Abstract
Nitration and oxidation of tyrosine, tryptophan, and methionine residues in proteins are potential markers of their interaction with peroxynitrite. This chapter describes the procedure for the detection of these nitro-oxidative modifications by tandem mass spectrometry. The peptide YGDLANWMIPGK, shown to contain a nitrohydroxytryptophan in the mitochondrial enzyme succinyl-CoA:3-ketoacid coenzyme A transferase (SCOT) in vivo, was synthesized and exposed to peroxynitrite in order to test whether an identical tryptophan derivative could be generated in vitro. Data show that the occurrence of specific fragmented ions corresponding to the oxidation of methionine, nitration of tyrosine, and nitration/oxidation of tryptophan residues can be used to identify the sites of the nitration and oxidation of proteins in vitro and in vivo. It is also demonstrated that a nitrohydroxy addition to the tryptophan, similar to that present in SCOT in vivo, can be produced in vitro.
1. Introduction
Several types of posttranslational modifications, induced by nitric oxide-derived reactive intermediates, can target specific amino acid residues in proteins (Beckman, 1996; Beckman et al., 1990; Yamakura and Ikeda, 2006). In vitro exposure of proteins, peptides, or amino acids to nitrating agents has been widely demonstrated to cause additions of a nitro group to the benzyl ring of aromatic amino acids and a hydroxyl group to cysteine, methionine, and tryptophan residues (Alvarez and Radi, 2003; Yamakura et al., 2005). Protein nitration can also occur in situ in cells under conditions of oxidative stress and inflammation, ostensibly by interaction with peroxynitrite (ONOO−), a potent nitrating and oxidizing agent produced by a reaction between superoxide anion and nitric oxide (Beckman, 1990). The presence of peroxynitrite in mitochondria is considered plausible due to the activity of a hypothetical isoform of nitric oxide synthase and a high rate of generation in this organelle (Elfering et al., 2002; Radi, 2004). The in vitro as well as in vivo detection of tyrosine nitration in proteins has been facilitated greatly by the availability of specific anti-3NT antibodies (Ye et al., 1996). In contrast, a similar detection of nitrated tryptophan residues remained elusive until the recent development of an appropriate antibody (Ikeda et al., 2007). Accumulation of tyrosine-nitrated proteins has been observed in association with several pathological conditions (Ischiropoulos, 1998; Turko and Murad, 2002), as well as during the normal aging process (Kanski et al., 2003, 2005a,b; Sharov et al., 2006; Viner et al., 1999). Thus, the identification of specific protein targets and the individual residues that are modified is desirable for establishing the relationship between oxidation/nitration and the functional effect on the protein.
The application of mass spectrometric (MS) methods has been a highly useful tool for the identification of structural modifications in proteins due to exposure to natural or artificial reactive agents. In particular, matrix-assisted laser desorption/ionization (MALDI) in combination with time-of-flight (TOF) and electrospray ionization mass spectrometry (ESI-MS) have been increasingly applied to investigate protein nitration and oxidation (Nielson and Pennington, 1995; Petersson et al., 2001; Sarver et al., 2001; Turko and Murad, 2005).
Using a combination of methods (Western blot with anti-3-nitrotyrosine monoclonal antibody, HPLC electrochemical detection, MALDI-TOF, and ESI-MS), we demonstrated a novel in vivo posttranslational modification in a mitochondrial protein in rat tissues, involving simultaneous additions of nitro and hydroxy groups to tryptophan 372 in succinyl-CoA:3-ketoacid coenzyme A transferase (SCOT) during aging (Rebrin et al., 2007). Thus the application of MS methods for the detection of distinctive modifications (nitro and hydroxy additions) of methionine, tyrosine, and tryptophan residues in proteins can facilitate the identification of cellular targets of oxidative/nitrative stress. This chapter describes procedures used for the (i) synthesis of a YGDLANWMIPGK peptide (tryptic peptide of SCOT, corresponding to amino acids 366–377, shown previously to contain nitro-hydroxytryptophan 372 residue in vivo); (ii) nitro-oxidative modification of this peptide by exposure to peroxynitrite; and (iii) tandem mass spectrometry for determination of the specific fragmentation patterns of this synthetic peptide, which are a characteristic of the peroxynitrite-induced modifications of tyrosine, tryptophan, and methionine residues. Such methods can be adapted easily for other proteins.
2. Experimental Procedures
2.1. Synthesis of SCOT peptide
Peptide synthesis should be carried out on a Rainin Symphony peptide synthesizer or some similar instrumentation capable of automated solid-phase peptide synthesis. Using fast-Fmoc chemistry, and starting from the carboxy-terminal residue, the amino acids will undergo deprotection/coupling cycles. The resultant peptide can be cleaved from the resin and deprotected using trifluoroacetic acid. The molecular weight of the neutral monoisotopic peptide YGDLANWMIPGK (1363.66) should be confirmed by MALDI-TOF spectrometry. The peptide is purified and desalted using a preparative-scale reversed-phase HPLC column and the purity confirmed by analytical HPLC analysis. The lyophilized peptide sample may be stored in a dark vial in an oxygen-free environment at −20 °C.
2.2. Nitration by peroxynitrite
Peroxynitrite can be procured from Upstate Biotechnology (Lake Placid, NY) or synthesized from sodium nitrite and hydrogen peroxide in the presence of an acid, according to the procedure described elsewhere (Koppenol et al., 1996). Peroxynitrite concentration in dilutions of the stock solution made with 0.3 N NaOH can be determined by measurement of absorbance at 303 nm (ε = 1670 M−1cm−1). To achieve efficient oxidation and nitration of different amino acid residues in the peptide, use of a 10-fold molar excess of peroxynitrite is recommended. Dissolve 5 mg peptide in 100 μl of 100 mM peroxynitrite while stirring the reaction mixture vigorously and incubate at 4 °C. The time course of reaction can be determined in aliquots of 10 μl every hour of the reaction, diluted 1000-fold in 0.1% formic acid in water, and analyzed immediately or stored at −80 °C in small aliquots in order to avoid repetitive freezing/thawing.
2.3. HPLC of the peptide
Following peroxynitrite treatment of the peptide YGDLANWMIPGK, products of the reaction can be separated by the BioBasic 18 reversed-phase capillary column (ThermoFinnigan, San Jose, CA, 100 × 0.18 mm) using a ThermoFinnigan Surveyor MS pump. Equilibrate the column for 5 min at a flow rate of 1.5 μl/min with 95% solution A and 5% solution B (A, 0.1% formic acid in water; B, 0.1% formic acid in acetonitrile). The sample (1–10 μl) can be injected via an autosample injector (Surveyor, ThermoFinnigan). After injecting the sample, hold solution A at 95% and solution B at 5%, followed by a linear gradient of 5 to 65% solution B over 45 min. Increase solution B to 80% over the subsequent 5 min and hold at 80% for an additional 5 min, after which the column should be reequilibrated back to 5% of solution B. Because the peptide YGDLANWMIPGK has no cysteine residues, there is no need for alkylation; however, if the source of the peptides is tryptic digests of proteins or synthetic peptides containing cysteine residues, an additional alkylation step should be carried out prior to injection of the samples on the column, as described previously (Yarian et al., 2005).
2.4. MS/MS spectrometry
Mass analysis can be performed using a ThermoFinnigan LCQ Deca XP Plus ion trap mass spectrometer equipped with a nanospray ion source (ThermoFinnigan) employing a 4-cm metal emitter (Proxeon, Odense, Denmark). Electrical contact and voltage application to the probe tip take place via the nanoprobe assembly. Spray voltage of the mass spectrometer is set to 2.9 kV and heated capillary temperature at 190°. Acquire mass spectra in the m/z 400 to 1800 range. A data-dependent acquisition mode should be used where each of the top five ions for a given scan is subjected to MS/MS analysis.
2.5. Data analysis
Protein identification may be carried out with the MS/MS search software Mascot (Matrix Science), with confirmatory or complementary analyses by TurboSequest (Bioworks Browser 3.2, build 41, from ThermoFinnigan and Sonar MS/MS from Genomics Solutions). Alkylated modification (carbami-domethyl) should be designated as fixed; in contrast, oxidation of methionine and tryptophan, as well as nitration of tyrosine and tryptophan, should be considered as variable modifications in the TurboSequest search. The NCBI rat genome database server complemented with the NCBI nonredundant protein database can be used for the search. Theoretical m/z values for the peptides and their fragmentation ions can be assessed using the “MS/MS fragment ion calculator” from The Institute for Systems Biology at http://db.systemsbiology.net:8080/proteomicsToolkit/. Acceptable cross-correlation scores (Xcorr) for positive identification of a modification (doubly charged ions) within the peptide should be greater than two. It is recommended that MS/MS spectra of particular interest should be inspected manually.
3. Anticipated Results
3.1. Synthesis, purification, and MS analysis of YGDLANWMIPGK peptide
A homogeneous product should be obtained after solid-phase peptide synthesis and the molecular weight confirmed using MALDI-TOF analysis. The observed m/z value of the singly protonated peptide [M+H]+ should correspond to the theoretical one, within deviation of <0.2 to 0.4 m/z unit (observed [M+H]+ of the peptide YGDLANWMIPGK at m/z 1365.0, calculated [M+H]+ = 1364.67). The purity of the synthetic peptide, indicated by HPLC analysis, should be >95%. After exposure of the synthetic peptide to peroxynitrite, HPLC separation of the reaction products should yield several peaks: peptides with oxidized methionine and nitrohydroxytryptophan should elute at earlier retention times, whereas nitrotyrosine-containing peptides should be retained longer than the unmodified peptide.
Electrospray ionization mass spectra of the reaction products would indicate synthetic peptide YGDLANWMIPGK to be either unmodified (doubly charged precursor ion 682.8) or modified by +16 (oxidation) at methionine (doubly charged precursor ion 690.8) or +45 (nitration) at tyrosine (doubly charged precursor ion 705.3) or +61 (oxidation and nitration) at tryptophan (doubly charged precursor ion 713.3) and +77 addition, corresponding to the oxidation of methionine combined with nitrohydroxytryptophan (doubly charged precursor ion 721.3). The theoretical m/z values of ions, expected from fragmentation of the synthetic peptide YGDLANWMIPGK, are indicated in Table 15.1. MS/MS spectra of the synthetic peptide YGDLANWMIPGK should indicate the distinctive fragmentation pattern, that is, relative dominance of the y3 and b9, but not of y4 or y5 ion peaks. Such fragmentation should be essentially identical to that obtained previously, using tryptic peptide from SCOT, purified from rat heart mitochondria (Rebrin et al., 2007). The occurrence of the indicated fragmentation pattern may be because of the presence of the proline residue (dominant peaks y3 and b9), which can cause suppression and/or disappearance of some of the fragments (including y4 and y5), as also shown in similar proline-containing peptides (Paizs and Suhai, 2005).
Table 15.1.
Theoretical m/z values for fragmented ions produced by high collision-induced dissociation of doubly protonated unmodified peptide (theoretical [M+2H]2+ at m/z 682.84)
| B ions | Y ions | |||
|---|---|---|---|---|
| Y | 1 | 164.07 | — | |
| G | 2 | 221.09 | 1201.6 | 11 |
| D | 3 | 336.12 | 1144.58 | 10 |
| L | 4 | 449.2 | 1029.55 | 9 |
| A | 5 | 520.24 | 916.47 | 8 |
| N | 6 | 634.28 | 845.43 | 7 |
| W | 7 | 820.36 | 731.39 | 6 |
| M | 8 | 951.4 | 545.31 | 5 |
| I | 9 | 1064.49 | 414.27 | 4 |
| P | 10 | 1161.54 | 301.19 | 3 |
| G | 11 | 1218.56 | 204.13 | 2 |
| K | — | 147.11 | 1 |
3.2. MS/MS analysis of unmodified peptide
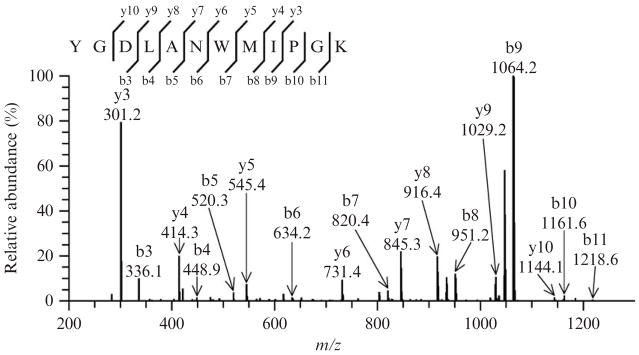
The MS/MS spectrum of the unmodified peptide YGDLANWMIPGK, as displayed in Fig. 15.1, should indicate the presence of 8 y and 9 b out of the potential 11 ions. The y3 and b9 ions at m/z 301.2 and 1064.2, respectively, are predominant, while the intensity of other ions is considerably lower. Such a fragmentation profile is virtually identical to the one reported for the native SCOT peptide (Rebrin et al., 2007).
Figure 15.1.
Tandem mass spectrum of the untreated synthetic peptide YGDLANW-MIPGK ([M+2H]2+ ion at m/z 683.4) displaying a distinctive fragmentation pattern, characterized by the dominance of two ions, y3 and b9. Such a profile was interpreted to be due to the “proline effect,” which favors fragmentation on its amino terminal bond and suppresses other ions. For all spectra, ion annotation is based on results presented in the Sequest’s “display ion view” window, corroborated by the dta file.
3.3. MS/MS analysis of methionine oxidation
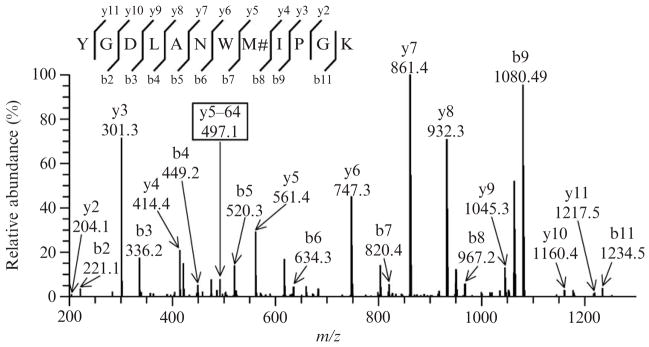
Exposure of peptide to peroxynitrite leads to the nearly ubiquitous presence of methionine sulfoxide containing peptides. The corresponding MS/MS spectrum (single charged precursor ion with m/z [M+H+16]+ = 1380.7 and cross-correlation score Xcorr = 4.3 in TurboSequest analysis), represented in Fig. 15.2, should indicate the presence of 10 y and 9 b out of the potential 11 ions. The presence of methionine sulfoxide dramatically alters the pattern of fragmentation of the synthetic peptide. The ions y5 (m/z 561.4), y6 (m/z 747.3), y7 (m/z 861.4), and y8 (m/z 932.3) become particularly abundant, whereas they are masked in the untreated peptide. The detection of oxidized methionine should also be facilitated by the presence of additional y and/or b ion fragments, which are 64 Da lower in mass than the methionine sulfoxide ion. Such a decrement in mass corresponds to the neutral loss of methane-sulfenic acid (CH3SOH, 64 Da) from the side chain of methionine sulfoxide. The corresponding y5–64 ion at m/z 497.4 is shown in Fig. 15.2. Indeed, this characteristic loss of the 64 Da has been suggested as a valuable “fingerprint” for the detection of methionine oxidation in proteins (Guan et al., 2003).
Figure 15.2.
Representative tandem mass spectrum of the synthetic peptide containing methionine sulfoxide ([M+2H]2+ ion at m/z 690.62). Multiple peptides containing methionine sulfoxide are obtained following peroxynitrite exposure. The presence of oxidized methionine alters the fragmentation profile of the peptide: the ions y5, y6, y7, and y8 become quite abundant. The low abundant ion at m/z 497.1, corresponding to a neutral mass loss of 64 Da fromthe y5 ion, constitutes a fingerprint of methionine sulfoxide and is indicated as y5–64. Such a mass decrement is known to be because of the elimination of methanesulfenic acid from the side chain of methionine sulfoxide. M#, methionine sulfoxide.
3.4. MS/MS analysis of tyrosine nitration
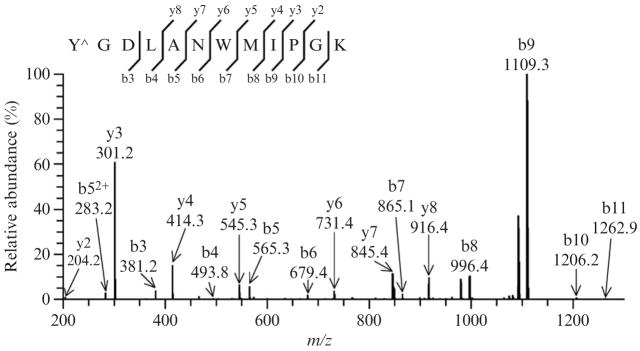
Several peptides containing an additional mass of 45 Da, indicative of the presence of a nitro group, can be detected in mass spectra. The program TurboSequest should unambiguously assign the +45 addition on the tyrosine residue of the peptide (singly charged precursor ion with m/z [M+H+45]+ = 1411.1 and cross-correlation score Xcorr = 3.83 in Turbo-Sequest analysis). The MS/MS spectrum, shown in Fig. 15.3, indicates the presence of 7 y and 9 b out of the potential 11 ions. The ions b3, b4, b5, and b6 at m/z 381.2, 493.8, 565.3, and 679.4, respectively, confirm the addition of a nitro group to the tyrosine residue, whereas the ions y6 (m/z = 731.4), y7 (m/z = 845.4), and y8 (m/z = 916.4) indicate that the tryptophan residue is not modified.
Figure 15.3.
Representative tandem mass spectrum of a nitrotyrosine containing peptide ([M+2H]2+ ion at m/z 705.49). Following exposure to peroxynitrite, a +45-Da mass increment of the peptide is detected and can be attributed to the presence of a nitro group on either tyrosine or tryptophan residues. The fragmented ions unambiguously indicate the addition of the nitro group to the tyrosine residue, as inferred from the presence of the ions b3, b4, b5, and b6 at m/z 381.2,493.8, 565.3, and 679.4, respectively, and the ions y6, y7, and y8 at m/z 731.4,845.4, and 916.4, respectively. Y^, nitrated tyrosine.
3.5. MS/MS analysis of tryptophan nitration and oxidation
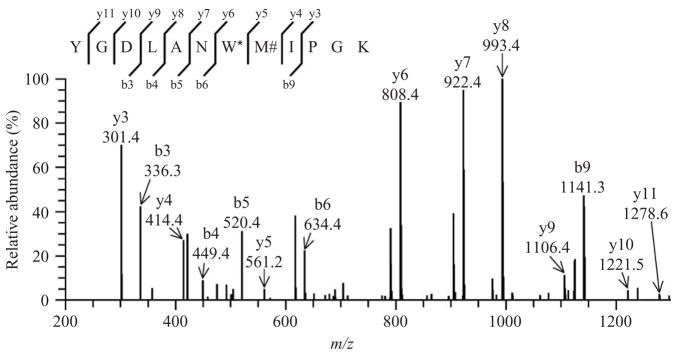
Peptides, containing tryptophan with an additional +61 mass (nitro and hydroxy additions) in association with methionine sulfoxide (+16), are observed with a m/z value of 721.3 (doubly charged precursor ion). In such cases, a relatively low abundance ion y5 corresponding to methionine sulfoxide at m/z = 561.31 can be detected. A representative MS/MS spectrum (cross-correlation score Xcorr = 3.62 in TurboSequest analysis) indicates the presence of 9 y and 5 b out of the potential 11 ions (Fig. 15.4). Such a fragmentation pattern is comparable to the one observed for the SCOT peptide in vivo (Rebrin et al., 2007). The presence of nitro and hydroxy groups on the tryptophan residue seemingly alters the fragmentation profile of this peptide, as indicated by the predominance of the ions y6 (m/z = 808.4), y7 (m/z = 922.4), and y9 (m/z = 1106.4). In some MS/MS spectra, the y5 (m/z = 561.2) ion, corresponding to methionine sulfoxide, is very weak or undetectable. Thus it appears that the double modification on the tryptophan disfavors the fragmentation of the residue located on its C-terminal side, which makes it difficult to detect the y5 ion corresponding to the methionine residue (or methionine sulfoxide derivative).
Figure 15.4.
Tandem mass spectrum showing the simultaneous presence of nitrohydroxytryptophan and methionine sulfoxide in the peptide ([M+2H]2+ ion at m/z 721.0) following peroxynitrite treatment. The fragmentation profile of the peptide is also modified considerably, with y6, y7, and y8 becoming particularly abundant. In contrast, y5 relative intensity is low, suggesting that nitrohydroxytryptophan might repress fragmentation on its C-terminal bond. M#, methionine sulfoxide; W*, nitrohydroxytryptophan.
3.6. MS/MS analysis of multiple nitration and oxidation products
Prolonged exposure of the synthetic peptide YGDLANWMIPGK to peroxynitrite should also yield peptides containing oxidation on both methionine [methionine sulfoxide (+16) or methionine sulfone (+32)] and tryptophan [hydroxytryptophan (+16) or N-formylkynurenine (+32) residues]. In addition, multiple peptides containing N-formylkynurenine, as well as nitrated tyrosine in combination with methionine sulfoxide or sulfone, can also be observed (data not shown). In contrast, nitration of tryptophan residues alone was not detected. Peptides, containing oxidized derivatives of tryptophan, mainly hydroxytryptophan (+16), but also kynurenine (+4) and N-formylkynurenine, (+32) are also observed.
4. Conclusion
Tandem mass spectrometry is highly informative for the mass determination of fragmented ions originating from peptides. Structural modifications of methionine, tyrosine, and tryptophan residues following exposure of the synthetic peptide YGDLANWMIPGK, derived from SCOT sequence, to peroxynitrite, can be detected by the additions of +16 or +32 (oxidation), +45 (nitration), and +61 (nitration and oxidation) to the mass/charge values of fragmentation ions at a high level of precision. Occurrence of nitrohydroxytryptophan can be demonstrated after in vitro exposure of the peptide to peroxynitrite. This novel modification of the tryptophan residue matches the one observed in SCOT in vivo during aging. Electrospray ionization MS/MS analysis of nitrative and oxidative modifications provides clear identification of the target amino acid residue(s) as well as estimation of the corresponding mass addition. This analytical approach can be applied to other proteins, which are targets of nitrative and/or oxidative stress.
Acknowledgments
This research was supported by Grant RO1 AG 13563 from National Institute on Aging–National Institutes of Health.
References
- Alvarez B, Radi R. Peroxynitrite reactivity with amino acids and proteins. Amino Acids. 2003;25:295–311. doi: 10.1007/s00726-003-0018-8. [DOI] [PubMed] [Google Scholar]
- Beckman JS. Ischaemic injury mediator. Nature. 1990;345:27–28. doi: 10.1038/345027b0. [DOI] [PubMed] [Google Scholar]
- Beckman JS. Oxidative damage and tyrosine nitration from peroxynitrite. Chem Res Toxicol. 1996;9:836–844. doi: 10.1021/tx9501445. [DOI] [PubMed] [Google Scholar]
- Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Apparent hydroxyl radical production by peroxynitrite: Implications for endothelial injury from nitric oxide and superoxide. Proc Natl Acad Sci USA. 1990;87:1620–1624. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfering SL, Sarkela TM, Giulivi C. Biochemistry of mitochondrial nitric-oxide synthase. J Biol Chem. 2002;277:38079–38086. doi: 10.1074/jbc.M205256200. [DOI] [PubMed] [Google Scholar]
- Guan Z, Yates NA, Bakhtiar R. Detection and characterization of methionine oxidation in peptides by collision-induced dissociation and electron capture dissociation. J Am Soc Mass Spectrom. 2003;14:605–613. doi: 10.1016/S1044-0305(03)00201-0. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Yukihiro Hiraoka B, Iwai H, Matsumoto T, Mineki R, Taka H, Takamori K, Ogawa H, Yamakura F. Detection of 6-nitrotryptophan in proteins by Western blot analysis and its application for peroxynitrite-treated PC12 cells. Nitric Oxide. 2007;16:18–28. doi: 10.1016/j.niox.2006.04.263. [DOI] [PubMed] [Google Scholar]
- Ischiropoulos H. Biological tyrosine nitration: A pathophysiological function of nitric oxide and reactive oxygen species. Arch Biochem Biophys. 1998;356:1–11. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- Kanski J, Alterman MA, Schoneich C. Proteomic identification of age-dependent protein nitration in rat skeletal muscle. Free Radic Biol Med. 2003;35:1229–1239. doi: 10.1016/s0891-5849(03)00500-8. [DOI] [PubMed] [Google Scholar]
- Kanski J, Behring A, Pelling J, Schoneich C. Proteomic identification of 3-nitrotyrosine-containing rat cardiac proteins: Effects of biological aging. Am J Physiol Heart Circ Physiol. 2005a;288:H371–H381. doi: 10.1152/ajpheart.01030.2003. [DOI] [PubMed] [Google Scholar]
- Kanski J, Hong SJ, Schoneich C. Proteomic analysis of protein nitration in aging skeletal muscle and identification of nitrotyrosine-containing sequences in vivo by nanoelectrospray ionization tandem mass spectrometry. J Biol Chem. 2005b;280:24261–24266. doi: 10.1074/jbc.M501773200. [DOI] [PubMed] [Google Scholar]
- Koppenol WH, Kissner R, Beckman JS. Syntheses of peroxynitrite: To go with the flow or on solid grounds? Methods Enzymol. 1996;269:296–302. doi: 10.1016/s0076-6879(96)69030-2. [DOI] [PubMed] [Google Scholar]
- Nielson KR, Pennington MW. Mass spectral analysis of peptides containing nitrobenzyl moieties. Lett Peptide Sci. 1995;2:301–305. [Google Scholar]
- Paizs B, Suhai S. Fragmentation pathways of protonated peptides. Mass Spectrom Rev. 2005;24:508–548. doi: 10.1002/mas.20024. [DOI] [PubMed] [Google Scholar]
- Petersson AS, Steen H, Kalume DE, Caidahl K, Roepstorff P. Investigation of tyrosine nitration in proteins by mass spectrometry. J Mass Spectrom. 2001;36:616–625. doi: 10.1002/jms.161. [DOI] [PubMed] [Google Scholar]
- Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci USA. 2004;101:4003–4008. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebrin I, Bregere C, Kamzalov S, Gallaher TK, Sohal RS. Nitration of tryptophan 372 in succinyl-CoA:3-ketoacid CoA transferase during aging in rat heart mitochondria. Biochemistry. 2007;46:10130–10144. doi: 10.1021/bi7001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarver A, Scheffler NK, Shetlar MD, Gibson BW. Analysis of peptides and proteins containing nitrotyrosine by matrix-assisted laser desorption/ionization mass spectrometry. J Am Soc Mass Spectrom. 2001;12:439–448. doi: 10.1016/S1044-0305(01)00213-6. [DOI] [PubMed] [Google Scholar]
- Sharov VS, Galeva NA, Kanski J, Williams TD, Schoneich C. Age-associated tyrosine nitration of rat skeletal muscle glycogen phosphorylase b: Characterization by HPLC-nanoelectrospray-tandem mass spectrometry. Exp Gerontol. 2006;41:407–416. doi: 10.1016/j.exger.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Turko IV, Murad F. Protein nitration in cardiovascular diseases. Pharmacol Rev. 2002;54:619–634. doi: 10.1124/pr.54.4.619. [DOI] [PubMed] [Google Scholar]
- Turko IV, Murad F. Mapping sites of tyrosine nitration by matrix-assisted laser desorption/ionization mass spectrometry. Methods Enzymol. 2005;396:266–275. doi: 10.1016/S0076-6879(05)96023-0. [DOI] [PubMed] [Google Scholar]
- Viner RI, Ferrington DA, Williams TD, Bigelow DJ, Schoneich C. Protein modification during biological aging: Selective tyrosine nitration of the SER-CA2a isoform of the sarcoplasmic reticulum Ca2+-ATPase in skeletal muscle. Biochem J. 1999;340(Pt 3):657–669. [PMC free article] [PubMed] [Google Scholar]
- Yamakura F, Ikeda K. Modification of tryptophan and tryptophan residues in proteins by reactive nitrogen species. Nitric Oxide. 2006;14:152–161. doi: 10.1016/j.niox.2005.07.009. [DOI] [PubMed] [Google Scholar]
- Yamakura F, Matsumoto T, Ikeda K, Taka H, Fujimura T, Murayama K, Watanabe E, Tamaki M, Imai T, Takamori K. Nitrated and oxidized products of a single tryptophan residue in human Cu, Zn-superoxide dismutase treated with either peroxynitrite-carbon dioxide or myeloperoxidase-hydrogen peroxide-nitrite. J Biochem (Tokyo) 2005;138:57–69. doi: 10.1093/jb/mvi095. [DOI] [PubMed] [Google Scholar]
- Yarian CS, Rebrin I, Sohal RS. Aconitase and ATP synthase are targets of malondialdehyde modification and undergo an age-related decrease in activity in mouse heart mitochondria. Biochem Biophys Res Commun. 2005;330:151–156. doi: 10.1016/j.bbrc.2005.02.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye YZ, Strong M, Huang ZQ, Beckman JS. Antibodies that recognize nitrotyrosine. Methods Enzymol. 1996;269:201–209. doi: 10.1016/s0076-6879(96)69022-3. [DOI] [PubMed] [Google Scholar]