Abstract
S-nitrosylation is an important mediator of multiple nitric oxide-dependent biological processes, including eukaryotic cellular events such as macrophage apoptosis and proinflammatory signaling. Many pathogenic bacteria possess NO detoxification mechanisms, such as the nitric oxide reductase (NorB) of Neisseria meningitidis and the flavohemoglobins (Hmp) of Salmonella enterica and Escherichia coli, which serve to protect the microorganism from nitrosative stress within the intracellular environment. In this study, we demonstrate that expression of meningococcal NorB increases the rate at which low-molecular-weight S-nitrosothiol (SNO) decomposes in vitro. To determine whether this effect occurs in cells during infection by bacteria, we induced SNO formation in murine macrophages by activation with lipopolysaccharide and γ-interferon and observed a reduced abundance of SNO during coincubation with N. meningitidis, S. enterica, or E. coli. In each case, this effect was shown to be dependent on bacterial NO detoxification genes, which act to prevent SNO formation through the removal of NO. This may represent a novel mechanism of host cell injury by bacteria.—Laver, J. R., Stevanin, T. M., Messenger, S. L., Dehn Lunn, A., Lee, M. E., Moir, J. W. B., Poole, R. K., Read, R. C. Bacterial nitric oxide detoxification prevents host cell S-nitrosothiol formation: a novel mechanism of bacterial pathogenesis.
Keywords: S-nitrosylation, infection, macrophage, nitrosative stress
Nitric oxide (NO) is a ubiquitous, biologically potent signaling molecule with functionally pleiotropic effects. NO, in the presence of a strongly oxidizing cofactor, can bond covalently with the thiol residues of proteins to form S-nitrosothiols (SNOs). The formation of SNOs, called S-nitrosylation, represents a form of reversible posttranslational modification of cellular proteins (1) that is critically involved in a number of cell signaling events, akin to the effect of tyrosine phosphorylation (2). Thus, S-nitrosylation has been implicated in regulating apoptosis (3,4,5,6), control of intracellular signaling cascades (7), neurotransmission (8), and regulation of gene expression (9,10,11). S-nitrosylation meets the criteria for validation as a cell signaling mechanism: it is stimulus evoked (12, 13), precisely targeted (14), reversible (15), spatiotemporally restricted (16, 17), and necessary for specific cell responses (18). The reverse process of S-nitrosylation is termed denitrosylation and involves the release of the NO moiety from the SNO bond. The rate of SNO decomposition in solution is affected by factors such as light, temperature, pH, and the presence of contaminating transition metal ions (19). Recent studies demonstrated that denitrosylation is also an enzymatically mediated process. In addition to S-nitrosoglutathione reductase (GSNOR), a ubiquitous and well-conserved SNO-metabolizing enzyme (20), denitrosylation has also been demonstrated as a function of the thioredoxin/thioredoxin reductase system (21).
Many bacteria can metabolize NO, for example, by reduction in denitrification and NO detoxification for defense against phagocyte-induced nitrosative stress. One example occurs in Neisseria meningitidis (the meningococcus), which causes meningococcal disease after colonization of its sole biological niche, the NO-rich human nasopharynx (22,23,24). Although N. meningitidis is incapable of anaerobic respiration (25), it can also grow under oxygen-limited conditions (26) and supplement its growth by partial denitrification, using the gene products of aniA (NMB1622) and norB (NMB1623) (26). The norB gene encodes an NO-reducing quinol oxidase (NorB, qNOR), which confers resistance to NO-mediated killing by monocyte-derived macrophages (MDMs) and enhances survival within human nasopharyngeal tissue (27). In addition, NorB exerts effects on the host response, modulating the cytokine and chemokine output of infected MDMs (28) and inhibiting MDM apoptosis (29). Expression of norB is controlled transcriptionally by the nitric oxide-sensitive protein, NsrR (30).
However, the best-characterized bacterial NO detoxification mechanism is that catalyzed by flavohemoglobins (Hmp), which have properties similar to erythrocyte hemoglobins and associated flavin-containing methemoglobin reductases (31, 32). Hmp consists of an N-terminal heme-binding domain integrated with a flavin-binding reductase domain, both of which are required to provide protection against nitrosative stress (33). The noncovalently bound FAD transfers electrons from NAD(P)H to the heme and thence to oxygen (34) to form an oxyhemoglobin species. However, in the presence of NO and physiological oxygen concentrations, Hmp preferentially binds NO (35) and reduces it to NO−, which aerobically forms nontoxic nitrate anion (NO3−) (36). Thus, Hmp catalyzes a denitrosylase, not a dioxygenase, reaction. Under anoxic conditions, Hmp catalyzes the slow reduction of NO to NO−, which can be detected as nitrous oxide (N2O) (37). In the enteric bacterium Escherichia coli, the efficacy of Hmp in resisting nitrosative stress has been demonstrated by assays of viability (38), cell respiration (39), and macrophage killing (40). In Salmonella, NsrR-dependent Hmp expression (41, 42) is an important determinant of NO detoxification and bacterial viability in macrophages (43).
We postulated that NorB-mediated alterations in host cell physiology are driven by bacterial perturbation of S-nitrosylation. If so, this would represent a mechanism of host cell septic injury and have far-reaching implications for all pathogens that consume NO. As proof of this principle, we tested the hypothesis that bacterial NO detoxification mechanisms can affect the abundance of host cell SNO, using a murine macrophage infection model. In response to cellular agonists, murine macrophage cell lines have a high natural output of NO and contain relatively high concentrations of SNO (44,45,46).
MATERIALS AND METHODS
Reagents and apparatus
Unless otherwise indicated, all chemicals were purchased either from Sigma-Aldrich (St. Louis, MO, USA) or BDH Laboratories (Poole, UK). Lipopolysaccharide (LPS) from E. coli 0127:B8 was purchased as a filter-sterilized stock solution (1 mg/ml) from Sigma-Aldrich, as were N-([3-(aminomethyl)phenyl]methyl)ethanimidamide dihydrochloride (1400W) and diethylene triamine pentaacetic acid (DTPA). Recombinant mouse interferon-γ (rmIFN-γ) was purchased from R&D Systems (Minneapolis, MN, USA). Dulbecco’s modified Eagle’s medium (DMEM) was purchased from Lonza (Basel, Switzerland). S-nitrosoglutathione (GSNO) was synthesized in-house, as described previously (47). Chemiluminescence measurements of nitrogen oxides were performed using a Bo 280i Nitric Oxide Analyzer (NOA; Sievers, Boulder, CO, USA) with output data analyzed using either the LIQUID program or OriginPro 8.0.
Bacterial strains and growth conditions
The bacterial strains used in this study are detailed in Table 1. Frozen stocks of viable bacteria were maintained in cryopreservation fluid (Protect™; TSC, Heywood, UK) at −80°C. Aliquots of frozen N. meningitidis stocks were streaked onto GC agar supplemented with 1% Vitox supplement (Oxoid, Basingstoke, UK) and spectinomycin (50 μg/ml) where appropriate. Plates were incubated overnight at 37°C, 5% CO2 and propagated daily on fresh medium. Cultures were produced using ≥3 meningococcal colonies from fresh overnight plates, to avoid problems of phase variability of surface structures in Neisseria. Meningococcal cultures were grown in 10 ml Mueller Hinton broth in 25 ml universal tubes at 37°C, 5% CO2 with agitation (<90 rpm) provided by a plate mixer. For infecting activated J774.2 macrophage cells, cultures were grown to midlog phase (OD600=0.25). For those experiments requiring a heat-killed wild-type control for meningococcal NorB activity, a suspension of wild-type bacteria, containing the same number of cells as the wild-type inoculum, was incubated at 80°C for 20 min.
TABLE 1.
Bacterial strains used in this work
| Strain | Relevant genotype | Source/ref. |
|---|---|---|
| Neisseria meningitidis | ||
| MC58 | Wild-type group B strain | McGuinness et al.(70) |
| norB | MC58, norB :: Ω | Anjum et al.(26) |
| nsrR | MC58, nsrR :: Ω | Rock et al.(51) |
| Salmonella entericaspp. Typhimurium | ||
| ATCC 14028s | Wild type | Crawford & Goldberg (71) |
| 14028sΔhmp | ATCC 14028s, hmp :: kanR | Crawford & Goldberg (71) |
| Escherichia coli | ||
| MG1665 | Wild type | Laboratory collection (R.K.P.) |
| MG1665Δhmp | MG1665, hmp :: Tn5 | Laboratory collection (R.K.P.) |
Salmonella strains were propagated from frozen stocks onto Columbia agar supplemented with horse blood (Oxoid) and kanamycin (50 μg/ml) where appropriate. Plates were incubated overnight at 37°C, 5% CO2 to produce single colonies. Cultures of Salmonella were produced by inoculating 100 ml of brain heart infusion broth in a 250 ml flask with 3 Salmonella colonies from fresh, overnight plates and incubating at 37°C with shaking at ∼200 rpm. For infecting activated J774.2 cells, cultures were grown to midlog phase (OD600=0.35).
Single colonies of each E. coli strain were grown on Luria-Bertani (LB) agar containing kanamycin (25 μg/ml) where appropriate. An overnight culture of the appropriate strain was used to inoculate (1:1000 v/v) 100 ml of fresh LB medium in a 250-ml flask, which was then incubated at 37°C with shaking at 200 rpm. For infection of activated J774.2 cells, cultures were grown to midlog phase (OD600=0.35). In experiments investigating the effect of NorB activity on the decomposition of GSNO, DMEM + 10% HI-FCS was supplemented with 100 μM GSNO before being inoculated with meningococci.
Tissue culture
The mouse [BALB/c] tumor monocyte-macrophage cell line, J774.2 (ECACC 85011428) was maintained in DMEM plus 10% heat-inactivated fetal calf serum (HI-FCS), in suspensions between 3 and 9 × 105 cells/ml at 37°C, 5% CO2. For use in the measurement of endogenous SNO, J774.2 cells were seeded at 1 × 106 cells/ml in 6-well plates and incubated at 37°C, 5% CO2 for 24 h before their use.
Activation of murine macrophages
A sterile preparation of LPS from E. coli 0127:B8 was added to DMEM + 10% HI-FCS at a final concentration of 1 μg/ml. rmIFN-γ was added to DMEM + 10% HI-FCS at a final concentration of 1000 U/ml. Where used, the specific inducible nitric oxide synthase (iNOS) inhibitor 1400W was added to medium at a final concentration of 100 μM. Following addition of agonists (t=0 h), cells were incubated at 37°C, 5% CO2 for 18 h.
Infection of activated macrophages
Aliquots of midlog phase culture containing the appropriate number of viable bacteria were centrifuged, washed, and resuspended in a suitable volume of DMEM + 10% HI-FCS. At t = 18 h, agonists were removed from wells, and the activated macrophages were infected at a multiplicity of infection (MOI) of 10 with suspensions of N. meningitidis, or at MOI 100 with suspensions of Salmonella or E. coli. The medium from uninfected (control) cells was replaced with fresh DMEM + 10% HI-FCS. Before infection, where appropriate, cells were treated for 30 min at 37°C with DMEM + 10% HI-FCS containing cytochalasin D (5 μg/ml). In experiments designed to disrupt the internalization of bacteria by the macrophage cells, bacterial suspensions also contained 2 μg/ml cytochalasin D. Where NO synthesis was inhibited at the same time as bacterial infection, suspensions were produced in medium containing 100 μM 1400W. Inocula were determined by 10-fold serial dilution followed by viable counting according to the Miles-Misra method (48). Infected macrophage cells were incubated at 37°C, 5% CO2 for either 2 h (Salmonella and E. coli) or 4 h (N. meningitidis). At the end of each infection, the number of viable bacteria in each supernatant was determined before removal of the supernatant.
Preparation of cell lysates
Washed, postinfection J774.2 cells were lysed using 300 μl SNO-compatible lysis buffer (50 mM phosphate buffer, pH 7.4+1 mM DTPA+protease inhibitor cocktail+50 mM NEM). For measurements using tri-iodide (I3−)-dependent chemiluminescence, lysis buffer also contained 2% saponin (from Quillaja bark). Plates were incubated at 37°C, 5% CO2 for 15 min to allow the alkylation of free thiol. Cell debris was scraped off each well and transferred to a fresh tube, where they were kept on ice and in the dark. In saponized lysates, cell debris and bacterial cells were removed immediately by centrifugation at 10,000 g for 5 min; otherwise, samples were first subjected to 3 freeze–thaw cycles using liquid N2. Supernatants were transferred carefully to fresh tubes, and where appropriate, aliquots were pretreated for measurement of SNO. The remainder of each lysate was stored for protein assay at −20°C. In some experiments, lysates were dialyzed (2000 Da cutoff) overnight against 50 mM phosphate buffer + 1 mM DTPA at 4°C, prior to measurement of SNO concentration.
Measurement of SNO in liquid samples
Measurement of SNO was performed using ozone-based chemiluminescence. To release NO from S-nitrosothiol, the purging vessel contained either I3− solution, as described previously (47), or a buffered solution of Cu(I) and cysteine, as described by Gaston and co-workers (49). Tri-iodide-dependent SNO measurements required samples to be treated with either 10% (v/v) 100 mM sulfanilamide in 2 N HCl or 10% (v/v) HgCl2, followed by 10 min incubation on ice and in the dark. Hg2+-treated samples were then treated with 10% (v/v) acidified sulfanilamide and incubated on ice and in the dark for another 10 min. No Hg2+ treatment was performed on GSNO-supplemented DMEM. SNO was measured by duplicate injection into the appropriate solution at either 30°C (I3−) or 50°C (Cu(I)/Cys). The mean AUC for duplicate injections was used to determine the SNO concentration, with reference to a GSNO standard curve. SNO-ablated (Hg2+-treated) signal was corrected for extra dilution (×1.1) and the adjusted concentration was deducted from each sulfanilamide-treated sample. SNO concentrations of lysates were normalized to the protein concentration of each lysate (pmol SNO/mg protein).
Protein assay
Determination of the protein concentration in lysates was performed using either the RC DC Protein Assay Kit (Bio-Rad, Hercules, CA, USA) or Bio-Rad Protein Assay Reagent, according to the manufacturer’s instructions.
Statistical analysis
Data were analyzed for skewness using SPSS 14.0 for Windows (SPSS Inc., Chicago, IL, USA). Data were considered significantly skewed—and therefore nonparametrically distributed—if the skewness was equal to or exceeded double the value of the se of the skewness. Values presented in the text are means ± sd of parametrically distributed data or the median and interquartile range of nonparametrically distributed data, unless otherwise specified. Details of specific statistical tests are detailed in the figure legends. Results of statistical tests were considered significant at derived values of P ≤ 0.05.
RESULTS
NorB accelerates the decomposition of SNO in vitro
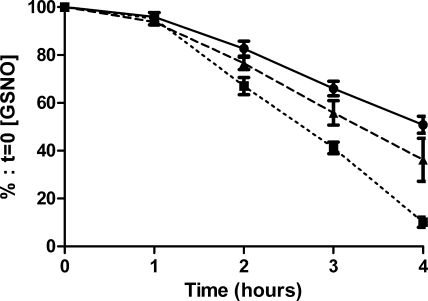
DMEM + 10% HI-FCS was spiked with the nitrosating compound GSNO to a final concentration of 100 μM and used to grow either wild-type N. meningitidis or the norB-mutant derivative. Inoculation of DMEM + 10% HI-FCS + 100 μM GSNO with wild-type N. meningitidis (Fig. 1) increased the rate of decomposition of SNO over 4 h, cf. uninfected medium. After 4 h, sterile medium retained 50.8 ± 10.7% of the initial SNO signal, while medium inoculated with wild-type bacteria retained only 10.1 ± 4.8%. In contrast, we found a nonsignificant increase in the rate of SNO decomposition in medium containing the norB strain (Fig. 1). After 4 h, norB-inoculated medium retained 36.2 ± 20.0% of the initial SNO signal. Both Neisseria strains had similar growth rates over the 4 h incubation period (data not shown).
Figure 1.
Effect of bacterial growth on the rate of degradation of GSNO in DMEM. Log phase wild-type N. meningitidis (squares; dotted line) or the norB-mutant derivative (triangles; broken line) were grown in DMEM + 10% HI-FCS supplemented with 100 μM GSNO (circles; solid line). SNO concentration was determined at intervals by duplicate injection of acidified sulfanilamide-treated medium into I3− reaction mixture and quantified by ozone-based chemiluminescence. Data are percentages of the starting SNO concentration. Individual points, representing means + se, were compared using unpaired samples t test. Area under curve for the three groups was compared by unpaired samples t test. n > 5. Error bars fall within the points where not visible.
Exposure to LPS and rmIFN-γ generates a prolonged enrichment of endogenously produced SNO in J774.2 murine macrophage cells by activating expression of iNOS
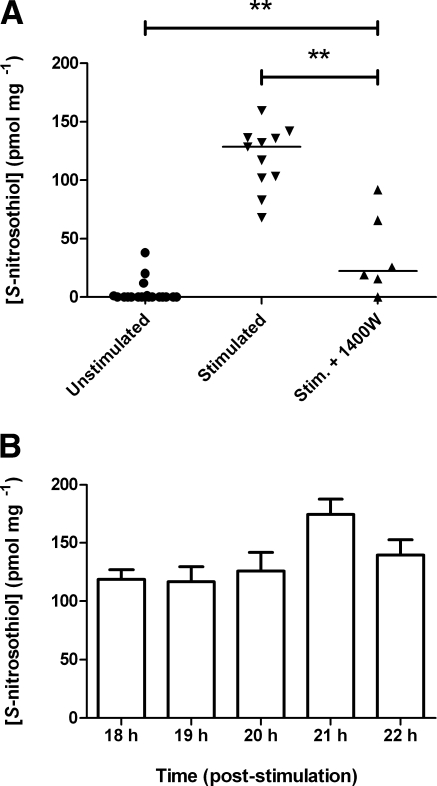
Stimulation of J774.2 murine macrophage cells with 1 μg/ml LPS + 1000 U/ml rmIFNγ in DMEM for 18 h increased the level of SNO in cell lysates (median 128.5 pmol/mg; interquartile range 101.7–136.3 pmol/mg), cf. lysates from untreated cells (median 0.00 pmol/mg; interquartile range 0.00–0.94 pmol/mg) (Fig. 2A). The iNOS inhibitor 1400W removed this effect (median 22.41 pmol/mg; interquartile range 11.7–72.3 pmol/mg). The high SNO concentration of stimulated J774.2 cell lysates remained detectable for 4 h after withdrawal of LPS and rmIFNγ (Fig. 2B). Throughout this period, we found no significant change in the SNO signal.
Figure 2.
Endogenous SNO generation is iNOS dependent, and SNO is stable for 4 h following removal of LPS and rmIFN-γ. A) Duplicate wells of J774.2 cells were incubated in either fresh DMEM (unstimulated) or medium containing 1 μg/ml LPS and 1000 U/ml rmIFN-γ (stimulated) for 18 h. iNOS inhibitor 1400 W (100 μM) was also added to medium (stim.+1400W). Horizontal bars denote medians; n > 6. **P < 0.001; Mann Whitney U test. B) SNO concentrations of stimulated J774.2 cells were measured hourly following removal of LPS and IFNγ and replacement of medium with fresh DMEM. Lysates were produced using SNO-compatible lysis buffer plus 2% saponin. SNO content was determined by duplicate injection into I3− reaction mixture linked to ozone-based chemiluminescence and normalized to the protein content of each lysate. Data are presented as SNO concentration (pmol SNO/mg protein). Bars denote means + se; n > 6; ANOVA with Tukey’s multiple comparisons test.
Infection of activated J774.2 murine macrophage cells with wild-type N. meningitidis results in a reduced abundance of endogenous SNO
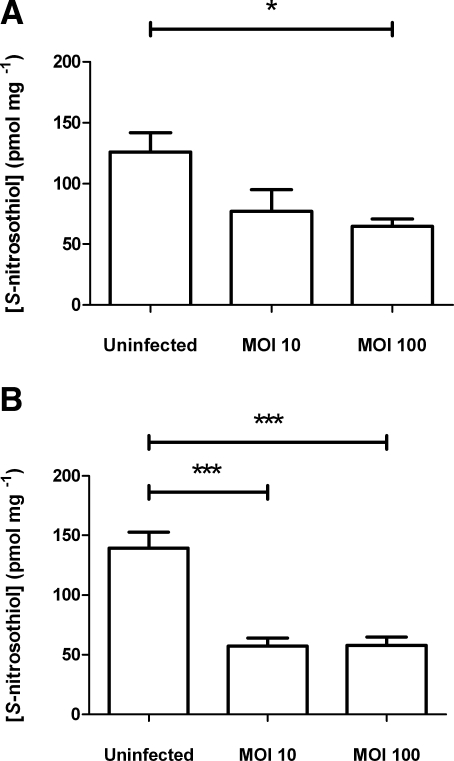
At 4 h after infection of J774.2 cells with N. meningitidis (Fig. 3B), the SNO concentration in stimulated but uninfected J774.2 cells (139.7±52.2 pmol/mg) was reduced significantly by coincubation with wild-type bacteria. Macrophages were infected at either MOI 10 or 100, but after 4 h, lysed macrophages contained similar concentrations of SNO (∼57 pmol/mg), in each case significantly lower than those of uninfected cells. After overnight dialysis of lysates produced from wild-type infected cells, the distribution of SNO was determined to be 35% low-molecular-weight SNO (<2000 Da) (data not shown). At 2 h after infection, while the sizes of the two wild-type populations (MOI 10 and 100) were similar (data not shown), cells infected at MOI 100 contained significantly lower concentrations of SNO (64.5±13.9 pmol/mg) than uninfected cells (126.0±47.4 pmol/mg), in contrast to cells infected at MOI 10 (77.3±39.5 pmol/mg) (Fig. 3A).
Figure 3.
Effect of infection by wild type N. meningitidis on endogenously produced SNO in activated J774.2 cells. At t = 0 h, duplicate wells of J774.2 cells were activated with 1 μg/ml LPS and 1000 U/ml rmIFN-γ for 18 h and then infected with a suspension of log-phase wild-type N. meningitidis in fresh medium at MOI 10 or 100 for 2 h (t=20 h) (A) or 4 h (t=22 h) (B). Lysates were produced using SNO-compatible lysis buffer plus 2% saponin. SNO content was determined by duplicate injection into I3− reaction mixture linked to ozone-based chemiluminescence and normalized to the protein concentration of the lysate. Data are presented as SNO concentration (pmol SNO/mg protein). Bars denote means + se. *P < 0.05, ***P < 0.001; ANOVA with Tukey’s multiple comparisons test.
NorB of N. meningitidis is associated with reduced abundance of SNO following infection of J774.2 murine macrophage cells
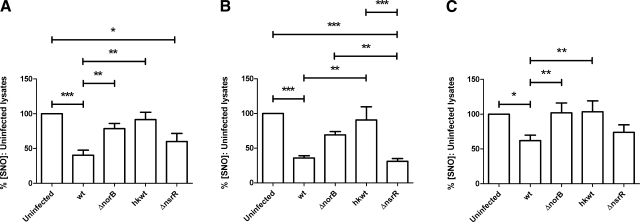
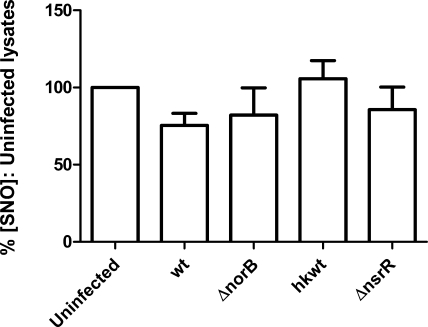
Infection of stimulated J774.2 cells with N. meningitidis strains expressing functional NorB (i.e., wild-type MC58 and the nsrR-mutant derivative) resulted in reduced abundance of host cell SNO (Fig. 4A). Stimulated but uninfected cells contained 139.7±52.2 pmol/mg SNO (100%), whereas wild-type infected and nsrR-infected cells contained only 40.5 ± 18.5 and 60.2 ± 28.6% as much SNO, respectively (Fig. 4A). Conversely, infection of stimulated cells with bacteria lacking functional NorB, either due to the effective deletion of the norB gene (i.e., norB-mutant derivative) or the metabolic inactivity of the bacterium (heat-killed wild-type MC58) did not result in significantly different concentrations of macrophage cell SNO. Cells infected with norB-mutant bacteria produced lysates containing 78.7 ± 22.3% of the SNO concentration measured in uninfected cells, while cells infected with heat-killed wild-type bacteria contained 91.6 ± 25.6% as much SNO as uninfected cells (Fig. 4A). In keeping with previous findings that the growth of the norB-mutant strain is attenuated in human MDMs (27), viable counts of bacteria at 4 h postinfection showed that significantly fewer of these bacteria were present per J774.2 cell than of either the wild-type or nsrR-mutant strain (data not shown). To overcome this confounding factor, we exposed cells to the f-actin polymerization inhibitor cytochalasin D, which itself had no effect on the concentration of cellular SNO, compared to uninfected cells without cytochalasin D treatment (data not shown). Inhibition of phagocytosis in this manner eliminated the difference in numbers of bacteria per J774.2 cell (data not shown). The concentration of SNO in uninfected, cytochalasin-treated cells was 106.8 pmol/mg (median; interquartile range 68.57–184.5 pmol/mg). Infection of cytochalasin-treated cells with wild-type or mutant meningococci resulted in a similar pattern of SNO abundance to that observed in untreated cells (Fig. 4B). From these findings, it is concluded that the differences in SNO depletion between NorB-expressing wild-type and the mutant are not due to intracellular attenuation of bacterial growth.
Figure 4.
Effect of NorB on endogenously produced SNO in J774.2 cells. A, B) At t = 0 h, J774.2 cells were activated with 1 μg/ml LPS and 1000 U/ml rmIFNγ for 18 h. Cells then remained untreated (A) or were treated with 5 μg/ml cytochalasin D for 30 min at 37°C (B). Duplicate wells of J774.2 cells were then incubated for 4 h with suspensions of log-phase N. meningitidis in fresh medium (A) or medium containing 2 μg/ml cytochalasin D (B) plus wild-type strain MC58 (wt), mutant lacking NorB (ΔnorB), heat-killed wild type strain MC58 (hkwt), and mutant lacking NsrR repressor protein (ΔnsrR). Uninfected samples were incubated with fresh medium or medium plus cytochalasin D only. Lysates were produced using SNO-compatible lysis buffer plus 2% saponin. SNO content was determined by duplicate injection into I3− reaction mixture linked to ozone-based chemiluminescence and normalized to the protein concentration of each lysate. C) Cells were treated as in B, but lysates were produced in SNO-compatible lysis buffer using 3 freeze-thaw cycles. SNO measurements were determined by injection into phosphate-buffered saline (pH 7.0) containing a saturating concentration of Cu(I) and 1 mM l-cysteine, linked to ozone-based chemiluminescence. Data are percentages of SNO concentration in uninfected lysates. Bars denote means + se; n > 6. *P < 0.05, **P < 0.01, ***P < 0.001; ANOVA with Tukey’s multiple comparisons test.
In an identical series of experiments using cytochalasin D, we corroborated our initial findings in I3− using a phosphate-buffered solution (pH 7.0) containing a saturating concentration of Cu(I) ion and 1 mM l-cysteine (Fig. 4C). Wild type-infected and nsrR-infected macrophage cells contained a reduced abundance of SNO compared to uninfected controls (62.0±20.4 and 73.8±28.6%, respectively), while cells infected with strains unable to detoxify NO contained SNO concentrations exceeding those of control cells (norB, 101.9±37.4%; heat-killed wild-type, 103.4±41.4%).
Bacterial NO detoxification prevents new SNO formation
To determine whether NorB activity degrades preformed SNO or prevents new SNO formation, we added the iNOS inhibitor 1400W to the macrophage cells at the point of infection (t=18 h). Exposure of stimulated J774.2 cells to 1400W resulted in a significant reduction in cellular SNO concentrations over 4 h cf. uninfected cells not exposed to the inhibitor (data not shown). The SNO concentration in 1400W-treated, uninfected cells was 32.5 ± 10.1 pmol/mg. Infection of 1400W-treated cells with any strain of N. meningitidis had no effect on the abundance of SNO in these cells (Fig. 5).
Figure 5.
Effect of inhibiting iNOS activity at the point of infection. J774.2 cells were activated with 1 μg/ml LPS and 1000 U/ml rmIFN-γ for 18 h, then infected with a suspension of log-phase N. meningitidis in fresh medium containing 100 μM 1400W. Strains were identical to those in Fig. 4. Lysates were produced using SNO-compatible lysis buffer plus 2% saponin. SNO content was determined by duplicate injection into I3− reaction mixture linked to ozone-based chemiluminescence and normalized to the protein concentration of each lysate. Data are percentages of SNO concentration in uninfected lysates. Bars denote means + se.
Hmp is also associated with reduced SNO abundance in infected J774.2 murine macrophage cells
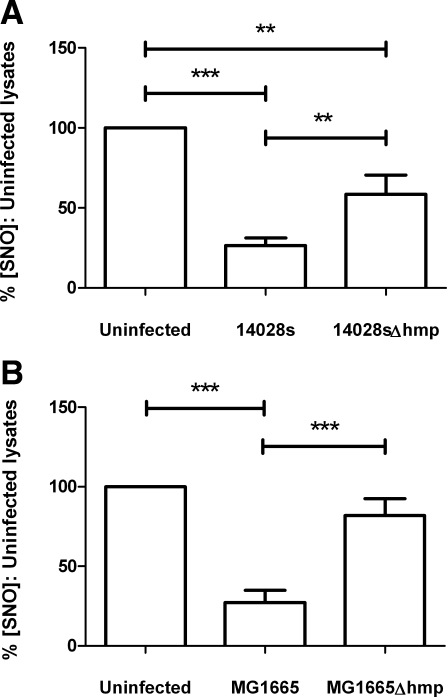
Infection of stimulated J774.2 cells with the gram-negative enteric bacteria Salmonella enterica spp. Typhimurium (strain 14028s) or E. coli (strain MG1655) also caused a reduced abundance of macrophage SNO (Fig. 6). 14028s-infected cells produced lysates containing only 26.4 ± 15.2% of the SNO concentrations measured in uninfected cells, which in this experimental series was determined to be 53.7 ± 19.2 pmol/mg (Fig. 6A). This activity was Hmp dependent; although 14028sΔhmp-infected cells yielded significantly less SNO than uninfected cells (58.5±36.2% of controls); these concentrations were significantly higher than those of cells infected with 14028s (Fig. 6A).
Figure 6.
Effect of Hmp on endogenously produced SNO in J774.2 cells following infection with S. enterica serovar Typhimurium (14028s) (A) or E. coli (MG1665) (B). At t = 0 h, duplicate wells of J774.2 cells were activated with 1 μg/ml LPS and 1000 U/ml rmIFN-γ for 18 h, then infected at MOI 100 with a suspension of either wild-type or Δhmp S. typhimurium serovar Typhimurium or E. coli for 2 h. Lysates were produced using SNO-compatible lysis buffer plus 2% saponin. SNO content was determined by duplicate injection into I3− reaction mixture linked to ozone-based chemiluminescence and normalized to the protein concentration of each lysate. Data are percentages of SNO concentration in uninfected lysates. Bars denote means + se;n = 9 (A), n = 7 (B). **P < 0.01, ***P < 0.001; repeated measures ANOVA with Tukey’s multiple comparisons test.
A different pattern was observed following infection of stimulated J774.2 cells with wild-type and mutant E. coli strains (Fig. 6B). Deletion of the hmp gene from strain MG1655 removed the ability of E. coli to influence macrophage SNO concentrations significantly. MG1655-infected cells produced lysates containing only 27.3 ± 20.2% of the SNO in uninfected cells, which were measured as 62.57 ± 27.97 pmol/mg in these experiments. However, no significant difference was found between the SNO concentration of lysates produced from uninfected and MG1665Δhmp-infected cells (Fig. 6B).
DISCUSSION
This work has shown that the process of bacterial NO detoxification, vital to the survival of pathogenic organisms under conditions of nitrosative stress, can prevent the formation of host cell SNO. Both the NorB of N. meningitidis and the Hmp of the enteric pathogens S. enterica and E. coli, which detoxify NO by different mechanisms in different cellular environments, have been demonstrated to inhibit the formation of SNO in infected macrophage cells significantly. To our knowledge, this represents the first observation of its kind: that bacterial energy metabolism can disrupt the formation of a specific chemical adduct inside host cells, one that is regulatory for a number of diverse host cell processes.
This study has demonstrated the capacity for a combination of LPS and IFN-γ to induce the formation of high concentrations of endogenously formed SNO in murine macrophage-like cells, through the expression of iNOS. Although the chemical species responsible for S-nitrosylation per se remains contentious, NO—as a precursor molecule—is necessary for the formation of these adducts. In this simplistic view, NO represents one of the two most important substrates for S-nitrosylation; the other is thiol. Our data support similar findings by Zhang and Hogg (46), who could generate SNO in the RAW 264.7 murine macrophage cell line following its exposure to LPS. The values measured in our study fall within the same range as those of Zhang and Hogg (46) (pmol SNO/mg protein) but exceed their average measurement (17±1.0 pmol/mg) between 4- and 8-fold in lysates produced from stimulated but uninfected cells. These differences are not unexpected, given the pleiotropic effects of IFN-γ on immune cells (50). Inclusion of the specific iNOS inhibitor 1400W demonstrated that the formation of SNO in our model was largely but not exclusively dependent on the activity of this enzyme (Fig. 2). In this situation, 1400W effectively removed one of the substrates necessary for SNO formation. With impaired NO synthesis, as expected, we found a reduced abundance of S-nitrosylating species. As such, S-nitrosylation was impaired, and intracellular SNO concentrations remained comparatively low.
Our evidence suggests that bacteria influence the concentration of host cell SNO by an analogous mechanism, i.e., the removal of S-nitrosylating substrate through the detoxification of NO. Infection of LPS and IFNγ-stimulated, SNO-loaded J774.2 cells with wild-type N. meningitidis significantly reduced the abundance of host cell SNO after either 2 or 4 h, dependent on the size of the bacterial inoculum (Fig. 3). This observation could reflect either depletion by the bacteria of preexisting SNO, where the presence of the infecting organism either directly or indirectly accelerates the breakdown of S-nitrosothiol bonds, or the prevention of the formation of new S-nitrosothiol. The data collected in this study support the latter hypothesis.
Figure 4 demonstrates the association of reduced SNO abundance with functional NorB, which was observed after infection with either wild-type N. meningitidis or a mutant strain overexpressing the nitric oxide reductase, nsrR (51). Conversely, identical J774.2 cells infected with bacteria lacking NorB function contained SNO concentrations similar to those found in uninfected cells (Fig. 4). Similarly, we demonstrated an association between reduced cellular SNO abundance and the bacterial flavohemoprotein Hmp, which is at least partially responsible for reducing SNO abundance during infection with either S. enterica or E. coli (Fig. 6). Both Δhmp strains exhibited impaired depletion of cellular SNO compared to wild-type, though in E. coli the effect of deleting this gene was more pronounced. SNO concentrations in MG1655Δhmp-infected cells were similar to those in uninfected cells (Fig. 6B). Conversely, Hmp-deficient Salmonella could reduce SNO abundance in J774.2 cells significantly, although Hmp appeared to account for ∼50% of this activity (Fig. 6A). This difference between bacterial species was unexpected given the similarities between Salmonella and E. coli Hmp, which have been shown to have similar NADH reductase activities and ligand-binding affinities in vitro (52). The differences measured in this study might, therefore, be due to differences in the relative expression levels of Hmp, the availability of oxygen for the denitrosylase reaction, or perhaps different contributions of alternative NO detoxification mechanisms, such as the NO reductase NorVW (41).
For both strains of enteric bacteria and for N. meningitidis, the association of NO detoxification machinery with a reduced abundance of host cell SNO suggests that the effect takes place through the detoxification of NO per se rather than through direct protein–protein interactions, akin to the mechanism shown for the thioredoxin/thioredoxin reductase system (21) and GSNO reductase (20). It is unlikely that these bacterial proteins physically interact with host cell SNO, given their subcellular localization to either the inner bacterial membrane (NorB) or the cytoplasm (Hmp). While NO is itself freely diffusible in biological systems, SNO requires transport across membranes (53).
By simultaneously inhibiting iNOS activity and infecting stimulated J774.2 cells with meningococci, we have shown that bacterial mechanisms for NO detoxification act to prevent de novo SNO formation rather than accelerate the decomposition of preexisting SNO (Fig. 5). Addition of 1400W at the start of the infection significantly impaired NO synthesis and removed one of the substrates necessary for SNO formation. In uninfected cells, the inhibition of NO synthesis prevented the replenishment of SNO, which continued to degrade and be degraded by cellular denitrosylation mechanisms. The result was a gradual decline in cellular SNO concentration, similar to observations made by Zhang and Hogg in RAW264.7 cells (46). If the activity of NorB were increasing the rate of degradation of preexisting SNO, then infection of cells with NorB-expressing strains would have caused further reduction in SNO concentrations, over and above the normal rate. Therefore, we would expect infection with the wild-type and nsrR-mutant strains to result in far less host cell SNO compared to the other samples. Instead, Fig. 5 shows no significant difference between the cellular SNO concentrations of J774.2 cells infected with any strain of N. meningitidis. This observation is consistent with the proposal that, because the rate of NO diffusion exceeds any rate of NO oxidation, cellular SNO formation involves NO that has first diffused out of, then back into a given cell (54). Addition of extracellular hemoglobin to LPS-stimulated RAW cells resulted in a reduced abundance of SNO (46), which was attributed to the failure of NO to return from the extracellular milieu after having been effectively scavenged (54). The activity of meningococcal NorB may be acting analogously to hemoglobin in our infection model, preventing NO that has diffused out of the cell from returning and forming SNO, by way of its reduction to N2O.
An alternative mechanism, not addressed experimentally in this work, may be that the SNO-depletion phenomenon is due to some effect of NO detoxification on amino acid transport. Since NO donation can enhance cystine transport across mammalian cell membranes (55), it is conceivable that bacterial modulation of NO concentration results in selective loss of SNO-associated amino acids (i.e., S-nitroso-l-cysteine). Transmembrane movement of S-nitroso-l-cysteine is mediated by system L amino acid transporters (56). However, in a rat model of bacterial sepsis, no significant difference was observed in the activity of amino acid transport system L between septic and control animals (57).
According to the data reported in Fig. 3, infection with wild-type meningococci results in the attainment of a lower, steady-state concentration of host cell SNO, the formation of which is not prevented by bacteria (Fig. 3). A lower inoculum of wild-type bacteria could not prevent a rise in host cell SNO over 2 h (Fig. 3A; MOI 10) but after 4 h had replicated to a population size sufficient to maintain this basal SNO concentration (Fig. 3B; MOI 10). The reduction in SNO concentration observed between 2 and 4 h represents the continued degradation/denitrosylation of SNO in the absence of new SNO formation, and a gradual return to basal levels. Larger inocula were sufficient to prevent any significant rises in SNO concentration (Fig. 3; MOI 100), but a detectable concentration of SNO remained in these cells (∼57 pmol/mg). This steady state may be evidence of a core subset of stable S-nitrosylated thiol, which corroborates the findings of Paige et al. (58). In a recent proteomic study, this group identified a subset of 10 proteins containing thiol-insensitive SNO, which they hypothesize to be the most likely mediators of the persistent cellular effects of NO (58). Their work divides SNOs into stable and unstable groups, the former of which shields the NO-cysteine bond from degradation. These observations are difficult to reconcile with the idea that bacteria metabolize NO before SNO formation can take place, especially given the demonstrated colocalization of NO synthesis with SNO formation (17) and the evidence that S-nitrosylation is compartmentalized (59). While we have postulated that bacterial NO detoxification prevents the formation of new SNO, it is possible that there is limited S-nitrosylation in the immediate vicinity of NOS to form so-called stable SNO. This is plausible if we assume that the qualities that stabilize the S-nitrosothiol bond also encourage its formation (60). In this scenario, therefore, bacterial NO detoxification is preventing the formation of “unstable” SNO.
Rapidly accumulating evidence indicates the critical importance of homeostatic regulation of SNO concentration in health and disease. Indeed, S-nitrosylation and its dysregulation are implicated in a number of chronic diseases, including amyotrophic lateral sclerosis (61), Parkinson’s disease (62, 63), multiple sclerosis (64), and Alzheimer’s disease (65). Deletion of GSNOR in mice is associated with higher systemic SNO concentrations and is protective against pathologies such as airway hyperresponsivity (66) but also increases mortality in endotoxemia (67). Conversely, humans with asthma have higher concentrations of GSNOR in bronchoalveolar lavage (BAL) fluid compared to nonasthmatic individuals and reduced abundance of pulmonary SNO (68). Our findings suggest a potential role for S-nitrosylation, or at least its dysregulation, in sepsis. Rather than regulated denitrosylation, the effect we have described results in wholesale SNO dysregulation, which we predict will have important consequences for the behavior of host cells, and therefore the natural history of infectious disease. Fine dissection of the physiological consequences of this phenomenon is likely to be difficult, given the apparent ubiquity of S-nitrosylation. While the S-nitrosylation of one protein may positively influence or stimulate a particular cellular process, the same process may be affected adversely or inhibited by S-nitrosylation of a different target. However, the evidence that SNO depletion is mediated by at least two NO detoxification mechanisms in three different mammalian pathogens supports the hypothesis that this phenomenon is a previously uncharacterized mechanism of septic disease. The bacterial burdens used in our system are physiologically relevant, given that studies of meningococcal septicemia, for example, have shown titers in the region of 108 bacterial genome copies/ml blood, occurring commonly in infected people (69). Similarly, the abundance of enteric bacteria in the intestine make it likely that, in these oxygen-poor, NO-rich environments, cells in intimate contact with bacteria under these conditions (i.e., endothelial cells and macrophages) are likely to experience SNO depletion and that soluble SNOs, such as plasma S-nitrosoalbumin, are also likely to be depleted.
We posit that bacterial proteins capable of detoxifying NO can remove freely diffusible NO from the cellular environment at a rate sufficient to prevent the formation of unstable SNO inside host cells. The potential pathophysiological consequences of this phenomenon are considerable and are currently under investigation.
Acknowledgments
We acknowledge the assistance of Prof. Neil Hogg for advice on the measurement of SNO using the Sievers NOA and of Dr. Jonathan Shaw for advice in the preparation of this manuscript. This work has been supported by grants to R.C.R. from the Wellcome Trust (grant 069791) and the Sheffield Hospitals Charitable Trust (grant 7866). J.R.L. and R.C.R. are supported by the Meningitis Research Foundation (project 0902.0).
References
- Stamler J S, Jaraki O, Osborne J, Simon D I, Keaney J, Vita J, Singel D, Valeri C R, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci U S A. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick J B, Schonhoff C M. Nitrosylation: the next phosphorylation? Arch Biochem Biophys. 2002;408:1–6. doi: 10.1016/s0003-9861(02)00490-3. [DOI] [PubMed] [Google Scholar]
- Hara M R, Snyder S H. Nitric. oxide-GAPDH-Siah: A novel cell death cascade. Cell Mol Neurobiol. 2006;26:527–538. doi: 10.1007/s10571-006-9011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y M, Talanian R V, Billiar T R. Nitric oxide inhibits apoptosis by preventing increases in caspase-3-like activity via two distinct mechanisms. J Biol Chem. 1997;272:31138–31148. doi: 10.1074/jbc.272.49.31138. [DOI] [PubMed] [Google Scholar]
- Li J, Billiar T R, Talanian R V, Kim Y M. Nitric oxide reversibly inhibits seven members of the caspase family via S-nitrosylation. Biochem Biophys Res Commun. 1997;240:419–424. doi: 10.1006/bbrc.1997.7672. [DOI] [PubMed] [Google Scholar]
- Mannick J B, Miao X Q, Stamler J S. Nitric oxide inhibits Fas-induced apoptosis. J Biol Chem. 1997;272:24125–24128. doi: 10.1074/jbc.272.39.24125. [DOI] [PubMed] [Google Scholar]
- Lander H M, Hajjar D P, Hempstead B L, Mirza U A, Chait B T, Campbell S, Quilliam L A. A molecular redox switch on p21(ras). Structural basis for the nitric oxide-p21(ras) interaction. J Biol Chem. 1997;272:4323–4326. doi: 10.1074/jbc.272.7.4323. [DOI] [PubMed] [Google Scholar]
- Jaffrey S R, Erdjument-Bromage H, Ferris C D, Tempst P, Snyder S H. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- Marshall H E, Stamler J S. Inhibition of NF-kappa B by S-nitrosylation. Biochemistry. 2001;40:1688–1693. doi: 10.1021/bi002239y. [DOI] [PubMed] [Google Scholar]
- Rabkin S W, Klassen S S. Nitric oxide differentially regulates the gene expression of caspase genes but not some autophagic genes. Nitric Oxide. 2007;16:339–347. doi: 10.1016/j.niox.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Reynaert N L, Ckless K, Korn S H, Vos N, Guala A S, Wouters E F, van der Vliet A, Janssen-Heininger Y M. Nitric oxide represses inhibitory kappaB kinase through S-nitrosylation. Proc Natl Acad Sci U S A. 2004;101:8945–8950. doi: 10.1073/pnas.0400588101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A J, Chen Q, Hess D T, Day B J, Ischiropoulos H, Stamler J S. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J Biol Chem. 2002;277:9637–9640. doi: 10.1074/jbc.C100746200. [DOI] [PubMed] [Google Scholar]
- Hoffmann J, Dimmeler S, Haendeler J. Shear stress increases the amount of S-nitrosylated molecules in endothelial cells: important role for signal transduction. FEBS Lett. 2003;551:153–158. doi: 10.1016/s0014-5793(03)00917-7. [DOI] [PubMed] [Google Scholar]
- Sun J H, Xin C L, Eu J P, Stamler J S, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci U S A. 2001;98:11158–11162. doi: 10.1073/pnas.201289098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett C M, Whorton A R. S-nitrosoglutathione reversibly inhibits GAPDH by S-nitrosylation. Am J Physiol. 1995;269:739–749. doi: 10.1152/ajpcell.1995.269.3.C739. [DOI] [PubMed] [Google Scholar]
- Fang M, Jaffrey S R, Sawa A, Ye K, Luo X, Snyder S H. Dexras1: a G protein specifically coupled to neuronal nitric oxide synthase via CAPON. Neuron. 2000;28:183–193. doi: 10.1016/s0896-6273(00)00095-7. [DOI] [PubMed] [Google Scholar]
- Iwakiri Y, Satoh A, Chatterjee S, Toomre D K, Chalouni C M, Fulton D, Groszmann R J, Shah V H, Sessa W C. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc Natl Acad Sci U S A. 2006;103:19777–19782. doi: 10.1073/pnas.0605907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D T, Matsumoto A, Kim S O, Marshall H E, Stamler J S. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- Singh R J, Hogg N, Joseph J, Kalyanaraman B. Mechanism of nitric oxide release from S-nitrosothiols. J Biol Chem. 1996;271:18596–18603. doi: 10.1074/jbc.271.31.18596. [DOI] [PubMed] [Google Scholar]
- Liu L, Hausladen A, Zeng M, Que L, Heitman J, Stamler J S. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- Benhar M, Forrester M T, Hess D T, Stamler J S. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch T, Kuhlen R, Knorr M, Kelly K, Lewandowski K, Rossaint R, Falke K J, Gerlach H. Nasal, pulmonary and autoinhaled nitric oxide at rest and during moderate exercise. Intensive Care Med. 2000;26:391–399. doi: 10.1007/s001340051172. [DOI] [PubMed] [Google Scholar]
- Gerlach H, Rossaint R, Pappert D, Knorr M, Falke K J. Autoinhalation of nitric oxide after endogenous synthesis in nasopharynx. Lancet. 1994;343:518–519. doi: 10.1016/s0140-6736(94)91465-6. [DOI] [PubMed] [Google Scholar]
- Kimberly B, Nejadnik B, Giraud G D, Holden W E. Nasal contribution to exhaled nitric oxide at rest and during breathholding in humans. Am J Resp Crit Care Med. 1996;153:829–836. doi: 10.1164/ajrccm.153.2.8564139. [DOI] [PubMed] [Google Scholar]
- Rock J D, Mahnane M R, Anjum M F, Shaw J G, Read R C, Moir J W. The pathogen Neisseria meningitidis requires oxygen, but supplements growth by denitrification. Nitrite nitric oxide and oxygen control respiratory flux at genetic and metabolic levels. Mol Microbiol. 2005;58:800–809. doi: 10.1111/j.1365-2958.2005.04866.x. [DOI] [PubMed] [Google Scholar]
- Anjum M F, Stevanin T M, Read R C, Moir J W. Nitric oxide metabolism in Neisseria meningitidis. J Bacteriol. 2002;184:2987–2993. doi: 10.1128/JB.184.11.2987-2993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin T M, Moir J W, Read R C. Nitric oxide detoxification systems enhance survival of Neisseria meningitidis in human macrophages and in nasopharyngeal mucosa. Infect Immun. 2005;73:3322–3329. doi: 10.1128/IAI.73.6.3322-3329.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin T M, Laver J R, Poole R K, Moir J W, Read R C. Metabolism of nitric oxide by Neisseria meningitidis modifies release of NO-regulated cytokines and chemokines by human macrophages. Microbes Infect. 2007;9:981–987. doi: 10.1016/j.micinf.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Tunbridge A J, Stevanin T M, Lee M, Marriott H M, Moir J W, Read R C, Dockrell D H. Inhibition of macrophage apoptosis by Neisseria meningitidis requires nitric oxide detoxification mechanisms. Infect Immun. 2006;74:729–733. doi: 10.1128/IAI.74.1.729-733.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurlier K, Thomson M J, Aziz N, Moir J W. The nitric oxide (NO)-sensing repressor NsrR of Neisseria meningitidis has a compact regulon of genes involved in NO synthesis and detoxification. J Bacteriol. 2008;190:2488–2495. doi: 10.1128/JB.01869-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R C. A brief history of hemoglobins: plant, animal, protist, and bacteria. Proc Natl Acad Sci U S A. 1996;93:5675–5679. doi: 10.1073/pnas.93.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Riggs A F. Yeast flavohemoglobin is an ancient protein related to globins and a reductase family. Proc Natl Acad Sci U S A. 1992;89:5015–5019. doi: 10.1073/pnas.89.11.5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Urzua E, Mills C E, White G P, Contreras-Zentella M L, Escamilla E, Vasudevan S G, Membrillo-Hernandez J, Poole R K. Flavohemoglobin Hmp, but not its individual domains, confers protection from respiratory inhibition by nitric oxide in Escherichia coli. J Biol Chem. 2003;278:34975–34982. doi: 10.1074/jbc.M303629200. [DOI] [PubMed] [Google Scholar]
- Poole R K, Ioannidis N, Orii Y. Reactions of the Escherichia coli flavohaemoglobin (Hmp) with oxygen and reduced nicotinamide adenine dinucleotide: evidence for oxygen switching of flavin oxidoreduction and a mechanism for oxygen sensing. Proc Biol Sci. 1994;255:251–258. doi: 10.1098/rspb.1994.0036. [DOI] [PubMed] [Google Scholar]
- Hausladen A, Gow A, Stamler J S. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc Natl Acad Sci U S A. 2001;98:10108–10112. doi: 10.1073/pnas.181199698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole R K, Hughes M N. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol Microbiol. 2000;36:775–783. doi: 10.1046/j.1365-2958.2000.01889.x. [DOI] [PubMed] [Google Scholar]
- Kim S O, Orii Y, Lloyd D, Hughes M N, Poole R K. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 1999;445:389–394. doi: 10.1016/s0014-5793(99)00157-x. [DOI] [PubMed] [Google Scholar]
- Membrillo-Hernandez J, Coopamah M D, Channa A, Hughes M N, Poole R K. A novel mechanism for upregulation of the Escherichia coli K-12 hmp (flavohaemoglobin) gene by the “NO releaser”, S-nitrosoglutathione: nitrosation of homocysteine and modulation of MetR binding to the glyA-hmp intergenic region. Mol Microbiol. 1998;29:1101–1112. doi: 10.1046/j.1365-2958.1998.01000.x. [DOI] [PubMed] [Google Scholar]
- Stevanin T M, Ioannidis N, Mills C E, Kim S O, Hughes M N, Poole R K. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo’ or bd, from nitric oxide. J Biol Chem. 2000;275:35868–35875. doi: 10.1074/jbc.M002471200. [DOI] [PubMed] [Google Scholar]
- Stevanin T M, Read R C, Poole R K. The hmp gene encoding the NO-inducible flavohaemoglobin in Escherichia coli confers a protective advantage in resisting killing within macrophages, but not in vitro: links with swarming motility. Gene. 2007;398:62–68. doi: 10.1016/j.gene.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Bang I S, Liu L, Vazquez-Torres A, Crouch M L, Stamler J S, Fang F C. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin hmp. J Biol Chem. 2006;281:28039–28047. doi: 10.1074/jbc.M605174200. [DOI] [PubMed] [Google Scholar]
- Gilberthorpe N J, Lee M E, Stevanin T M, Read R C, Poole R K. NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-γ-stimulated J774.2 macrophages. Microbiology. 2007;153:1756–1771. doi: 10.1099/mic.0.2006/003731-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin T M, Poole R K, Demoncheaux E A, Read R C. Flavohemoglobin Hmp protects Salmonella enterica serovar typhimurium from nitric oxide-related killing by human macrophages. Infect Immun. 2002;70:4399–4405. doi: 10.1128/IAI.70.8.4399-4405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Keshive M, Deen WM. Diffusion and reaction of nitric oxide in suspension cell cultures. Biophys J. 1998;75:745–754. doi: 10.1016/S0006-3495(98)77564-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eu J P, Liu L, Zeng M, Stamler J S. An apoptotic model for nitrosative stress. Biochemistry. 2000;39:1040–1047. doi: 10.1021/bi992046e. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hogg N. Formation and stability of S-nitrosothiols in RAW 264.7 cells. Am J Physiol Lung Cell Mol Physiol. 2004;287:L467–L474. doi: 10.1152/ajplung.00350.2003. [DOI] [PubMed] [Google Scholar]
- Laver J R, Stevanin T M, Read R C. Chemiluminescence quantification of NO and its derivatives in liquid samples. Methods Enzymol. 2008;436:113–127. doi: 10.1016/S0076-6879(08)36007-8. [DOI] [PubMed] [Google Scholar]
- Miles A A, Misra S S. The estimation of the bactericidal power of the blood. J Hygiene. 1938;38:732–749. doi: 10.1017/s002217240001158x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang K, Ragsdale N V, Carey R M, MacDonald T, Gaston B. Reductive assays for S-nitrosothiols: implications for measurements in biological systems. Biochem Biophys Res Commun. 1998;252:535–540. doi: 10.1006/bbrc.1998.9688. [DOI] [PubMed] [Google Scholar]
- Boehm U, Klamp T, Groot M, Howard J C. Cellular responses to interferon-gamma. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- Rock J D, Thomson M J, Read R C, Moir J W. Regulation of denitrification genes in Neisseria meningitidis by nitric oxide and the repressor. NsrR J Bacteriol. 2007;189:1138–1144. doi: 10.1128/JB.01368-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farres J, Rechsteiner M P, Herold S, Frey A D, Kallio P T. Ligand binding properties of bacterial hemoglobins and flavohemoglobins. Biochemistry. 2005;44:4125–4134. doi: 10.1021/bi047389d. [DOI] [PubMed] [Google Scholar]
- Broniowska K A, Zhang Y, Hogg N. Requirement of transmembrane transport for S-nitrosocysteine-dependent modification of intracellular thiols. J Biol Chem. 2006;281:33835–33841. doi: 10.1074/jbc.M603248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster J R, Jr, Gaston B. NO and nitrosothiols: spatial confinement and free diffusion. Am J Physiol Lung Cell Mol Physiol. 2004;287:L465–L466. doi: 10.1152/ajplung.00151.2004. [DOI] [PubMed] [Google Scholar]
- Li H, Marshall Z M, Whorton A R. Stimulation of cystine uptake by nitric oxide: regulation of endothelial cell glutathione levels. Am J Physiol. 1999;276:C803–C811. doi: 10.1152/ajpcell.1999.276.4.C803. [DOI] [PubMed] [Google Scholar]
- Li S, Whorton A R. Functional characterization of two S-nitroso-L-cysteine transporters, which mediate movement of NO equivalents into vascular cells. Am J Physiol Cell Physiol. 2007;292:C1263–C1271. doi: 10.1152/ajpcell.00382.2006. [DOI] [PubMed] [Google Scholar]
- James J H, Hasselgren P O, Hummel R P, 3rd, Warner B W, Fischer J E. Effect of sepsis on amino acid transport system A and its response to insulin in incubated rat skeletal muscle. Metabolism. 1990;39:335–340. doi: 10.1016/0026-0495(90)90245-8. [DOI] [PubMed] [Google Scholar]
- Paige J S, Xu G, Stancevic B, Jaffrey S R. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannick J B, Schonhoff C, Papeta N, Ghafourifar P, Szibor M, Fang K, Gaston B. S-Nitrosylation of mitochondrial caspases. J Cell Biol. 2001;154:1111–1116. doi: 10.1083/jcb.200104008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamler J S, Toone E J, Lipton S A, Sucher N J. (S)NO. signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–696. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- Schonhoff C M, Matsuoka M, Tummala H, Johnson M A, Estevez A G, Wu R, Kamaid A, Ricart K C, Hashimoto Y, Gaston B, Macdonald T L, Xu Z, Mannick J B. S-nitrosothiol depletion in amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2006;103:2404–2409. doi: 10.1073/pnas.0507243103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung K K, Thomas B, Li X, Pletnikova O, Troncoso J C, Marsh L, Dawson V L, Dawson T M. S-nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- Fang J, Nakamura T, Cho D H, Gu Z, Lipton S A. S-nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson’s disease. Proc Natl Acad Sci U S A. 2007;104:18742–18747. doi: 10.1073/pnas.0705904104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzozero O A, DeJesus G, Bixler H A, Pastuszyn A. Evidence of nitrosative damage in the brain white matter of patients with multiple sclerosis. Neurochem Res. 2005;30:139–149. doi: 10.1007/s11064-004-9695-2. [DOI] [PubMed] [Google Scholar]
- Benhar M, Forrester M T, Stamler J S. Nitrosative stress in the ER: a new role for S-nitrosylation in neurodegenerative diseases. ACS Chem Biol. 2006;1:355–358. doi: 10.1021/cb600244c. [DOI] [PubMed] [Google Scholar]
- Que L G, Liu L, Yan Y, Whitehead G S, Gavett S H, Schwartz D A, Stamler J S. Protection from experimental asthma by an endogenous bronchodilator. Science. 2005;308:1618–1621. doi: 10.1126/science.1108228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Yan Y, Zeng M, Zhang J, Hanes M A, Ahearn G, McMahon T J, Dickfeld T, Marshall H E, Que L G, Stamler J S. Essential roles of S-nitrosothiols in vascular homeostasis and endotoxic shock. Cell. 2004;116:617–628. doi: 10.1016/s0092-8674(04)00131-x. [DOI] [PubMed] [Google Scholar]
- Que L G, Yang Z, Stamler J S, Lugogo N L, Kraft M. S-Nitrosoglutathione reductase—an important regulator in human asthma. Am J Resp Crit Care Med. 2009;180:226–231. doi: 10.1164/rccm.200901-0158OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darton T, Guiver M, Naylor S, Jack D L, Kaczmarski E B, Borrow R, Read R C. Severity of meningococcal disease associated with genomic bacterial load. Clin Infect Dis. 2009;48:587–594. doi: 10.1086/596707. [DOI] [PubMed] [Google Scholar]
- McGuinness B T, Clarke I N, Lambden P R, Barlow A K, Poolman J T, Jones D M, Heckels J E. Point mutation in meningococcal por A gene associated with increased endemic disease. Lancet. 1991;337:514–517. doi: 10.1016/0140-6736(91)91297-8. [DOI] [PubMed] [Google Scholar]
- Crawford M J, Goldberg D E. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J Biol Chem. 1998;273:12543–12547. doi: 10.1074/jbc.273.20.12543. [DOI] [PubMed] [Google Scholar]