Summary
Osteoporosis, a low bone mass disease, is associated with decreased osteoblast numbers and increased levels of oxidative stress in these cells. Since the FoxO family of transcription factors, confers stress resistance, we investigated their potential impact on skeletal integrity. We show here through cell-specific deletion and molecular analyses that, among the 3 FoxO proteins, only FoxO1 is required for proliferation and redox balance in osteoblasts, and as a result controls bone formation. FoxO1 regulation of osteoblast proliferation occurs because of its interaction with ATF4, a transcription factor regulating amino acid import; and of its regulation of a stress-dependent pathway influencing p53 signaling. Accordingly, decreasing oxidative stress levels or increasing protein intake normalizes bone formation and bone mass in mice lacking FoxO1 in osteoblasts only. These results identify FoxO1 as a crucial regulator of osteoblast physiology and provide a direct mechanistic link between oxidative stress and the regulation of bone remodeling.
Introduction
In adult vertebrates, bones are constantly renewed by a physiological process called bone remodeling, which includes two cellular events occurring in succession. The first one is resorption, or destruction of the mineralized bone matrix, by osteoclasts, and is followed by de novo bone formation by osteoblasts (Harada and Rodan, 2003; Teitelbaum and Ross, 2003). Bone remodeling is affected in the most frequent degenerative disease of bones, osteoporosis, a low bone mass disease resulting from an imbalance between bone formation and resorption (Rodan and Martin, 2000; Raisz, 2005). Starting in their mid-40s, both men and women experience a progressive decline in bone mass and strength (Riggs et al., 2006; Bouxsein et al., 2006) which in women is accelerated at menopause because of the decline of estrogens. Hence, osteoporosis can be viewed also as a disease of aging.
A growing number of evidence has linked aging and the development of age-related diseases to increased levels of oxidative stress, indicating that oxidative stress plays a significant role in their pathogenesis (Finkel and Holbrook, 2000; Quarrie and Riabowol, 2004). Similar to other aging-related diseases, the development of osteoporosis, has been associated with increased levels of oxidative stress in osteoblasts, suggesting that this may be one critical component of the pathophysiology of bone loss (Levasseur et al., 2003; Bai et al., 2004; Lean et al., 2003; Almeida et al., 2007). Consistent with this idea, an osteoporotic phenotype has been observed in mouse models of premature aging associated with oxidative damage (Tyner et al., 2002; De Boer et al., 2002).
Oxidative stress is the result of elevated levels of reactive oxygen species (ROS), the most important of which are superoxide anions, hydroxyl radicals, and hydrogen peroxide. A rise in the level of ROS can damage proteins, lipids, and DNA, eventually leading to cell death. Alternatively, it can trigger the activation of specific physiologic signaling pathways. As a matter of fact, physiological levels of stress activate defense signaling mechanisms that maintain cellular and organismal functionality. Both the damage of various cell components and the triggering of the activation of specific signaling pathways by ROS can influence numerous cellular processes which have been correlated with overall longevity in invertebrates and vertebrates (Quarrie and Riabowol, 2004; Finkel and Holbrook, 2000).
Cells counteract the adverse effects of ROS by up-regulating enzymatic scavengers or DNA-damage repair genes. This response involves dephosphorylation and subsequent activation of a small family of ubiquitous transcription factors known as FoxOs (Liu et al., 2005; Lehtinen et al., 2006; Nemoto and Finkel, 2002). The 3 FoxO molecules, FoxO1, FoxO3 and FoxO4, are encoded by different genes and they all affect differentiation, proliferation and survival of a variety of cells including adipocytes, hepatocytes, β-cells, myoblasts, thymocytes and cancer cells (reviewed in (Accili and Arden, 2004; Greer and Brunet, 2005; Arden, 2006; Murakami, 2006)). To cite one example, analysis of mice lacking each of the FoxO proteins in all cells have established their role in the resistance of hematopoietic stem cells to physiologic oxidative stress (Tothova et al., 2007). Yet, the putative role of any of the members of this small family of transcription factors in bone cells is unknown for now.
We show here that among the 3 FoxO proteins, FoxO1 is the main regulator of redox balance and function in osteoblasts and the only one that overtly controls bone mass. Deletion of FoxO1 specifically from osteoblasts (Foxo1ob-/- mice) decreases osteoblast numbers, bone formation rate and bone volume. These effects stem from molecular alterations, presented below, in both oxidative stress and amino acid import in the FoxO1 mutant mice; the latter being reverted by a high protein diet. These observations identify FoxO1 as a crucial regulator of osteoblast physiology and provide a direct mechanistic link between oxidative stress and the control of bone mass.
Results
FoxO1, the most highly expressed and regulated FoxO protein in osteoblasts
As a first step toward understanding the role, if any, of FoxO proteins in the skeleton we studied their pattern of expression in osteoblasts and osteoclasts. As shown in Figures 1A-1C primary osteoblasts as well as whole bone and osteoclasts express all three FoxO isoforms, with FoxO1 being, by far, the most abundant member of this family of proteins. To understand the role of FoxO proteins in skeletogenesis we focused on studying first the function that they may exert on osteoblasts. Osteoblast-specific expression of FoxO1 was confirmed by immunohistochemical analysis in bone sections obtained from femurs of newborn wild type mice (Figures 1D-1L). A comparison between sections stained with FoxO1 (Figures 1D and 1G) and sections stained with 4′,6-diamidino-2-phenylindole (DAPI), a fluorescent stain that binds strongly to DNA (Figures 1E and 1H), indicated that FoxO1 is expressed in the nucleus of osteoblastic cells embedded in the endosteal area of the cortex or present in the periosteum (Figures 1D-1I). Consistent with the notion that in the absence of stress stimuli these transcription factors can shuttle between the nucleus and the cytoplasm, FoxO1 was also present in the cytoplasm of osteoblasts. The localization of FoxO1 with respect to the nucleus, can be viewed clearer by overlaying the images of FoxO1 and DAPI staining (Figures 1F and 1I). In addition, cytoplasmic and nuclear localization of FoxO1 was further confirmed in adjacent bone sections stained for FoxO1 and counterstained with either the cytoplasmic stain eosin (Figure 1K) or the nuclear stain hematoxylin (Figure 1L).
Figure 1. Expression and regulation of FoxO family members in bone.
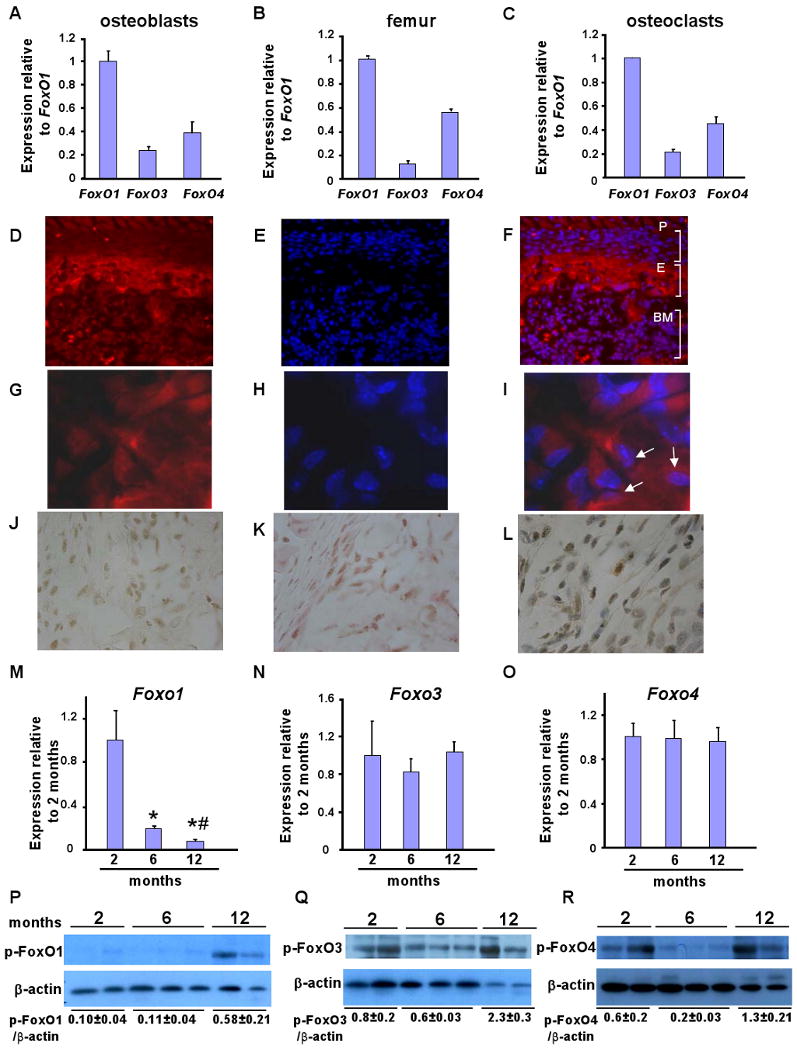
A-C) Expression of all the 3 FoxO genes in primary calvarial osteoblasts, femurs and osteoclasts of WT mice by real-time PCR (n=4 mice/group and duplicates were performed for cell extracts). Bars indicate means ± sem. Expression levels are relative to FoxO1. FoxO1 expression has been considered 1. Mice were 2 months old.
D-L) Immunohistochemical localization of FoxO1 in femoral sections of newborn WT mice. (D,and G) Images of bone sections depicting FoxO1 staining at 40× and 100× magnification. (E and H) Sections were counterstained with DAPI. (F and I) FoxO1 and DAPI images were overlaid to visualize nuclear and cytoplasmic localization of FoxO1 at 40× and 100× magnification. Arrows indicate representative cells showing nuclear localization of FoxO1 (purple). (J) DAB staining of FoxO1 in femoral sections. Adjacent sections were stained with FoxO1 and counterstained with (K) eosin or L) hematoxylin (100× magnification). P indicates periosteal surface; E indicates endosteal surface; and, BM indicates bone marrow. The 100× magnification images are obtained from the endosteal surface.
M, N and O) Expression analysis of all the 3 FoxO isoforms in murine bones collected from different ages (n=4 mice/group). * p < 0.05 (6 months vs. 2 months); # p < 0.05 (12 months vs. 6 months). Expression levels are relative to FoxO expression at 2 months of age. Expression at 2 months of age has been considered 1.
P, Q and R) Activity assessment by phosphorylation status of FoxO1, FoxO3 and FoxO4 in bones collected from mice of different ages. See also Figures S1 and S2.
We next asked whether accumulation or activity of FoxO proteins are altered with aging or under conditions of increased oxidative stress. Aging mice fulfill both requirements as they show a progressive decrease in bone formation which correlates with a parallel increase in oxidative stress levels in bone (Almeida et al., 2007). In this setting, FoxO1 expression progressively decreased from 2 to 12 months of age whereas FoxO3 and FoxO4 levels remained stable in 2-, 6- and 12-month old animals (Figures 1M, 1N and 1O). Potential changes in the activity of FoxO1, FoxO3 and FoxO4 were assessed by measuring the phosphorylation status of Ser256 of FoxO1 and Foxo3 or Ser193/258 of Foxo4 –sites that are phosphorylated by the phosphoinositide 3-(PI3) kinase/Akt pathway resulting in the inactivation of FoxO. FoxO1 phosphorylation/inactivation increased in the bone of aging mice (Figure 1P). FoxO3 activation status was unaffected by age despite the progressive decrease in bone formation and the concomitant increase in oxidative stress (Figure 1Q). Hence, it is unlikely that FoxO3 will be essential in maintaining bone homeostasis under stressful conditions. On the other hand, FoxO4 phosphorylation/inactivation increased progressively in aging animals (Figure 1R). Taken together, the results of these experiments along with evidence from several reports identifying the pivotal role of FoxO1 in angiogenesis, insulin action, organismal growth, and tumorigenesis (reviewed in (Accili and Arden, 2004)), identify FoxO1 as the best candidate among all FoxO proteins to be involved in osteoblast biology. This led us to analyze its function in vivo.
FoxO1 Controls bone mass through its actions on osteoblasts
In view of the results presented above, we generated mice lacking FoxO1 specifically in osteoblasts by crossing a floxed allele of this gene (Paik et al., 2007; Dacquin et al., 2002) with mice expressing Cre only in osteoblasts (Figure S1). As a control of specificity we also deleted FoxO3 specifically in osteoblasts using the same strategy (Figure S2).
FoxO1ob +/- or FoxO3ob +/- mice were intercrossed and animals homozygous for FoxO1 (FoxO1ob -/-) or FoxO3 (FoxO3ob -/-) deletion in osteoblasts were obtained. α1(I) Collagen-Cre-mediated deletion of FoxO1 or FoxO3 was specific to bone and was accompanied by a marked, 75%, reduction in FoxO1 expression in osteoblasts derived from FoxO1ob -/- mice (Figure S1). Similarly, FoxO3 deletion was bone-specific and its expression levels were reduced by 65% in osteoblasts from FoxO3ob -/- mice (Figure S2). FoxO1 as well as FoxO3 expression was unaffected in a variety of different tissues examined, including liver, gut, pancreas, brown and white adipose tissue, skeletal muscle and brainstem (Figures S1 and S2).
At two months of age, all mutant Foxo1 mice had a body weight similar to that of wild type littermates. In contrast, bone mineral density (BMD) measured by peripheral dual-energy X-ray absorptiometry (DEXA, PIXImus) was decreased in the spine and femur of both Foxo1ob+/- and Foxo1ob-/- mice as compared to wild type (WT) controls (Figure 2A). At the cellular level, Foxo1ob-/- mice were characterized by decreased osteoblast numbers (N.Ob./T.Ar.), bone formation rate (BFR) and bone volume (BV/TV) (Figure 2B). Cortical thickness in the midshaft femur, although not statistically significant, was decreased by 11% by FoxO1 deletion in osteoblasts (Figure 2C). Additionally, femoral BV/TV was decreased by 40-50%, an effect similar to that observed in the lumbar verterbrae (Figure 2C). And, although trabecular bone loss and osteoblast numbers in FoxO1ob-/- mice persisted with age, they did not progress with aging and there were no age – related changes in their bone phenotype (Figure 2D). Thus, FoxO1ob-/- mice showed lack of any age-related changes in their bone phenotype. Loss of FoxO1 expression was stable in the bone of aging FoxO1ob-/- mice (Figure S1D). There was no statistically significant difference in BFR or any of the other measured bone parameters between 2, 6 and 12 month-old FoxO1ob-/- mice. In addition, the relative suppression in BFR between FoxO1ob-/- and WT mice was not statistically significant different at 2, 6 or 12 months of age. Finally, bone resorption increased as indicated by the increase in osteoclast surface in FoxO1ob-/- as compared to WT control mice (Figure 2B). There were no age-related changes in the resorption phenotype of FoxO1ob-/- mice (data not shown). Expression of FoxO3 and FoxO4 was not affected in the bone of mice with FoxO1 deletion in osteoblasts (data not shown). Similarly, we found no changes in FoxO4 activity in the bones of FoxO1ob-/- as compared to wild type control mice (Figure 2G). Thus, the bone phenotypic abnormalities in FoxO1ob-/- mice are independent of changes in the expression or activity of the other FoxO isoforms.
Figure 2. Low bone formation in FoxO1ob-/- mice.
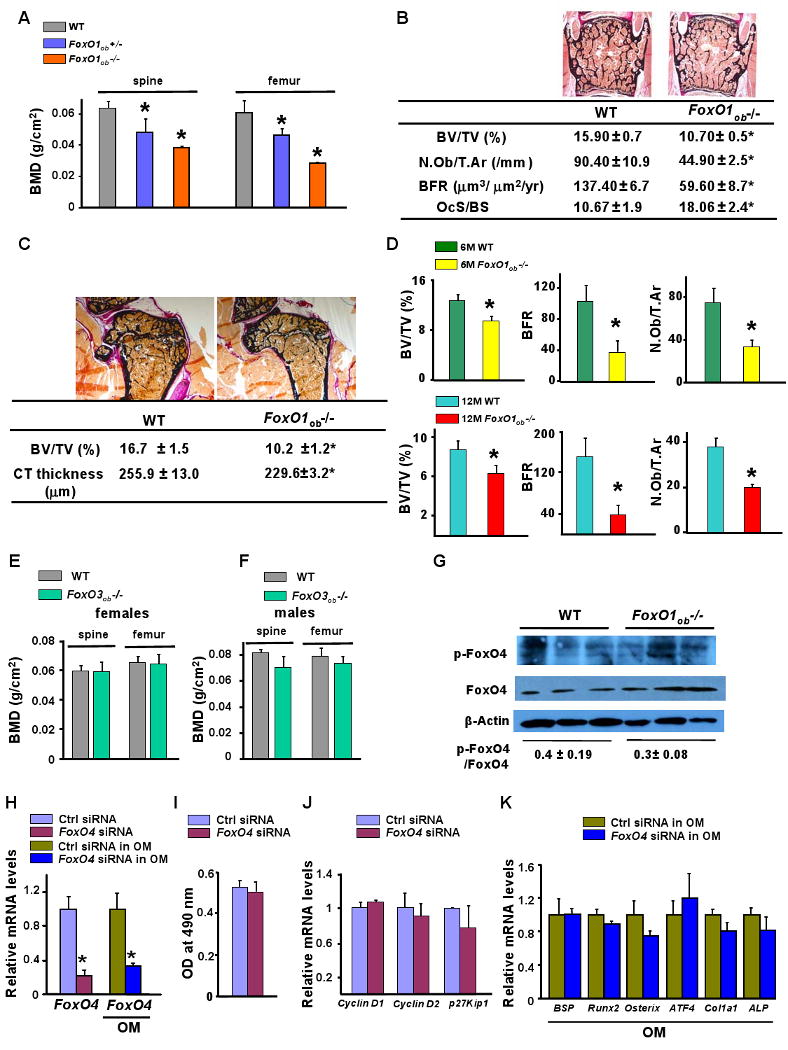
A) Bone mineral density (BMD) measured by DEXA in spine and femur of 2 month-old WT, FoxO1ob+/- and FoxO1ob-/- mice (n=10 mice/group). * p < 0.05 vs WT.
B) BV/TV, bone volume over trabecular volume; N.Ob/T.Ar, number of osteoblasts per trabecular area; BFR, bone formation rate, of 2 month-old and OcS/BS, osteoclast surface per bone surface in verterbrae of 1 month-old WT and FoxO1ob-/- mice (n=10 mice/group). * p < 0.05 vs WT.
C) BV/TV in femoral head and midshaft cortical thickness in the femurs of 2 month-old WT and FoxO1ob-/- mice (n=10 mice/group). * p < 0.05 vs WT.
D) BV/TV, BFR and N.Ob/T.Ar in verterbrae of 6 and 12 month-old WT and FoxO1ob-/- mice (n=5-10 mice/group). * p < 0.05 vs WT.
E, F) BMD measured by DEXA in spine and femur of 2 month-old WT and FoxO3ob-/- mice (n=10 mice/group).
G) Immunoblotting analysis of FoxO4 activity in the bone of WT and FoxO1ob-/- mice.
H) RT-PCR analysis of FoxO4 expression in osteoblasts transfected with siRNA oligos for FoxO4 or control (Ctrl), scrambled oligos. * p < 0.05 vs control (Ctrl) siRNA and Ctrl siRNA in OM. OM denotes Osteogenic Medium.
I) Proliferation in cultures of osteoblasts transfected with siRNA oligos for FoxO4 or control (Ctrl), scrambled oligos. OM denotes Osteogenic Medium.
J) Real-time PCR (RT-PCR) analysis of CyclinD1, D2 and p27Kip1 expression in osteoblasts with silenced FoxO4.
K) RT-PCR analysis of osteoblast differentiation markers in osteoblasts with silenced FoxO4. In all panels values are means ± sem.
The effect of FoxO1 deletion in osteoblasts was specific since a similar deletion of FoxO3 did not affect BMD in the spine or long bones of female or male mice (Figure 2E and 2F). Bone volume, bone formation rate, and osteoblast numbers as well as osteoclast surface were also not affected in FoxO3ob-/- mice (data not shown). Confirming the notion that FoxO4 does not regulate bone formation, siRNA of FoxO4 in osteoblasts (Figure 2H) had no effect on osteoblast proliferation as well as on cyclin D1 and D2 and p27Kip expression (Figure 2I and 2J). Differentiation was also not altered by FoxO4 silencing as evidenced by the lack of an effect on osteoblast differentiation markers (Figure 2K). These results support the notion that FoxO1 plays an important and unique role among all FoxO proteins as a regulator of bone mass.
Molecular bases of the low bone formation phenotype of FoxO1ob -/- mice
To elucidate the underlying reason for the low bone formation phenotype of the FoxO1ob -/- mice we searched for alterations in osteoblast function. In vivo analysis of osteoblast proliferation in FoxO1 mutant mice showed that the decrease in osteoblast numbers correlated with decreased osteoblast proliferation as measured by BrdU incorporation (Figures 3A and 3B) in 5.5 day-old pups. Osteoblast proliferation, expressed either per trabecular or per bone perimeter was decreased by approximately 40% in FoxO1ob-/- mice as compared to WT littermates. FoxOs control cell cycle through regulation of cyclins D1 and D2 and the cell cycle inhibitor p27Kip1 (Arden, 2006). Similar to adult mice, FoxO1 is the most abundant of the 3 FoxO isoforms in the bone of 5.5 day-old pups; and, its inactivation in FoxO1ob-/- mice at this age has no effect on the expression levels of FoxO3 or FoxO4 (Figure 3C). Accordingly, the decline in osteoblast proliferation was correlated with a decrease in the expression of cyclin D1 and cyclin D2 and an increase in the expression of p27kip1 in FoxO1ob-/- bones (Figure 3D). Consistent with the RNA analysis, protein levels of Cyclins D1 and D2 decreased in the bone of FoxO1ob-/- mice (Figures 3E and 3F).
Figure 3. Decreased osteoblast proliferation in bones of FoxO1Ob-/- mice.
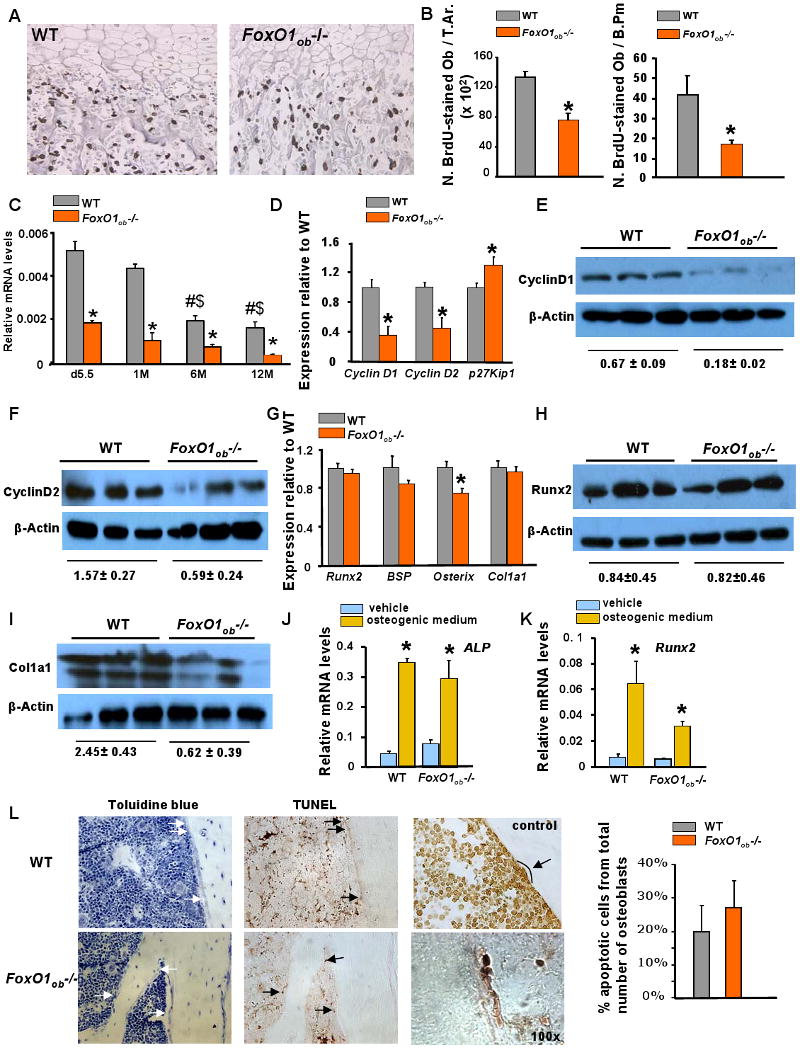
A) Sections of femurs from d5.5 WT and FoxO1ob-/- pups stained with BrdU (Image, 40× magnification)
B) Osteoblast proliferation expressed as number of BrdU stained osteoblasts per Trabecular Area (T.Ar.) and per Bone Perimeter (B.Pm) (n=4 mice/group). * p < 0.05 vs WT.
C) RT-PCR analysis of FoxO1, FoxO3 and FoxO4 expression in the bone of WT and FoxO1ob-/- mice at indicated ages. d denotes day and M demotes months (n=3 mice per group). * p < 0.05 vs WT; # p < 0.05 vs WT at d5.5; $ p < 0.05 vs WT at 1M.
D) RT-PCR analysis of CyclinD1, D2 and p27Kip1 expression in femurs of WT and FoxO1ob-/- mice at 2 months of age (n=6 mice/group).
E and F) Immunoblotting of cyclins D1 and D2 in bones of WT and FoxO1ob -/- mice.
G) RT-PCR analysis of Runx2, BSP, Osterix and Col1a1 expression in bones of WT and FoxO1ob -/- mice (n=6 mice/group). * p < 0.05 vs WT.
H and I) Immunoblotting of Runx2 and Type I collagen in bones of WT and FoxO1ob -/- mice.
J and K) RT-PCR analysis of Runx2 and Alkaline phosphatase (ALP) expression in cultured primary calvarial osteoblasts of WT and FoxO1ob-/- mice (n=3) treated with vehicle or osteogenic medium. * p < 0.05 vs vehicle
L) Apoptosis in bone sections from the femurs of 3 month-old WT and FoxO1ob-/- mice (n=4 mice per group). Arrows indicate osteoblasts (left panel) or apoptotic osteoblasts (right panel). Control indicates TUNEL staining of a bone section treated with DNAse. Magnifications are 40× unless otherwise stated. The 100× magnification panel shows an example of a group of apoptotic osteoblasts.
In all panels values are means ± sem.
Contrasting with the changes in proliferation and expression of cell cycle genes, expression of osteoblast differentiation markers such as Runx2, bone sialoprotein (Bsp) and type 1 collagen was not altered in FoxO1ob-/- mice (Figure 3G). Expression of osterix was decreased suggesting that osterix may be a target of FoxO1. However, whereas Runx2 protein levels, like Runx2 RNA, were not affected, protein levels of type I collagen were decreased in the bone of FoxO1ob-/- mice (Figure 3H and 3I). In agreement with the lack of changes in the expression of most osteoblast differentiation markers, calvaria-derived osteoblastic cells from Foxo1ob-/- mice were able to differentiate as efficiently as WT cells when cultured in osteogenic medium (Figures 3J and 3K). This was evidenced by normal expression of Runx2 and alkaline phosphatase (ALP) in primary osteoblast cultures from FoxO1ob -/- as compared to WT mice.
Finally, we searched for potential effects of FoxO1 deletion on osteoblast apoptosis. Using TUNEL staining of bone sections we found that osteoblast apoptosis is not affected in FoxO1ob-/- mice as compared to wild type control animals (Figure 3L).
Increased Oxidative stress suppresses osteoblast proliferation in FoxO1ob -/- mice
As the first step in elucidating the molecular basis of defective osteoblast proliferation in FoxO1ob -/-mice we examined whether FoxO1 deletion alters redox balance in osteoblasts. One of the well described transcriptional targets of the anti-oxidant properties of FoxO1 is Sod2, the gene encoding the anti-oxidant defense enzyme Superoxide Dismutase 2. Mitochondrial SOD2 activity in whole femur extracts was decreased in Foxo1ob-/- mice (Figure 4A). Consistent with this observation, the expression of two redox-sensitive genes, Gadd45 and FasL, was altered. Consistent with the lack of an effect on osteoblast apoptosis expression of the stress-activated DNA repair gene Gadd45 was increased whereas expression of the pro-apoptotic gene FasL was decreased, in the bone of the Foxo1ob-/- mice (Figure 4B). The expression and activity pattern of FoxO1-regulated genes indicate a decrease in anti-oxidant defense responses in FoxO1ob-/- mice.
Figure 4. N-acetyl L-cysteine (NAC) rescues the increased oxidative stress and the bone phenotype of FoxO1ob-/- mice.
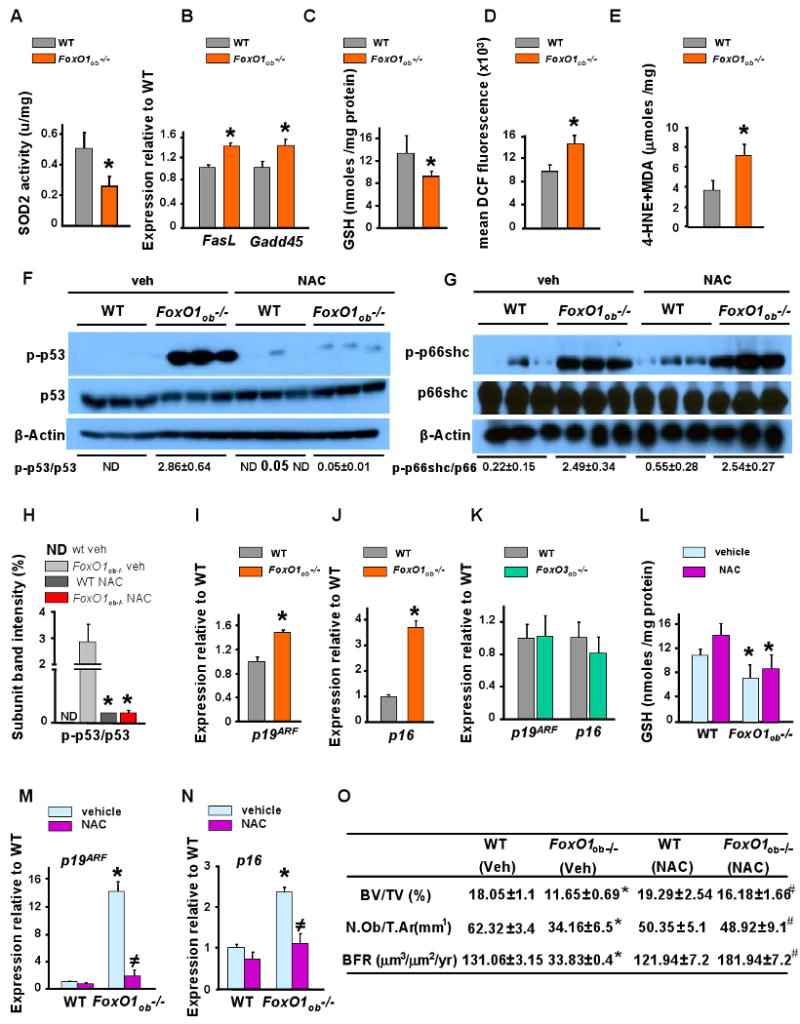
A) SOD2 activity in femurs of WT and FoxO1ob-/- mice (u/mg) (n=6 mice/group). * p < 0.05 vs WT.
B) RT-PCR analysis of FasL and Gadd45 expression in femurs of WT and FoxO1ob-/- mice (n=5 mice/group). * p < 0.05 vs WT.
C) Glutathione (GSH) levels in bones of WT and FoxO1ob-/- mice (n=5 mice/group). * p < 0.05 vs WT.
D) Flow cytometry analysis of reactive oxygen species (ROS) levels in osteoblasts from bone marrow cells of WT and FoxO1ob-/- mice (n=5 mice/group). * p < 0.05 vs WT.
E) Lipid peroxidation levels in bones of WT and FoxO1ob-/- mice (n=5 mice/group). * p < 0.05 vs WT.
F and G) p53 and p66shc activity measured by immunoblotting in bones of WT and FoxO1ob-/- mice (n=2 mice/group).
H) Densitometric analysis of the ratio of p-p53 / p53. ND denotes not determined.
I and J) RT-PCR analysis of p19ARF and p16 expression in bones of WT and FoxO1ob-/- mice (n=5 mice/group). * p < 0.05 vs WT.
K) RT-PCR analysis of p19ARF and p16 expression in bones of WT and FoxO3ob-/- mice (n=5 mice/group).
L) GSH levels in osteoblasts of vehicle- or NAC-treated WT and FoxO1ob-/- mice (n=5 mice/group). * p < 0.05 vs WT vehicle and WT NAC.
M and N) RT-PCR analysis of p19ARF and p16 expression in bones of vehicle- or NAC-treated WT and FoxO1ob-/- mice (n=5 mice/group). * p < 0.05 vs WT vehicle; # p < 0.05 vs WT vs FoxO1ob-/- vehicle.
O) BV/TV, N.Ob./T.Ar and BFR in spines of vehicle or NAC-treated WT and FoxO1ob-/- mice (n=5 mice/group). * P < 0.05 (FoxO1ob-/- vehicle vs. WT); # P < 0.05 (FoxO1ob-/- NAC vs FoxO1ob-/- vehicle).
In all panels, except I and J where mice were 5.5 days of age, animals were 3 months of age; and, values are means ± sem.
See also Figures S3 and S4.
More important, accumulation of glutathione (GSH), a small protein with redox-active sulfhydryl moieties which scavenges and neutralizes ROS thereby detoxifying cells, was reduced in the bone of FoxO1ob-/- mice (Figure 4C). The reduction in GSH levels in FoxO1-deficient osteoblasts should lead to increased oxidative stress in bone. Indeed, we determined that production of reactive oxygen species (ROS) was increased in osteoblasts from FoxO1ob-/- mice (Figure 4D). Elevated levels of lipid peroxidation end products, a measure of cellular injury, were also observed in the bone of FoxO1ob-/- mice (Figure 4E). Taken together these results indicate that FoxO1 functions in osteoblasts to limit oxidative stress and its deleterious consequences on the cells.
Two of the most powerful signal transduction pathways activated by ROS are those involving p53 and p66shc signaling. Whereas p53 mediates ROS-induced anti-proliferative actions and early senescence or apoptosis, p66shc is an adapter protein mediating specifically pro-apoptotic actions of ROS (Migliaccio et al., 1999). We found that the activity of p53 and p66shc increased in bone from FoxO1ob-/- mice (Figures 4F-4H). Two proteins, the products of p19ARF and p16, activate a p53 pathway which leads to anti-proliferative effects (Satyanarayana and Rudolph, 2004). Thus, consistent with defective osteoblast proliferation of FoxO1-deficient osteoblasts, and concomitant with elevated p53 activity, expression of its upstream regulators p19ARF and p16 increased in osteoblasts from FoxO1ob-/- mice as compared to wild type control animals (Figures 4I and 4J). Similar to the lack of a bone phenotype in the FoxO3ob-/- mice, these perturbations were specific of FoxO1 deletion since expression of p19ARF and p16 in bone was unaffected by FoxO3 deletion (Figure 4K). Similarly, knockdown of FoxO4 in osteoblasts had no effect on Sod2 as well as p19ARF and p16 expression (Figure S3).
Rescue of the low bone mass phenotype of FoxO1ob-/- mice
To determine whether increased oxidative stress in osteoblasts could be a causative factor for the low bone formation phenotype of the FoxO1ob-/- mice we normalized redox levels by supplying the anti-oxidant N- acetyl L- cysteine (NAC). The anti-oxidant actions of NAC are due to its ability to induce either one of 2 independent effects: neutralize ROS products by supplying a pool of amino acids utilized for glutathione synthesis; or block ROS signaling by suppressing the activity of p53 (Nakamura et al., 1997; Nogueira et al., 2008). While administration of NAC had no effect on GSH levels in FoxO1ob-/- mice (Figure 4L), or p66shc activity (Figure 4G) it suppressed p53 activity as well as the expression of p19ARF and p16 (Figures 4F, 4M and 4N). Consistent with normalization of p53 signaling, NAC rescued the phenotypic bone abnormalities of FoxO1ob-/- mice such as osteoblast numbers, bone formation rate and bone volume (Figure 4O).
To further confirm that the defect in osteoblast proliferation and the increase in oxidative stress in osteoblasts results from FoxO1 deficiency, we overexpressed FoxO1 in osteoblasts derived from FoxO1ob-/- mice. FoxO1 overexpression, confirmed by RT-PCR and immunoblotting, rescued the decrease in osteoblast proliferation and, similar to NAC, obviated the increase in ROS levels, normalized GSH levels and inhibited the increase in p19ARF and p16 expression (Figure S4). These results demonstrate that increased levels of oxidative stress in the bone of FoxO1ob-/- mice decrease osteoblast proliferation by a pathway that controls p19ARF/p16/p53 signaling thus providing a mechanistic link between pathways regulating oxidative stress and bone homeostasis under the control of FoxO1.
FoxO1 promotes protein synthesis in osteoblasts by interacting with ATF4
The observations that GSH levels were reduced in the bone of FoxO1ob-/- mice and that NAC administration failed to replenish them raised the hypothesis that, in addition to activation of p53 signaling presented above, there was also a defect in amino acid import and protein synthesis in FoxO1-deficient osteoblasts. This notion was further supported by the observation that whereas type I collagen expression was not altered, its production was decreased in the bone of FoxO1ob-/- mice.
To examine this possibility we measured the activity of the translation initiation factor 2 (eIF2) which integrates amino acid metabolism and resistance to oxidative stress (Harding et al., 2003). Eukaryotic cells respond to oxidants by phosphorylating the alpha subunit of eIF2. In turn, activation of eIF2α initiates the pathway controlling amino acid import and protection against oxidative stress (Harding et al., 2002). We found that the phosphorylation levels of eIF2α were increased in FoxO1ob-/- osteoblasts (Figure 5A) indicating a normal response of eIF2α. Thus, we reasoned that another component of the protein synthesis pathway, downstream of eIF2α may be affected in FoxO1ob-/- osteoblasts. The transcription factor ATF4 is an integral component of a negative-feedback pathway controlling amino acid import, leading to glutathione synthesis, and, its expression or activity is dependent on eIF2α (Harding et al., 2003; Yang et al., 2004). Although ATF4 mRNA expression remained unchanged in FoxO1ob-/- bones as compared to WT control animals (Figure 5B) ATF4 was found to physically associate with FoxO1 in the nuclei of osteoblasts as well as in bone nuclear extracts from WT mice (Figures 5C and 5D). Immunohistochemical analysis in bone sections confirmed that FoxO1 and ATF4 co-localize in the nucleus (Figures 5E and 5F). However, both proteins can also be found in the cytoplasm as they are known to shuttle between the two subcellular compartments. In the absence of any stimuli, and in particular stress-related stimulus, the two transcription factors are predominantly located in the cytoplasm. Stress signals stimulate their translocation to the nucleus, where they actively operate to initiate transcriptional events that protect cellular functionality.
Figure 5. Altered amino acid metabolism in osteoblasts of FoxO1ob-/- mice.
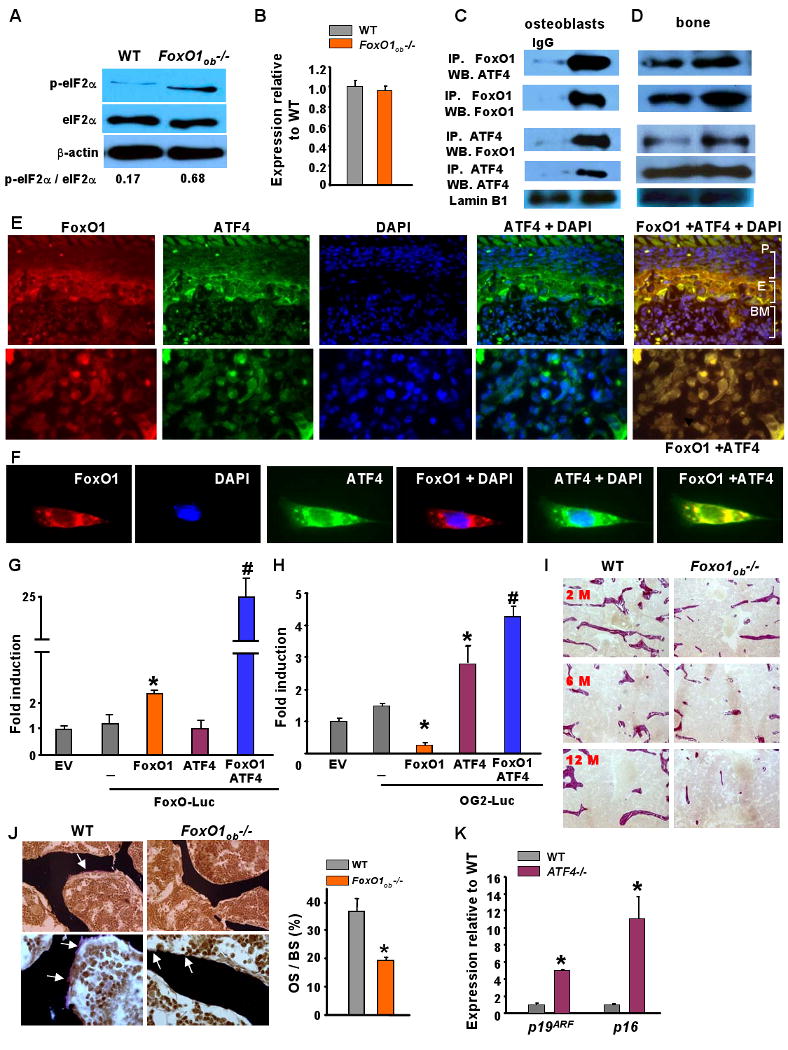
A) Immunoblotting analysis of the phosphorylation status of eIF2α̣ in bone from WT and FoxO1ob-/- mice (n=5 mice/group).
B) RT-PCR analysis of ATF4 expression in bone from WT and FoxO1ob-/- mice (n=5 mice/group).
C and D) Immumoprecipitation (IP) and immunoblotting of FoxO1 and ATF4 in nuclear extracts from primary osteoblasts and bones of WT mice.
E) Immunohistochemical localization of FoxO1 and ATF4 in femoral sections of newborn, WT mice. Images of bone sections depicting FoxO1, ATF4, DAPI and combination of ATF4 with DAPI or FoxO1 with ATF4 stainings. The top 5 panels show 40× and the 5 lower panels show 100× magnifications. P indicates periosteal surface; E indicates endosteal surface; and, BM indicates bone marrow. The 100× magnification images are obtained from the endosteal surface.
F) Immunohistochemical localization of FoxO1 and ATF4 in primary osteoblasts. Single cell images at 100× magnification show staining with the indicated antibodies.
G) Co-transfection of FoxO1, ATF4 and FoxO-Luc reporter construct in COS-7 cells. EV denotes empty vector. Results are presented as fold induction over EV. (EV=1). * p < 0.05 vs.FoxO-luc; # P < 0.05 vs FoxO1/FoxO-luc.
H) Co-transfection of FoxO1, ATF4 and OG2-Luc reporter construct in COS-7 cells. EV denotes empty vector. Results are presented as fold induction over EV. (EV=1). * P < 0.05 vs. OG2-luc; # P < 0.05 vs ATF4/OG2-Luc and vs FoxO1/OG2-luc).
I) Von Gieson staining indicating collagen content (stained red) on vertebral sections from 2, 6 and 12 month-old WT and FoxO1ob-/- mice. Representative results from n = 5 mice per group.
J) Osteoid surface by Von Kossa staining in vertebral sections from 1 month-old WT and FoxO1ob-/- mice (n= 6 mice per group). OS and BS denote osteoid surface and bone surface, respectively. Arrows indicate osteoid deposition. * p < 0.05 vs WT.
K) RT-PCR analysis of p19ARF and p16 expression in primary osteoblasts derived from Atf4-/- mice. * p < 0.05 vs WT.
In all panels values are means ± sem.
To examine whether the FoxO1-ATF4 interaction influences the activity of either of the 2 transcription factors we used COS-7 cells. Cotransfection experiments indicated that ATF4 stimulates FoxO1 activity as measured on a FoxO1 reporter (Figure 5G). Since osteocalcin is an ATF4 target gene in osteoblasts (Yang et al., 2004), activation of ATF4 was measured using a reporter construct that carries 147bp of the osteocalcin promoter fused to the Luciferase gene (pOG-2-Luc). This construct contains one binding site for ATF4 (Ducy and Karsenty, 1995). Forced expression of FoxO1 along with ATF4, enhanced the transactivating ability of ATF4 on pOG2-Luc (Figure 5H). These observations raised the prospect that FoxO1 deletion in osteoblasts compromises amino acid synthesis by interfering with the activity of ATF4. This event leads to a reduction in both GSH levels and collagen production. Indeed, collagen content was decreased in the bones of the FoxO1ob-/- mice (Figure 5I) supporting the notion that protein synthesis by osteoblasts is compromised in these animals. Consistent with the low collagen content, we found a decrease in osteoid surface, the amount of bone extracellular matrix not mineralized (Figure 5J). Additionally, a reduction in GSH synthesis will result in a subsequent increase in ROS and oxidative stress. Elevated ROS do in turn activate p53 signaling.
An implication of this model is that induction of p53 signaling may be, at least in part, ATF4-dependent. To determine if this was the case, we measured expression levels of p19ARF and p16 in Atf4-/- osteoblasts. ATF4 deletion increased the expression of both p53 regulators (Figure 5K) suggesting that the FoxO1-ATF4 pathway is a determinant of p53 signaling in osteoblasts. Collectively these observations suggest that FoxO1 regulates protein synthesis and oxidative stress in osteoblasts by interacting with ATF4.
High protein diet rescues the low bone formation phenotype of FoxO1ob-/- mice
The finding that FoxO1 is required for protein synthesis in osteoblasts and its ability to interact with ATF4 suggested that a high-protein diet could rescue the skeletal manifestations observed in FoxO1ob-/- mice as it does for the Atf4-/- mice (Elefteriou et al., 2006). To test this hypothesis we used the same diet that had been used to correct the bone phenotype of the Atf4-/- mice and examined whether it could affect the bone phenotype of the FoxO1ob-/- mice. WT and FoxO1ob-/- mothers were fed from the first day of gestation with a normal (ND) or a high-protein diet (HPD). After weaning, pups were kept on this same diet up to 28 days of age. At this stage, histomorphometric analysis showed that HPD fully rescued the low osteoblast numbers in FoxO1ob-/- mice, but had no effect on the increase in osteoclast surface (Figure 6A). In agreement with these observations and the notion that osteoblast recruitment is compromised in FoxO1ob-/- mice, BV/TV was rescued and bone formation rate, although not statistically significant, showed a trend towards partial rescue in high protein diet - fed FoxO1ob-/- mice (Figure 6A). This latter effect may indicate that additional pathways, independent of protein synthesis, may compromise osteoblast function in the FoxO1ob-/- mice. HPD also normalized the upregulation of p53 activity and the increase in the expression of p19ARF and p16 in bones of Foxo1ob-/- mice (Figures 6B, 6C and 6D). Taken together, these results demonstrate that FoxO1 regulation of osteoblast proliferation and bone formation occurs mainly through its ability to favor amino acid import and protein synthesis through interacting with ATF4 and subsequently maintaining redox balance in osteoblasts.
Figure 6. High protein diet (HPD) rescues the bone phenotype of FoxO1ob-/- mice.
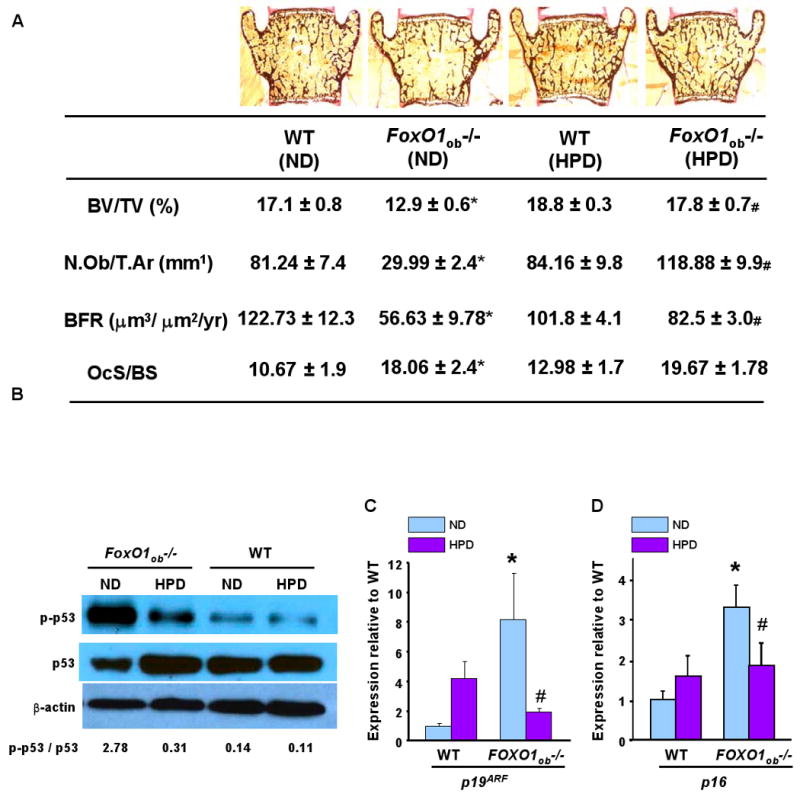
A) BV/TV, N.Ob/T.Ar., BFR and osteoclast surface in the verterbrae of WT and FoxO1ob-/- on normal (ND) or HPD (n=5 mice/group). * P < 0.05 (FoxO1ob-/- ND vs. WT ND); # P < 0.05 (FoxO1ob-/- HPD vs FoxO1ob-/- ND). The OcS/BS data shown for WT and FoxO1ob-/- mice on normal diet, are identical to those shown in Figure 2B.
B) Immunoblotting of phospho-p53 in bones of WT and FoxO1ob-/- on ND or HPD. C and D) RT-PCR analysis of p19ARF and p16 expression in bones of WT and FoxO1ob-/- on ND or HPD (n=5 mice/group). * P < 0.05 (FoxO1ob-/- vs. WT); # P < 0.05 (FoxO1ob-/- HPD vs FoxO1ob-/- ND).
In all panels, mice were 1 month of age.
Discussion
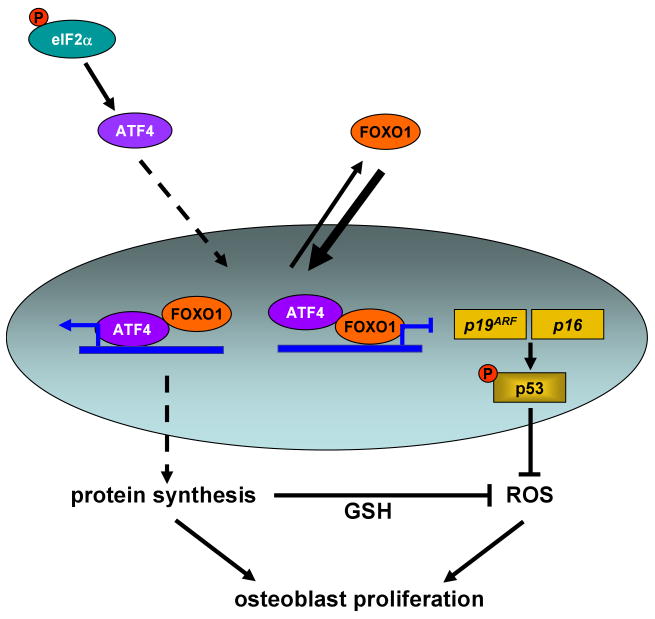
This study reveals that FoxO1 is a major regulator of osteoblast proliferation and as result of bone homeostasis. The effects of FoxO1 stem from its ability to maintain redox balance in osteoblasts in a p53-dependent manner. FoxO1 also affects osteoblast function by regulating amino acid import and thus protein synthesis. This function results from the physical interaction of FoxO1 with ATF4, a transcription factor regulating amino acid import and collagen synthesis by osteoblasts. These results establish a sequence of molecular events that mediate the protective effects of FoxO1 in osteoblasts (Figure 7). More important, they provide proof of principle evidence that oxidative stress does indeed regulate bone mass.
Figure 7. Model depicting the mechanism of FoxO1 action in osteoblasts.
Under physiological levels of stress, FoxO1 shuttles between the nucleus and the cytoplasm. In the nucleus, FoxO1 interacts with ATF4. This interaction promotes the transcriptional activity of FoxO1 and is required for amino acid import and protein synthesis. Normal protein synthesis allows FoxO1 to orchestrate an anti-oxidant defense mechanism that maintains redox balance by suppressing expression of p19ARF and p16 and downstream activation of their target protein p53. Transcriptional repression of the p19ARF/p16/p53 pathway prevents cell cycle arrest in osteoblasts and maintains their normal proliferation and bone homeostasis
ROS to bone remodeling: FoxO1 delineates a signaling cascade mediating the effects of oxidative stress on osteoblast proliferation
The oxidative hypothesis of senescence, since its origin in 1956, has garnered significant evidence and growing support for the notion that free radicals play an important role in aging, either as “damaging” molecules or as signaling molecules. Age-increasing oxidative injuries induced by free radicals, higher susceptibility to oxidative stress, genetic manipulations that alter oxidative resistance and the anti-aging effect of caloric restriction are a few examples documenting the implication of oxidative stress in the aging process and the development of aging-associated diseases. However, to this time, the mechanisms and mediators that implement the effects of oxidative stress in different tissues are not completely understood due to the complex “network” of redox regulatory systems. The identification of FoxO1 as a crucial mediator of ROS signaling in osteoblasts leads to the delineation of the molecular events that mediate the effects of oxidative stress on osteoblast proliferation (Figure 7). Our observations that a) FoxO1 interacts with ATF4 and this interaction promotes both FoxO1 and ATF4 activity, b) protein synthesis is compromised in osteoblasts from Foxo1ob-/- mice and c) high protein diet rescues the low bone formation phenotype of Foxo1ob-/- mice, indicate that FoxO1 controls osteoblast proliferation by interacting with ATF4 and promoting protein synthesis.
The p19/p16/p53 signaling cascade is a novel, nontranscriptional target of FoxO1 mediating its anti-oxidant properties
Increased levels of ROS and lipid peroxidation products and activation of a stress-evoked, p53-dependent signaling cascade were observed in the bones of Foxo1ob-/- mice. Administration of the antioxidant N-acetyl L-cysteine (NAC), rescues the low bone formation phenotype of Foxo1ob-/- mice and restores redox balance in FoxO1-deficient osteoblasts. These studies reveal that FoxO1 in osteoblasts utilizes a previously unrecognized mechanism to organize anti-oxidant responses, which involves inhibition of p53-dependent signaling. This is particularly important for the biology of FoxO1 action in anti-oxidant defense which to this time had identified only Sod2, catalase, Gadd45, Bim and FasL as the direct transcriptional targets of FoxO1 during stress (Lehtinen et al., 2006). In our hands high protein diet along with rescuing the bone phenotype of Foxo1ob-/- mice also suppresses p53 signaling in bone, thus indicating that protein synthesis is required for redox balance in osteoblasts. Thus, FoxO1 controls osteoblast proliferation by actions that involve interaction with ATF4; and, downstream regulation of a stress-dependent pathway that controls p53 signaling (Figure 7).
Interaction of FoxO1 with Insulin and IGF-1 signaling pathways
In work carried out in our laboratoty, we have found that FoxO1ob-/- mice have a metabolic phenotype characterized by improved glucose tolerance and insulin sensitivity and increased insulin secretion, secondary to an increase in β-cell proliferation. Thus, it is possible that increased insulin levels in the FoxO1ob-/- mice may affect osteoblast proliferation. In addition to insulin, IGF-1 signaling suppresses FoxO activity and is known to promote osteoblast function (Zhang et al., 2002). However, we have found that proliferation is decreased in ex vivo cultures of osteoblasts obtained from FoxO1ob-/- mice as compared to wild type littermates (data not shown), suggesting that an intrinsic factor is not the main cause for the low osteoblast numbers. These observations, along with a plethora of evidence in the literature that FoxO1 is the main mediator of insulin actions (Accili and Arden, 2004), suggest that the defect in osteoblast proliferation and its rescue with high protein diet is most likely independent of changes in insulin levels. However, further gene inactivation experiments will be required to obtain a definitive answer.
Decreased collagen content but intact mineralization in by FoxO1 deletion in osteoblasts
We have found that although Osterix expression was suppressed by FoxO1 deletion, osteoblast differentiation was not overtly affected as determined by the lack of changes in the expression of Alp and Bsp in the bone and in ex vivo cultures of osteoblasts from FoxO1ob-/- mice. However, as reported for ATF4-/- mice (Yang et al., 2004), type I collagen expression is not affected but its synthesis and bone collagen content are decreased in the FoxO1ob-/- mice supporting the notion that protein synthesis by osteoblasts is compromised in these animals. Consistent with the low collagen content, we found a decrease in osteoid surface, the amount of bone extracellular matrix not mineralized. However, because Alp expression is not affected and serum calcium and phosphate levels are normal in the serum of FoxO1ob-/- mice (data not shown) mineralization appears to be overall unaffected in FoxO1ob-/- mice.
The role of broadly expressed transcription factors in osteoblast function
Until recently the prevailing view of the transcriptional control of osteoblast biology was that it involves either one of 3 proteins that are expressed specifically in osteoblasts: Runx2, Osterix and/or ATF4. However, FoxO1 has a potent effect on bone formation in spite of its ubiquitous expression. The paradigm of FoxO1 action on osteoblast adds to the growing list of broadly expressed proteins and transcription factors that are found to be crucial for the control of bone remodeling. LRP5 is now recognized to control bone formation by inhibiting serotonin synthesis in the duodenum (Yadav et al., 2008). CREB, another ubiquitous transcription factor, is a transcriptional effector of serotonin action on bone (Yadav et al., 2008). Scnurri-3 mostly known for its functions as an adapter protein in the immune system is an important regulator of adult bone formation (Jones et al., 2006). And, the broadly expressed AP-1 family members (Fos/Jun) have an essential role in bone development and bone formation (Wagner, 2002; Fu et al., 2005). These observations suggest that in addition to the osteoblast-specific transcription factors, ubiquitously expressed ones can influence the function of these cells. Whether this is achieved by means of interaction with the osteoblast-specific proteins or by means of activating novel signaling cascades (such as FoxO1) it remains to be examined. In any case, the transition from a broadly expressed protein to a tissue specific pathway highlights the importance of the cross talk between different organs and illustrates the importance of an integrative approach to bone physiology.
Experimental Procedures
Generation of mutant mice
All the protocols and animal treatment procedures described have been approved by the institute of comparative medicine, Columbia University. FoxO1Ob-/- and FoxO3Ob-/- were generated by crossing α1(I)Collagen-Cre mice with FoxO1 or FoxO3 floxed mice that have been previously described (Paik J.H et al. 2007 and Dacquin R. et al. 2002). In all experiments, littermates of FoxO1ob-/- mice were used as wild type control animals. Those mice were homozygous for the floxed FoxO1 allele.
Animal treatments
N-acetyl-l cysteine (Sigma) was administrated IP, 50mg/kg twice a day for 3 weeks (Almeida M. 2007). 1× PBS was given to sham mice. Normal diet (ND) and high protein diet (HPD) were obtained from Harlan Teklad (USA). The high protein diet contained 50% protein, 31.6% carbohydrate and 5.5% fat. The normal diet contained 20.3% protein, 61.6% carbohydrate and 5.5% fat. They are matched for calcium (0.7%) and phosphorus (0.54%) content. For all animal experiments littermates were used as wild type control animals. Genotyping was performed at 3 weeks of age by PCR analysis of genomic DNA.
Histology, Protein Expression, and Proliferation Assays
Immunohistochemistry was performed according to standard protocols on undecalcified specimens embedded in paraffin and sectioned at 5 μm. Immune complexes were visualized using anti–FOXO1 (Cell Signaling) or anti-ATF4 (Santa Cruz). FoxO1 and ATF4 were visualized with CY3-conjugated (red) and CY2-conjugated (green) secondary antibodies (Jackson Immuno.), respectively. Sections were counterstained with DAPI to visualize the cell nuclei (blue). Alternatively, FoxO1 expression was detected using the stable chromogen DAB (Diaminobenzidine) and counterstaining with hematoxylin or eosin. Images were acquired with a Nikon 80i Eclipse Microscope using a Retiga digital camera. Osteoblast or bone extracts (50-80 μg) from WT and FoxO1ob-/- mice were anayzed on a SDS-polyacrylamide gel, transferred to a PVDF membrane and probed with the appropriate antibodies. Immunoblotting primary antibodies were against total or Phospho-p66Shc (BD biosciences), phospho-FoxO1, phospho-FoxO3, phospho-FoxO4, phospho-53 and eIF2α (Cell Signaling, Beverly, MA), and FoxO1, FoxO3 and FoxO4 (Santa Cruz). Antibodies against β-actin and Lamin B1 were obtained from Santa Cruz. The intensity of the bands was measured by densitometry (Image J software). The ratios of phospho/total proteins was calculated based on the intensity of the bands determined by densitometry. In vivo osteoblast proliferation assays (Zymed Laboratories) were performed on femurs of d5.5 mice pups injected with BrdU (0.4 mg) and sacrificed 4 hr later. In vitro osteoblast proliferation was assessed using the CellTiter One System (Promega) according to the manufacturer's instructions. Static and dynamic histomorphometric analyses were performed on vertebral column specimens collected from 1, 2 and 3, 6 and 12 month-old mice using undecalcified sections according to standard protocols using the Osteomeasure analysis system (Osteometrics). Type I collagen content was analyzed by Von Gieson staining in undecalcified sections. Analysis of bone mineral density (BMD) was performed as previously described (Kousteni et al., 2002). Six to 12 animals were analyzed for each group.
Cell culture and treatments
Calvaria-derived primary osteoblastic cells from WT and FoxO1ob-/- mice were plated in αMEM medium containing 1% FBS. Treatments with NAC were performed at 2 mM. Differentiation to the osteogenic lineage was induced in osteogenic medium containing 50 μM ascorbic acid and 50 μM β-glycerol phosphate. Co-tranfection was carried out in COS-7 cells as described previously (Kousteni et al., 2001). Renilla luciferase control vector (Promega) was cotransfected as an internal standard to normalize for transfection efficiency. Normalized luciferase activity is presented as fold induction over the empty vector control (EV, considered 1).
Biochemical studies
Intracellular ROS levels were measured in BM cells by means of flow cytometry using an oxidation sensitive fluorescent probe dye, 2′,7′-dichlorodihydrofluorescin diacetate (H2DCFDA). BM cells flushed from femurs were incubated with goat anti-mouse Osteocalcin (OCN) antibody for 20 mins at 4°C followed by incubation with donkey anti-goat alexa-680 secondary antibody for 20mins at 4°C. After 2 washes, cells were incubated with 10 μM H2DCFDA for 30 min at 37°C and fluorescence was measured using an LSR II Flow cytometer (BD biosciences). Osteoblastic cells were identified as a CD45low, OCN+ population. OCN positive cells were first gated and the mean fluorescence intensity (MFI) of the DCF is measured. Analysis was performed using FloJo software (TreeStar). Lipid peroxidation end product (4-hydroxy-2(E) nonenal: 4-HNE + malondialdehyde: MDA) levels were measured in femurs by using lipid peroxidation assay kit (Calbiochem, San Diego, CA, USA). The levels of 4-HNE + MDA were determined from standard calibration curve constructed using MDA according to the manufacturer's instructions. The values were expressed as μmol 4-HNE+MDA/mg protein. Reduced glutathione levels in the tibia extracts were measured by the 5,5′-dithiobis-2-nitrobenzoic acid-glutathione disulfide reductase recycling method as described previously (Tietz F, Anal Biochem. 1969 and Rahman I et al, Nature Protocols 2006). Mitochondrial Superoxide Dismutase (SOD-2) activity was measured in femurs using Superoxide Dismutase Assay Kit II (Calbiochem).
Molecular studies
RNA isolation and real-time PCR was performed following standard protocols. Primer sequences are shown in Table S1.
siRNA knockdown
Primary osteoblasts were transfected with the FoxO4 siRNA oligos or control scrambled siRNA oligos (Santa Cruz) using siRNA transfection reagents (Santa Cruz) as described by the manufacturer.
Adenoviral infection
High- titer virus stocks were produced by infecting FoxO1 and empty vector (EV) adenoviral expression vectors into HEK 293- based packaging cell line. After 72 hours, virus was collected and viral titer was determined. Calvaria-derived osteoblastic cells were infected with FoxO1 or EV control adenoviral constructs. Forty eight hours later, cells were harvested and used for different assays.
Apoptosis in bone sections
Apoptosis was assessed in decalcified, paraffin-embedded femoral sections from 3 month-old WT and FoxO1ob-/- mice using the DeadEnd Colorimetric TUNEL System (Promega) according to the manufacturer's instructions. Apoptotic osteoblasts and total osteoblasts were counted and the % of apoptotic cells was calculated.
Statistical Analysis
Results are given as means ± standard deviations from the mean (s.e.m). Statistical analysis was performed by Student's t test where p<0.05 is considered significant.
Acknowledgments
The authors are grateful to Dr Gerard Karsenty for helpful discussions, critical reading of the manuscript and for providing ATF4-deficient osteoblasts. We are also thankful to Dr John P. Bilezikian and Dr Patricia Ducy for helpful discussions, Dr John Manavalan for help with measurements of ROS levels in osteoblasts, Dr Li Qiang for help with the adenoviral infections and Charles Duncan for technical assistance. We are thankful to the histology facility of the Diabetes And Endocrinology Research Center (DERC) of Columbia University Medical Center (supported by NIDDK DK063608-07) for help with histological analysis. Dr Kousteni is indebted to Dr Vijay Yadav for his generous help and constant availability throughout this project. This work was supported by the National Institutes of Health (R01 AR055931 and R01 AR054447 to SK). The authors do not have any conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–426. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- Almeida M, Han L, Martin-Millan M, Plotkin LI, Stewart SA, Roberson PK, Kousteni S, O'Brien CA, Bellido T, Parfitt AM, Weinstein RS, Jilka RL, Manolagas SC. Skeletal involution by age-associated oxidative stress and its acceleration by loss of sex steroids. J Biol Chem. 2007;282:27285–27297. doi: 10.1074/jbc.M702810200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arden KC. Multiple roles of FOXO transcription factors in mammalian cells point to multiple roles in cancer. Exp Gerontol. 2006;41:709–717. doi: 10.1016/j.exger.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Bai XC, Lu D, Bai J, Zheng H, Ke ZY, Li XM, Luo SQ. Oxidative stress inhibits osteoblastic differentiation of bone cells by ERK and NF-kappaB. Biochem Biophys Res Commun. 2004;314:197–207. doi: 10.1016/j.bbrc.2003.12.073. [DOI] [PubMed] [Google Scholar]
- Bouxsein ML, Melton LJ, III, Riggs BL, Muller J, Atkinson EJ, Oberg AL, Robb RA, Camp JJ, Rouleau PA, McCollough CH, Khosla S. Age- and sex-specific differences in the factor of risk for vertebral fracture: a population-based study using QCT. J Bone Miner Res. 2006;21:1475–1482. doi: 10.1359/jbmr.060606. [DOI] [PubMed] [Google Scholar]
- Dacquin R, Starbuck M, Schinke T, Karsenty G. Mouse alpha1(I)-collagen promoter is the best known promoter to drive efficient Cre recombinase expression in osteoblast. Dev Dyn. 2002;224:245–251. doi: 10.1002/dvdy.10100. [DOI] [PubMed] [Google Scholar]
- De Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, Weeda G, van der Horst GT, van Leeuwen W, Themmen AP, Meradji M, Hoeijmakers JH. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- Ducy P, Karsenty G. Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol Cell Biol. 1995;15:1858–1869. doi: 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefteriou F, Benson MD, Sowa H, Starbuck M, Liu X, Ron D, Parada LF, Karsenty G. ATF4 mediation of NF1 functions in osteoblast reveals a nutritional basis for congenital skeletal dysplasiae. Cell Metab. 2006;4:441–451. doi: 10.1016/j.cmet.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Fu L, Patel MS, Bradley A, Wagner EF, Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene. 2005;24:7410–7425. doi: 10.1038/sj.onc.1209086. [DOI] [PubMed] [Google Scholar]
- Harada S, Rodan GA. Control of osteoblast function and regulation of bone mass. Nature. 2003;423:349–355. doi: 10.1038/nature01660. [DOI] [PubMed] [Google Scholar]
- Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, Stojdl DF, Bell JC, Hettmann T, Leiden JM, Ron D. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- Hosaka T, Biggs WH, III, Tieu D, Boyer AD, Varki NM, Cavenee WK, Arden KC. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc Natl Acad Sci U S A. 2004;101:2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Wein MN, Oukka M, Hofstaetter JG, Glimcher MJ, Glimcher LH. Regulation of adult bone mass by the zinc finger adapter protein Schnurri-3. Science. 2006;312:1223–1227. doi: 10.1126/science.1126313. [DOI] [PubMed] [Google Scholar]
- Kousteni S, Bellido T, Plotkin LI, O'Brien CA, Bodenner DL, Han K, DiGregorio G, Katzenellenbogen JA, Katzenellenbogen BS, Roberson PK, Weinstein RS, Jilka RL, Manolagas SC. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity. Cell. 2001;104:719–730. [PubMed] [Google Scholar]
- Kousteni S, Chen JR, Bellido T, Han L, Ali AA, O'Brien C, Plotkin LI, Fu Q, Mancino AT, Wen Y, Vertino AM, Powers CC, Stewart SA, Ebert R, Parfit AM, Weinstein RS, Jilka RL, Manolagas SC. Reversal of bone loss in mice by nongenotropic signaling of sex steroids. Science. 2002;298:843–846. doi: 10.1126/science.1074935. [DOI] [PubMed] [Google Scholar]
- Lean JM, Davies JT, Fuller K, Jagger CJ, Kirstein B, Partington GA, Urry ZL, Chambers TJ. A crucial role for thiol antioxidants in estrogen-deficiency bone loss. J Clin Invest. 2003;112:915–923. doi: 10.1172/JCI18859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de l I, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Levasseur R, Barrios R, Elefteriou F, Glass DA, Lieberman MW, Karsenty G. Reversible skeletal abnormalities in gamma-glutamyl transpeptidase-deficient mice. Endocrinology. 2003;144:2761–2764. doi: 10.1210/en.2002-0071. [DOI] [PubMed] [Google Scholar]
- Liu JW, Chandra D, Rudd MD, Butler AP, Pallotta V, Brown D, Coffer PJ, Tang DG. Induction of prosurvival molecules by apoptotic stimuli: involvement of FOXO3a and ROS. Oncogene. 2005;24:2020–2031. doi: 10.1038/sj.onc.1208385. [DOI] [PubMed] [Google Scholar]
- Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- Murakami S. Stress resistance in long-lived mouse models. Exp Gerontol. 2006;41:1014–1019. doi: 10.1016/j.exger.2006.06.061. [DOI] [PubMed] [Google Scholar]
- Nakamura H, Nakamura K, Yodoi J. Redox regulation of cellular activation. Annu Rev Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- Nemoto S, Finkel T. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science. 2002;295:2450–2452. doi: 10.1126/science.1069004. [DOI] [PubMed] [Google Scholar]
- Nogueira V, Park Y, Chen CC, Xu PZ, Chen ML, Tonic I, Unterman T, Hay N. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell. 2008;14:458–470. doi: 10.1016/j.ccr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, Jiang S, Gilliland DG, Chin L, Wong WH, Castrillon DH, DePinho RA. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarrie JK, Riabowol KT. Murine models of life span extension. Sci Aging Knowledge Environ . 2004;2004:re5. doi: 10.1126/sageke.2004.31.re5. [DOI] [PubMed] [Google Scholar]
- Raisz LG. Clinical practice. Screening for osteoporosis. N Engl J Med. 2005;353:164–171. doi: 10.1056/NEJMcp042092. [DOI] [PubMed] [Google Scholar]
- Riggs BL, Melton LJ, III, Robb RA, Camp JJ, Atkinson EJ, Oberg AL, Rouleau PA, McCollough CH, Khosla S, Bouxsein ML. Population-based analysis of the relationship of whole bone strength indices and fall-related loads to age- and sex-specific patterns of hip and wrist fractures. J Bone Miner Res. 2006;21:315–323. doi: 10.1359/JBMR.051022. [DOI] [PubMed] [Google Scholar]
- Rodan GA, Martin TJ. Therapeutic approaches to bone diseases. Science. 2000;289:1508–1514. doi: 10.1126/science.289.5484.1508. [DOI] [PubMed] [Google Scholar]
- Satyanarayana A, Rudolph KL. p16 and ARF: activation of teenage proteins in old age. J Clin Invest. 2004;114:1237–1240. doi: 10.1172/JCI23437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teitelbaum SL, Ross FP. Genetic regulation of osteoclast development and function. Nat Rev Genet. 2003;4:638–649. doi: 10.1038/nrg1122. [DOI] [PubMed] [Google Scholar]
- Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, Armstrong SA, Passegue E, DePinho RA, Gilliland DG. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, Jones S, Ghebranious N, Igelmann H, Lu X, Soron G, Cooper B, Brayton C, Hee PS, Thompson T, Karsenty G, Bradley A, Donehower LA. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Wagner EF. Functions of AP1 (Fos/Jun) in bone development. Ann Rheum Dis. 2002;61 2:ii40–ii42. doi: 10.1136/ard.61.suppl_2.ii40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VK, Ryu JH, Suda N, Tanaka KF, Gingrich JA, Schutz G, Glorieux FH, Chiang CY, Zajac JD, Insogna KL, Mann JJ, Hen R, Ducy P, Karsenty G. Lrp5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell. 2008;135:825–837. doi: 10.1016/j.cell.2008.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xuan S, Bouxsein ML, von SD, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, Clemens TL. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277:44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]