Abstract
The measurement of circulating nucleic acids has transformed the management of chronic viral infections such as HIV. The development of analogous markers for individuals with cancer could similarly enhance the management of their disease. DNA containing somatic mutations is highly tumor specific and thus, in theory, can provide optimum markers. However, the number of circulating mutant gene fragments is small compared to the number of normal circulating DNA fragments, making it difficult to detect and quantify them with the sensitivity required for meaningful clinical use. In this study, we applied a highly sensitive approach to quantify circulating tumor DNA (ctDNA) in 162 plasma samples from 18 subjects undergoing multimodality therapy for colorectal cancer. We found that ctDNA measurements could be used to reliably monitor tumor dynamics in subjects with cancer who were undergoing surgery or chemotherapy. We suggest that this personalized genetic approach could be generally applied to individuals with other types of cancer.
Cancers arise through the sequential alteration of genes that control cell growth. In solid tumors such as those of the colon or breast, it has been shown that, on average, approximately 80 genes harbor subtle mutations that are present in virtually every tumor cell but are not present in normal cells1. These somatic mutations thereby have the potential to serve as highly specific biomarkers. They are, in theory, much more specific indicators of neoplasia than any other biomarker yet described. One challenge for modern cancer research is therefore to exploit somatic mutations as tools to improve the detection of disease and, ultimately, to positively affect individual outcomes.
Tumor cells can often be found in the circulation of individuals with advanced cancers2,3. It has been shown that tumor-derived mutant DNA can also be detected in the cell-free fraction of the blood of individuals with cancer4–6. Most of this mutant DNA is not derived from circulating tumor cells4–6 and, in light of the specificity of mutations, raises the possibility that the circulating mutant DNA fragments themselves can be used to track disease. However, the reliable detection of such mutant DNA fragments is challenging7. In particular, the circulating mutant DNA represents only a tiny fraction of the total circulating DNA, sometimes less than 0.01% (ref. 8).
In the current study, we developed modifications of a technique called BEAMing (beads, emulsion, amplification and magnetics)8,9 to quantify ctDNA in serially collected plasma samples from subjects with colorectal cancers. We were interested in determining whether such measurements provided information about the dynamics of tumor burden in these subjects during the course of their disease.
RESULTS
Measurement of ctDNA
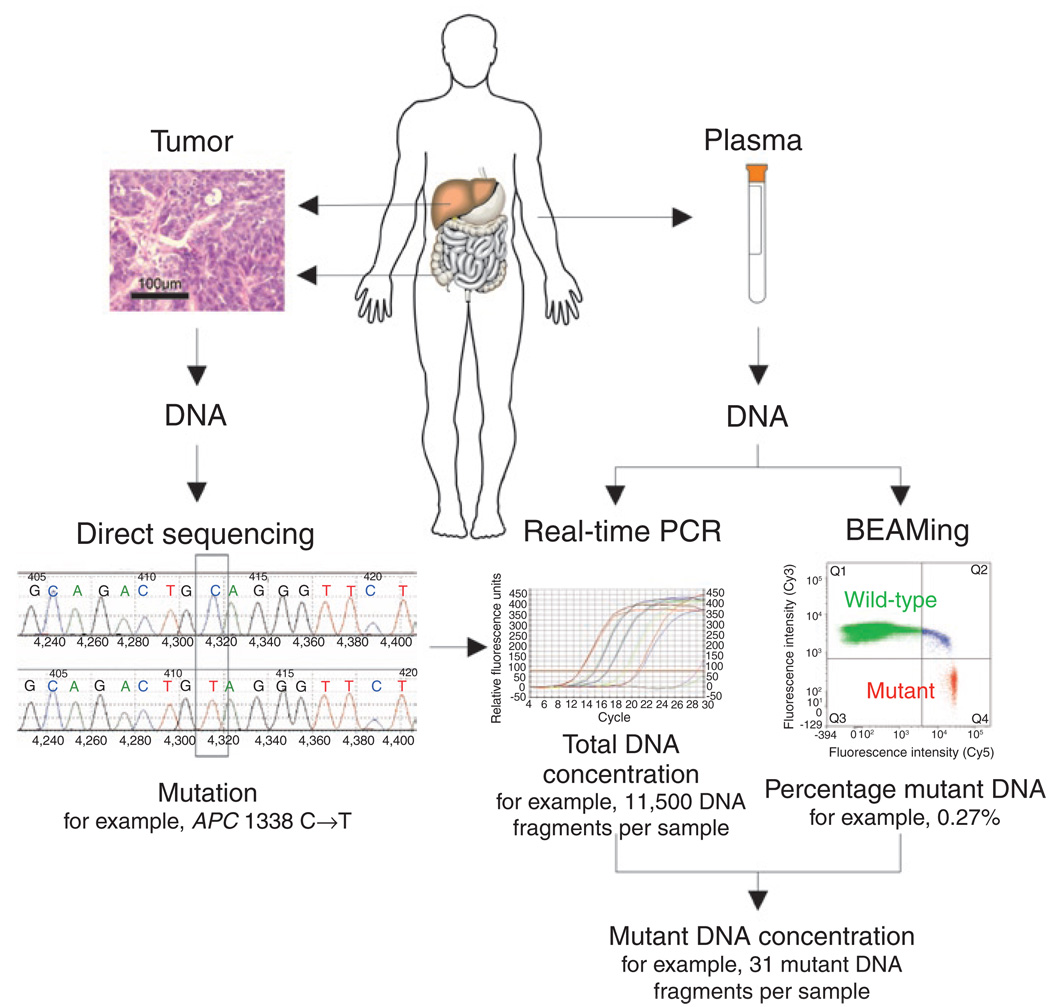
Quantification of circulating mutant ctDNA by BEAMing represents a personalized approach for assessing disease in subjects with cancer. The first step in this process is the identification of a somatic mutation in the subject’s tumor (Fig. 1). Supplementary Table 1 online lists the characteristics of the subjects with colorectal cancer evaluated in this study. Four genes were assessed by direct sequencing in tumors from 18 subjects, and each of the tumors was found to have at least one mutation (Supplementary Table 2 online).
Figure 1. Measurement of ctDNA.
The left side of the schematic depicts conventional Sanger sequencing of DNA derived from the subject’s tumor, representing the first step of the analysis. The approach for quantifying tumor-derived DNA in plasma samples is shown on the right. Real-time PCR is used to measure the total number of DNA fragments in the plasma, whereas BEAMing measures the ratio of mutant to wild-type fragments labeled with the fluorescent probes Cy5 and Cy3, respectively.
The second step in the process is the estimation of the total number of DNA fragments in the plasma by real-time PCR (Fig. 1). Before surgery (day 0), there was a median of 4,000 fragments per milliliter of plasma in the 18 subjects described above (range between 10th and 90th percentiles, 1,810–12,639 DNA fragments ml−1).
The third and final step is the determination of the fraction of DNA fragments of a given gene that contains the queried mutation. Such mutant DNA fragments are expected to represent only a small fraction of the total DNA fragments in the circulation. To achieve the sensitivity required for detection of such rare tumor-derived DNA fragments, we developed an improved version of BEAMing (detailed in Supplementary Methods online). These improvements achieved high signal-to-noise ratios and permitted detection of many different mutations via simple hybridization probes under identical conditions. We attempted to design 28 assays, at least one for each of the 18 subjects, and were successful in every case. The median percentage of mutant DNA fragments in the 95 positive samples evaluated in this study was 0.18% (range between 10th and 90th percentiles, 0.005–11.7%). Examples of typical assays from plasma serially collected from a representative subject are shown in Figure 2.
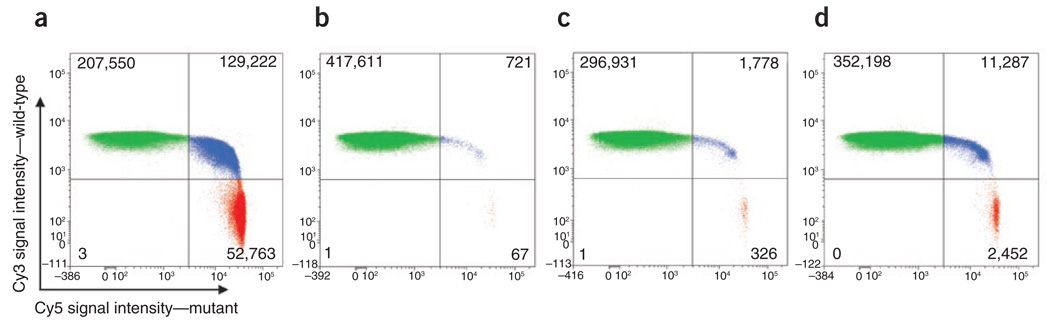
Figure 2. Representative flow cytometric data obtained from BEAMing.
The four graphs illustrate the data obtained from subject 6 (APC G4189T) at different time points during treatment. The green and red dots represent beads bound to wild-type and mutant fragments, respectively. The blue dots represent beads bound to both wild-type and mutant fragments resulting from their inclusion in an emulsion microdroplet that contained both wild-type and mutant DNA templates15. Numbers in each quadrant represent events for each population measured. (a) Before surgery, the fraction of mutant DNA fragments was 13.4%. (b) After surgery (day 3), the fraction of mutant DNA fragments dropped to 0.015%. (c) After surgery (day 48), the fraction of mutant DNA fragments increased to 0.11%, suggesting disease recurrence. (d) On day 244, the subject had progressive disease and the fraction mutant DNA fragments increased further to 0.66%.
Multiplying the total number of DNA fragments of a gene in the analyzed volume of plasma (as determined by real-time PCR) by the fraction of mutant fragments (as determined by BEAMing) yields the number of mutant fragments (ctDNA number) in that volume of plasma (Fig. 1). The median number of mutant DNA fragments in the 95 positive samples evaluated in this study was 39 (range between 10th and 90th percentiles, 1.3–1833.0).
The accuracy of these assays was assessed by measurements of the number of mutant DNA fragments derived from two different genes in the same subject. We were able to assay mutations in two different genes in 43 samples derived from nine study subjects. The ctDNA levels corresponding to the two mutant genes were found to be remarkably similar (correlation coefficient R2 = 0.95, Supplementary Fig. 1a online).
ctDNA dynamics in subjects with cancer undergoing therapy
We evaluated 18 subjects after a total of 22 surgeries during the course of this study (Supplementary Table 1). The ctDNA level determined before surgery (day 0) varied widely, ranging from 1.3 to 23,000 mutant templates per sample (median 99 mutant templates per sample; range between 10th and 90th percentiles, 3–2,837). Seventeen of these surgeries involved complete resection of all evident tumor tissue, whereas five were incomplete resections. A sharp drop in the ctDNA level by the day of discharge (two to ten days after surgery) was observed in all subjects who underwent complete resections, with a 99.0% median decrease in ctDNA (range between 10th and 90th percentiles, 58.9–99.8%; Table 1). This decrease was already evident 24 h after surgery (96.7% median decrease, range between 10th and 90th percentiles, 31.4–100.0%). Through evaluation of a subject whose plasma was sampled at multiple early times after complete resection, we estimated the half-life of ctDNA after surgery as 114 min (Supplementary Fig. 2 online).
Table 1.
ctDNA in subjects before and after surgery
| ctDNA level (ctDNA molecules in 2 ml)a | CEA level (ng ml−1) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Subject number |
Stage | Surgery number |
Surgical procedure | Resection | Gene | Pre-surgery day 0 |
Post-operative day 1 |
Post-operative days 2–10 |
Post-operative follow-up days 13–56 |
Pre-surgery day 0 |
Post-operative day 1 |
Post-operative days 2–10 |
Post-operative follow-up days 13–56 |
Disease recurrence |
| 1 | IV | 1 | Hepatic metastectomy | Complete | APC | 99 ± 38 | 4.0 ± 2.0 | 0.9 ± 0.3 | 16 ± 2.3 | 69 | 53 | 24 | 22 | Yes |
| KRAS | 79 ± 35 | 2.6 ± 1.3 | Neg | 5.6 ± 0.9 | ||||||||||
| 2 | IV | 1 | Hepatic metastectomy (first-stage) |
Incomplete | APC | 2,951 ± 773 | 1,336 ± 408 | 1,508 ± 156 | 1,049 ± 254 | 22 | 14 | 10 | 12 | Yes |
| 3 | IV | 1 | Right hemicolectomy with hepatic metastectomy |
Incomplete | APC | 58 ± 18 | 421 ± 85 | 404 ± 121 | 166 ± 63 | 6.8 | 7.2 | 6.5 | 12 | Yes |
| KRAS | 46 ± 15 | 485 ± 98 | 393 ± 118 | 136 ± 51 | ||||||||||
| 4 | IV | 1 | Hepatic metastectomy with RFA |
Complete | APC | 16 ± 4 | Neg | Neg | Neg | 1.6 | 1.4 | 0.8 | 1.8 | No |
| KRAS | 15 ± 4 | Neg | Neg | Neg | ||||||||||
| 5 | IV | 1 | Hepatic metastectomy | Complete | PIK3CA | 213 ± 90 | 4.9 ± 2.1 | 7.7 ± 2.3 | 9.8 ± 3.8 | 9.4 | 6.8 | 4.4 | 2.1 | Yes |
| TP53 | 275 ± 116 | 4.5 ± 2.0 | 2.6 ± 0.8 | 5.7 ± 1.7 | ||||||||||
| 6 | IV | 1 | Pulmonary and hepatic metastectomy | Complete | APC | 3,650 ± 1,313 | 1.3 ± 0.4 | 3.3 ± 1.3 | 2.2 ± 0.6 | 422 | 149 | 61 | 7.2 | Yes |
| KRAS | 2,375 ± 917 | 1.2 ± 0.5 | 4.4 ± 1.7 | 1.2 ± 0.4 | ||||||||||
| 7 | IV | 1 | Hepatic metastectomy with RFA |
Complete | KRAS | 856 ± 394 | 117 ± 24 | - | 220 ± 9 | 5.3 | 2.7 | - | 3.4 | Yes |
| 8 | IV | 1 | Sigmoid colectomy with hepatic metastectomy (first-stage) |
Incomplete | PIK3CA | 2,575 ± 1,027 | 1,370 ± 551 | 6,459 ± 2,305 | 1,420 ± 408 | 4.5 | 2.4 | 1.6 | 3.8 | Yes |
| TP53 | 2,352 ± 937 | 802 ± 248 | 4,171 ± 1,484 | 944 ± 258 | ||||||||||
| 2 | Right hepatectomy | Complete | PIK3CA | 644 ± 281 | 26.5 ± 7.9 | 3.7 ± 1.2 | 0.9 ± 0.1 | 1.4 | 0.6 | 0.5 | 0.7 | Yes | ||
| TP53 | 389 ± 169 | 5.9 ± 1.7 | Neg | Neg | ||||||||||
| 9 | IV | 1 | Right hepatectomy | Complete | TP53 | 91 ± 33 | 2.6 ± 3.3 | 1.2 ± 1.3 | Neg | 4.4 | 1.1 | 0.7 | 0.5 | No |
| 10 | IV | 1 | Right hemicolectomy with hepatic metastectomy |
Complete | APC | 6 ± 1.6 | Neg | Neg | Neg | 0.9 | 0.8 | 0.7 | 1.2 | Yes |
| KRAS | 3 ± 1.0 | Neg | 2.4 ± 0.5 | 1.5 ± 0.7 | ||||||||||
| 2 | Hepatic metastectomy with RFA |
Complete | APC | 128 ± 34 | 37 ± 14 | - | 316 ± 64 | 2.0 | 0.9 | - | 1.8 | Yes | ||
| KRAS | 42 ± 13 | 11 ± 5 | - | 131 ± 32 | ||||||||||
| 11 | IV | 1 | Sigmoid colectomy with hepatic metastectomy with RFA |
Incomplete | TP53 | 14 ± 3 | 59 ± 22 | - | 295 ± 71 | 93 | 55 | - | 32 | Yes |
| 2 | Hepatic metastectomy | Complete | TP53 | 2,713 ± 775 | 10 ± 5 | 14 ± 7 | 1.8 ± 0.4 | 74 | 43 | 22 | 2.8 | Yes | ||
| 3 | RFA to hepatic metastases | Complete | TP53 | 1,267 ± 243 | 465 ± 8 | - | 39 ± 2 | 52 | 44 | - | 9.1 | Yes | ||
| 12 | IV | 1 | Hepatic metastectomy with RFA |
Incomplete | KRAS | 28 ± 8 | 67 ± 9 | 27 ± 9 | 120 ± 42 | 1.6 | 0.8 | 0.8 | 0.7 | Yes |
| 13 | IV | 1 | Hepatic metastectomy with partial gastrectomy and omentectomy |
Complete | APC | 1 ± 0.3 | 1.2 ± 0.9 | - | - | 2.0 | 1.5 | - | - | Yes |
| 14 | II | 1 | Sigmoid colectomy | Complete | APC | 2 ± 0.7 | Neg | Neg | Neg | 3.9 | 2.7 | 1.7 | 1.3 | No |
| 15 | IV | 1 | Right hepatectomy | Complete | KRAS | 519 ± 125 | 612 ± 60 | 35 ± 11 | 20 ± 4 | 17 | 14 | 11 | 4.5 | Yes |
| 16 | III | 1 | Sigmoid colectomy | Complete | APC | 5 ± 1.7 | - | 3.5 ± 0.9 | Neg | <0.5 | <0.5 | <0.5 | <0.5 | No |
| TP53 | 2 ± 0.6 | - | 1.6 ± 0.2 | Neg | ||||||||||
| 17 | IV | 1 | Right hepatectomy | Complete | APC | 20,144 ± 5,504 | 58 ± 22 | 13 ± 3 | - | 25 | 17 | 0.8 | - | Yes |
| KRAS | 17,482 ± 4,777 | 97 ± 36 | 108 ± 27 | - | ||||||||||
| 18 | IV | 1 | Hepatic metastectomy | Complete | APC | 29 ± 8 | 4 ± 1.28 | - | 0.7 ± 0.4 | 24 | 16 | - | 2.2 | No |
ctDNA level ±s.d. RFA, radiofrequency ablation; APC, adenomatous polyposis coli; KRAS, Kirsten rat sarcoma viral oncogene homolog; PIK3CA; phosphoinositide-3-kinase, catalytic, αpolypeptide; TP53, tumor protein p53; Neg, undetectable ctDNA; –, plasma sample not collected.
In the five cases with incomplete resections, the change in ctDNA was quite different. In two of these cases, the number of mutant fragments decreased only slightly at 24 h (55–56%), whereas in the other three cases, the number actually increased (141%, 329% and 794%). This increase was perhaps due to injury of remnant tumor tissue during the surgical procedure, with subsequent release of DNA. Surgically induced tissue injury is consistent with the observation that the total amount of DNA in the plasma (mutant plus normal) increased immediately after surgery in all subjects (Supplementary Fig. 3 online).
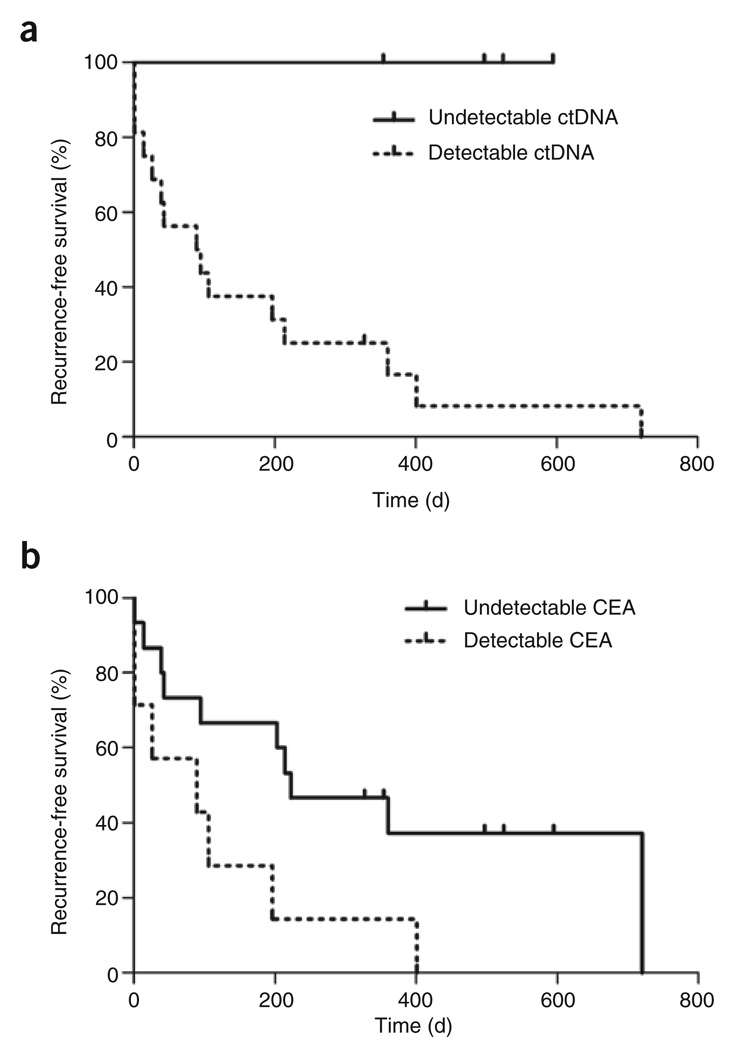
Though the amount of ctDNA generally decreased after surgery, it did not decrease to undetectable levels in most cases. Plasma samples were available from the first follow-up visit, 13–56 d after surgery, in 20 instances. ctDNA was still detectable in 16 of these 20 instances, and recurrences occurred in all but one of these 16 (Table 1). In a marked contrast, no recurrence occurred in the four subjects in whom ctDNA was undetectable at the first follow-up visit (Table 1). The difference in recurrence rate between subjects with and without detectable ctDNA at the first follow-up was significant (P = 0.006 by Mantel-Cox log-rank test, Fig. 3a).
Figure 3. Recurrence-free survival, as detected by ctDNA and CEA.
(a) The difference in recurrence-free survival in subjects with detectable versus undetectable post-operative ctDNA levels (P = 0.006 by Mantel-Cox log-rank test). (b) The difference in recurrence-free survival in subjects with detectable versus undetectable post-operative CEA levels (P = 0.03 by Mantel-Cox log-rank test).
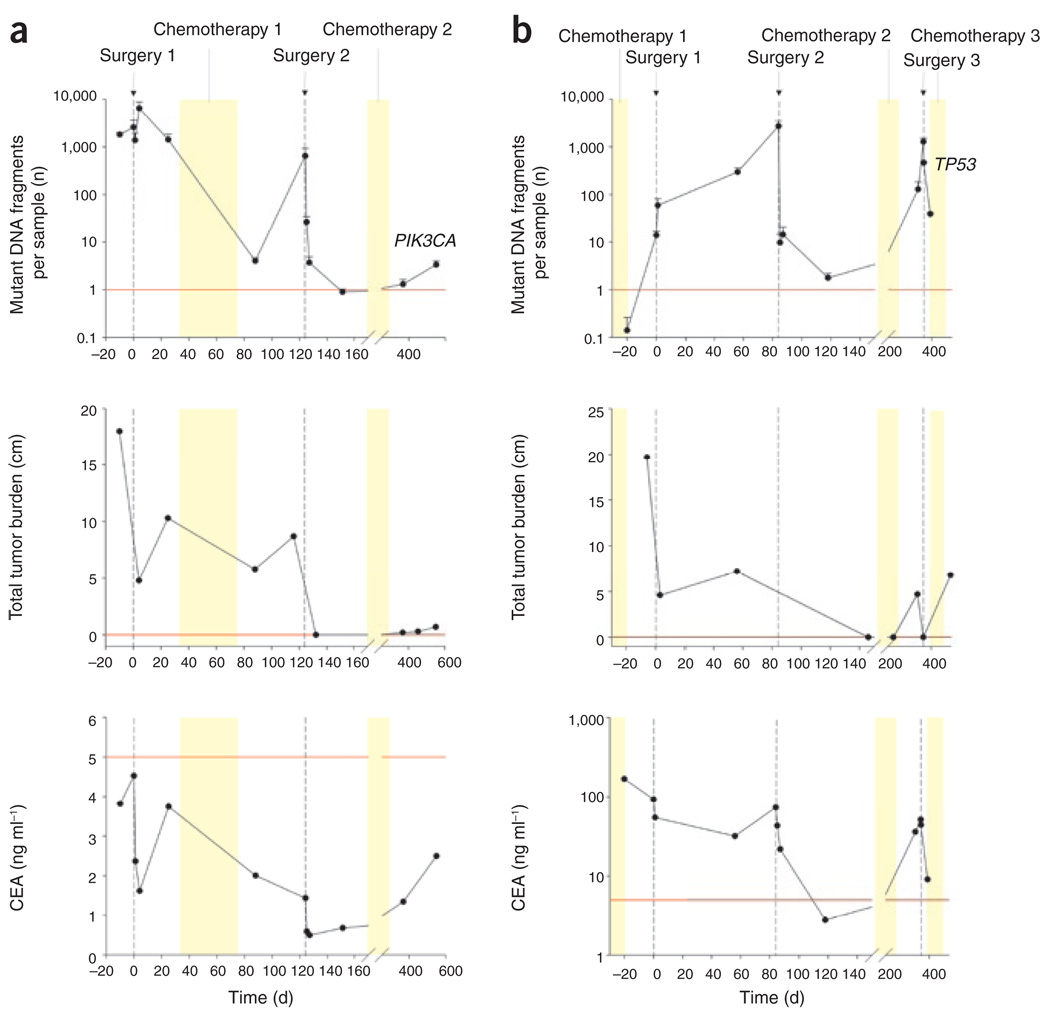
Representative time courses of ctDNA along with clinical and radiologic data on two subjects are provided in Figure 4, and similar data on all other subjects are shown in Supplementary Figure 4 online. Subjects 8 and 11 had more than one surgical procedure during the study, providing special opportunities to assess changes in ctDNA after repeated, controlled manipulation of tumor burden. Both of these subjects had incomplete resections in their initial surgery, and their ctDNA levels did not decrease (Fig. 4). They had complete resections in their second surgery, and the ctDNA abundance dropped precipitously thereafter. The ctDNA abundance then climbed back to higher levels over the next several months (Fig. 4).
Figure 4. Comparison of ctDNA, CEA and imaging dynamics in individual study subjects.
For each subject, the top, middle, and bottom graphs represent ctDNA level, tumor volume as assessed by imaging, and CEA level. The red lines represent the upper bound of the normal levels: one mutant DNA fragment per sample for ctDNA levels, 0.0 cm for tumor diameter, and 5.0 ng ml−1 for CEA abundance. (a) Subject 8 had a sigmoid adenocarcinoma and solitary metastases in both hepatic lobes. The subject underwent a sigmoidectomy and left lateral hepatic sectorectomy (Surgery 1). A right-sided liver metastasis was left in place while the subject was treated with systemic chemotherapy (Chemotherapy 1). On day 120, a right hepatectomy was performed (Surgery 2). After surgery, the subject was treated for 4 months with systemic chemotherapy (Chemotherapy 2). (b) Subject 11 had a sigmoid adenocarcinoma and two liver metastases that were treated with systemic chemotherapy before surgery (Chemotherapy 1). The subject underwent a sigmoid colectomy, left hepatic lobectomy and RFA of a solitary right hepatic lesion (Surgery 1). Imaging studies at 2 months showed recurrence in the liver, and the subject underwent a right hepatectomy (Surgery 2). Given the high risk of recurrence, chemotherapy was reinitiated (Chemotherapy 2). At 8 months, imaging showed three recurrent liver lesions and a suspicious celiac lymph node. The subject underwent RFA of these lesions and resection of the celiac node (Surgery 3). After surgery, the subject received additional chemotherapy (Chemotherapy 3); however, later imaging revealed multiple pulmonary metastases.
Eleven of the subjects in our cohort received chemotherapy during the course of the study. In three of these subjects, ctDNA levels declined during the treatment (Fig. 4 and Supplementary Fig. 4). An example is provided by subject 8: ctDNA decreased by more than 99.9%, whereas tumor volume (composed of live and dead neoplastic cells in addition to stromal cells) decreased only slightly (Fig. 4). In six subjects, there was an immediate rise in ctDNA after discontinuation of chemotherapy, as is evident in subjects 8 and 11 after the first chemotherapy (Fig. 4) and in subjects 1, 4, 10 and 12 (Supplementary Fig. 4).
Comparison with carcinoembryonic antigen
Carcinoembryonic antigen (CEA) is the standard biomarker for following disease in subjects with colorectal cancer and is routinely used in the management of the disease10. Only ten of the eighteen subjects had CEA levels >5 ng ml−1 (the boundary of the normal range) before study entry (Table 1). This difference in sensitivity between the two assays (ctDNA versus CEA) was statistically significant; 56% versus 100%, respectively (P = 0.008, McNemar test). Moreover, even in those subjects with positive CEA levels before surgery, complete tumor resection resulted in a much less marked decrease in CEA than that observed with ctDNA (median decrease of 99.0% versus 32.5% in ctDNA versus CEA, respectively; P < 0.001, Student’s t-test). There was a modest overall correlation between CEA abundance and ctDNA levels after correcting for clustering within subjects (R2 = 0.20, P < 0.001, Supplementary Fig. 1b). Finally, when measured at the first post-operative follow-up visit on days 24–48, the ability of CEA levels to predict recurrent disease was less impressive than that of ctDNA levels (P = 0.03 by Mantel-Cox log-rank test, Fig. 3b).
DISCUSSION
The results reported herein show that ctDNA is a promising biomarker for following the course of therapy in patients with metastatic colorectal cancer. ctDNA was detectable in all subjects before surgery, and serial blood sampling revealed oscillations in the level of ctDNA that correlated with the extent of surgical resection. Subjects who had detectable ctDNA after surgery generally relapsed within 1 year. The ctDNA seemed to be a much more reliable and sensitive indicator than the current standard biomarker (CEA) in this cohort of subjects.
Our studies are consistent with others that have shown that ctDNA can be detected in subjects with cancer, particularly in advanced tumors6. However, most such previous studies have not used techniques sufficiently sensitive to detect the low levels of ctDNA found in many of the subjects evaluated in the current study. Moreover, one of the crucial and distinguishing features of our approach lies in the ability to precisely measure the level of ctDNA rather than to simply determine whether or not ctDNA is detectable.
The results of our study suggest that ctDNA levels reflect the total systemic tumor burden, in that ctDNA levels decreased upon complete surgery and generally increased as new lesions became apparent upon radiological examination. However, whether ctDNA levels are exactly proportional to systemic tumor burden cannot be definitively determined, because there is no independent way to measure systemic total burden at this time. Radiographs are inaccurate, because lesions that are observed upon imaging are composed of live neoplastic cells, dead neoplastic cells and varying amounts of non-neoplastic cells (stromal fibroblasts, inflammatory cells, vasculature, and the like)11. The proportion of these cell types in any lesion is unknown. Additionally, micrometastatic lesions that are smaller than a few millimeters, which in aggregate may make a large contribution to the total tumor burden, are not detectable by positron emission tomography, computed tomography or magnetic resonance imaging scans.
The approach used in our study can be considered a form of ‘personalized genomics’. As such, it has both advantages and disadvantages. The advantage over other biomarkers lies in its specificity, as the queried mutation should never be found in the circulation unless residual tumor cells are present somewhere in the subject’s body. The disadvantage is that a marker specific for each subject must be developed. This entails the identification of mutations in the subject’s tumor as a preliminary step (Fig. 1). Though we have performed this step with direct sequencing of DNA from paraffin-embedded tissues, it could be performed with simpler technologies, such as microarrays querying mutation hotspots12,13. The second step—designing and testing a mutation-specific probe—is also time consuming at this stage of technological development. But it, too, could be simplified, in that a stock of probes, representing the most common mutations, could easily be prepared in advance. This strategy may also be particularly useful for a different application of the approach, that is, cancer screening in a healthy population where mutational status is not known in advance.
In sum, we present a framework for using circulating tumor DNA as a measure of tumor dynamics. The rationale is similar to that employed in the care of patients with HIV, in whom viral nucleic acids are quantitatively assessed to monitor asymptomatic disease and used to tailor therapy to the individual’s needs. We envision that ctDNA could be used to noninvasively monitor many types of cancer and, as in the treatment of individuals with HIV, help influence clinical decision-making. As sequencing technologies improve, it will become relatively simple to identify such mutations in virtually any cancer. Indeed, such diagnostic applications are one of the major goals of the Cancer Genome Atlas Project (http://cancergenome.nih.gov/index.asp/).
METHODS
Subjects and study design
This study was approved by the Institutional Review Board of the Johns Hopkins Medical Institutions. Subjects were eligible if they had primary or metastatic colorectal cancer that was being treated surgically at The Johns Hopkins Sidney Kimmel Comprehensive Cancer Center. Between October 2005 and July 2006, 31 subjects diagnosed with colorectal cancer were screened during preoperative evaluation for possible surgery. Twenty-eight subjects consented for the study, but seven of these were found not to be candidates for therapy, two subjects were lost during follow-up and one subject was found to have a medical condition other than colorectal cancer, leaving eighteen participants. Each subject agreed to have ctDNA assessed in plasma samples obtained before and after surgery and during prespecified intervals during their post-operative course (Supplementary Fig. 5 online) through October 2007. We prospectively collected 162 plasma samples from the 18 subjects. Formalin-fixed paraffin-embedded tumor tissue was obtained from each subject and processed by the Surgical Pathology Laboratory at The Johns Hopkins Medical Institutes using routine procedures. We performed the analyses of the tumor tissues and the plasma samples in a blinded fashion once the clinical assessment was complete. We measured tumor sizes radiographically with computed tomography, and we used cross-sectional measurements in centimeters to estimate tumor burden.
Isolation and quantification of DNA from plasma
We drew peripheral blood into EDTA tubes (Becton Dickinson). Within one hour, we subjected the tubes to centrifugation at 820g for 10 min. We transferred 1-ml aliquots of the plasma to 1.5-ml tubes and centrifuged at 16,000g for 10 min to pellet any remaining cellular debris. We transferred the supernatant to fresh tubes and stored them at −80 °C. We purified total genomic DNA from 2 ml of the plasma aliquots using the QIAamp MinElute virus vacuum kit (Qiagen) according to the manufacturer’s instructions. We quantified the amount of total DNA isolated from plasma with a modified version of a human LINE-1 quantitative real-time PCR assay, as described previously14. Details are provided in Supplementary Methods online.
Mutation analysis of DNA from tumor tissue
We determined the mutation status of four genes in DNA purified from paraffin-embedded tumor tissue. We cut 10-µm sections and stained them with H&E. We used laser-capture microdissection to acquire neoplastic cells from these sections. We digested the dissected material overnight with proteinase K (Invitrogen) and purified genomic DNA from it with the QIAamp Micro Kit (Qiagen). We analyzed a total of 26 PCR products by direct sequencing. Further details concerning DNA amplification and sequencing are provided in the Supplementary Methods.
Mutation analysis of DNA from plasma
We queried at least one mutation identified by sequencing of each subject’s tumor tissue in plasma. In brief, we designed primers that could amplify the region containing the mutation for an initial amplification step with a high-fidelity DNA polymerase (New England BioLabs). We used the amplified product as a template in the subsequent BEAMing assay. The sequences of the primers and probes used for each test are listed in the Supplementary Methods. The basic experimental features of BEAMing have been previously described15, and the modifications used in the current study are described in the Supplementary Methods. We used the DNA purified from 2 ml plasma for each BEAMing assay. We repeated each measurement at least two times.
We used DNA purified from each subject’s tumor as a positive control. We also included negative controls, performed with DNA from subjects without cancer, in each assay. Depending on the mutation being queried, the percentage of beads bound to mutant-specific probes in these negative control samples varied from 0.0061% to 0.00023%. This fraction represented sequence errors introduced by the high-fidelity DNA polymerase during the first PCR step, as explained in detail previously16. To be scored as positive in an experimental sample, the fraction of beads bound to mutant fragments had to be higher than the fraction found in the negative control, and the mean value of mutant DNA fragments per sample plus one standard deviation had to be >1.0. We analyzed bead populations generated by BEAMing at least twice for each plasma sample.
Carcinoembryonic antigen measurement
We analyzed CEA abundance by a two-step chemiluminescent microparticle immunoassay with the Abbott ARCHITECT i2000 instrument (Abbott Laboratories) at the Johns Hopkins Medical Institutions Clinical Chemistry Research Laboratory.
Statistical analyses
We quantified post-operative changes in ctDNA as a mean percentage decrease after surgery, with its standard error. We compared relative changes in CEA to ctDNA values with Student’s unpaired t-test. We assessed changes from baseline with a one-sample t-test. We calculated the correlation between CEA and ctDNA levels with partial residuals from linear regression, taking into account within-subject clustering. Recurrence was defined on the basis of radiographic and clinical findings. We calculated all confidence intervals at the 95% level. We performed computations using JMP 6.0 software (SAS Institute) and SigmaPlot 10.0.1 (Systat Software).
Supplementary Material
Note: Supplementary information is available on the Nature Medicine website.
ACKNOWLEDGMENTS
We thank G.Traverso for useful discussions; D. Heslop, C. Vaughn and D. Edelstein for their help with plasma collection; and L. Fayad for help with the interpretation of imaging studies. This work was supported by the Virginia and D.K. Ludwig Fund for Cancer Research, the Miracle Foundation, the US National Colorectal Cancer Research Alliance and US National Institutes of Health grants CA43460, CA57345, CA62924 and CA129825.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemedicine/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 2.Nagrath S, et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristofanilli M, et al. Circulating tumor cells, disease progression and survival in metastatic breast cancer. N. Engl. J. Med. 2004;351:781–791. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 4.Sidransky D. Emerging molecular markers of cancer. Nat. Rev. Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 5.Goebel G, Zitt M, Zitt M, Muller HM. Circulating nucleic acids in plasma or serum (CNAPS) as prognostic and predictive markers in patients with solid neoplasias. Dis. Markers. 2005;21:105–120. doi: 10.1155/2005/218759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer—a survey. Biochim. Biophys. Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat. Res. 2007;635:105–117. doi: 10.1016/j.mrrev.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Diehl F, et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA. 2005;102:16368–16373. doi: 10.1073/pnas.0507904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstein MJ, Mitchell EP. Carcinoembryonic antigen in the staging and follow-up of patients with colorectal cancer. Cancer Invest. 2005;23:338–351. doi: 10.1081/cnv-58878. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Fan X, Houghton J. Tumor microenvironment: the role of the tumor stroma in cancer. J. Cell. Biochem. 2007;101:805–815. doi: 10.1002/jcb.21159. [DOI] [PubMed] [Google Scholar]
- 12.Hacia JG, Collins FS. Mutational analysis using oligonucleotide microarrays. J. Med. Genet. 1999;36:730–736. doi: 10.1136/jmg.36.10.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shendure J, Mitra RD, Varma C, Church GM. Advanced sequencing technologies: methods and goals. Nat. Rev. Genet. 2004;5:335–344. doi: 10.1038/nrg1325. [DOI] [PubMed] [Google Scholar]
- 14.Rago C, et al. Serial Assessment of human tumor burdens in mice by the analysis of circulating DNA. Cancer Res. 2007;67:9364–9370. doi: 10.1158/0008-5472.CAN-07-0605. [DOI] [PubMed] [Google Scholar]
- 15.Diehl F, et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Diehl F, Dressman D, Vogelstein B, Kinzler KW. BEAMing up for detection and quantification of rare sequence variants. Nat. Methods. 2006;3:95–97. doi: 10.1038/nmeth850. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Medicine website.