Abstract
BACKGROUND
A recent genomewide mutational analysis of glioblastomas (World Health Organization [WHO] grade IV glioma) revealed somatic mutations of the isocitrate dehydrogenase 1 gene (IDH1) in a fraction of such tumors, most frequently in tumors that were known to have evolved from lower-grade gliomas (secondary glioblastomas).
METHODS
We determined the sequence of the IDH1 gene and the related IDH2 gene in 445 central nervous system (CNS) tumors and 494 non-CNS tumors. The enzymatic activity of the proteins that were produced from normal and mutant IDH1 and IDH2 genes was determined in cultured glioma cells that were transfected with these genes.
RESULTS
We identified mutations that affected amino acid 132 of IDH1 in more than 70% of WHO grade II and III astrocytomas and oligodendrogliomas and in glioblastomas that developed from these lower-grade lesions. Tumors without mutations in IDH1 often had mutations affecting the analogous amino acid (R172) of the IDH2 gene. Tumors with IDH1 or IDH2 mutations had distinctive genetic and clinical characteristics, and patients with such tumors had a better outcome than those with wild-type IDH genes. Each of four tested IDH1 and IDH2 mutations reduced the enzymatic activity of the encoded protein.
CONCLUSIONS
Mutations of NADP+-dependent isocitrate dehydrogenases encoded by IDH1 and IDH2 occur in a majority of several types of malignant gliomas.
GLIOMAS, THE MOST COMMON TYPE OF primary brain tumors, are classified as grade I to grade IV on the basis of histopathological and clinical criteria established by the World Health Organization (WHO).1 This group of tumors includes specific histologic subtypes, the most common of which are astrocytomas, oligodendrogliomas, and ependymomas. WHO grade I gliomas, often considered to be benign, are generally curable with complete surgical resection and rarely, if ever, evolve into higher-grade lesions.2 By contrast, gliomas of WHO grade II or III are invasive, progress to higher-grade lesions, and have a poor outcome. WHO grade IV tumors (glioblastomas), the most invasive form, have a dismal prognosis.3,4 On the basis of histopathological criteria, it is impossible to distinguish a secondary glioblastoma, defined as a tumor that was previously diagnosed as a lower-grade glioma, from a primary tumor.5,6
Several genes, including TP53, PTEN, CDKN2A, and EGFR, are altered in gliomas.7-12 These alterations tend to occur in a defined order during the progression to a high-grade tumor. The TP53 mutation appears to be a relatively early event during the development of an astrocytoma, whereas the loss or mutation of PTEN and amplification of EGFR are characteristic of higher-grade tumors.6,13,14 In oligodendrogliomas, allelic losses of 1p and 19q occur in many WHO grade II tumors, whereas losses of 9p21 are largely confined to WHO grade III tumors.15
In a recent genomewide analysis, we identified somatic mutations at codon 132 of the isocitrate dehydrogenase 1 gene (IDH1) in approximately 12% of glioblastomas.16 These mutations were also found in five of six secondary glioblastomas. The results suggested that IDH1 mutations might occur after formation of a low-grade glioma and drive the progression of the tumor to a glioblastoma. To evaluate this possibility, we analyzed a large number of gliomas of various types.
METHODS
DNA SAMPLES
DNA was extracted from samples of primary brain tumor and xenografts and from patient-matched normal blood lymphocytes obtained from the Tissue Bank at the Preston Robert Tisch Brain Tumor Center at Duke University and collaborating centers, as described previously.17 All analyzed brain tumors were subjected to consensus review by two neuropathologists. Table 1 lists the types of brain tumors we analyzed. The samples from glioblastomas included 138 primary tumors and 13 secondary tumors. Of the 138 primary tumors, 15 were from patients under the age of 21 years. Secondary glioblastomas were categorized as WHO grade IV on the basis of histologic criteria but had been categorized as WHO grade II or III at least 1 year earlier. Of the 151 tumors, 63 had been analyzed in our previous genomewide mutation analysis of glioblastomas. None of the lower-grade tumors were included in that analysis.16
Table 1.
Summary of Genetic and Clinical Characteristics of Brain Tumors in the Study.*
| Tumor Classification† | No. of Tumors Analyzed | Median Age of Patient‡ | Male Sex | Median Survival | Tumors with IDH Mutations | Median Age of Patient | Tumors with Other Alterations§ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IDH1 | IDH2 | Combined | Mutated IDH | Wild-Type IDH | TP53 | 1p and 19q | PTEN | EGFR | CDKN2A or CDKN2B | |||||
| yr | % | mo | no. | % | yr | % | ||||||||
| Astrocytic tumors | ||||||||||||||
| Pilocytic astrocytoma (grade I) | 21 | 5 | 48 | ND | 0 | 0 | 0 | ND | 5 | 0 | NA | 0 | 0 | NA |
| Subependymal giant-cell astrocytoma (grade I) | 2 | 16 | 100 | ND | 0 | NA | 0 | ND | ND | NA | NA | NA | NA | NA |
| Diffuse astrocytoma (grade II) | 30 | 34 | 53 | 132 | 25 | 2 | 90 | 35 | 5 | 74 | 0 | 0 | 0 | 0 |
| Pleomorphic xanthoastrocytoma (grade II) | 7 | 11 | 14 | 44 | 1 | NA | 14 | 20 | 11 | NA | NA | NA | 0 | NA |
| Anaplastic astrocytoma (grade III) | 52 | 38 | 67 | 51 | 36 | 2 | 73 | 34 | 56 | 65 | 10 | 9 | 2 | 9 |
| Secondary glioblastoma (grade IV)¶ | 13 | 33 | 70 | 16 | 11 | 0 | 85 | 32 | 62 | 62 | NA | 0 | 0 | 20 |
| Primary adult glioblastoma (grade IV) | 123 | 59 | 60 | 15 | 6 | 0 | 5 | 32 | 59 | 23 | 4 | 23 | 38 | 42 |
| Primary pediatric glioblastoma (grade IV) | 15 | 5 | 60 | 8 | 0 | 0 | 0 | ND | 5 | 33 | NA | NA | NA | 20 |
| Oligodendroglial tumors | ||||||||||||||
| Oligodendroglioma (grade II) | 51 | 37 | 63 | 135 | 41 | 2 | 84 | 37 | 13.5 | 16 | 60 | 0 | 0 | 4 |
| Anaplastic oligodendroglioma (grade III) | 36 | 45 | 64 | 84 | 31 | 3 | 94 | 45 | ND | 10 | 84 | 0 | 0 | 14 |
| Oligoastrocytic tumors | ||||||||||||||
| Oligoastrocytoma (grade II) | 3 | 38 | 67 | ND | 3 | NA | 100 | 38 | ND | 33 | NA | 0 | 0 | 0 |
| Anaplastic oligoastrocytoma (grade III) | 7 | 30 | 57 | ND | 7 | NA | 100 | 30 | ND | 71 | 50 | 0 | 0 | 0 |
| Ependymoma (grade II) | 30 | 5.5 | 45 | ND | 0 | 0 | 0 | ND | 5.5 | 0 | NA | 0 | NA | NA |
| Medulloblastoma (grade IV) | 55 | 7 | 65 | 27 | 0 | 0 | 0 | ND | 7 | NA | NA | NA | NA | NA |
Of the indicated tumors, 6 secondary and 60 primary glioblastomas were previously described in Parsons et al.16 Copy-number changes in EGFR, CDKN2A, and CDKN2B were determined by quantitative real-time polymerase chain reaction. For such assays, copy-number levels of more than 6 or less than 0.3 were considered amplifications or losses, respectively. NA denotes not analyzed, and ND not determined because of limited sample size and status of data censoring.
Tumors were graded according to histopathological and clinical criteria established by the World Health Organization.
Patient age refers to age at which the study sample was obtained.
Alterations included mutations in TP53 and PTEN, loss of heterozygosity in 1p and 19q, amplification in EGFR, and deletion in CDKN2A or CDKN2B.
Secondary glioblastoma designates a tumor that was resected more than 1 year after a previous diagnosis of a lower-grade glioma (grade II or grade III).
In addition to brain tumors, we analyzed 35 lung cancers, 57 gastric cancers, 27 ovarian cancers, 96 breast cancers, 114 colorectal cancers, 95 pancreatic cancers, and 7 prostate cancers, along with 4 samples from patients with chronic myelogenous leukemia, 7 from patients with chronic lymphocytic leukemia, 7 from patients with acute lymphoblastic leukemia, and 45 from patients with acute myelogenous leukemia. All samples were obtained in accordance with the Health Insurance Portability and Accountability Act. Acquisition of tissue specimens was approved by the institutional review board at the Duke University Health System and at each of the participating institutions.
Exon 4 of the IDH1 gene was amplified with the use of a polymerase-chain-reaction (PCR) assay and sequenced in DNA from the tumor and lymphocytes from each patient, as described previously.16 In all gliomas and medulloblastomas without an R132 IDH1 mutation, exon 4 of the IDH2 gene (which contains the IDH2 residue equivalent to R132 of IDH1) was sequenced and analyzed for somatic mutations. In addition, we evaluated all astrocytomas and oligodendrogliomas of WHO grade I to grade III, all secondary glioblastomas, and 96 primary glioblastomas without R132 IDH1 mutations or R172 IDH2 mutations for alterations in the remaining coding exons of IDH1 and IDH2. All coding exons of TP53 and PTEN were also sequenced in the panel of diffuse astrocytomas, oligodendrogliomas, anaplastic oligodendrogliomas, anaplastic astrocytomas, and glioblastomas. EGFR amplification and the CDKN2A-CDKN2B deletion were analyzed with the use of quantitative real-time PCR in the same tumors.18 We evaluated samples of oligodendrogliomas and anaplastic oligodendrogliomas for loss of heterozygosity at 1p and 19q, as described previously.15,19
ENZYMATIC ACTIVITY
To assess the enzymatic activity of wild-type and mutant IDH1 and IDH2 proteins, a human oligodendroglioma line without IDH1 or IDH2 mutations was transfected with a vector (pCMV6, Invitrogen) containing the coding sequences of the wild-type IDH1, wild-type IDH2, or mutant IDH genes (corresponding to the most common IDH1 mutation, R132H, or the IDH2 mutations R172G, R172K, and R172M). Clones of the wild-type IDH1 and IDH2 genes were obtained from Origene, and mutations were introduced by standard methods.
Cells were collected 48 hours after transfection, subjected to centrifugation at 1000×g for 10 minutes at 4°C, washed once with cold phosphate-buffered saline, and lysed in buffer containing 0.1% Triton X-100. They were then disrupted by ultrasonication and centrifuged at 12,000×g for 30 minutes. The supernatants were used to measure IDH activity. Expression levels of wild-type and mutant IDH proteins were determined by Western blotting with the use of an antibody against FLAG, a polypeptide protein tag. For each enzymatic reaction, a volume of cell lysate containing the same amount of IDH protein was added to 1 ml of assay solution containing 33 mM of Tris buffer, 0.33 mM of EDTA, 0.1 mM of NADP+, 1.33 mM of manganese chloride, and 1.3 mM of isocitrate. The activity of IDH was analyzed through the reduction of NADP+ to NADPH, which was measured at 25°C by spectrophotometry at 340 nm 5 times a second for 300 seconds.20
CLINICAL DATA AND SURVIVAL
Clinical information included the date of birth, the date the study sample was obtained, the date of pathological diagnosis, the date and pathology of any preceding diagnosis of a lower-grade glioma, the use or nonuse of radiation therapy or chemotherapy before the date that the study sample was obtained, the date of the last contact with the patient, and the patient's status at the time of the last contact. We calculated overall survival for patients with anaplastic astrocyomas, including 38 patients with mutations in IDH1 or IDH2 and 14 with wild-type genes, and for adult patients (≥21 years of age) with glioblastomas, including 14 patients with mutations in IDH1 or IDH2 and 115 with wild-type genes, using the date of histologic diagnosis and the date of the last contact with the patient or death. For patients with secondary glioblastomas, survival was calculated from the date of secondary diagnosis. Seven patients with glioblastomas were not included in the statistical analysis because of insufficient survival data.
STUDY DESIGN
The authors designed the study, gathered and analyzed the data, wrote the manuscript, and made the decision to publish the findings. Gene sequencing was performed by Agencourt Bioscience, a subsidiary of Beckman Coulter. The lead academic authors vouch for the completeness and accuracy of the data and the analyses.
STATISTICAL ANALYSIS
We examined the association between the occurrence of mutations in IDH1 or IDH2 and other genetic alterations using Fisher's exact test. Kaplan-Meier survival curves were plotted and the survival distributions were compared with the use of the Mantel-Cox log-rank test and the Wilcoxon test. All reported P values are two-sided, and P values of less than 0.01 were considered to indicate statistical significance.
RESULTS
SEQUENCE ANALYSIS
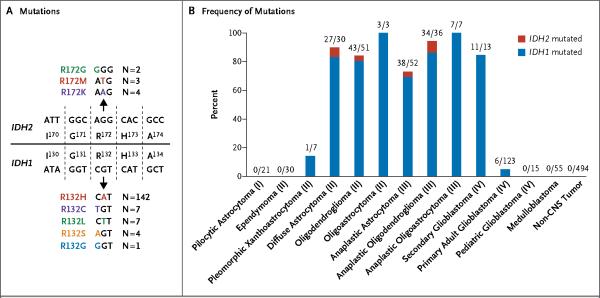
Sequence analysis of IDH1 in 939 tumor samples revealed 161 somatic mutations at residue R132, including R132H (142 tumors), R132C (7 tumors), R132S (4 tumors), R132L (7 tumors), and R132G (1 tumor) (Fig. 1A; and Fig. 1 in the Supplementary Appendix, available with the full text of this article at NEJM.org). Table 1 and Figure 1B show the tumors with somatic R132 mutations. No other somatic mutations of IDH1 in the remaining IDH1 exons of R132-negative tumors were found in all WHO grade I to grade III astrocytomas and oligodendrogliomas, in all secondary glioblastomas, and in 96 primary glioblastomas. No R132 mutations were observed in 21 pilocytic astrocytomas (WHO grade I), 2 subependymal giant-cell astrocytomas (WHO grade I), 30 ependymomas (WHO grade II), 55 medulloblastomas, or any of the 494 non-central nervous system tumor samples.
Figure 1. IDH1 and IDH2 Mutations in Human Gliomas.
Panel A shows mutations at codon R132 in IDH1 and R172 in IDH2 that were identified in human gliomas, along with the number of patients who carried each mutation. Codons 130 to 134 of IDH1 and 170 to 174 of IDH2 are shown. Panel B shows the number and frequency of IDH1 and IDH2 mutations in gliomas and other types of tumors. The roman numerals in parentheses are the tumor grades, according to histopathological and clinical criteria established by the World Health Organization. CNS denotes central nervous system.
We also sought alterations in other genes with functions similar to those of IDH1 in tumors without IDH1 mutations. For this purpose, we analyzed the IDH2 gene, which encodes the only human protein homologous to IDH1 that uses NADP+ as an electron acceptor. Sequence evaluation of all IDH2 exons in these glioma samples revealed nine somatic mutations of IDH2, all at residue R172: R172G in two tumors, R172M in three tumors, and R172K in four tumors (Fig. 1A, and Fig. 1 in the Supplementary Appendix). The R172 residue in IDH2 is the exact analogue of the R132 residue in IDH1, which is located in the active site of the enzyme and forms hydrogen bonds with the isocitrate substrate.21
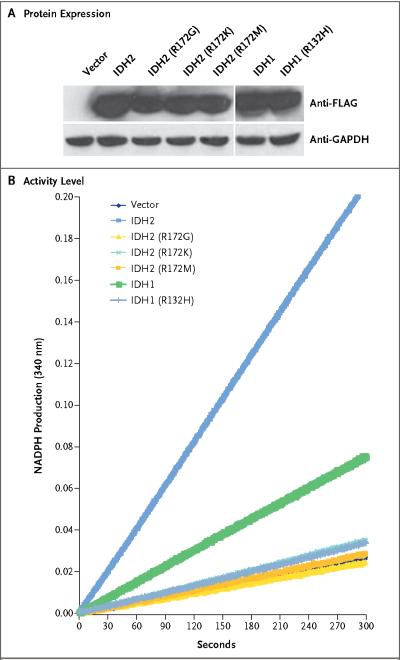
To determine whether the mutations in IDH1 and IDH2 disturb the function of the corresponding proteins, we measured the enzymatic activity (reduction of NADP+ to NADPH) of IDH1 and IDH2 proteins in an oligodendroglioma line that had been transfected with wild-type or mutant IDH1 or IDH2 genes. These mutants represented 88% of the IDH1 mutations and 100% of the IDH2 mutations found in patients. Figure 2 shows that exogenous expression of wild-type IDH1 or IDH2 significantly increased the production of NADPH, whereas only endogenous IDH activity was observed in cells that had been transfected with mutant IDH1 or IDH2 genes.
Figure 2. Enzymatic Activity of Wild-Type and Mutant IDH1 and IDH2 Proteins.
Cell lysates were extracted from a human oligodendroglioma cell line without IDH1 or IDH2 mutations that had been transfected with vectors encoding the indicated proteins. Panel A shows the expression of proteins encoded by wild-type and mutant IDH1 and IDH2, as determined by Western blotting, with the use of an anti-FLAG antibody. Panel B shows the activity levels of these proteins, as analyzed by monitoring the production of NADPH. GAPDH denotes glyceraldehyde 3-phosphate dehydrogenase.
To further evaluate IDH alterations during glioma progression, we assessed IDH1 mutations in seven progressive gliomas in which both lowgrade and high-grade tumor samples were available. Sequence analysis identified IDH1 mutations in both the low-grade and high-grade tumors in all seven cases (Table 1, and Fig. 2 in the Supplementary Appendix). These results demonstrate that IDH1 alterations in high-grade tumors are derived from the earlier lesions.
We also examined diffuse astrocytomas, oligodendrogliomas, anaplastic oligodendrogliomas, anaplastic astrocytomas, and a subgroup of glioblastomas for mutations in TP53 and PTEN, amplification of EGFR, deletion of CDKN2A-CDKN2B, and allelic losses of 1p and 19q (Table 1). TP53 mutations were more common in diffuse astrocytomas (74%), anaplastic astrocytomas (65%), and secondary glioblastomas (62%) than in oligodendrogliomas (16%) or anaplastic oligodendrogliomas (9%) (P<0.001 for all comparisons by Fisher's exact test). Conversely, deletions of 1p and 19q were found more often in oligodendrocytic than in astrocytic tumors, as expected.15
Most (80%) of the anaplastic astrocytomas and glioblastomas with mutated IDH1 or IDH2 genes also had a mutation of TP53, but only 3% had alterations in PTEN, EGFR, CDKN2A, or CDKN2B (Table 2). Conversely, anaplastic astrocytomas and glioblastomas with wild-type IDH1 and IDH2 genes had few TP53 mutations (18%) and more frequent alterations of PTEN, EGFR, CDKN2A, or CDKN2B (74%) (P<0.001 for both comparisons by Fisher's exact test). Loss of 1p and 19q was observed in 45 of 53 (85%) of the oligodendrocytic tumors with mutated IDH1 or IDH2 but in none of the tumors with wild-type IDH genes (P<0.001 by Fisher's exact test).
Table 2.
Frequency of Common Genetic Alterations in Gliomas with Mutated or Wild-Type IDH1 and IDH2 Genes.*
| Tumor Type and IDH1 or IDH2 Mutational Status† | Total Patients | Location of Other Alterations‡ | ||||
|---|---|---|---|---|---|---|
| TP53 | PTEN | EGFR | CDKN2A or CDKN2B | 1p and 19q | ||
| no. | no./total no. (%) | |||||
| Astrocytic tumors | ||||||
| Diffuse astrocytoma (grade II) | ||||||
| Mutant | 27 | 17/20 (85) | 0/19 | 0/23 | 0/23 | 1/19 (5) |
| Wild-type | 3 | 0/3 | 0/2 | 0/3 | 0/3 | 0/3 |
| Total | 30 | 17/23 (74) | 0/21 | 0/26 | 0/26 | 1/22 (5) |
| Anaplastic astrocytoma (grade III) | ||||||
| Mutant | 38 | 23/28 (82) | 0/22 | 0/35 | 0/33 | 1/22 (5) |
| Wild-type | 14 | 3/12 (25) | 3/11 (27) | 1/12 (8) | 4/12 (33) | 1/6 (17) |
| Total | 52 | 26/40 (65) | 3/33 (9) | 1/47 (2) | 4/45 (9) | 1/28 (4) |
| Primary adult glioblastoma (grade IV) | ||||||
| Mutant | 6 | 5/6 (83) | 0/5 | 0/6 | 0/6 | 1/2 (50) |
| Wild-type | 117 | 23/117 (20) | 21/88 (24) | 35/86 (41) | 39/87 (45) | 0/24 |
| Total | 123 | 28/123 (23) | 21/93 (23) | 35/92 (38) | 39/93 (42) | 1/26 (4) |
| Secondary adult glioblastoma (grade IV) | ||||||
| Mutant | 11 | 8/11 (73) | 0/6 | 0/10 | 1/8 (12) | NA |
| Wild-type | 2 | 0/2 | 1/2 (50) | 0/2 | 1/2 (50) | NA |
| Total | 13 | 8/13 (62) | 1/8 (12) | 0/12 | 2/10 (20) | NA |
| Oligodendroglial tumors | ||||||
| Oligodendroglioma (grade II) | ||||||
| Mutant | 43 | 5/24 (21) | 0/20 | 0/43 | 2/40 (5) | 18/23 (78) |
| Wild-type | 8 | 0/8 | 0/7 | 0/8 | 0/8 | 0/7 |
| Total | 51 | 5/32 (16) | 0/27 | 0/51 | 2/48 (4) | 18/30 (60) |
| Anaplastic oligodendroglioma (grade III) | ||||||
| Mutant | 34 | 3/30 (10) | 0/28 | 0/33 | 3/33 (9) | 27/30 (90) |
| Wild-type | 2 | 0/2 | 0/2 | 0/2 | 2/2 (100) | 0/2 |
| Total | 36 | 3/32 (9) | 0/30 | 0/35 | 5/35 (14) | 27/32 (84) |
All tumors were analyzed for IDH1 R132 and IDH2 R172 mutations. In addition, all pilocytic astrocytomas, diffuse astrocytomas, oligodendrogliomas, anaplastic oligodendrogliomas, anaplastic astrocytomas, secondary glioblastomas, and 96 primary glioblastomas were evaluated for mutations in the remaining coding exons of IDH1 and IDH2. NA denotes not analyzed.
Tumors were graded according to histopathological and clinical criteria established by the World Health Organization.
Alterations included mutations in TP53 and PTEN, loss of heterozygosity in 1p and 19q, amplification in EGFR, and deletion in CDKN2A or CDKN2B.
Patients with anaplastic astrocytomas or glioblastomas with IDH1 or IDH2 mutations were significantly younger than were patients with tumors carrying wild-type IDH1 and IDH2 genes (median age, 34 years vs. 56 years for patients with anaplastic astrocytomas and 32 years vs. 59 years for those with glioblastomas; P<0.001 for both comparisons by Student's t-test). Despite the lower median age of patients with IDH1 or IDH2 mutations, no mutations were identified in glioblastomas from the 15 patients who were under the age of 21 (Fig. 3 in the Supplementary Appendix). In patients with oligodendrogliomas or anaplastic oligodendrogliomas, the median age of the patients with IDH1 or IDH2 mutation was 39 years; IDH1 mutations were identified in two teenagers (14 and 16 years) but not in four younger patients.
Our previous observation of improved outcome for patients whose glioblastomas carried the IDH1 mutation16 was confirmed in this larger data set and extended to include such patients with mutations in IDH2. Patients with a glioblastoma carrying an IDH1 or IDH2 mutation had a median overall survival of 31 months, which was significantly longer than the 15-month survival in patients with wild-type IDH1 (P = 0.002 by the log-rank test) (Fig. 3A). Mutations of IDH genes were also associated with improved outcome in patients with anaplastic astrocytomas; the median overall survival was 65 months for patients with mutations and 20 months for those without mutations (P<0.001 by the log-rank test) (Fig. 3B). Differential survival analyses could not be performed in patients with diffuse astrocytomas, oligodendrogliomas, or anaplastic oligodendrogliomas because there were too few tumors of these types without IDH gene mutations.
Figure 3. Survival of Adult Patients with Malignant Gliomas with or without IDH Gene Mutations.
For patients with glioblastomas, the median survival was 31 months for the 14 patients with mutated IDH1 or IDH2, as compared with 15 months for the 115 patients with wild-type IDH1 or IDH2 (Panel A). For patients with anaplastic astrocytomas, the median survival was 65 months for the 38 patients with mutated IDH1 or IDH2, as compared with 20 months for the 14 patients with wild-type IDH1 or IDH2 (Panel B). Patients with both primary and secondary tumors were included in the analysis. For patients with secondary glioblastomas, survival was calculated from the date of the secondary diagnosis. Survival distributions were compared with the use of the logrank test.
DISCUSSION
Our findings implicate mutations in the NADP+-dependent isocitrate dehydrogenase genes, IDH1 and IDH2, in the pathogenesis of malignant gliomas. Gliomas with IDH mutations were clinically and genetically distinct from gliomas with wild-type IDH genes. Notably, two subtypes of gliomas of WHO grade II or III (astrocytomas and oligodendrogliomas) often carried IDH mutations but not other genetic alterations that are detectable relatively early during the progression of gliomas. This finding suggests that IDH mutations occur early in the development of a glioma from a stem cell that can give rise to both astrocytes and oligodendrocytes. The identification of IDH1 mutations in 10 of 10 oligoastrocytomas and anaplastic oligoastrocytomas, tumors with morphologic features of both cell types, supports this conjecture.
Mutations in IDH1 or IDH2 were not identified in any pilocytic astrocytomas of WHO grade I, indicating that these tumors arise through a different mechanism. This conclusion is consistent with clinical observations that pilocytic astrocytomas rarely if ever undergo malignant transformation2 and with recent data indicating that a duplication at 7q34 producing a BRAF fusion gene occurs frequently in pilocytic astrocytomas but not higher-grade gliomas.22
In each of the tested mutations, the enzymatic activity of the IDH proteins was eliminated. A previous study showed that in vitro substitution of glutamate for arginine at residue 132 of IDH1 (an alteration not observed in patients) resulted in a catalytically inactive enzyme.23 Although our results demonstrate an effect of the mutations on the function of the IDH1 protein, they do not necessarily mean that the mutations are inactivating. For example, the mutant proteins that preclude the use of isocitrate as substrate could allow other, asyet-unknown substrates to be used by the enzyme, thereby conferring a gain rather than a loss of activity. If future studies confirm this possibility, mutant IDH could become a target for therapeutic intervention.
Our results have important practical implications. Historically, glioblastomas have been divided into cancers that arise from low-grade gliomas (secondary tumors) and those without such an antecedent (primary tumors).5,6 Secondary tumors account for only 5% of all glioblastomas. The finding that IDH1 or IDH2 is mutated in the vast majority of WHO grade II or III gliomas and in the secondary glioblastomas that develop from these precursors provides a biologic explanation for this clinical categorization: tumors with mutated NADP+-dependent isocitrate dehydrogenases comprise a specific subgroup of glioblastomas.
The localization of IDH1 and IDH2 mutations to a single amino acid (R132 and R172, respectively) simplifies the use of this genetic alteration for diagnostic purposes. For example, IDH mutation tests could help distinguish pilocytic astrocytomas (WHO grade I) from diffuse astrocytomas (WHO grade II), since these lesions can sometimes be difficult to categorize solely on the basis of histopathological criteria.2
Supplementary Material
Acknowledgments
Supported by a grant from the Pediatric Brain Tumor Foundation Institute, a Damon Runyon Foundation Scholar Award, a grant from the Southeastern Brain Tumor Foundation, Alex's Lemonade Stand Foundation, a grant from the V Foundation for Cancer Research, the Virginia and D.K. Ludwig Fund for Cancer Research, the Pew Charitable Trusts, the American Brain Tumor Association, the Brain Tumor Research Fund at Johns Hopkins, grants (R01CA118822, NS20023-21, R37CA11898-34, CA121113, CA43460, CA57345, 5P50-CA-108786, 5P50-NS-20023, 5R37-CA-11898, and 2P30-CA-14236) from Beckman Coulter, and grants from the Accelerate Brain Cancer Cure Foundation.
Drs. Yan, Parsons, Jones, Kinzler, Velculescu, Vogelstein, and Bigner report being eligible for royalties received by Johns Hopkins University on sales of products related to research described in this article, under licensing agreements between the university and Beckman Coulter. No other potential conflict of interest relevant to this article was reported.
The views expressed in this article are those of the authors and do not necessarily represent the official views of the Food and Drug Administration.
We thank Melissa J. Ehinger, Diane L. Satterfield, Eric Lipp, Michael Leonard, Jennifer D. Funkhouser, and Patrick Killela for their assistance with the clinical information; P. Buckhaults, S. Powell, S. Kern, J. Eshleman, C. Civin, I.-M. Shih, and A. Gazdar for providing DNA from non-central nervous system cancers; and Dr. A.T. Campagnoni at the University of California, Los Angeles, for donating the human oligodendroglioma cell line.
REFERENCES
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, editors. WHO classifcation of tumours of the central nervous system. 4th ed. IARC Press; Lyon, France: 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burger PC, Scheithauer BW, Paulus W, et al. Pilocytic astrocytoma. In: Kleihues P, Cavenee WK, editors. Pathology and genetics of tumours of the nervous system. IARC Press; Lyon, France: 2000. pp. 45–51. [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359:492–507. doi: 10.1056/NEJMra0708126. [Erratum, N Engl J Med 2008;359:877] [DOI] [PubMed] [Google Scholar]
- 5.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–9. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 6.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–53. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 9.Nigro JM, Baker SJ, Preisinger AC, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705–8. doi: 10.1038/342705a0. [DOI] [PubMed] [Google Scholar]
- 10.Ueki K, Ono Y, Henson JW, Efird JT, von Deimling A, Louis DN. CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res. 1996;56:150–3. [PubMed] [Google Scholar]
- 11.Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84:6899–903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong AJ, Ruppert JM, Bigner SH, et al. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A. 1992;89:2965–9. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furnari FB, Fenton T, Bachoo RM, et al. Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev. 2007;21:2683–710. doi: 10.1101/gad.1596707. [DOI] [PubMed] [Google Scholar]
- 14.Weber RG, Sabel M, Reifenberger J, et al. Characterization of genomic alterations associated with glioma progression by comparative genomic hybridization. Oncogene. 1996;13:983–94. [PubMed] [Google Scholar]
- 15.Bigner SH, Matthews MR, Rasheed BK, et al. Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am J Pathol. 1999;155:375–86. doi: 10.1016/S0002-9440(10)65134-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sjöblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–74. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 18.Wang TL, Diaz LA, Jr, Romans K, et al. Digital karyotyping identifies thymidylate synthase amplification as a mechanism of resistance to 5-fluorouracil in metastatic colorectal cancer patients. Proc Natl Acad Sci USA. 2004;101:3089–94. doi: 10.1073/pnas.0308716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reifenberger J, Reifenberger G, Liu L, James CD, Wechsler W, Collins VP. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145:1175–90. [PMC free article] [PubMed] [Google Scholar]
- 20.Batinic-Haberle I, Benov LT. An SOD mimic protects NADP+-dependent isocitrate dehydrogenase against oxidative inactivation. Free Radic Res. 2008;42:618–24. doi: 10.1080/10715760802209639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu X, Zhao J, Xu Z, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel selfregulatory mechanism of activity. J Biol Chem. 2004;279:33946–57. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 22.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–7. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jennings GT, Minard KI, McAlister-Henn L. Expression and mutagenesis of mammalian cytosolic NADP+-specific isocitrate dehydrogenase. Biochemistry. 1997;36:13743–7. doi: 10.1021/bi970916r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.