are either blocked or precluded from arising
- Photodamage
an umbrella term describing photo-induced toxicity to the sample, sometimes also including the bleaching of fluorescent markers
- Numerical aperture (NA)
a dimensionless measure that characterizes the light gathering and resolving power of a lens
- AOD
acousto-optic deflector
- Hologram
interferometric encoding of a 3D image within a 2D representation
- Super-resolution fluorescence microscopy
techniques that circumvent the diffraction limit to achieve higher resolution; many of these techniques use photophysical or photochemical switching effects to transiently darken some fluorophores
- Optical fiber
an optical light guide, typically made of glass or plastic, often used in microscopy to route light from remote sources and to remote detectors
- Diffraction limit
classical resolution limit of light microscopy, based on the physical restrictions imposed by light diffraction, which scale with wavelength
- Point-spread function
functional description of a microscope’s image of an ideal point source of light, often used to characterize the microscope’s resolving power
- STED
stimulated emission depletion
- SIM
structured illumination microscopy
- STORM
stochastic optical reconstruction microscopy
- PALM
photoactivated localization microscopy
- Intrinsic contrast mechanism
mechanism for generating image contrast based on tissue’s inherent optical qualities, without need for application of exogenous dyes or contrast agents
- OCT
optical coherence tomography
- CARS
coherent anti-Stokes Raman scattering
- SHG
second-harmonic generation
- MADM
mosaic analysis with double markers
- SLICK
single-neuron labeling with inducible Cre-mediated knockout
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- Adelsberger H, Garaschuk O, Konnerth A. Cortical calcium waves in resting newborn mice. Nat Neurosci. 2005;8:988–90. doi: 10.1038/nn1502. [DOI] [PubMed] [Google Scholar]
- Aguirre AD, Chen Y, Fujimoto JG, Ruvinskaya L, Devor A, Boas DA. Depth-resolved imaging of functional activation in the rat cerebral cortex using optical coherence tomography. Opt Lett. 2006a;31:3459–61. doi: 10.1364/ol.31.003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AD, Nishizawa N, Fujimoto JG, Seitz W, Lederer M, Kopf D. Continuum generation in a novel photonic crystal fiber for ultrahigh resolution optical coherence tomography at 800 nm and 1300 nm. Opt Express. 2006b;14:1145–60. doi: 10.1364/oe.14.001145. [DOI] [PubMed] [Google Scholar]
- Akkin T, Davé DP, Milner TE, Rylander HG. Detection of neural activity using phase-sensitive optical low-coherence reflectometry. Opt Express. 2004;12:2377–86. doi: 10.1364/opex.12.002377. [DOI] [PubMed] [Google Scholar]
- Akkin T, Joo C, de Boer JF. Depth-resolved measurement of transient structural changes during action potential propagation. Biophys J. 2007;93:1347–53. doi: 10.1529/biophysj.106.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert O, Sherman L, Mourou G, Norris TB, Vdovin G. Smart microscope: an adaptive optics learning system for aberration correction in multiphoton confocal microscopy. Opt Lett. 2000;25:52–54. doi: 10.1364/ol.25.000052. [DOI] [PubMed] [Google Scholar]
- Allen PB, Sgro AE, Chao DL, Doepker BE, Scott Edgar J, et al. Single-synapse ablation and long-term imaging in live C. elegans. J Neurosci Methods. 2008;173:20–26. doi: 10.1016/j.jneumeth.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose WP, Moerner WE. Fluorescence spectroscopy and spectral diffusion of single impurity molecules in a crystal. Nature. 1991;349:225–27. [Google Scholar]
- Andresen M, Stiel AC, Fölling J, Wenzel D, Schönle A, et al. Photoswitchable fluorescent proteins enable monochromatic multilabel imaging and dual color fluorescence nanoscopy. Nat Biotechnol. 2008;26:1035–40. doi: 10.1038/nbt.1493. [DOI] [PubMed] [Google Scholar]
- Andresen V, Egner A, Hell SW. Time-multiplexed mutifocal multiphoton microscope. Opt Lett. 2001;26:7577. doi: 10.1364/ol.26.000075. [DOI] [PubMed] [Google Scholar]
- Anger EM, Unterhuber A, Hermann B, Sattmann H, Schubert C, et al. Ultrahigh resolution optical coherence tomography of the monkey fovea. Identification of retinal sublayers by correlation with semithin histology sections. Exp Eye Res. 2004;78:1117–25. doi: 10.1016/j.exer.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Araya R, Jiang J, Eisenthal KB, Yuste R. The spine neck filters membrane potentials. Proc Natl Acad Sci USA. 2006;103:17961–66. doi: 10.1073/pnas.0608755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–10. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Bahlmann K, So PTC, Kirber M, Reich R, Kosicki B, et al. Multifocal multiphoton microscopy (MMM) at a frame rate beyond 600 Hz. Opt Express. 2007;15:10991–98. doi: 10.1364/oe.15.010991. [DOI] [PubMed] [Google Scholar]
- Bailey B, Farkas DL, Taylor DL, Lanni F. Enhancement of axial resolution in fluorescence microscopy by standing-wave excitation. Nature. 1993;366:44–48. doi: 10.1038/366044a0. [DOI] [PubMed] [Google Scholar]
- Bates M, Blosser TR, Zhuang X. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys Rev Lett. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates M, Huang B, Dempsey GT, Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 2007;317:1749–53. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaurepaire E, Oheim M, Mertz J. Ultra-deep two-photon fluorescence excitation in turbid media. Opt Commun. 2001;188:25–29. [Google Scholar]
- Betzig E, Chichester RJ. Single molecules observed by near-field scanning optical microscopy. Science. 1993;262:1422–25. doi: 10.1126/science.262.5138.1422. [DOI] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–45. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Bewersdorf J, Pick R, Hell SW. Multifocal multiphoton microscopy. Opt Lett. 1998;23:655–57. doi: 10.1364/ol.23.000655. [DOI] [PubMed] [Google Scholar]
- Bishop DL, Misgeld T, Walsh MK, Gan W-B, Lichtman JW. Axon branch removal at developing synapses by axosome shedding. Neuron. 2004;44:651–61. doi: 10.1016/j.neuron.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. Super-resolution imaging in live Caulobacter crescentus cells using photoswitchable EYFP. Nat Methods. 2008;5:947–49. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizheva K, Pflug R, Hermann B, Považay B, Sattmann H, et al. Optophysiology: depth-resolved probing of retinal physiology with functional ultrahigh-resolution optical coherence tomography. Proc Natl Acad Sci USA. 2006;103:5066–71. doi: 10.1073/pnas.0506997103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobroff N. Position measurement with a resolution and noise-limited instrument. Rev Sci Instrum. 1986;57:1152–57. [Google Scholar]
- Bock H, Geisler C, Wurm CA, von Middendorff C, Jakobs S, et al. Two-color far-field fluorescence nanoscopy based on photoswitchable emitters. Appl Phys B. 2007;88:161–65. [Google Scholar]
- Boot MJ, Westerberg CH, Sanz-Ezquerro J, Cotterell J, Schweitzer R, et al. In vitro whole-organ imaging: 4D quantification of growing mouse limb buds. Nat Methods. 2008;5:609–12. doi: 10.1038/nmeth.1219. [DOI] [PubMed] [Google Scholar]
- Booth MJ, Neil MAA, Juškaitis R, Wilson T. Adaptive aberration correction in a confocal microscope. Proc Natl Acad Sci USA. 2002;99:5788–92. doi: 10.1073/pnas.082544799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakenhoff GJ, Squier J, Norris T, Bliton AC, Wade MH, Athey B. Real-time two-photon confocal microscopy using a femtosecond, amplified Ti:sapphire system. J Microsc. 1996;181:253–59. doi: 10.1046/j.1365-2818.1996.97379.x. [DOI] [PubMed] [Google Scholar]
- Brendza RP, Bacskai BJ, Cirrito JR, Simmons KA, Skoch JM, et al. Anti-Aβ antibody treatment promotes the rapid recovery of amyloid-associated neuritic dystrophy in PDAPP transgenic mice. J Clin Invest. 2005;115:428–33. doi: 10.1172/JCI23269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretschneider S, Eggeling C, Hell SW. Breaking the diffraction barrier in fluorescence microscopy by optical shelving. Phys Rev Lett. 2007;98:218103. doi: 10.1103/PhysRevLett.98.218103. [DOI] [PubMed] [Google Scholar]
- Brown E, McKee T, diTomaso E, Pluen A, Seed B, et al. Dynamic imaging of collagen and its modulation in tumors in vivo using second-harmonic generation. Nat Med. 2003;9:796–800. doi: 10.1038/nm879. [DOI] [PubMed] [Google Scholar]
- Brown GD, Yamada S, Sejnowski TJ. Independent component analysis at the neural cocktail party. Trends Neurosci. 2001;24:54–63. doi: 10.1016/s0166-2236(00)01683-0. [DOI] [PubMed] [Google Scholar]
- Buist AH, Müller M, Squier J, Brakenhoff GJ. Real time two-photon absorption microscopy using multi point excitation. J Microsc. 1998;192:217–26. [Google Scholar]
- Bullen A, Patel SS, Saggau P. High-speed, random-access fluorescence microscopy: I. High-resolution optical recording with voltage-sensitive dyes and ion indicators. Biophys J. 1997;73:477–91. doi: 10.1016/S0006-3495(97)78086-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajal SR. The structure and connexions of neurons. Nobel lecture. 1906 Dec 12; [Google Scholar]
- Campagnola PJ, Loew LM. Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms. Nat Biotechnol. 2003;21:1356–60. doi: 10.1038/nbt894. [DOI] [PubMed] [Google Scholar]
- Campagnola PJ, Millard AC, Terasaki M, Hoppe PE, Malone CJ, Mohler WA. Three-dimensional high-resolution second-harmonic generation imaging of endogenous structural proteins in biological tissues. Biophys J. 2002;82:493–508. doi: 10.1016/S0006-3495(02)75414-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cense B, Nassif NA, Chen TC, Pierce MC, Yun S-H, et al. Ultrahigh-resolution high-speed retinal imaging using spectral-domain optical coherence tomography. Opt Express. 2004;12:2435–47. doi: 10.1364/opex.12.002435. [DOI] [PubMed] [Google Scholar]
- Chalasani SH, Chronis N, Tsunozaki M, Gray JM, Ramot D, et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450:63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–5. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Chen CC, Zappe S, Sahin O, Zhang XJ, Fish M, et al. Design and operation of a microfluidic sorter for Drosophila embryos. Sens Actuators B. 2004;102:59–66. [Google Scholar]
- Chen T-S, Zeng S-Q, Luo Q-M, Zhang Z-H, Zhou W. High-order photobleaching of green fluorescent protein inside live cells in two-photon excitation microscopy. Biochem Biophys Res Commun. 2002;291:1272–75. doi: 10.1006/bbrc.2002.6587. [DOI] [PubMed] [Google Scholar]
- Chen Y, Aguirre AD, Ruvinskaya L, Devor A, Boas DA, Fujimoto JG. Optical coherence tomography (OCT) reveals depth-resolved dynamics during functional brain activation. J Neurosci Methods. 2008a;178:162–73. doi: 10.1016/j.jneumeth.2008.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Geisler WS, Seidemann E. Optimal decoding of correlated neural population responses in the primate visual cortex. Nat Neurosci. 2006;9:1412–20. doi: 10.1038/nn1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Geisler WS, Seidemann E. Optimal temporal decoding of neural population responses in a reaction-time visual detection task. J Neurophysiol. 2008b;99:1366–79. doi: 10.1152/jn.00698.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Milner TE, Srinivas S, Wang X, Malekafzali A, et al. Noninvasive imaging of in vivo blood flow velocity using optical Doppler tomography. Opt Lett. 1997;22:1119–21. doi: 10.1364/ol.22.001119. [DOI] [PubMed] [Google Scholar]
- Cheng J-X. Coherent anti-Stokes Raman scattering microscopy. Appl Spectrosc. 2007;61:197–208. doi: 10.1366/000370207781746044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J-X, Volkmer A, Book LD, Xie XS. An epi-detected coherent anti-Stokes Raman scattering (E-CARS) microscope with high spectral resolution and high sensitivity. J Phys Chem B. 2001;105:1277–80. [Google Scholar]
- Chow DK, Groszer M, Pribadi M, Machniki M, Carmichael ST, et al. Laminar and compartmental regulation of dendritic growth in mature cortex. Nat Neurosci. 2009;12:116–18. doi: 10.1038/nn.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis N, Zimmer M, Bargmann CI. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat Methods. 2007;4:727–31. doi: 10.1038/nmeth1075. [DOI] [PubMed] [Google Scholar]
- Chu S-W, Chen I-H, Liu T-M, Chen PC, Sun C-K, Lin B-L. Multimodal nonlinear spectral microscopy based on a femtosecond Cr:forsterite laser. Opt Lett. 2001;26:1909–11. doi: 10.1364/ol.26.001909. [DOI] [PubMed] [Google Scholar]
- Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods. 2008;5:637–43. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- Conley NR, Biteen JS, Moerner WE. Cy3-Cy5 covalent heterodimers for single-molecule photoswitching. J Phys Chem B. 2008;112:11878–80. doi: 10.1021/jp806698p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crépel V, Aronov D, Jorquera I, Represa A, Ben-Ari Y, Cossart R. A parturition-associated nonsynaptic coherent activity pattern in the developing hippocampus. Neuron. 2007;54:105–20. doi: 10.1016/j.neuron.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Cui X, Lee LM, Heng X, Zhong W, Sternberg PW, et al. Lensless high-resolution on-chip optofluidic microscopes for Caenorhabditis elegans and cell imaging. Proc Natl Acad Sci USA. 2008;105:10670–75. doi: 10.1073/pnas.0804612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagani GT, Monzo K, Fakhoury JR, Chen C-C, Sisson JC, Zhang X. Microfluidic self-assembly of live Drosophila embryos for versatile high-throughput analysis of embryonic morphogenesis. Biomed Microdevices. 2007;9:681–94. doi: 10.1007/s10544-007-9077-z. [DOI] [PubMed] [Google Scholar]
- Davalos D, Lee JK, Smith WB, Brinkman B, Ellisman MH, et al. Stable in vivo imaging of densely populated glia, axons and blood vessels in the mouse spinal cord using two-photon microscopy. J Neurosci Methods. 2008;169:1–7. doi: 10.1016/j.jneumeth.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedecker P, Hotta J, Flors C, Sliwa M, Uji-i H, et al. Subdiffraction imaging through the selective donut-mode depletion of thermally stable photoswitchable fluorophores: numerical analysis and application to the fluorescent protein Dronpa. J Am Chem Soc. 2007;129:16132–41. doi: 10.1021/ja076128z. [DOI] [PubMed] [Google Scholar]
- Deerinck TJ, Martone ME, Lev-Ram V, Green DPL, Tsien RY, et al. Fluorescence photooxidation with eosin: a method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J Cell Biol. 1994;126:901–10. doi: 10.1083/jcb.126.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska AK, Miesenböck G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–86. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- De Paola V, Holtmaat A, Knott G, Song S, Wilbrecht L, et al. Cell type-specific structural plasticity of axonal branches and boutons in the adult neocortex. Neuron. 2006;49:861–75. doi: 10.1016/j.neuron.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Dodt H-U, Leischner U, Schierloh A, Jährling N, Mauch CP, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4:331–36. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- Dombeck DA, Blanchard-Desce M, Webb WW. Optical recording of action potentials with second-harmonic generation microscopy. J Neurosci. 2004;24:999–1003. doi: 10.1523/JNEUROSCI.4840-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56:43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombeck DA, Sacconi L, Blanchard-Desce M, Webb WW. Optical recording of fast neuronal membrane potential transients in acute mammalian brain slices by second-harmonic generation microscopy. J Neurophysiol. 2005;94:3628–36. doi: 10.1152/jn.00416.2005. [DOI] [PubMed] [Google Scholar]
- Donnert G, Eggeling C, Hell SW. Major signal increase in fluorescence microscopy through dark-state relaxation. Nat Methods. 2007a;4:81–86. doi: 10.1038/nmeth986. [DOI] [PubMed] [Google Scholar]
- Donnert G, Keller J, Medda R, Andrei MA, Rizzoli SO, et al. Macromolecular-scale resolution in biological fluorescence microscopy. Proc Natl Acad Sci USA. 2006;103:11440–45. doi: 10.1073/pnas.0604965103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnert G, Keller J, Wurm CA, Rizzoli SO, Westphal V, et al. Two-color far-field fluorescence nanoscopy. Biophys J. 2007b;92:L67–69. doi: 10.1529/biophysj.107.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler W, Fujimoto JG. State-of-the-art retinal optical coherence tomography. Prog Retin Eye Res. 2008;27:45–88. doi: 10.1016/j.preteyeres.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Drexler W, Morgner U, Kärtner FX, Pitris C, Boppart SA, et al. In vivo ultrahigh-resolution optical coherence tomography. Opt Lett. 1999;24:1221–23. doi: 10.1364/ol.24.001221. [DOI] [PubMed] [Google Scholar]
- Duncan MD, Reintjes J, Manuccia TJ. Scanning coherent anti-Stokes Raman microscope. Opt Lett. 1982;7:350–52. doi: 10.1364/ol.7.000350. [DOI] [PubMed] [Google Scholar]
- Durst ME, Zhu G, Xu C. Simultaneous spatial and temporal focusing for axial scanning. Opt Express. 2006;14:12243–54. doi: 10.1364/oe.14.012243. [DOI] [PubMed] [Google Scholar]
- Durst ME, Zhu G, Xu C. Simultaneous spatial and temporal focusing in nonlinear microscopy. Opt Commun. 2008;281:1796–805. doi: 10.1016/j.optcom.2007.05.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggeling C, Ringemann C, Medda R, Schwarzmann G, Sandhoff K, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2008;457:1159–62. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- Egner A, Geisler C, von Middendorff C, Bock H, Wenzel D, et al. Fluorescence nanoscopy in whole cells by asynchronous localization of photoswitching emitters. Biophys J. 2007;93:3285–90. doi: 10.1529/biophysj.107.112201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egner A, Hell SW. Time multiplexing and parallelization in multifocal multiphoton microscopy. J Opt Soc Am A. 2000;17:1192–201. doi: 10.1364/josaa.17.001192. [DOI] [PubMed] [Google Scholar]
- Engelbrecht CJ, Johnston RS, Seibel EJ, Helmchen F. Ultra-compact fiber-optic two-photon microscope for functional fluorescence imaging in vivo. Opt Express. 2008;16:5556–64. doi: 10.1364/oe.16.005556. [DOI] [PubMed] [Google Scholar]
- Espinosa JS, Luo L. Timing neurogenesis and differentiation: insights from quantitative clonal analyses of cerebellar granule cells. J Neurosci. 2008;28:2301–12. doi: 10.1523/JNEUROSCI.5157-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CL, Xu X, Kesari S, Xie XS, Wong STC, Young GS. Chemically-selective imaging of brain structures with CARS microscopy. Opt Express. 2007;15:12076–87. doi: 10.1364/oe.15.012076. [DOI] [PubMed] [Google Scholar]
- Fang-Yen C, Chu MC, Seung HS, Dasari RR, Feld MS. Noncontact measurement of nerve displacement during action potential with a dual-beam low-coherence interferometer. Opt Lett. 2004;29:2028–30. doi: 10.1364/ol.29.002028. [DOI] [PubMed] [Google Scholar]
- Fee MS. Active stabilization of electrodes for intracellular recording in awake behaving animals. Neuron. 2000;27:461–68. doi: 10.1016/s0896-6273(00)00057-x. [DOI] [PubMed] [Google Scholar]
- Feng G, Mellor RH, Bernstein M, Keller-Peck C, Nguyen QT, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Ferezou I, Bolea S, Petersen CCH. Visualizing the cortical representation of whisker touch: voltage-sensitive dye imaging in freely moving mice. Neuron. 2006;50:617–29. doi: 10.1016/j.neuron.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Ferezou I, Haiss F, Gentet LJ, Aronoff R, Weber B, Petersen CCH. Spatiotemporal dynamics of cortical sensorimotor integration in behaving mice. Neuron. 2007;56:907–23. doi: 10.1016/j.neuron.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Fernández-Suárez M, Baruah H, Martinez-Hernandez L, Xie KT, Baskin JM, et al. Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. Nat Biotechnol. 2007;25:1483–87. doi: 10.1038/nbt1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittinghoff DN, Squier JA. Time-decorrelated multifocal array for multiphoton microscopy and micro-machining. Opt Lett. 2000;25:1213–15. doi: 10.1364/ol.25.001213. [DOI] [PubMed] [Google Scholar]
- Fittinghoff DN, Wiseman PW, Squier JA. Widefield multiphoton and temporally decorrelated multifocal multiphoton microscopy. Opt Express. 2000;7:273–79. doi: 10.1364/oe.7.000273. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Cocker ED, Piyawattanametha W, Jung JC, Cheung EL, Schnitzer MJ. Fiber-optic fluorescence imaging. Nat Methods. 2005a;2:941–50. doi: 10.1038/nmeth820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, Jung JC, Cocker ED, Anderson EP, Schnitzer MJ. In vivo brain imaging using a portable 3.9 gram two-photon fluorescence microendoscope. Opt Lett. 2005b;30:2272–74. doi: 10.1364/ol.30.002272. [DOI] [PubMed] [Google Scholar]
- Flusberg BA, Nimmerjahn A, Cocker ED, Mukamel EA, Barretto RPJ, et al. High-speed, miniaturized fluorescence microscopy in freely moving mice. Nat Methods. 2008;5:935–38. doi: 10.1038/nmeth.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fölling J, Belov V, Kunetsky R, Medda R, Schönle A, et al. Photochromic rhodamines provide nanoscopy with optical sectioning. Angew Chem Int Ed Engl. 2007;46:6266–70. doi: 10.1002/anie.200702167. [DOI] [PubMed] [Google Scholar]
- Fölling J, Belov V, Riedel D, Schönle A, Egner A, et al. Fluorescence nanoscopy with optical sectioning by two-photon induced molecular switching using continuous-wave lasers. Chem Phys Chem. 2008a;9:321–26. doi: 10.1002/cphc.200700655. [DOI] [PubMed] [Google Scholar]
- Fölling J, Bossi M, Bock H, Medda R, Wurm CA, et al. Fluorescence nanoscopy by ground-state depletion and single-molecule return. Nat Methods. 2008b;5:943–45. doi: 10.1038/nmeth.1257. [DOI] [PubMed] [Google Scholar]
- Freudiger CW, Min W, Saar BG, Lu S, Holtom GR, et al. Label-free biomedical imaging with high sensitivity by stimulated Raman scattering microscopy. Science. 2008;322:1857–61. doi: 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke M, Nielsen T. Two-dimensional imaging without scanning by multifocal multiphoton microscopy. Appl Opt. 2005;44:2984–88. doi: 10.1364/ao.44.002984. [DOI] [PubMed] [Google Scholar]
- Fu L, Gu M. Fibre-optic nonlinear optical microscopy and endoscopy. J Microsc. 2007;226:195–206. doi: 10.1111/j.1365-2818.2007.01777.x. [DOI] [PubMed] [Google Scholar]
- Fu Y, Huff TB, Wang H-W, Cheng J-X, Wang H. Ex vivo and in vivo imaging of myelin fibers in mouse brain by coherent anti-Stokes Raman scattering microscopy. Opt Express. 2008;16:19396–409. doi: 10.1364/oe.16.019396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wang H, Huff TB, Shi R, Cheng J-X. Coherent anti-Stokes Raman scattering imaging of myelin degradation reveals a calcium-dependent pathway in lyso-PtdCho-induced demyelination. J Neurosci Res. 2007;85:2870–81. doi: 10.1002/jnr.21403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs E, Jaffe JS, Long RA, Azam F. Thin laser light sheet microscope for microbial oceanography. Opt Express. 2002;10:145–54. doi: 10.1364/oe.10.000145. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Mitteregger G, Kretzschmar H, Herms J. Dendritic pathology in prion disease starts at the synaptic spine. J Neurosci. 2007;27:6224–33. doi: 10.1523/JNEUROSCI.5062-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor D. A new microscopic principle. Nature. 1948;161:777–78. doi: 10.1038/161777a0. [DOI] [PubMed] [Google Scholar]
- Gaietta GM, Deerinck TJ, Adams SR, Bouwer J, Tour O, et al. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–7. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- Gaietta GM, Giepmans BNG, Deerinck TJ, Smith WB, Ngan L, et al. Golgi twins in late mitosis revealed by genetically encoded tags for live cell imaging and correlated electron microscopy. Proc Natl Acad Sci USA. 2006;103:17777–82. doi: 10.1073/pnas.0608509103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galison PL. Image and Logic: A Material Culture of Microphysics. Chicago: Univ. Chicago Press; 1997. p. 982. [Google Scholar]
- Gan W-B, Kwon E, Feng G, Sanes JR, Lichtman JW. Synaptic dynamism measured over minutes to months: age-dependent decline in an autonomic ganglion. Nat Neurosci. 2003;6:956–60. doi: 10.1038/nn1115. [DOI] [PubMed] [Google Scholar]
- Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–23. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- Giepmans BNG, Deerinck TJ, Smarr BL, Jones YZ, Ellisman MH. Correlated light and electron microscopic imaging of multiple endogenous proteins using Quantum dots. Nat Methods. 2005;2:743–49. doi: 10.1038/nmeth791. [DOI] [PubMed] [Google Scholar]
- Gloesmann M, Hermann B, Schubert C, Sattmann H, Ahnelt PK, Drexler W. Histologic correlation of pig retina radial stratification with ultrahigh-resolution optical coherence tomography. Invest Ophthalmol Vis Sci. 2003;44:1696–703. doi: 10.1167/iovs.02-0654. [DOI] [PubMed] [Google Scholar]
- Göbel W, Helmchen F. In vivo calcium imaging of neural network function. Physiology. 2007;22:358–65. doi: 10.1152/physiol.00032.2007. [DOI] [PubMed] [Google Scholar]
- Göbel W, Kampa BM, Helmchen F. Imaging cellular network dynamics in three dimensions using fast 3D laser scanning. Nat Methods. 2007;4:73–79. doi: 10.1038/nmeth989. [DOI] [PubMed] [Google Scholar]
- Göbel W, Kerr JND, Nimmerjahn A, Helmchen F. Miniaturized two-photon microscope based on a flexible coherent fiber bundle and a gradient-index lens objective. Opt Lett. 2004;29:2521–23. doi: 10.1364/ol.29.002521. [DOI] [PubMed] [Google Scholar]
- Goldin M, Epsztein J, Jorquera I, Represa A, Ben-Ari Y, et al. Synaptic kainate receptors tune orienslacunosum moleculare interneurons to operate at theta frequency. J Neurosci. 2007;27:9560–72. doi: 10.1523/JNEUROSCI.1237-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golgi C. Nobel lecture. 1906. Dec 11, The neuron doctrine—theory and facts. [Google Scholar]
- Gorostiza P, Isacoff EY. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–99. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz KG, Wenking H. Visual control of locomotion in the walking fruitfly Drosophila. J Comp Physiol A. 1973;85:235–66. [Google Scholar]
- Gould TJ, Gunewardene MS, Gudheti MV, Verkhusha VV, Yin S-R, et al. Nanoscale imaging of molecular positions and anisotropies. Nat Methods. 2008;5:1027–30. doi: 10.1038/nmeth.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NW, Weimer RM, Bureau I, Svoboda K. Rapid redistribution of synaptic PSD-95 in the neocortex in vivo. PLoS Biol. 2006;4:e370. doi: 10.1371/journal.pbio.0040370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg DS, Houweling AR, Kerr JND. Population imaging of ongoing neuronal activity in the visual cortex of awake rats. Nat Neurosci. 2008;11:749–51. doi: 10.1038/nn.2140. [DOI] [PubMed] [Google Scholar]
- Greenberg DS, Kerr JND. Automated correction of fast motion artifacts for two-photon imaging of awake animals. J Neurosci Methods. 2009;176:1–15. doi: 10.1016/j.jneumeth.2008.08.020. [DOI] [PubMed] [Google Scholar]
- Griffin BA, Adams SR, Tsien RY. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–72. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- Grinvald A, Frostig RD, Siegel RM, Bartfeld E. High-resolution optical imaging of functional brain architecture in the awake monkey. Proc Natl Acad Sci USA. 1991;88:11559–63. doi: 10.1073/pnas.88.24.11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutzendler J, Kasthuri N, Gan W-B. Long-term dendritic spine stability in the adult cortex. Nature. 2002;420:812–16. doi: 10.1038/nature01276. [DOI] [PubMed] [Google Scholar]
- Gugel H, Bewersdorf J, Jakobs S, Engelhardt J, Storz R, Hell SW. Cooperative 4Pi excitation and detection yields sevenfold sharper optical sections in live-cell microscopy. Biophys J. 2004;87:4146–52. doi: 10.1529/biophysj.104.045815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SX, Bourgeois F, Chokshi T, Durr NJ, Hilliard MA, et al. Femtosecond laser nanoaxotomy lab-on-a-chip for in vivo nerve regeneration studies. Nat Methods. 2008;5:531–33. doi: 10.1038/nmeth.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MGL. Extended resolution fluorescence microscopy. Curr Opin Struct Biol. 1999;9:627–34. doi: 10.1016/s0959-440x(99)00016-0. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J Microsc. 2000;198:82–87. doi: 10.1046/j.1365-2818.2000.00710.x. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL. Nonlinear structured-illumination microscopy: wide-field fluorescence imaging with theoretically unlimited resolution. Proc Natl Acad Sci USA. 2005;102:13081–86. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson MGL, Agard DA, Sedat JW. I5M: 3D widefield light microscopy with better than 100 nm axial resolution. J Microsc. 1999;195:10–16. doi: 10.1046/j.1365-2818.1999.00576.x. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL, Shao L, Carlton PM, Wang CJR, Golubovskaya IN, et al. Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 2008;94:4957–70. doi: 10.1529/biophysj.107.120345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harke B, Keller J, Ullal CK, Westphal V, Schönle A, Hell SW. Resolution scaling in STED microscopy. Opt Express. 2008;16:4154–62. doi: 10.1364/oe.16.004154. [DOI] [PubMed] [Google Scholar]
- Hayworth KJ, Kasthuri N, Hartwieg E, Lichtman JW. Automating the collection of ultrathin brain sections for electron microscopic volume imaging. SfN Poster #534.6/III27 Abstr; Presented at Annu. Meet. Soc. Neurosci., 37th; San Diego, CA. 2007. [Google Scholar]
- Heilemann M, van de Linde S, Schüttpelz M, Kasper R, Seefeldt B, et al. Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew Chem Int Ed Engl. 2008;47:6172–76. doi: 10.1002/anie.200802376. [DOI] [PubMed] [Google Scholar]
- Hein B, Willig KI, Hell SW. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc Natl Acad Sci USA. 2008;105:14271–76. doi: 10.1073/pnas.0807705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzmann R, Jovin TM, Cremer C. Saturated patterned excitation microscopy—a concept for optical resolution improvement. J Opt Soc Am A. 2002;19:1599–609. doi: 10.1364/josaa.19.001599. [DOI] [PubMed] [Google Scholar]
- Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–58. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- Hell SW, Kroug M. Ground-state-depletion fluorescence microscopy: a concept for breaking the diffraction resolution limit. Appl Phys B. 1995;60:495–97. [Google Scholar]
- Hell SW, Schrader M, van der Voort HTM. Far-field fluorescence microscopy with three-dimensional resolution in the 100-nm range. J Microsc. 1997;187:1–7. doi: 10.1046/j.1365-2818.1997.2410797.x. [DOI] [PubMed] [Google Scholar]
- Hell SW, Stelzer EHK. Properties of a 4Pi confocal fluorescence microscope. J Opt Soc Am A. 1992;9:2159–66. [Google Scholar]
- Hell SW, Wichmann J. Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt Lett. 1994;19:780–82. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–40. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Fee MS, Tank DW, Denk W. A miniature head-mounted two-photon microscope: high-resolution brain imaging in freely moving animals. Neuron. 2001;31:903–12. doi: 10.1016/s0896-6273(01)00421-4. [DOI] [PubMed] [Google Scholar]
- Helmchen F, Imoto K, Sakmann B. Ca2+ buffering and action potential-evoked Ca2+ signaling in dendrites of pyramidal neurons. Biophys J. 1996;70:1069–81. doi: 10.1016/S0006-3495(96)79653-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng X, Erickson D, Baugh LR, Yaqoob Z, Sternberg PW, et al. Optofluidic microscopy–method for implementing a high resolution optical microscope on a chip. Lab Chip. 2006;6:1274–76. doi: 10.1039/b604676b. [DOI] [PubMed] [Google Scholar]
- Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–72. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hübener M. Experience leaves a lasting structural trace in cortical circuits. Nature. 2009;457:313–17. doi: 10.1038/nature07487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M, Eggeling C, Jakobs S, Hell SW. Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci USA. 2005;102:17565–69. doi: 10.1073/pnas.0506010102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holekamp TF, Turaga D, Holy TE. Fast three-dimensional fluorescence imaging of activity in neural populations by objective-coupled planar illumination microscopy. Neuron. 2008;57:661–72. doi: 10.1016/j.neuron.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Holtmaat AJGD, Bonhoeffer T, Chow D, Chuckowree J, De Paola V. Long-term high resolution imaging in the mouse neocortex through a chronic cranial window. Nat Protoc. 2009 doi: 10.1038/nprot.2009.89. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat AJGD, Wilbrecht L, Knott GW, Welker E, Svoboda K. Experience-dependent and cell-type-specific spine growth in the neocortex. Nature. 2006;441:979–83. doi: 10.1038/nature04783. [DOI] [PubMed] [Google Scholar]
- Holtmaat AJGD, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, et al. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–91. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Hopt A, Neher E. Highly nonlinear photodamage in two-photon fluorescence microscopy. Biophys J. 2001;80:2029–36. doi: 10.1016/S0006-3495(01)76173-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoy CL, Durr NJ, Chen P, Piyawattanametha W, Ra H, et al. Miniaturized probe for femtosecond laser microsurgery and two-photon imaging. Opt Express. 2008;16:9996–10005. doi: 10.1364/oe.16.009996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008a;5:1047–52. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008b;319:810–13. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Swanson EA, Lin CP, Schuman JS, Stinson WG, et al. Optical coherence tomography. Science. 1991;254:1178–81. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, Stelzer EHK. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–9. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- Hulme SE, Shevkoplyas SS, Apfeld J, Fontana W, Whitesides GM. A microfabricated array of clamps for immobilizing and imaging C. elegans. Lab Chip. 2007;7:1515–23. doi: 10.1039/b707861g. [DOI] [PubMed] [Google Scholar]
- Izatt JA, Kulkami MD, Yazdanfar S, Barton JK, Welch AJ. In vivo bidirectional color Doppler flow imaging of picoliter blood volumes using optical coherence tomograghy. Opt Lett. 1997;22:1439–41. doi: 10.1364/ol.22.001439. [DOI] [PubMed] [Google Scholar]
- Ji N, Magee JC, Betzig E. High-speed, low-photodamage nonlinear imaging using passive pulse splitters. Nat Methods. 2008;5:197–202. doi: 10.1038/nmeth.1175. [DOI] [PubMed] [Google Scholar]
- Jiang J, Eisenthal KB, Yuste R. Second harmonic generation in neurons: electro-optic mechanism of membrane potential sensitivity. Biophys J. 2007;93:L26–28. doi: 10.1529/biophysj.107.111021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–29. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- Jung JC, Mehta AD, Aksay E, Stepnoski R, Schnitzer MJ. In vivo mammalian brain imaging using one-and two-photon fluorescence microendoscopy. J Neurophysiol. 2004;92:3121–33. doi: 10.1152/jn.00234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JC, Schnitzer MJ. Multiphoton endoscopy. Opt Lett. 2003;28:902–4. doi: 10.1364/ol.28.000902. [DOI] [PubMed] [Google Scholar]
- Jung W, Tang S, McCormic DT, Xie T, Ahn Y-C, et al. Miniaturized probe based on a microelectromechanical system mirror for multiphoton microscopy. Opt Lett. 2008;33:1324–26. doi: 10.1364/ol.33.001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao HP, Verkman AS. Tracking of single fluorescent particles in three dimensions: use of cylindrical optics to encode particle position. Biophys J. 1994;67:1291–300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EHK. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–69. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- Keller-Peck CR, Walsh MK, Gan W-B, Feng G, Sanes JR, Lichtman JW. Asynchronous synapse elimination in neonatal motor units: studies using GFP transgenic mice. Neuron. 2001;31:381–94. doi: 10.1016/s0896-6273(01)00383-x. [DOI] [PubMed] [Google Scholar]
- Kellner RR, Baier CJ, Willig KI, Hell SW, Barrantes FJ. Nanoscale organization of nicotinic acetyl-choline receptors revealed by stimulated emission depletion microscopy. Neuroscience. 2007;144:135–43. doi: 10.1016/j.neuroscience.2006.08.071. [DOI] [PubMed] [Google Scholar]
- Kerr JND, Greenberg DS, Helmchen F. Imaging input and output of neocortical networks in vivo. Proc Natl Acad Sci USA. 2005;102:14063–68. doi: 10.1073/pnas.0506029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschensteiner M, Schwab ME, Lichtman JW, Misgeld T. In vivo imaging of axonal degeneration and regeneration in the injured spinal cord. Nat Med. 2005;11:572–77. doi: 10.1038/nm1229. [DOI] [PubMed] [Google Scholar]
- Kim KH, Buehler C, Bahlmann K, Ragan T, Lee W-CA. Multifocal multiphoton microscopy based on multianode photomultiplier tubes. Opt Express. 2007;15:11658–78. doi: 10.1364/oe.15.011658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittel RJ, Wichmann C, Rasse TM, Fouquet W, Schmidt M, et al. Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science. 2006;312:1051–54. doi: 10.1126/science.1126308. [DOI] [PubMed] [Google Scholar]
- Klar TA, Hell SW. Subdiffraction resolution in far-field fluorescence microscopy. Opt Lett. 1999;24:954–56. doi: 10.1364/ol.24.000954. [DOI] [PubMed] [Google Scholar]
- Klar TA, Jakobs S, Dyba M, Egner A, Hell SW. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA. 2000;97:8206–10. doi: 10.1073/pnas.97.15.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinfeld D, Mitra PP, Helmchen F, Denk W. Fluctuations and stimulus-induced changes in blood flow observed in individual capillaries in layers 2 through 4 of rat neocortex. Proc Natl Acad Sci USA. 1998;95:15741–46. doi: 10.1073/pnas.95.26.15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittel J, Schnieder L, Buess G, Messerschmidt B, Possner T. Endoscope-compatible confocal microscope using a gradient index-lens system. Opt Commun. 2001;188:267–73. [Google Scholar]
- Knott GW, Holtmaat A. Dendritic spine plasticity—current understanding from in vivo studies. Brain Res Rev. 2008;58:282–89. doi: 10.1016/j.brainresrev.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Knott GW, Holtmaat A, Wilbrecht L, Welker E, Svoboda K. Spine growth precedes synapse formation in the adult neocortex in vivo. Nat Neurosci. 2006;9:1117–24. doi: 10.1038/nn1747. [DOI] [PubMed] [Google Scholar]
- Koenigsknecht-Talboo J, Meyer-Luehmann M, Parsadanian M, Garcia-Alloza M, Finn MB, et al. Rapid microglial response around amyloid pathology after systemic anti-Aβ antibody administration in PDAPP mice. J Neurosci. 2008;28:14156–64. doi: 10.1523/JNEUROSCI.4147-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz R, Fricke M, Kalb J, Tinnefeld P, Sauer M. Application of multiline two-photon microscopy to functional in vivo imaging. J Neurosci Methods. 2006;151:276–86. doi: 10.1016/j.jneumeth.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Laing NG, Nowak KJ. When contractile proteins go bad: the sarcomere and skeletal muscle disease. BioEssays. 2005;27:809–22. doi: 10.1002/bies.20269. [DOI] [PubMed] [Google Scholar]
- Larson DR, Johnson MC, Webb WW, Vogt VM. Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci USA. 2005;102:15453–58. doi: 10.1073/pnas.0504812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazebnik M, Marks DL, Potgieter K, Gillette R, Boppart SA. Functional optical coherence tomography for detecting neural activity through scattering changes. Opt Lett. 2003;28:1218–20. doi: 10.1364/ol.28.001218. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22:451–61. doi: 10.1016/s0896-6273(00)80701-1. [DOI] [PubMed] [Google Scholar]
- Lee W-CA, Chen JL, Huang H, Leslie JH, Amitai Y, et al. A dynamic zone defines interneuron remodeling in the adult neocortex. Proc Natl Acad Sci USA. 2008;105:19968–73. doi: 10.1073/pnas.0810149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-CA, Huang H, Feng G, Sanes JR, Brown EN, et al. Dynamic remodeling of dendritic arbors in GABAergic interneurons of adult visual cortex. PLoS Biol. 2006;4:e29. doi: 10.1371/journal.pbio.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Harzic R, Weinigel M, Riemann I, König K, Messerschmidt B. Nonlinear optical endoscope based on a compact two axes piezo scanner and a miniature objective lens. Opt Express. 2008;16:20588–96. doi: 10.1364/oe.16.020588. [DOI] [PubMed] [Google Scholar]
- Leitgeb RA, Drexler W, Unterhuber A, Hermann B, Bajraszewski T, et al. Ultrahigh resolution Fourier domain optical coherence tomography. Opt Express. 2004;12:2156–65. doi: 10.1364/opex.12.002156. [DOI] [PubMed] [Google Scholar]
- Leitgeb RA, Schmetterer L, Drexler W, Fercher AF, Zawadzki RJ, Bajraszewski T. Real-time assessment of retinal blood flow with ultrafast acquisition by color Doppler Fourier domain optical coherence tomography. Opt Express. 2003;11:3116–21. doi: 10.1364/oe.11.003116. [DOI] [PubMed] [Google Scholar]
- Levene MJ, Dombeck DA, Kasischke KA, Molloy RP, Webb WW. In vivo multiphoton microscopy of deep brain tissue. J Neurophysiol. 2004;91:1908–12. doi: 10.1152/jn.01007.2003. [DOI] [PubMed] [Google Scholar]
- Lévêque-Fort S, Fontaine-Aupart MP, Roger G, Georges P. Fluorescence-lifetime imaging with a multifocal two-photon microscope. Opt Lett. 2004;29:2884–86. doi: 10.1364/ol.29.002884. [DOI] [PubMed] [Google Scholar]
- Levoy M, Ng R, Adams A, Footer M, Horowitz M. Light field microscopy. ACM Trans Graph (Proc SIGGRAPH) 2006;25:924–34. [Google Scholar]
- Levoy M. Stanford Light Field Microscope Project. 2009 http://graphics.stanford.edu/projects/lfmicroscope/
- Levoy M, Zhang Z, McDowall I. Recording and controlling the 4D light field in a microscope. J Microsc. 2009 doi: 10.1111/j.1365-2818.2009.03195.x. In press. [DOI] [PubMed] [Google Scholar]
- Lichtman JW, Hayworth K. ATLUM Project. 2009 http://mcb.harvard.edu/lichtman/ATLUM/ATLUM_web.htm.
- Lichtman JW, Livet J, Sanes JR. A technicolour approach to the connectome. Nat Rev Neurosci. 2008;9:417–22. doi: 10.1038/nrn2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtman JW, Magrassi L, Purves D. Visualization of neuromuscular junctions over periods of several months in living mice. J Neurosci. 1987;7:1215–22. doi: 10.1523/JNEUROSCI.07-04-01215.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Margolskee R, Donnert G, Hell SW, Restrepo D. Olfactory neurons expressing transient receptor potential channel M5 (TRPM5) are involved in sensing semiochemicals. Proc Natl Acad Sci USA. 2007;104:2471–76. doi: 10.1073/pnas.0610201104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindek S, Stefany T, Stelzer EHK. Single-lens theta microscopy—a new implementation of confocal theta microscopy. J Microsc. 1997;188:280–84. [Google Scholar]
- Lindek S, Swoger J, Stelzer EHK. Single-lens theta microscopy: resolution, efficiency and working distance. J Mod Opt. 1999;46:843–58. [Google Scholar]
- Liu TM, Chu SW, Sun CK, Lin BL, Cheng PC, Johnson I. Multiphoton confocal microscopy using a femtosecond Cr:forsterite laser. Scanning. 2001;23:249–54. doi: 10.1002/sca.4950230405. [DOI] [PubMed] [Google Scholar]
- Livet J, Weissman TA, Kang H, Draft RW, Lu J, et al. Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system. Nature. 2007;450:56–62. doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- Llewellyn ME, Barretto RPJ, Delp SL, Schnitzer MJ. Minimally invasive high-speed imaging of sarcomere contractile dynamics in mice and humans. Nature. 2008;454:784–88. doi: 10.1038/nature07104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord SJ, Conley NR, Lee HD, Samuel R, Liu N, et al. A photoactivatable push-pull fluorophore for single-molecule imaging in live cells. J Am Chem Soc. 2008;130:9204–5. doi: 10.1021/ja802883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Lee JH, Fu LA, Patel NH, Ismagilov RF. Dynamics of Drosophila embryonic patterning network perturbed in space and time using microfluidics. Nature. 2005;434:1134–38. doi: 10.1038/nature03509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukosz W, Marchand M. Optischen Abbildung unter Überschreitung der beugungsbedingten Auflösungsgrenze. Opt Acta. 1963;10:241–55. [Google Scholar]
- Lutz C, Otis TS, DeSars V, Charpak S, DiGregorio DA, Emiliani V. Holographic photolysis of caged neurotransmitters. Nat Methods. 2008;5:821–27. doi: 10.1038/nmeth.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maheswari RU, Takaoka H, Homma R, Kadono H, Tanifuji M. Implementation of optical coherence tomography (OCT) in visualization of functional structures of cat visual cortex. Opt Commun. 2002;202:47–54. [Google Scholar]
- Maheswari RU, Takaoka H, Kadono H, Homma R, Tanifuji M. Novel functional imaging technique from brain surface with optical coherence tomography enabling visualization of depth resolved functional structure in vivo. J Neurosci Methods. 2003;124:83–92. doi: 10.1016/s0165-0270(02)00370-9. [DOI] [PubMed] [Google Scholar]
- Majewska AK, Newton JR, Sur M. Remodeling of synaptic structure in sensory cortical areas in vivo. J Neurosci. 2006;26:3021–29. doi: 10.1523/JNEUROSCI.4454-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–57. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- Maranto AR. Neuronal mapping: a photooxidation reaction makes Lucifer yellow useful for electron microscopy. Science. 1982;217:953–55. doi: 10.1126/science.7112109. [DOI] [PubMed] [Google Scholar]
- Martin C, Berwick J, Johnston D, Zheng Y, Martindale J, et al. Optical imaging spectroscopy in the unanaesthetised rat. J Neurosci Methods. 2002;120:25–34. doi: 10.1016/s0165-0270(02)00185-1. [DOI] [PubMed] [Google Scholar]
- Martin C, Martindale J, Berwick J, Mayhew J. Investigating neural-hemodynamic coupling and the hemodynamic response function in the awake rat. NeuroImage. 2006;32:33–48. doi: 10.1016/j.neuroimage.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Mayerich D, Abbott L, McCormick B. Knife-edge scanning microscopy for imaging and reconstruction of three-dimensional anatomical structures of the mouse brain. J Microsc. 2008;231:134–43. doi: 10.1111/j.1365-2818.2008.02024.x. [DOI] [PubMed] [Google Scholar]
- McGurk L, Morrison H, Keegan LP, Sharpe J, O’Connell MA. Three-dimensional imaging of Drosophila melanogaster. PLoS ONE. 2007;2:e834. doi: 10.1371/journal.pone.0000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–33. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melder RJ, Salehi HA, Jain RK. Interaction of activated natural killer cells with normal and tumor vessels in cranial windows in mice. Microvasc Res. 1995;50:35–44. doi: 10.1006/mvre.1995.1036. [DOI] [PubMed] [Google Scholar]
- Meyer L, Wildanger D, Medda R, Punge A, Rizzoli SO, et al. Dual-color STED microscopy at 30-nm focal-plane resolution. Small. 2008;4:1095–100. doi: 10.1002/smll.200800055. [DOI] [PubMed] [Google Scholar]
- Meyer-Luehmann M, Spires-Jones T, Prada C, Garcia-Alloza M, de Calignon A, et al. Rapid appearance and local toxicity of amyloid-β plaques in a mouse model of Alzheimer’s disease. Nature. 2008;451:720–24. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Weiss S. Using photon statistics to boost microscopy resolution. Proc Natl Acad Sci USA. 2006;103:4797–98. doi: 10.1073/pnas.0600808103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheva KD, Smith SJ. Array tomography: a new tool for imaging the molecular architecture and ultrastructure of neural circuits. Neuron. 2007;55:25–36. doi: 10.1016/j.neuron.2007.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misgeld T, Kerschensteiner M. In vivo imaging of the diseased nervous system. Nat Rev Neurosci. 2006;7:449–63. doi: 10.1038/nrn1905. [DOI] [PubMed] [Google Scholar]
- Miyawaki A. Innovations in the imaging of brain functions using fluorescent proteins. Neuron. 2005;48:189–99. doi: 10.1016/j.neuron.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Mizrahi A. Dendritic development and plasticity of adult-born neurons in the mouse olfactory bulb. Nat Neurosci. 2007;10:444–52. doi: 10.1038/nn1875. [DOI] [PubMed] [Google Scholar]
- Mizrahi A, Lu J, Irving R, Feng G, Katz LC. In vivo imaging of juxtaglomerular neuron turnover in the mouse olfactory bulb. Proc Natl Acad Sci USA. 2006;103:1912–17. doi: 10.1073/pnas.0506297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerner WE. A dozen years of single-molecule spectroscopy in physics, chemistry, and biophysics. J Phys Chem B. 2002;106:910–27. [Google Scholar]
- Moerner WE. New directions in single-molecule imaging and analysis. Proc Natl Acad Sci USA. 2007;104:12596–602. doi: 10.1073/pnas.0610081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moerner WE, Kador L. Optical detection and spectroscopy of single molecules in a solid. Phys Rev Lett. 1989;62:2535–38. doi: 10.1103/PhysRevLett.62.2535. [DOI] [PubMed] [Google Scholar]
- Monfared A, Blevins NH, Cheung ELM, Jung JC, Popelka G, Schnitzer MJ. In vivo imaging of mammalian cochlear blood flow using fluorescence microendoscopy. Otol Neurotol. 2006;27:144–52. doi: 10.1097/01.mao.0000190708.44067.b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movshon JA, Albright TD, Stoner GR, Majaj NJ, Smith MA. Cortical responses to visual motion in alert and anesthetized monkeys. Nat Neurosci. 2003;6:3. doi: 10.1038/nn0103-3a. [DOI] [PubMed] [Google Scholar]
- Mrsic-Flogel TD, Hofer SB, Ohki K, Reid RC, Bonhoeffer T, Hübener M. Homeostatic regulation of eye-specific responses in visual cortex during ocular dominance plasticity. Neuron. 2007;54:961–72. doi: 10.1016/j.neuron.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, Schnitzer MJ. Automated sorting of intracellular calcium signals for large-scale imaging studies of neuronal and glial populations. SfN Poster #252.2/K18 Abstr; Presented at Annu. Meet. Soc. Neurosci., 37th; San Diego, CA. 2007. [Google Scholar]
- Murayama M, Pérez-Garci E, Lüscher H-R, Larkum ME. Fiberoptic system for recording dendritic calcium signals in layer 5 neocortical pyramidal cells in freely moving rats. J Neurophysiol. 2007;98:1791–805. doi: 10.1152/jn.00082.2007. [DOI] [PubMed] [Google Scholar]
- Murayama M, Pérez-Garci E, Nevian T, Bock T, Senn W, Larkum ME. Dendritic encoding of sensory stimuli controlled by deep cortical interneurons. Nature. 2009;457:1137–41. doi: 10.1038/nature07663. [DOI] [PubMed] [Google Scholar]
- Nägerl UV, Willig KI, Hein B, Hell SW, Bonhoeffer T. Live-cell imaging of dendritic spines by STED microscopy. Proc Natl Acad Sci USA. 2008;105:18982–87. doi: 10.1073/pnas.0810028105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neil MAA, Juškaitis R, Booth MJ, Wilson T, Tanaka T, Kawata S. Adaptive aberration correction in a two-photon microscope. J Microsc. 2000;200:105–8. doi: 10.1046/j.1365-2818.2000.00770.x. [DOI] [PubMed] [Google Scholar]
- Nemet BA, Nikolenko V, Yuste R. Second harmonic imaging of membrane potential of neurons with retinal. J Biomed Opt. 2004;9:873–81. doi: 10.1117/1.1783353. [DOI] [PubMed] [Google Scholar]
- Nguyen QT, Sanes JR, Lichtman JW. Pre-existing pathways promote precise projection patterns. Nat Neurosci. 2002;5:861–67. doi: 10.1038/nn905. [DOI] [PubMed] [Google Scholar]
- Niell CM, Smith SJ. Functional imaging reveals rapid development of visual response properties in the zebrafish tectum. Neuron. 2005;45:941–51. doi: 10.1016/j.neuron.2005.01.047. [DOI] [PubMed] [Google Scholar]
- Nielsen T, Fricke M, Hellweg D, Andresen P. High efficiency beam splitter for multifocal multiphoton microscopy. J Microsc. 2001;201:368–76. doi: 10.1046/j.1365-2818.2001.00852.x. [DOI] [PubMed] [Google Scholar]
- Niesner R, Andresen V, Neumann J, Spiecker H, Gunzer M. The power of single and multibeam two-photon microscopy for high-resolution and high-speed deep tissue and intravital imaging. Biophys J. 2007;93:2519–29. doi: 10.1529/biophysj.106.102459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolenko V, Watson BO, Araya R, Woodruff A, Peterka DS, Yuste R. SLM microscopy: scanless two-photon imaging and photostimulation with spatial light modulators. Front Neural Circuits. 2008;2:5. doi: 10.3389/neuro.04.005.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A. Astrocytes going live: advances and challenges. J Physiol. 2009 doi: 10.1113/jphysiol.2008.167171. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A, Kirchhoff F, Kerr JND, Helmchen F. Sulforhodamine 101 as a specific marker of astroglia in the neocortex in vivo. Nat Methods. 2004;1:31–37. doi: 10.1038/nmeth706. [DOI] [PubMed] [Google Scholar]
- Nimmerjahn A, Mukamel EA, Schnitzer MJ. Two-photon imaging of neuronal and glial network activity in the cerebellar cortex of awake mice. SfN Poster #252.2/K21 Abstr; Presented at Annu. Meet. Soc. Neurosci., 37th; San Diego, CA. 2007. [Google Scholar]
- Nimmerjahn A, Mukamel EA, Schnitzer MJ. Motor behavior activates Bergmann glial networks. Neuron. 2009 doi: 10.1016/j.neuron.2009.03.019. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama H, Fukaya M, Watanabe M, Linden DJ. Axonal motility and its modulation by activity are branch-type specific in the intact adult cerebellum. Neuron. 2007;56:472–87. doi: 10.1016/j.neuron.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuriya M, Jiang J, Nemet B, Eisenthal KB, Yuste R. Imaging membrane potential in dendritic spines. Proc Natl Acad Sci USA. 2006;103:786–90. doi: 10.1073/pnas.0510092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober RJ, Ram S, Ward ES. Localization accuracy in single-molecule microscopy. Biophys J. 2004;86:1185–200. doi: 10.1016/S0006-3495(04)74193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oheim M, Beaurepaire E, Chaigneau E, Mertz J, Charpak S. Two-photon microscopy in brain tissue: parameters influencing the imaging depth. J Neurosci Methods. 2001;111:29–37. doi: 10.1016/s0165-0270(01)00438-1. [DOI] [PubMed] [Google Scholar]
- Ohki K, Chung S, Kara P, Hübener M, Bonhoeffer T, Reid RC. Highly ordered arrangement of single neurons in orientation pinwheels. Nature. 2006;442:925–28. doi: 10.1038/nature05019. [DOI] [PubMed] [Google Scholar]
- Orger MB, Kampff AR, Severi KE, Bollmann JH, Engert F. Control of visually guided behavior by distinct populations of spinal projection neurons. Nat Neurosci. 2008;11:327–33. doi: 10.1038/nn2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oron D, Tal E, Silberberg Y. Scanningless depth-resolved microscopy. Opt Express. 2005;13:1468–76. doi: 10.1364/opex.13.001468. [DOI] [PubMed] [Google Scholar]
- Ozden I, Lee HM, Sullivan MR, Wang SS-H. Identification and clustering of event patterns from in vivo multiphoton optical recordings of neuronal ensembles. J Neurophysiol. 2008;100:495–503. doi: 10.1152/jn.01310.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack CC, Berezovskii VK, Born RT. Dynamic properties of neurons in cortical area MT in alert and anaesthetized macaque monkeys. Nature. 2001;414:905–8. doi: 10.1038/414905a. [DOI] [PubMed] [Google Scholar]
- Pan F, Gan W-B. Two-photon imaging of dendritic spine development in the mouse cortex. Dev Neurobiol. 2008;68:771–78. doi: 10.1002/dneu.20630. [DOI] [PubMed] [Google Scholar]
- Papagiakoumou E, de Sars V, Oron D, Emiliani V. Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt Express. 2008;16:22039–47. doi: 10.1364/oe.16.022039. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Piston DW. Photobleaching in two-photon excitation microscopy. Biophys J. 2000;78:2159–62. doi: 10.1016/S0006-3495(00)76762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavani SRP, Piestun R. Three dimensional tracking of fluorescent microparticles using a photon-limited double-helix response system. Opt Express. 2008;16:22048–57. doi: 10.1364/oe.16.022048. [DOI] [PubMed] [Google Scholar]
- Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci USA. 2009;106:2995–99. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. Bioimage informatics: a new area of engineering biology. Bioinformatics. 2008;24:1827–36. doi: 10.1093/bioinformatics/btn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perentes JY, McKee TD, Ley CD, Mathiew H, Dawson M, et al. In vivo imaging of extracellular matrix remodeling by tumor-associated fibroblasts. Nat Methods. 2009;6:143–45. doi: 10.1038/nmeth.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CCH. Genetic manipulation, whole-cell recordings and functional imaging of the sensorimotor cortex of behaving mice. Acta Physiol. 2009;195:91–99. doi: 10.1111/j.1748-1716.2008.01925.x. [DOI] [PubMed] [Google Scholar]
- Petersen CCH, Hahn TTG, Mehta M, Grinvald A, Sakmann B. Interaction of sensory responses with spontaneous depolarization in layer 2/3 barrel cortex. Proc Natl Acad Sci USA. 2003;100:13638–43. doi: 10.1073/pnas.2235811100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielage J, Cheng L, Fetter RD, Carlton PM, Sedat JW, Davis GW. A presynaptic giant ankyrin stabilizes the NMJ through regulation of presynaptic microtubules and transsynaptic cell adhesion. Neuron. 2008;58:195–209. doi: 10.1016/j.neuron.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyawattanametha W, Barretto RPJ, Ko TH, Flusberg BA, Cocker ED, et al. Fast-scanning two-photon fluorescence imaging based on a microelectromechanical systems two-dimensional scanning mirror. Opt Lett. 2006;31:2018–20. doi: 10.1364/ol.31.002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyawattanametha W, Cocker ED, Barretto RPJ, Jung JC, Flusberg BA, et al. A portable two-photon fluorescence microendoscope based on a two-dimensional scanning mirror. Presented at Opt. MEMS Nanophotonics; Hualien, Taiwan. 2007. [Google Scholar]
- Plotnikov SV, Kenny AM, Walsh SJ, Zubrowski B, Joseph C, et al. Measurement of muscle disease by quantitative second-harmonic generation imaging. J Biomed Opt. 2008;13:044018. doi: 10.1117/1.2967536. [DOI] [PubMed] [Google Scholar]
- Plotnikov SV, Millard AC, Campagnola PJ, Mohler WA. Characterization of the myosin-based source for second-harmonic generation from muscle sarcomeres. Biophys J. 2006;90:693–703. doi: 10.1529/biophysj.105.071555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poe GR, Kristensen MP, Rector DM, Harper RM. Hippocampal activity during transient respiratory events in the freely behaving cat. Neuroscience. 1996;72:39–48. doi: 10.1016/0306-4522(95)00525-0. [DOI] [PubMed] [Google Scholar]
- Pons T, Mertz J. Membrane potential detection with second-harmonic generation and two-photon excited fluorescence: a theoretical comparison. Opt Commun. 2006;258:203–9. [Google Scholar]
- Pons T, Moreaux L, Mongin O, Blanchard-Desce M, Mertz J. Mechanisms of membrane potential sensing with second-harmonic generation microscopy. J Biomed Opt. 2003;8:428–31. doi: 10.1117/1.1581871. [DOI] [PubMed] [Google Scholar]
- Pontén E, Gantelius S, Lieber RL. Intraoperative muscle measurements reveal a relationship between contracture formation and muscle remodeling. Muscle Nerve. 2007;36:47–54. doi: 10.1002/mus.20780. [DOI] [PubMed] [Google Scholar]
- Poon T-C, Doh KB, Schilling BW, Wu MH, Shinoda KK, Suzuki Y. Three-dimensional microscopy by optical scanning holography. Opt Eng. 1995;34:1338–44. [Google Scholar]
- Popp MW, Antos JM, Grotenbreg GM, Spooner E, Ploegh HL. Sortagging: a versatile method for protein labeling. Nat Chem Biol. 2007;3:707–8. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- Portera-Cailliau C, Weimer RM, De Paola V, Caroni P, Svoboda K. Diverse modes of axon elaboration in the developing neocortex. PLoS Biol. 2005;3:e272. doi: 10.1371/journal.pbio.0030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potsaid B, Gorczynska I, Srinivasan VJ, Chen Y, Jiang J, et al. Ultrahigh speed spectral/Fourier domain OCT ophthalmic imaging at 70,000 to 312,500 axial scans per second. Opt Express. 2008;16:15149–69. doi: 10.1364/oe.16.015149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Považay B, Bizheva K, Unterhuber A, Hermann B, Sattmann H, et al. Submicrometer axial resolution optical coherence tomography. Opt Lett. 2002;27:1800–2. doi: 10.1364/ol.27.001800. [DOI] [PubMed] [Google Scholar]
- Prabhat P, Ram S, Ward ES, Ober RJ. Simultaneous imaging of different focal planes in fluorescence microscopy for the study of cellular dynamics in three dimensions. IEEE Trans Nanobiosci. 2004;3:237–42. doi: 10.1109/tnb.2004.837899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purves D, Hadley RD. Changes in the dendritic branching of adult mammalian neurones revealed by repeated imaging in situ. Nature. 1985;315:404–6. doi: 10.1038/315404a0. [DOI] [PubMed] [Google Scholar]
- Purves D, Hadley RD, Voyvodic JT. Dynamic changes in the dendritic geometry of individual neurons visualized over periods of up to three months in the superior cervical ganglion of living mice. J Neurosci. 1986;6:1051–60. doi: 10.1523/JNEUROSCI.06-04-01051.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quraishi S, Heider B, Siegel RM. Attentional modulation of receptive field structure in area 7a of the behaving monkey. Cereb Cortex. 2007;17:1841–57. doi: 10.1093/cercor/bhl093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffi M, Siegel RM. A functional architecture of optic flow in the inferior parietal lobule of the behaving monkey. PLoS ONE. 2007;2:e200. doi: 10.1371/journal.pone.0000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan UM, Tanifuji M. Functional optical coherence tomography reveals localized layer-specific activations in cat primary visual cortex in vivo. Opt Lett. 2007;32:2614–16. doi: 10.1364/ol.32.002614. [DOI] [PubMed] [Google Scholar]
- Ram S, Prabhat P, Chao J, Ward ES, Ober RJ. High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells. Biophys J. 2008;95:6025–43. doi: 10.1529/biophysj.108.140392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram S, Ward ES, Ober RJ. Beyond Rayleigh’s criterion: a resolution measure with application to single-molecule microscopy. Proc Natl Acad Sci USA. 2006;103:4457–62. doi: 10.1073/pnas.0508047103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector DM, Harper RM. Imaging of hippocampal neural activity in freely behaving animals. Behav Brain Res. 1991;42:143–49. doi: 10.1016/s0166-4328(05)80005-1. [DOI] [PubMed] [Google Scholar]
- Rector DM, Poe GR, Harper RM. Imaging of hippocampal and neocortical neural activity following intravenous cocaine administration in freely behaving cats. Neuroscience. 1993;54:633–41. doi: 10.1016/0306-4522(93)90234-7. [DOI] [PubMed] [Google Scholar]
- Reddy GD, Kelleher K, Fink R, Saggau P. Three-dimensional random access multiphoton microscopy for functional imaging of neuronal activity. Nat Neurosci. 2008;11:713–20. doi: 10.1038/nn.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GD, Saggau P. Fast three-dimensional laser scanning scheme using acousto-optic deflectors. J Biomed Opt. 2005;10:064038. doi: 10.1117/1.2141504. [DOI] [PubMed] [Google Scholar]
- Reidl J, Starke J, Omer DB, Grinvald A, Spors H. Independent component analysis of high-resolution imaging data identifies distinct functional domains. NeuroImage. 2007;34:94–108. doi: 10.1016/j.neuroimage.2006.08.031. [DOI] [PubMed] [Google Scholar]
- Rinberg D, Koulakov A, Gelperin A. Sparse odor coding in awake behaving mice. J Neurosci. 2006;26:8857–65. doi: 10.1523/JNEUROSCI.0884-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittweger E, Han KY, Irvine SE, Eggeling C, Hell SW. STED microscopy reveals crystal colour centers with nanometric resolution. Nat Photonics. 2009;3:144–47. [Google Scholar]
- Rohde CB, Zeng F, Gonzalez-Rubio R, Angel M, Yanik MF. Microfluidic system for on-chip high-throughput whole-animal sorting and screening at subcellular resolution. Proc Natl Acad Sci USA. 2007;104:13891–95. doi: 10.1073/pnas.0706513104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen J, Brooker G. Non-scanning motionless fluorescence three-dimensional holographic microscopy. Nat Photonics. 2008;2:190–95. [Google Scholar]
- Rueckel M, Mack-Bucher JA, Denk W. Adaptive wavefront correction in two-photon microscopy using coherence-gated wavefront sensing. Proc Natl Acad Sci USA. 2006;103:17137–42. doi: 10.1073/pnas.0604791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–95. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabharwal YS, Rouse AR, Donaldson L, Hopkins MF, Gmitro AF. Slit-scanning confocal microendoscope for high-resolution in vivo imaging. Appl Opt. 1999;38:7133–44. doi: 10.1364/ao.38.007133. [DOI] [PubMed] [Google Scholar]
- Sacconi L, Dombeck DA, Webb WW. Overcoming photodamage in second-harmonic generation microscopy: real-time optical recording of neuronal action potentials. Proc Natl Acad Sci USA. 2006;103:3124–29. doi: 10.1073/pnas.0511338103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacconi L, Mapelli J, Gandolfi D, Lotti J, O’Connor RP, et al. Optical recording of electrical activity in intact neuronal networks with random access second-harmonic generation microscopy. Opt Express. 2008;16:14910–21. [PubMed] [Google Scholar]
- Sasaki T, Takahashi N, Matsuki N, Ikegaya Y. Fast and accurate detection of action potentials from somatic calcium fluctuations. J Neurophysiol. 2008;100:1668–76. doi: 10.1152/jn.00084.2008. [DOI] [PubMed] [Google Scholar]
- Sawinski J, Denk W. Miniature random-access fiber scanner for in vivo multiphoton imaging. J Appl Phys. 2007;102:034701. [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, et al. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science. 2008;320:1332–36. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Wurm CA, Jakobs S, Engelhardt J, Egner A, Hell SW. Spherical nanosized focal spot unravels the interior of cells. Nat Methods. 2008;5:539–44. doi: 10.1038/nmeth.1214. [DOI] [PubMed] [Google Scholar]
- Schneider A, Rajendran L, Honsho M, Gralle M, Donnert G, et al. Flotillin-dependent clustering of the amyloid precursor protein regulates its endocytosis and amyloidogenic processing in neurons. J Neurosci. 2008;28:2874–82. doi: 10.1523/JNEUROSCI.5345-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwentker MA, Bock H, Hofmann M, Jakobs S, Bewersdorf J, et al. Wide-field subdiffraction RESOLFT microscopy using fluorescent protein photoswitching. Microsc Res Tech. 2007;70:269–80. doi: 10.1002/jemt.20443. [DOI] [PubMed] [Google Scholar]
- Seidemann E, Arieli A, Grinvald A, Slovin H. Dynamics of depolarization and hyperpolarization in the frontal cortex and saccade goal. Science. 2002;295:862–65. doi: 10.1126/science.1066641. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–9. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- Shao L, Isaac B, Uzawa S, Agard DA, Sedat JW, Gustafsson MGL. I5S: wide-field light microscopy with 100-nm-scale resolution in three dimensions. Biophys J. 2008;94:4971–83. doi: 10.1529/biophysj.107.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J. Optical projection tomography. Annu Rev Biomed Eng. 2004;6:209–28. doi: 10.1146/annurev.bioeng.6.040803.140210. [DOI] [PubMed] [Google Scholar]
- Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, et al. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296:541–45. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Sherman L, Ye JY, Albert O, Norris TB. Adaptive correction of depth-induced aberrations in multi-photon scanning microscopy using a deformable mirror. J Microsc. 2002;206:65–71. doi: 10.1046/j.1365-2818.2002.01004.x. [DOI] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, Betzig E. Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat Methods. 2008;5:417–23. doi: 10.1038/nmeth.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H, Galbraith CG, Galbraith JA, White H, Gillette J, et al. Dual-color superresolution imaging of genetically expressed probes within individual adhesion complexes. Proc Natl Acad Sci USA. 2007;104:20308–13. doi: 10.1073/pnas.0710517105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci USA. 2009;106:3125–30. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber JJ, Willig KI, Heintzmann R, Hell SW, Lang T. The SNARE motif is essential for the formation of syntaxin clusters in the plasma membrane. Biophys J. 2006;90:2843–51. doi: 10.1529/biophysj.105.079574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber JJ, Willig KI, Kutzner C, Gerding-Reimers C, Harke B, et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–76. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- Slovin H, Arieli A, Hildesheim R, Grinvald A. Long-term voltage-sensitive dye imaging reveals cortical dynamics in behaving monkeys. J Neurophysiol. 2002;88:3421–38. doi: 10.1152/jn.00194.2002. [DOI] [PubMed] [Google Scholar]
- Smith SJ. Circuit reconstruction tools today. Curr Opin Neurobiol. 2007;17:601–8. doi: 10.1016/j.conb.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosinsky GE, Giepmans BNG, Deerinck TJ, Gaietta GM, Ellisman MH. Markers for correlated light and electron microscopy. Methods Cell Biol. 2007;79:575–91. doi: 10.1016/S0091-679X(06)79023-9. [DOI] [PubMed] [Google Scholar]
- Spires TL, Meyer-Luehmann M, Stern EA, McLean PJ, Skoch J, et al. Dendritic spine abnormalities in amyloid precursor protein transgenic mice demonstrated by gene transfer and intravital multiphoton microscopy. J Neurosci. 2005;25:7278–87. doi: 10.1523/JNEUROSCI.1879-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell JM, Wokosin DL, White JG, Bavister BD. Long-term two-photon fluorescence imaging of mammalian embryos without compromising viability. Nat Biotechnol. 1999;17:763–67. doi: 10.1038/11698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan VJ, Chen Y, Duker JS, Fujimoto JG. In vivo functional imaging of intrinsic scattering changes in the human retina with high-speed ultrahigh resolution OCT. Opt Express. 2009;17:3861–77. doi: 10.1364/oe.17.003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan VJ, Wojtkowski M, Fujimoto JG, Duker JS. In vivo measurement of retinal physiology with high-speed ultrahigh-resolution optical coherence tomography. Opt Lett. 2006;31:2308–10. doi: 10.1364/ol.31.002308. [DOI] [PubMed] [Google Scholar]
- Stelzer EHK, Lindek S. Fundamental reduction of the observation volume in far-field light microscopy by detection orthogonal to the illumination axis: confocal theta microscopy. Opt Commun. 1994;111:536–47. [Google Scholar]
- Stiel AC, Andresen M, Bock H, Hilbert M, Schilde J, et al. Generation of monomeric reversibly switchable red fluorescent proteins for far-field fluorescence nanoscopy. Biophys J. 2008;95:2989–97. doi: 10.1529/biophysj.108.130146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stosiek C, Garaschuk O, Holthoff K, Konnerth A. In vivo two-photon calcium imaging of neuronal networks. Proc Natl Acad Sci USA. 2003;100:7319–24. doi: 10.1073/pnas.1232232100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subach FV, Patterson GH, Manley S, Gillette JM, Lippincott-Schwartz J, Verkhusha VV. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat Methods. 2009;6:153–59. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MR, Nimmerjahn A, Sarkisov DV, Helmchen F, Wang SS-H. In vivo calcium imaging of circuit activity in cerebellar cortex. J Neurophysiol. 2005;94:1636–44. doi: 10.1152/jn.01013.2004. [DOI] [PubMed] [Google Scholar]
- Sumbre G, Muto A, Baier H, Poo M. Entrained rhythmic activities of neuronal ensembles as perceptual memory of time interval. Nature. 2008;456:102–6. doi: 10.1038/nature07351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun MG, Williams J, Munoz-Pinedo C, Perkins GA, Brown JM, et al. Correlated three-dimensional light and electron microscopy reveals transformation of mitochondria during apoptosis. Nat Cell Biol. 2007;9:1057–65. doi: 10.1038/ncb1630. [DOI] [PubMed] [Google Scholar]
- Tal E, Oron D, Silberberg Y. Improved depth resolution in video-rate line-scanning multiphoton microscopy using temporal focusing. Opt Lett. 2005;30:1686–88. doi: 10.1364/ol.30.001686. [DOI] [PubMed] [Google Scholar]
- Testa I, Schönle A, von Middendorff C, Geisler C, Medda R, et al. Nanoscale separation of molecular species based on their rotational mobility. Opt Express. 2008;16:21093–104. doi: 10.1364/oe.16.021093. [DOI] [PubMed] [Google Scholar]
- Theer P, Hasan MT, Denk W. Two-photon imaging to a depth of 1000 μm in living brains by use of a Ti:Al2O3 regenerative amplifier. Opt Lett. 2003;28:1022–24. doi: 10.1364/ol.28.001022. [DOI] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophys J. 2002;82:2775–83. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni N, Teng EM, Bushong EA, Aimone JB, Zhao C, et al. Synapse formation on neurons born in the adult hippocampus. Nat Neurosci. 2007;10:727–34. doi: 10.1038/nn1908. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, et al. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–94. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Tsai J, Grutzendler J, Duff K, Gan W-B. Fibrillar amyloid deposition leads to local synaptic abnormalities and breakage of neuronal branches. Nat Neurosci. 2004;7:1181–83. doi: 10.1038/nn1335. [DOI] [PubMed] [Google Scholar]
- Tsai PS, Friedman B, Ifarraguerri AI, Thompson BD, Lev-Ram V, et al. All-optical histology using ultrashort laser pulses. Neuron. 2003;39:27–41. doi: 10.1016/s0896-6273(03)00370-2. [DOI] [PubMed] [Google Scholar]
- van de Linde S, Sauer M, Heilemann M. Subdiffraction-resolution fluorescence imaging of proteins in the mitochondrial inner membrane with photoswitchable fluorophores. J Struct Biol. 2008;164:250–54. doi: 10.1016/j.jsb.2008.08.002. [DOI] [PubMed] [Google Scholar]
- van Oijen AM, Köhler J, Schmidt J, Müller M, Brakenhoff GJ. 3-dimensional super-resolution by spectrally selective imaging. Chem Phys Lett. 1998;292:183–87. [Google Scholar]
- van Rijnsoever C, Oorschot V, Klumperman J. Correlative light-electron microscopy (CLEM) combining live-cell imaging and immunolabeling of ultrathin cryosections. Nat Methods. 2008;5:973–80. doi: 10.1038/nmeth.1263. [DOI] [PubMed] [Google Scholar]
- Vaziri A, Tang J, Shroff H, Shank CV. Multilayer three-dimensional super resolution imaging of thick biological samples. Proc Natl Acad Sci USA. 2008;105:20221–26. doi: 10.1073/pnas.0810636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voie AH, Burns DH, Spelman FA. Orthogonal-plane fluorescence optical sectioning: three-dimensional imaging of macroscopic biological specimens. J Microsc. 1993;170:229–36. doi: 10.1111/j.1365-2818.1993.tb03346.x. [DOI] [PubMed] [Google Scholar]
- Volkmer A, Cheng J-X, Xie XS. Vibrational imaging with high sensitivity via epidetected coherent anti-Stokes Raman scattering microscopy. Phys Rev Lett. 2001;87:023901. [Google Scholar]
- von Middendorff C, Egner A, Geisler C, Hell SW, Schönle A. Isotropic 3D nanoscopy based on single emitter switching. Opt Express. 2008;16:20774–88. doi: 10.1364/oe.16.020774. [DOI] [PubMed] [Google Scholar]
- Vučinić D, Bartol TM, Sejnowski TJ. Hybrid reflecting objectives for functional multiphoton microscopy in turbid media. Opt Lett. 2006;31:2447–49. doi: 10.1364/ol.31.002447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vučinić D, Sejnowski TJ. A compact multiphoton 3D imaging system for recording fast neuronal activity. PLoS ONE. 2007;2:e699. doi: 10.1371/journal.pone.0000699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MK, Lichtman JW. In vivo time-lapse imaging of synaptic takeover associated with naturally occurring synapse elimination. Neuron. 2003;37:67–73. doi: 10.1016/s0896-6273(02)01142-x. [DOI] [PubMed] [Google Scholar]
- Wang H, Fu Y, Zickmund P, Shi R, Cheng J-X. Coherent anti-Stokes Raman scattering imaging of axonal myelin in live spinal tissues. Biophys J. 2005;89:581–91. doi: 10.1529/biophysj.105.061911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Huff TB, Fu Y, Jia KY, Cheng J-X. Increasing the imaging depth of coherent anti-Stokes Raman scattering microscopy with a miniature microscope objective. Opt Lett. 2007a;32:2212–14. doi: 10.1364/ol.32.002212. [DOI] [PubMed] [Google Scholar]
- Wang RK, Hurst S. Mapping of cerebro-vascular blood perfusion in mice with skin and skull intact by optical micro-angiography at 1.3 μm wavelength. Opt Express. 2007;15:11402–12. doi: 10.1364/oe.15.011402. [DOI] [PubMed] [Google Scholar]
- Wang RK, Jacques SL, Ma Z, Hurst S, Hanson SR, Gruber A. Three dimensional optical angiography. Opt Express. 2007b;15:4083–97. doi: 10.1364/oe.15.004083. [DOI] [PubMed] [Google Scholar]
- Wang TD, Mandella MJ, Contag CH, Kino GS. Dual-axis confocal microscope for high-resolution in vivo imaging. Opt Lett. 2003;28:414–16. doi: 10.1364/ol.28.000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins LP, Yang H. Information bounds and optimal analysis of dynamic single molecule measurements. Biophys J. 2004;86:4015–29. doi: 10.1529/biophysj.103.037739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb RH, Rogomentich F. Confocal microscope with large field and working distance. Appl Opt. 1999;38:4870–75. doi: 10.1364/ao.38.004870. [DOI] [PubMed] [Google Scholar]
- Weiss S. Fluorescence spectroscopy of single biomolecules. Science. 1999;283:1676–83. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- Westphal V, Hell SW. Nanoscale resolution in the focal plane of an optical microscope. Phys Rev Lett. 2005;94:143903. doi: 10.1103/PhysRevLett.94.143903. [DOI] [PubMed] [Google Scholar]
- Westphal V, Rizzoli SO, Lauterbach MA, Kamin D, Jahn R, Hell SW. Video-rate far-field optical nanoscopy dissects synaptic vesicle movement. Science. 2008;320:246–49. doi: 10.1126/science.1154228. [DOI] [PubMed] [Google Scholar]
- White BR, Pierce MC, Nassif N, Cense B, Park BH, et al. In vivo dynamic human retinal blood flow imaging using ultra-high-speed spectral domain optical Doppler tomography. Opt Express. 2003;11:3490–97. doi: 10.1364/oe.11.003490. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc London Ser B. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Wildanger D, Rittweger E, Kastrup L, Hell SW. STED microscopy with a supercontinuum laser source. Opt Express. 2008;16:9614–21. doi: 10.1364/oe.16.009614. [DOI] [PubMed] [Google Scholar]
- Willig KI, Harke B, Medda R, Hell SW. STED microscopy with continuous wave beams. Nat Methods. 2007;4:915–18. doi: 10.1038/nmeth1108. [DOI] [PubMed] [Google Scholar]
- Willig KI, Rizzoli SO, Westphal V, Jahn R, Hell SW. STED microscopy reveals that synaptotagmin remains clustered after synaptic vesicle exocytosis. Nature. 2006;440:935–39. doi: 10.1038/nature04592. [DOI] [PubMed] [Google Scholar]
- Winship IR, Murphy TH. In vivo calcium imaging reveals functional rewiring of single somatosensory neurons after stroke. J Neurosci. 2008;28:6592–606. doi: 10.1523/JNEUROSCI.0622-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtkowski M, Srinivasan VJ, Ko TH, Fujimoto JG, Kowalczyk A, Duker JS. Ultrahigh-resolution, high-speed, Fourier domain optical coherence tomography and methods for dispersion compensation. Opt Express. 2004;12:2404–22. doi: 10.1364/opex.12.002404. [DOI] [PubMed] [Google Scholar]
- Wright AJ, Poland SP, Girkin JM, Freudiger CW, Evans CL, Xie XS. Adaptive optics for enhanced signal in CARS microscopy. Opt Express. 2007;15:18209–19. doi: 10.1364/oe.15.018209. [DOI] [PubMed] [Google Scholar]
- Xu H-T, Pan F, Yang G, Gan W-B. Choice of cranial window type for in vivo imaging affects dendritic spine turnover in the cortex. Nat Neurosci. 2007;10:549–51. doi: 10.1038/nn1883. [DOI] [PubMed] [Google Scholar]
- Yaksi E, Friedrich RW. Reconstruction of firing rate changes across neuronal populations by temporally deconvolved Ca2+ imaging. Nat Methods. 2006;3:377–83. doi: 10.1038/nmeth874. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Kobayashi M, Mitsui S, Ishida Y, van der Horst GTJ, et al. View of a mouse clock gene ticking. Nature. 2001;409:684. doi: 10.1038/35055628. [DOI] [PubMed] [Google Scholar]
- Yang Z, Heeger DJ, Seidemann E. Rapid and precise retinotopic mapping of the visual cortex obtained by voltage-sensitive dye imaging in the behaving monkey. J Neurophysiol. 2007;98:1002–14. doi: 10.1152/jn.00417.2007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X-C, Yamauchi A, Perry B, George JS. Rapid optical coherence tomography and recording functional scattering changes from activated frog retina. Appl Opt. 2005;44:2019–23. doi: 10.1364/ao.44.002019. [DOI] [PubMed] [Google Scholar]
- Yaroslavsky AN, Schulze PC, Yaroslavsky IV, Schober R, Ulrich F, Schwarzmaier H-J. Optical properties of selected native and coagulated human brain tissues in vitro in the visible and near infrared spectral range. Phys Med Biol. 2002;47:2059–73. doi: 10.1088/0031-9155/47/12/305. [DOI] [PubMed] [Google Scholar]
- Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–65. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- Young P, Qiu L, Wang D, Zhao S, Gross J, Feng G. Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nat Neurosci. 2008;11:721–28. doi: 10.1038/nn.2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F, Salehi HA, Boucher Y, Vasthare US, Tuma RF, Jain RK. Vascular permeability and microcirculation of gliomas and mammary carcinomas transplanted in rat and mouse cranial windows. Cancer Res. 1994;54:4564–68. [PubMed] [Google Scholar]
- Zawadzki RJ, Cense B, Zhang Y, Choi SS, Miller DT, Werner JS. Ultrahigh-resolution optical coherence tomography with monochromatic and chromatic aberration correction. Opt Express. 2008;16:8126–43. doi: 10.1364/oe.16.008126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Rohde CB, Yanik MF. Sub-cellular precision on-chip small-animal immobilization, multiphoton imaging and femtosecond-laser manipulation. Lab Chip. 2008;8:653–56. doi: 10.1039/b804808h. [DOI] [PubMed] [Google Scholar]
- Zeng S, Lv X, Zhan C, Chen WR, Xiong W, et al. Simultaneous compensation for spatial and temporal dispersion of acousto-optical deflectors for two-dimensional scanning with a single prism. Opt Lett. 2006;31:1091–93. doi: 10.1364/ol.31.001091. [DOI] [PubMed] [Google Scholar]
- Zhang F, Aravanis AM, Adamantidis A, de Lecea L, Deisseroth K. Circuit-breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci. 2007;8:577–81. doi: 10.1038/nrn2192. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhou X, Lu J, Lichtman JW, Adjeroh D, Wong STC. 3D axon structure extraction and analysis in confocal fluorescence microscopy images. Neural Comput. 2008;20:1899–927. doi: 10.1162/neco.2008.05-07-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZG, Zhang L, Ding G, Jiang Q, Zhang RL, et al. A model of mini-embolic stroke offers measurements of the neurovascular unit response in the living mouse. Stroke. 2005;36:2701–4. doi: 10.1161/01.STR.0000190007.18897.e3. [DOI] [PubMed] [Google Scholar]
- Zhu G, van Howe J, Durst M, Zipfel W, Xu C. Simultaneous spatial and temporal focusing of femtosecond pulses. Opt Express. 2005;13:2153–59. doi: 10.1364/opex.13.002153. [DOI] [PubMed] [Google Scholar]
- Zhu L, Sun P-C, Bartsch D-U, Freeman WR, Fainman Y. Adaptive control of a micromachined continuous-membrane deformable mirror for aberration compensation. Appl Opt. 1999;38:168–76. doi: 10.1364/ao.38.000168. [DOI] [PubMed] [Google Scholar]
- Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–92. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Zumbusch A, Holtom GR, Xie XS. Three-dimensional vibrational imaging by coherent anti-Stokes Raman scattering. Phys Rev Lett. 1999;82:4142–45. [Google Scholar]
- Zuo Y, Lin A, Chang P, Gan W-B. Development of long-term dendritic spine stability in diverse regions of cerebral cortex. Neuron. 2005a;46:181–89. doi: 10.1016/j.neuron.2005.04.001. [DOI] [PubMed] [Google Scholar]
- Zuo Y, Yang G, Kwon E, Gan W-B. Long-term sensory deprivation prevents dendritic spine loss in primary somatosensory cortex. Nature. 2005b;436:261–65. doi: 10.1038/nature03715. [DOI] [PubMed] [Google Scholar]