PLK4 is a key regulator of centriole duplication. Here, we show that PLK4 is active beyond the initiation of centriole duplication with the abundance of active kinase increasing to a peak in mitosis. Importantly, we show that differences in PLK4 abundance exist between mother and daughter centrioles and that active PLK4 is restricted to the centrosome.
Abstract
Centrosome duplication occurs once every cell cycle in a strictly controlled manner. Polo-like kinase 4 (PLK4) is a key regulator of this process whose kinase activity is essential for centriole duplication. Here, we show that PLK4 autophosphorylation of serine S305 is a consequence of kinase activation and enables the active fraction to be identified in the cell. Active PLK4 is detectable on the replicating mother centriole in G1/S, with the proportion of active kinase increasing through interphase to reach a maximum in mitosis. Activation of PLK4 at the replicating daughter centriole is delayed until G2, but a level equivalent to the replicating mother centriole is achieved in M phase. Active PLK4 is regulated by the proteasome, because either proteasome inhibition or mutation of the degron motif of PLK4 results in the accumulation of S305-phosphorylated PLK4. Autophosphorylation probably plays a role in the process of centriole duplication, because mimicking S305 phosphorylation enhances the ability of overexpressed PLK4 to induce centriole amplification. Importantly, we show that S305-phosphorylated PLK4 is specifically sequestered at the centrosome contrary to the nonphosphorylated form. These data suggest that PLK4 activity is restricted to the centrosome to prevent aberrant centriole assembly and sustained kinase activity is required for centriole duplication.
INTRODUCTION
The centrosome consists of two centrioles, attached to one another by a flexible linker, that are associated with a matrix of proteins known as pericentriolar material (Bornens, 2002; Doxsey et al., 2005). Centrioles are barrel-like structures ∼200 nm in diameter and 500 nm in length formed from nine sets of microtubules, which are triplet at the proximal ends and doublet at the distal ends (Rattner and Phillips, 1973; Kuriyama and Borisy, 1981; Vorobjev and Chentsov Yu, 1982; Paintrand et al., 1992). The two centrioles present in the centrosome differ from one another in both structure and age. One was formed one cell cycle later than the other, is slightly shorter, and does not possess a set of subdistal and distal appendages (Paintrand et al., 1992). To reflect the fact that this centriole is younger than the other centriole, it is referred to as the daughter centriole and the older centriole is known as the mother.
Centriole duplication occurs once per cell cycle and is thought to be initiated at the G1/S boundary with the formation of procentrioles at each parental centriole (Robbins et al., 1968). This process is dependent upon several proteins required for the formation of the centriole structure, including γ-tubulin, Cep135, HsSAS-6, CP110, CPAP (SAS-4), hPOC5, and POC1 (Chen et al., 2002; Leidel and Gonczy, 2003; Leidel et al., 2005; Kleylein-Sohn et al., 2007; Azimzadeh et al., 2009; Keller et al., 2009). The initiation of procentriole assembly is governed by a growing number of regulatory proteins such as the phosphatase Cdc14B and the kinases Mps1, polo-like kinase (PLK)2, PLK4, and Cdk2 cyclinA/E (Meraldi et al., 1999; Fisk et al., 2003; Warnke et al., 2004; Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Wu et al., 2008). PLK4 was identified as a key regulator of centriole duplication because its overexpression resulted in centriole amplification; conversely, RNA interference (RNAi) of the Plk4 gene caused the sequential reduction of centriole number (Bettencourt-Dias et al., 2005; Habedanck et al., 2005). The kinase domain of PLK4 is similar to that of PLK1-3, but its C terminus differs in that it possesses a single Polo-box and a crypto Polo-box domain instead of the tandem Polo-box domains that are present in PLK1-3 (Fode et al., 1994; Leung et al., 2002). The crypto Polo-box and Polo-box domains of PLK4 contain motifs that are responsible for targeting the kinase to centrosomes and the cleavage furrow during cytokinesis (Hudson et al., 2001; Leung et al., 2002; Habedanck et al., 2005).
A clear link has been established between PLK4 and cell proliferation. The kinase was first described in mice as SNK/PLK akin kinase (SAK) due to structural similarity with these kinases (Fode et al., 1994). In situ hybridization studies showed that PLK4/SAK mRNA transcripts were present in proliferating cells of multiple organs during embryogenesis and at the base of colonic crypts in the small intestine of adults (Fode et al., 1994). Deletion of the kinase is lethal, causing null embryos to arrest at stage embryonic day 7.5, with increased numbers of late mitotic and apoptotic cells (Hudson et al., 2001). The loss of a single copy is detrimental as embryonic fibroblasts from PLK4+/− mice exhibit increased centrosomal amplification, multipolar spindle formation, and aneuploidy (Ko et al., 2005). In addition, aging heterozygous mice have an increased incidence of spontaneous liver and lung cancers (Ko et al., 2005). These data, coupled with evidence from RNAi and overexpression studies, suggest that the abundance of the kinase must be tightly regulated within the cell to ensure proper centriole duplication (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Ko et al., 2005).
PLK4 expression has been shown to be controlled at transcriptional and posttranslational levels. The kinase is rapidly degraded, with a half-life of ∼2–3 h, and a substrate of both the anaphase-promoting complex/cyclosome and SCF/Slimb ubiquitin ligases (Fode et al., 1996; Cunha-Ferreira et al., 2009; Rogers et al., 2009). Ubiquitinylation by the SCF/Slimb complex is dependent upon the recognition, by Slimb, of a phosphorylated degron motif that is located in the N terminus of PLK4 (Cunha-Ferreira et al., 2009; Rogers et al., 2009). There is evidence showing that expression is controlled at the transcriptional level with PLK4 mRNA being detectable in late G1 and increasing to reach a maximum in mitosis (Fode et al., 1996).
In this article, we address one of the previously unanswered questions in the role of PLK4 in centriole duplication: When is the kinase active in the cell cycle? Current models would suggest that the kinase acts once in the cell cycle to initiate procentriole assembly; however, our data indicate that this is not the case. By generating tools to identify active PLK4, we show the kinase is active for most of the cell cycle. Furthermore, we find that PLK4 is differentially regulated at replicating centrioles, becoming active at the mother centriole first in S phase and then later at the daughter centriole in G2. We also show that more PLK4 is associated with the mother centriole than the daughter. These data suggest that PLK4 has other functions, in addition to initiating centriole duplication, at centrosomes as the kinase is active in S, G2, and M phase. Importantly, we find that active PLK4 is sequestered at centrosomes, thereby restricting its activity to this site. The possible implications of PLK4 kinase activity upon centriole structure maintenance and centriole maturation are discussed.
MATERIALS AND METHODS
Recombinant DNA Technology, Production of PLK4 Proteins, and Site-directed Mutagenesis
All SUMO fusion plasmids were created by polymerase chain reaction (PCR) by using the full-length PLK4 as a template and cloned into a modified version of pSUMO (Invitrogen, Carlsbad, CA). After culture upon agar plates containing kanamycin and 1% glucose, a single colony was resuspended in 2YT media containing kanamycin and 0.1% glucose for 3 h. One milliliter of the preculture was then incubated in 400 ml of autoinduction medium (Novagen, Madison, WI) without glucose at 37°C for 3 h to reach an OD (600 nm) of 0.6. At this point, the temperature was reduced to 23°C for another 16 h. The samples were spun down at 3500 rpm for 25 min at 4°C. The pellets were then resuspended in 10 ml of ice-cold Milli-Q water (Millipore, Billerica, MA) and 10 ml of 2× lysis buffer containing 100 mM Tris, pH 8.0, 600 mM NaCl, 0.015% β mercaptoethanol, 2% NP-40, 40% glycerol, and protease inhibitors (Complete; Roche Diagnostics, Mannheim, Germany). Sonication for 5 s, 12 times at low output was performed to complete lysis. The proteins, recuperated after centrifugation at 25,000 rpm for 20 min at 4°C, were bound to HIS-Select HF nickel affinity gel (Sigma Chemie, Deisenhofen, Germany) for 2.5 h under agitating conditions at 4°C. The HIS beads were spun down at 5000 rpm for 5 min, washed twice with 20 mM Tris, pH 8.0, 300 mM NaCl, 10 mM imidazole, pH 8.0, 0.2% NP-40, 20% glycerol, and 0.007% β mercaptoethanol for 10 min under agitating conditions at 4°C. Finally, the proteins were eluted from the beads with 20 mM Tris, pH 8.0, 300 mM NaCl, 500 mM imidazole, pH 8.0, 0.2% NP-40, 20% glycerol, and 0.007% β mercaptoethanol for 15 min under agitating conditions at 4°C and collecting the supernatant after centrifugation at 5000 rpm for 5 min at 4°C.
Site-directed mutagenesis was performed using a Quick-change XL mutagenesis kit (Stratagene, La Jolla, CA) according to manufacturer's protocol. The enhanced green fluorescent protein (EGFP)-PLK4 wild-type and kinase dead (K41M) plasmids were used as templates to produce S305 to alanine (S305A), S305 to glutamic acid (S305E) mutants. Nondegradable PLK4 (PLK ND) was made by replacing S285 and T289 with alanine. The primers used for sited-directed mutagenesis were as follows: S305A FW, 5′-CCAGTACCAGTATAAGTGGTGCGTTATTTGACAAAAGAAGAC-3′ and S305A RV, 5′-GTCTTCTTTTGTCAAATAACGCACCACTTATACTGGTACTGG-3′; S305E FW, 5′-CAGTACCAGTATAAGTGGTGAATTATTTGACAAAAGAAGAC-3′ and S305E RV, 5′-GTCTTCTTTTGTCAAATAATTCACCACTTATACTGGTACTG-3′; and PLK4 ND FW, 5′-CTGTGGAAGACTCAATTGATGCTGGGCATGCCGCAATTTCTACTGCAATTACAGC-3′ and PLK4 ND RV, 5′-GCTGTAATTGCAGTAGAAATTGCGGCATGCCCAGCATCAATTGAGTCTTCCACAG-3′.
To generate a small interfering RNA (siRNA)-resistant form of PLK4 four nucleotide mutations, silent at the amino acid level, were introduced to the target sequence of duplex 2 by site-directed mutagenesis. The sequences of the primers used were PLK4 siRes Fw 5′-GGTTACAAATGAAGGACTGGGCTTGACAACTACAGCTTCTG-3′ and PLK4 siRes Rv 5′-CAGAAGCTGTAGTTGTCAAGCCCAGTCCTTCATTTGTAACC-3′. The mutated nucleotides are bold in the PLK4 siRes Fw primer.
Cells, Transfections, and Protein Detection
HCT116 colorectal carcinoma cells were cultured in McCoy's 5A and Ham's F-12, RPE1 cells in DMEM/F-12, PC3 cells in Ham's F-12, KE37 cells in RPMI 1640 medium, and HeLa green fluorescent protein (GFP)-centrin-1 cells in DMEM (Invitrogen). All media were supplemented with 1% l-glutamine (Invitrogen), 1% penicillin/streptomycin, and 10% fetal calf serum (Invitrogen). Gentamicin was added to HCT116 cell culture medium to a final concentration of 1%. PC3 cell medium also contained 1 mM sodium pyruvate, 0.1 mM nonessential amino acids and 1.5 g/l sodium bicarbonate. PLK4 plasmids were transfected with either Lipofectamine 2000 (Invitrogen) or FuGENE 6 (Roche Diagnostics), whereas siRNAs were transfected using either Lipofectamine 2000 (Invitrogen) or Interferin (Polyplus-transfection, Illkirch, France). Three PLK4-specific siRNAs were used the target sequences of the duplexes were duplex 1, 5′-AGGAGTGTCTTCTATCAGTTA-3′ (QIAGEN, Hilden, Germany); duplex 2, 5′-AAGGACTTGGTCTTACAACTA-3′ (QIAGEN); and duplex 3, 5′-AAGGACCTTATTCACCAGTTA-3′ and a Mismatch control (4611; Ambion, Austin, TX) at either 10 or 50 nM depending on the transfection reagent used. For myc-tagged and EPFG-tagged PLK4 experiments, cells were lysed in triple detergent buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% SDS, 1% NP-40, 0.5% Na-deoxycholate, and protease and phosphatase inhibitor cocktail (Roche Diagnostics). For experiments including the detection of phosphorylated S305, cells were lysed in boiling buffer (1% SDS, 100 mM Tris pH 7.4, and 1 mM Na3VO4). The proteasome inhibitors AdaAhx3L3VS (Calbiochem, San Diego, CA), MG132 (Calbiochem), and MG115 (Sigma Chemie) were dissolved in DMSO. Proteins were transferred to nitrocellulose membrane after SDS-polyacrylamide gel electrophoresis (PAGE) and detected with appropriate antibodies.
Antibodies
Primary antibodies were obtained from the following sources: anti-FLAG M2 antibody (Sigma Chemie), anti-γ-tubulin monoclonal antibody (mAb) GTU-88 (Sigma Chemie), anti-α-tubulin antibody (clone 3A2; a gift from L. Lafanechére, Institut National de la Santé et de la Recherche Medicale Unité 836, Institut des Neurosciences de Grenoble, Grenoble, France), anti-PLK4 antibody (Abcam, Cambridge, United Kingdom), anti-glyceraldehyde-3-phosphate dehydrogenase (Abcam), and anti-HsSAS-6 (Santa Cruz Biotechnology, Santa Cruz, CA). Anti-PLK4 antibodies, detecting the kinase domain (KD), crypto Polo-box (CPB), or phosphorylated S305 (pS305), were produced and affinity purified by Agro-Bio (La Ferte Saint Aubin, France) by using recombinant PLK4 domains and synthetic peptide SISGpSLFDKRRLLC coupled to keyhole limpet hemocyanin. Humanized anti-ninein single chain antibody was produced by screening a phage display library with a N-terminal fragment of human ninein encoding residues 1–246 and subsequently cloning cDNAs into the vector into pFuse-hFc2 (InvivoGen, San Diego, CA) for expression in Chinese hamster ovary cells. Horseradish peroxidase (HRP)-secondary antibodies were obtained from Santa Cruz Biotechnology. Alexa488 and Alexa680 secondary antibodies were obtained from Invitrogen. Cyanine (Cy)3, Cy5, and aminomethylcoumarin acetate secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Affinity purified PLK4 kinase domain antibody was directly labeled with Dylight 549 fluorophore using a microscale antibody coupling kit according to the protocol provided (Thermo Fisher Scientific, Rockford, IL).
Immunoprecipitations
Cells were washed once with ice-cold Tris-buffered saline (TBS) and lysed in triple detergent buffer containing protease and phosphatase inhibitor cocktails (Roche Diagnostics). Lysates were passed through a 25-gauge needle several times to shear genomic DNA and cleared by centrifuging at 14,000 rpm for 10 min at 4°C. For each immunoprecipitation, a 30-μl volume of protein G agarose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) was washed once with triple detergent buffer, 5 μg of antibody was added, and then the beads were incubated in ice for 30 min. Lysates were added and proteins immunoprecipitated by incubating at 4°C for 1 h with end-over end mixing. Immunoprecipitates were washed five times with triple detergent buffer and three times with HEPES-buffered saline (20 mM HEPES and 150 mM NaCl, pH 7.4).
Preparation of Soluble/Insoluble Extracts
Soluble and insoluble extracts were prepared using a previously published protocol (Tassin et al., 1997). In brief, cells were sequentially washed with phosphate-buffered saline (PBS) and PHEM buffer (45 mM PIPES, pH 6.9, 45 mM HEPES, pH 6.9, 10 mM EGTA, and 5 mM MgCl2) before being lysed in PHEM buffer containing 1% Triton X-100. Lysates were incubated on ice for 10 min, insoluble material was precipitated by centrifugation at 300 × g for 10 min at 4°C, and the soluble material was transferred to a clean tube. The insoluble material was washed twice with PHEM buffer before solubilizing in SDS sample buffer. Soluble proteins were precipitated by adding 9 volumes of methanol and incubating on ice for 1 h. The precipitated proteins were pelleted by centrifugation at 4000 × g for 10 min at 4°C and solubilized in SDS sample buffer. Equivalent volumes of SDS sample buffer were used to solubilize the soluble and insoluble extracts.
Kinase Reactions, SDS-PAGE, Protein Determination, and Radioactive Analysis
Kinase reactions were incubated for 1 h at 22°C using the following buffer: 50 mM HEPES, 50 mM NaCl, 10 mM MgCl2, 1 mM NaF, 5 μM xylene cyanole, 1 μM ATP, 1 mM dithiothreitol, 0.3 μCi of [γ-33P]ATP, and PLK4 recombinant protein. The samples were separated on a 4–12% Bis Tris NuPage gel (Invitrogen), stained with Sypro Ruby (Invitrogen,) and imaged using a LumiImager (Roche Diagnostics). Gels were destained, dried on 3MM paper (Whatman, Maidstone, United Kingdom), exposed to a phosphorimager screen, and imaged using Typhoon ImageQuant software (GE Healthcare) was used to quantify the intensity of the bands.
Peptide Synthesis and Phosphorylation Analysis
A proprietary peptide library containing putative phosphorylation sites in PLK4 was prepared by Jerini Peptide Technology (Berlin, Germany) by using their proprietary microscale technology. The peptides, corresponding locations in PLK4 and mutations of each peptide scanned are displayed in the Supplemental Table 1. The peptides were subjected to a kinase reaction using purified SUMO-PLK41-285 and processed as described above.
Isolation of Centrosomes
Centrosomes were isolated from KE37 cells according to a previously published protocol (Moudjou and Bornens, 1994).
Fabrication of Micropatterned Coverslips
Micropatterned coverslips were made according to published protocols (Fink et al., 2007; Azioune et al., 2009). Ethanol-washed 18-mm glass coverslips were activated by exposing to deep UV (185 nm) light for 5 min. To create a nonadherent surface, the activated coverslips were coated with 0.1 mg/ml poly-l-lysine-graft-polyethylene glycol (Surface Solutions, Zurich, Switzerland) in 10 mM HEPES, pH 7.3, for 1 h at ambient temperature and the washed twice with distilled water. A quartz mask (Delta Mask B.V., Enschede, The Netherlands), printed with 37-μm-diameter disks, was activated by exposing to deep UV light for 5 min. The coverslips were attached to the quartz mask with a drop of water and exposed to deep UV light for 5 min. After air drying, the micropatterned surface of the coverslip was inverted onto a 30-μl drop of 50 μg/ml fibronectin (Sigma Chemie), incubated at ambient temperature for 1 h, and washed twice with PBS.
Immunofluorescence Imaging
HeLa centrin-1 GFP or RPE1 cells growing on fibronectin/collagen-coated coverslips were washed once with PBS and fixed with methanol at −20°C for 20 min. Coverslips were sequentially washed with TBS and antibody blocking buffer (TBS containing 1% bovine serum albumin fraction V [Sigma Chemie] and 0.5% Triton X-100 [Sigma Chemie] before adding primary antibodies diluted in antibody blocking buffer. Cells were stained with primary antibodies for 1 h at ambient temperature and washed extensively with antibody blocking buffer before incubating with secondary antibodies, diluted in the same buffer, for 30 min at ambient temperature. The coverslips were washed with antibody blocking buffer, DNA stained with a 0.2 μg/ml solution of 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma Chemie), and sequentially washed with antibody dilution buffer and distilled water. After allowing to air dry, the coverslips were mounted onto glass slides using Mowiol mounting medium. Images were captured on a DMRA2 microscope (Leica, Wetzlar, Germany), fitted with a CoolSNAP camera (Princeton Instruments, Princeton, NJ), by using a 100 × 1.4 numerical aperture objective lens (Leica) and MetaMorph software (Molecular Devices, Sunnyvale, CA). Processing of images and fluorescence intensity measurements were carried out using MetaMorph software. A 16-pixel-diameter disk was placed over the centrosome of maximal projections and the average fluorescence intensity measured using a journal that automatically subtracted background fluorescence. Exposure settings, for the quantitative analysis of PLK4 levels, were the same except for the images of mitotic cells were it was necessary to reduce exposure times to avoid saturating and exceeding the dynamic range of the digital camera. Values were subsequently corrected by multiplying by a factor corresponding to the reduction in exposure time.
RESULTS
PLK4 Autophosphorylates
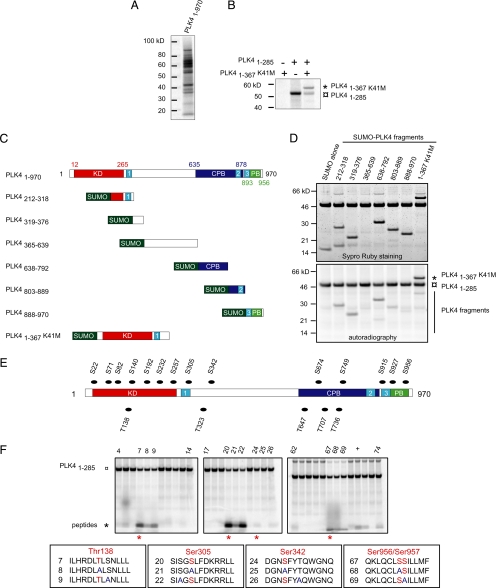
We sought to elucidate when PLK4 is active during the cell cycle and speculated that autophosphorylation, which has been suggested to occur previously (Yamashita et al., 2001), could be used as a means of identifying active PLK4. To verify that the kinase autophosphorylates, we carried out an in vitro kinase assay using human PLK4 purified from bacteria as a SUMO fusion protein. Autoradiography demonstrated that PLK4 is able to autophosphorylate in vitro, and the presence of multiple bands indicated that it is subject to degradation (Figure 1A). To investigate whether intermolecular autophosphorylation could occur, an N-terminal fragment of PLK4 made up of residues 1–367 with a mutation in the kinase domain to render it inactive (PLK41-367 K41M) was used as a substrate in an in vitro phosphorylation assay with the kinase domain of PLK4 (PLK41-285). PLK41-367 K41M was phosphorylated in the presence, but not absence, of PLK41-285 demonstrating that intermolecular autophosphorylation can occur (Figure 1B). It is possible that PLK4 is able to autophosphorylate in an intramolecular manner, although we have not addressed this issue. Three different techniques, based on in vitro phosphorylation assays, were used to identify the autophosphorylation sites within PLK4: phosphorylation of PLK4 fragments, screening peptide libraries to determine the consensus PLK4 phosphorylation sequence, and phosphorylation of PLK4-derived peptides containing predicted phosphorylation sites. In vitro phosphorylation assays carried out using purified fragments of PLK4 fused to SUMO demonstrated that several domains of the kinase were phosphorylated with sites being present in the kinase, central and crypto Polo-box domains of PLK4 (Figure 1, C and D). After determining the consensus phosphorylation sequence (Supplemental Data), which agrees with a previously published sequence (Leung et al., 2007), 12–13 amino acid-long peptides spanning predicted PLK4 phosphorylation sites were used as substrates in in vitro phosphorylation assays. A complete list of all the peptides synthesized is given in Supplemental Table 1. From a total of 19 predicted phosphorylation sites, six residues were found to be phosphorylated: T138, S140, S305, S342, S956, and S957. However, variation in the intensity of signal was observed between peptides, with the peptide containing residue S305 possessing the highest signal (Figure 1, E and F). Mutation of this residue to an alanine (S305A) to generate a nonphosphorylatable form resulted in a dramatic decrease in incorporation of phosphate, indicating that this residue was the major phosphorylation site within this peptide that contains three serine residues. Because S305 was readily phosphorylated, we decided to determine whether this phosphorylation event occurred in cells.
Figure 1.
PLK4 autophosphorylates. (A) Results of an in vitro kinase assay using full-length human PLK4 purified from bacteria showing that it autophosphorylates. It should be noted that PLK4 is readily degraded, explaining the presence of multiple bands. (B) An N-terminal fragment of PLK4 consisting of residues 1–367 that possessed a mutation within the catalytic domain rendering the kinase inactive (PLK41-367 K41M) was used as a substrate in an in vitro kinase assay using the kinase domain of PLK4 (PLK41-285). Autoradiography showed that PLK41-367 k41M was phosphorylated when mixed with the kinase domain of PLK4, indicating that intermolecular phosphorylation had occurred as well as autophosphorylation of the kinase domain. (C) Schematic showing the location of the three PEST sequences (numbered light blue boxes), kinase (red box), crypto Polo-box (dark blue box), and Polo-box (green box) domains in PLK4. The fragments of PLK4 used as substrates in in vitro phosphorylation assays with the kinase domain of PLK4 are also shown. (D) Results of in vitro kinase assays using as substrates the different fragments of PLK4 depicted in C. The kinase and central and crypto Polo-box domains of PLK4 were all found to be phosphorylated. (E) Diagram showing the location of predicted autophosphorylation sites in PLK4. (F) Results of in vitro phosphorylation assays using peptides spanning predicted autophosphorylation sites showed that S305 was a major site within PLK4. Peptide numbers are noted on top of the gels and sequences are given in the boxes with potential phosphorylation sites marked in red with S/T to A mutations marked in blue. Serine residue 305 seemed to be the major autophosphorylation site within PLK4. The plus sign indicates a positive control carried out using a 12 amino acid peptide from RAF containing serine 338 closely matching the consensus phosphorylation sequence of PLK4.
PLK4 Localizes Asymmetrically to Centrioles
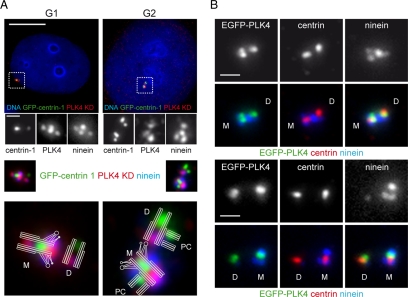
To study the cell cycle dynamics of PLK4, we raised antibodies against two different domains of PLK4; the kinase domain made up of residues 1–285 (PLK4 KD) and the crypto Polo-box domain 635–878 (PLK4 CPB). Data demonstrating the specificity of these antibodies can be found in Supplemental Figure S1. We used the PLK4 KD antibody in conjunction with an anti-ninein antibody to stain HeLa cells stably expressing GFP-tagged centrin-1 (Figure 2A). The GFP-centrin-1 protein identified the distal ends of centrioles (Paoletti et al., 1996), and the anti-ninein antibody labeling enabled us to distinguish between mother and daughter centrioles (Mogensen et al., 2000). The anti-PLK4 antibody clearly labeled centrioles, but we observed a staining pattern that has not been reported before, with the mother centriole being more intensely stained than the daughter centriole. PLK4 seemed to localize to two sites on the mother centriole, with one site lying at the proximal end, as has been reported previously (Kleylein-Sohn et al., 2007), and the other close to the subdistal appendages. This second site was present in both G1 and G2 phase cells, often appearing as an elongated spot spanning two ninein positive-staining foci, suggesting that it was at the base of the subdistal appendages. We next sought to determine whether this localization pattern could be observed by another means. We transfected RPE1 cells with an EGFP-PLK4 construct, fixed the cells, and stained them with antibodies against centrin and ninein to label and distinguish the mother and daughter centrioles. The localization of EGFP-PLK4 to centrioles was not totally identical to that of the endogenous protein, but asymmetry, in terms of the amount of protein associated, was consistently observed between the mother and daughter centrioles (Figure 2B). The mother centriole often possessed two foci of EGFP-PLK4 lying close to the distal end of the centriole but lacked exogenous protein at the proximal end of the centriole. In comparison, the daughter centriole possessed a single focus of EGFP-PLK4.
Figure 2.
PLK4 localizes asymmetrically to centrioles. (A) HeLa cells stably expressing GFP-centrin-1 (green) were fixed and stained with DAPI (blue) to label DNA, anti-PLK4 KD (red), and anti-ninein (blue insets) antibodies. PLK4 was found to localize to two sites on the mother centriole, with one site at the proximal end and the other site close to the subdistal appendages between two ninein-positive staining foci. In contrast, the daughter centriole possessed a single focus of PLK4 staining at the proximal end of the centriole, which often colocalized with ninein staining. A cartoon depicting the arrangement of mother (M), daughter (D), and procentrioles (PC) is superimposed on the enlargements of the merged centriole images presented. (B) RPE1 cells transiently transfected with EGFP-PLK4 (green) and stained with centrin (red) and ninein (blue) antibodies. Mother centrioles consistently possessed more EGFP-PLK4 than daughter centrioles. Bar, 10 μm; inset, 1 μm.
PLK4 Serine 305 Autophosphorylation Occurs in Cultured Cells
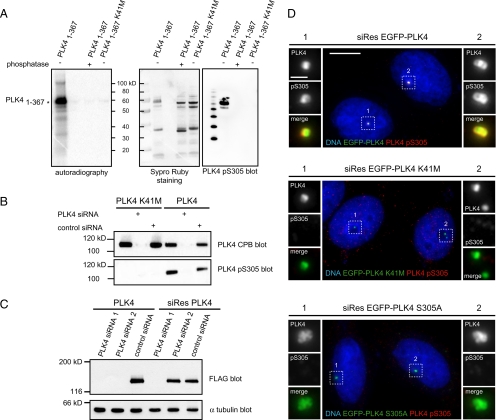
To determine whether PLK4 S305 autophosphorylation occurred in cultured cells as well as in vitro an antibody against the phosphorylated S305 residue was raised. The specificity of this antibody was verified by subjecting PLK41-367 to an in vitro kinase assay and treating a sample with lambda phosphatase (Figure 3A). PLK41-367 K41M, lacking kinase activity, was included as a second control in this experiment. Autoradiography demonstrated that the PLK41-367 had autophosphorylated, treatment of PLK41-367 with lambda phosphatase resulted in a loss of signal and PLK41-367 K41M was unable to autophosphorylate. Western blotting of these samples with the PLK4 pS305 antibody showed that it specifically recognized the phosphorylated form of PLK41-367, whereas neither the lambda phosphatase-treated PLK41-367 nor the PLK41-367 K41M forms were detected. The results of this experiment showed that the PLK4 pS305 antibody specifically recognized the S305 phosphorylated form of PLK4. We next sought to determine whether it was possible to detect PLK4 S305 autophosphorylation in cells. To address this question, we transiently transfected HCT116 cells with EGFP-PLK4 or -PLK4 K41M in conjunction with control siRNA or siRNA targeting PLK4. Lysates were prepared from these cells, and Western blotting was carried out using the PLK4 CPB and PLK4 pS305 antibodies (Figure 3B). The results showed that PLK4 S305 autophosphorylation occurs in cells, because the PLK4 pS305 antibody recognized only wild-type PLK4 in untreated or control siRNA-treated samples. Treatment with PLK4 siRNA resulted in a complete loss of this signal, and it could not be detected in the PLK4 K41M samples despite PLK4 CPB blotting showing that more of the inactive kinase was present than the wild-type form. To determine whether the PLK4 pS305 antibody was able to detect S305 phosphorylated PLK4 by immunofluorescent staining, and confirm its specificity, an siRNA rescue experiment was carried out, whereby HeLa cells were simultaneously transfected with siRNA against PLK4- and siRNA-resistant forms of PLK4, PLK4 K41M, or PLK4 S305A, where serine 305 had been mutated to an alanine to prevent phosphorylation. The siRNA-resistant forms of PLK4 were generated by introducing four silent nucleotide changes into the sequence of PLK4 siRNA 2 (see Materials and Methods). The efficacy of this mutation was verified by simultaneously transfecting HeLa cells with FLAG-tagged wild-type or siRNA-resistant PLK4 and either control or PLK4 siRNAs and carrying out Western blotting with an anti-FLAG antibody (Figure 3C). Both PLK4 siRNAs effectively targeted the wild-type PLK4 mRNA, because overexpressed wild-type PLK4 could not be detected in lysates from cells transfected with these duplexes, whereas it was clearly present in lysate from control siRNA-transfected cells. The siRNA-resistant form of PLK4 was still targeted by PLK4 siRNA 1, because no overexpressed protein could be detected but was resistant to PLK4 siRNA 2 with the level of overexpressed PLK4 being similar to the control siRNA-treated sample. Immunofluorescent staining of HeLa cells, which had been simultaneously transfected with PLK4 siRNA 2 and wild-type siRNA-resistant PLK4, with the PLK4 pS305 antibody showed that it was possible to detect S305 phosphorylated PLK4 (Figure 3D). In contrast, no signal could be detected in either PLK4 K41M or PLK4 S305A-transfected cells, in the absence of the endogenous kinase, indicating the PLK4 pS305 antibody specifically recognized its epitope (further information demonstrating the specificity of this antibody can be found in Supplemental Figure S2). Together, these results showed that PLK4 S305 phosphorylation occurs in cultured cells and the PLK4 pS305 antibody could be useful for determining when PLK4 autophosphorylates and becomes active.
Figure 3.
Serine 305 of PLK4 is autophosphorylated in vitro and in cells. (A) PLK41-367 and PLK41-367 K41M were subjected to in vitro phosphorylation and then treated with or without lambda phosphatase. Sypro Ruby staining was used to detect the PLK4 fragments, and autoradiography was used to determine whether they had been phosphorylated. Only the untreated PLK41-367 fragment was found to have autophosphorylated, and this form was specifically recognized by the PLK4 pS305 antibody. HRP-conjugated molecular weight markers were loaded onto the PLK4 pS305 blot (left lane). (B) Lysates were prepared from HCT116 cells that had been transiently transfected with EGFP-PLK4 or -PLK4 K41M in conjunction with control or PLK4 siRNA and Western blotted with anti-PLK4 CPB and anti-PLK4 pS305 antibodies. The anti-PLK4 pS305 antibody was found to recognize specifically the active form of the kinase in cells. (C) Western blotting of lysates from HeLa cells simultaneously transfected with siRNA and FLAG-tagged PLK4 expression constructs. The results show that both PLK4 siRNA duplexes are efficacious to the wild-type PLK4 mRNA, whereas PLK4 siRNA 2 is no longer effective against the siRNA-resistant form of PLK4 (siRes PLK4). α-Tubulin blotting was carried out as a loading control. (D) Images of HeLa cells transfected with siRNA-resistant EGFP-PLK4, -PLK4 K41M, or -PLK4 S305A (all green) and PLK4 siRNA 2, stained with DAPI (blue) to label DNA and PLK4 pS305 antibody (red). The PLK4 pS305 antibody recognizes wild-type siRes EGFP-PLK4, but neither siRes EGFP-PLK4 K41M nor siRes EGFP-PLK4 S305A in the absence of endogenous PLK4. Bar, 10 μm; inset, 1 μm.
PLK4 Autophosphorylation Occurs in a Cell Cycle-dependent Manner
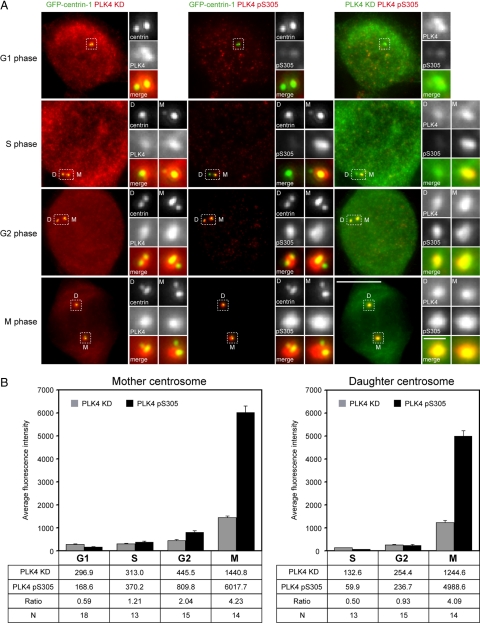
Because above-mentioned data demonstrated that it was possible to detect S305-phosphorylated overexpressed PLK4 with the PLK4 pS305 antibody, we next sought to determine whether it could recognize the endogenous protein at the centrosome. HeLa cells stably expressing GFP-centrin-1 were stained with PLK4 KD and PLK4 pS305 antibodies, and cells were compared at various phases of the cell cycle; mother and daughter centrioles could be clearly distinguished by the intensity of the GFP-centrin-1 signal (Figure 4A; Piel et al., 2000; White et al., 2000). Fluorescence intensity measurements were carried out to determine the amounts of PLK4 and pS305 PLK4 at each centriole pair, and the ratio of the average PLK4 pS305-to-PLK4 fluorescence intensities were calculated for each cell cycle phase to estimate the level of PLK4 autophosphorylation (Figure 4B).
Figure 4.
PLK4 autophosphorylation peaks in mitosis. (A) HeLa cells stably expressing GFP-centrin (green) were stained with anti-PLK4 KD (red) and anti-PLK4 pS305 (red) antibodies. GFP-tagged centrin served as a marker for centrioles. Staining with the anti-PLK4 KD antibody enabled detection of all the PLK4 present within the cell, whereas the anti-PLK4 pS305 antibody detected only the phosphorylated form of the kinase. In G1, the abundance of S305 phosphorylated PLK4 at centrioles is extremely low, being barely detectable, whereas in S phase levels increase to produce a clearly discernible signal that rises to reach a maximum in mitosis. Strikingly, more active PLK4 seemed to be present at mother centriole (marked as D, daughter; and M, mother). Bar, 10 μm; inset, 1 μm. (B) Results of fluorescence intensity measurements of PLK4 and PLK4 pS305 at the mother and daughter centriole pairs. The table below the graphs lists the average fluorescence intensity at each centriole pair at G1, S, G2, and M phases of the cell cycle. The ratio of PLK4 pS305 to PLK4 is also given to enable comparison of the level of PLK4 autophosphorylation between the cell cycle phases. The acquisition settings for mitotic cells were not the same as those for interphasic cells, because the exposure times were reduced to avoid saturating and exceeding the dynamic range of the digital camera. Fluorescence intensity measurements were multiplied by a factor corresponding to the reduction in exposure time to produce the final results for the mitotic cells.
In agreement with the above-mentioned observations (Figure 2), fluorescence intensity measurements showed that more PLK4 was associated with the replicating mother centriole than the replicating daughter centriole in interphasic cells. The amount of PLK4 at the centrosome was low in G1, increased slightly in S and G2 phases, and rose dramatically in mitosis to reach a peak. The replicating daughter centriole possessed even less PLK4 than the replicating mother centriole in S and G2 phases of the cell cycle, but a similar dramatic increase was observed in mitosis. At this point in the cell cycle, there was no significant difference in the amount of PLK4 present at the mother and daughter centrosomes.
Staining with the PLK4 pS305 antibody revealed that little S305 phosphorylated PLK4 was present at centrioles in G1. In S phase, S305-phosphorylated PLK4 could be detected at the replicating mother centriole, with the signal increasing in G2 to reach a maximum in mitosis. Comparison of the ratio of PLK4 pS305-to-PLK4 fluorescence intensities showed an approximately exponential increase in the amount of S305 phosphorylated PLK4 at the replicating mother centriole through the cell cycle. This exponential increase reflects an augmentation in the amount of PLK4 protein as well as an increase in the proportion of active kinase at the centrosome. The replicating daughter centriole possessed extremely low levels of S305 phosphorylated PLK4 in S phase although this approximately doubled in G2, with a clearly detectable signal being present. Interestingly, in mitosis there was almost as much S305 phosphorylated PLK4 at the daughter centrosome as at the mother centrosome. The finding that PLK4 abundance is low in G1, S, and G2 phases and reaches a maximum in mitosis coinciding with an increase in the amount of S305-phosphorylated (active) kinase suggests that PLK4 might have a function at the centrosome at this point in the cell cycle.
Inhibition of the Proteasome Causes the Accumulation of S305-phosphorylated PLK4
We speculated that S305-phosphorylated PLK4 might be targeted to the proteasome for degradation during interphase to maintain its abundance at a low level. We tested this hypothesis by treating RPE1 cells that had been transiently transfected with EGFP-PLK4 with the proteasome inhibitor MG115 for 3 h. Western blotting with the PLK4 KD and PLK4 pS305 antibodies demonstrated that proteasome inhibition resulted in the accumulation of S305-phosphorylated PLK4 but had little effect upon the overall amount of PLK4 (Figure 5A). Similar results were obtained using the proteasome inhibitors MG132 and AdaAhx3L3VS (Supplemental Figure S3). These results show that S305-phosphorylated PLK4 accumulates in response to proteasome inhibition, suggesting that it is a substrate of the proteasome. To determine whether the S305-phosphorylated form of PLK4 was active, PC3 cells, which are easily transfected, were transiently transfected with either FLAG-tagged-PLK4 or -PLK4 K41M and treated with MG115 for 3 h to induce the accumulation of S305-phosphorylated PLK4. The cells were subsequently lysed and PLK4 immunoprecipitated using either anti-FLAG or anti-PLK4 pS305 antibodies. A fraction of each immunoprecipitate was subjected to an in vitro phosphorylation assay before fractionating on a gradient gel and staining with silver (Figure 5B). The anti-FLAG antibody immunoprecipitated similar amounts of PLK4 and PLK4 K41M kinase, whereas the PLK4 pS305 antibody only immunoprecipitated wild-type PLK4. Autoradiography demonstrated that only wild-type PLK4 immunoprecipitated by the anti-FLAG and anti-PLK4 pS305 antibodies had been phosphorylated in vitro. These results suggest, as the PLK4 K41M mutant was not found to be phosphorylated, that PLK4 S305 phosphorylation is a result of autophosphorylation, and that S305 phosphorylated PLK4 is active. In agreement with this, we found that there was as much phosphorylated PLK4 in the PLK4 pS305 immunoprecipitate as the anti-FLAG, despite less kinase being pulled down by the PLK4 pS305 antibody (as judged by silver staining). This result suggested that only a fraction of the PLK4 immunoprecipitated with the anti-FLAG antibody was active. To confirm that the immunoprecipitated protein was PLK4, Western blotting was carried out on the remaining fraction of immunoprecipitate with the anti-PLK4 KD antibody (Figure 5C). This conclusively demonstrated that the immunoprecipitated protein was PLK4 and that the PLK4 pS305 antibody had specifically recognized S305 phosphorylated PLK4.
Figure 5.
Proteasome inhibition causes the accumulation of S305 autophosphorylated PLK4. (A) RPE1 cells were transfected with EGFP-PLK4 and treated with either DMSO, as a control, or 1 μM MG115 for 3 h before preparing total cell lysates. Western blotting was carried out using the anti-PLK4 kinase domain and PLK4 pS305 antibodies to determine the relative quantities of PLK4- and S305-autophosphorylated PLK4. This demonstrated that proteasome inhibition specifically causes the accumulation of autophosphorylated PLK4. (B) PC3 cells were transiently transfected with either PLK4 or PLK4 K41M, tagged at their C termini with a 3xFLAG motif and treated with 1 μM MG115 for 3 h. Immunoprecipitations were carried out using either anti-FLAG or anti-PLK4 pS305 antibodies. Approximately 70% of the immunoprecipitated material was subjected to an in vitro phosphorylation assay, migrated on a gradient gel, stained with silver, and exposed to a phosphorimager screen. The open arrowhead points to immunoprecipitated PLK4 and PLK4 K41M. Autoradiography demonstrated that phosphorylated PLK4 was present in the anti-FLAG and anti-PLK4 pS305 immunoprecipitates from PC3 cells transfected with wild-type PLK4 but not the kinase inactive mutant PLK4 K41M. (C) Western blotting was carried out on the remaining immunoprecipitated material with the PLK4 KD antibody. This showed that approximately equal amounts of PLK4 and PLK4 K41M had been immunoprecipitated with the anti-FLAG antibody, whereas the PLK4 pS305 antibody only immunoprecipitated wild-type PLK4.
Proteasome Inhibition Increases the Amount of Endogenous S305-autophosphorylated PLK4 at Centrioles in Interphase
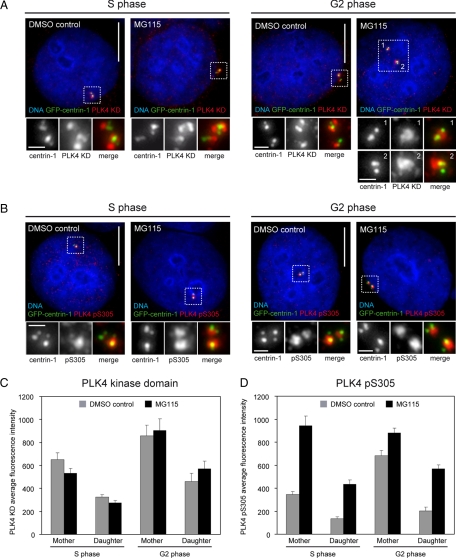
Inhibition of the proteasome caused an increase in the amount of S305-phosphorylated overexpressed PLK4. To determine whether proteasome inhibition would cause a similar increase in endogenous S305-phosphorylated PLK4, we treated HeLa GFP-centrin-1 cells with MG115 for 3 h and then stained them with either the PLK4 KD or PLK4 pS305 antibody (Figure 6, A and B). Fluorescence intensity measurements were carried out as described above to determine the abundance of PLK4- and S305-phosphorylated PLK4 at the replicating mother and daughter centrioles in S and G2 phase cells. In S phase cells, inhibition of the proteasome resulted in an approximately threefold increase in the amount of S305-phosphorylated PLK4 at both replicating centrioles (Figure 6C). A similar increase in S305-phosphorylated PLK4 levels was observed at the replicating daughter centriole in G2 cells upon MG115 treatment, but levels at the replicating mother centriole were only slightly augmented. This result is intriguing, and suggests that there is a maximum level of S305 phosphorylation that can be attained in interphase. In contrast, the overall amount of PLK4 at the replicating mother and daughter centrioles in S and G2 phases remained unchanged upon proteasome inhibition, indicating that only the proportion of active kinase had increased (Figure 6C). It has previously been shown that proteasome inhibitor treatment induces centriole over duplication (Duensing et al., 2007), and the above-mentioned data offer an explanation as to why this occurs. Proteasome inhibition seems to allow the accumulation of S305-autophosphorylated PLK4 at centrioles, which could trigger centriole over duplication because the levels of active kinase are higher than normal.
Figure 6.
PLK4 S305 autophosphorylation increases at centrioles upon proteasome inhibition. (A and B) HeLa GFP-centrin-1 cells (green) were treated with either DMSO, as a control, or 1 μM MG115 for 3 h; fixed; and stained with DAPI (blue) to label DNA and either anti-PLK4 kinase domain (A) or anti-PLK4 pS305 antibody (B) (both red). Inhibition of the proteasome in S phase cells caused an increase in the abundance of S305-phosphorylated PLK4 at both the mother and daughter centriole pairs. In G2 cells, an increase was observed only at the daughter centriole pair. Bar, 10 μm; inset, 1 μm. (C and D) Fluorescence intensity measurements showing PLK4 and S305 phosphorylated PLK4 levels at mother and daughter centriole pairs in S and G2 phase with and without proteasome inhibitor treatment. At least 19 centrosomes were quantified in each group.
In mitosis, PLK4 S305 autophosphorylation at centrosomes increases to much higher level than in G2 phase, reflecting an increase in the overall amount of kinase at the centrosome (Figure 4B) and possibly a change in its stability or activity.
Serine 305 Autophosphorylation Plays a Role in Centriole Duplication
The above-mentioned data indicated that S305-phosphorylated PLK4 represents the active form of the kinase, which is a potential proteasomal substrate because it accumulates within the cell upon treatment with a proteasome inhibitor. Recently published data has shown that the turnover of Drosophila PLK4 is regulated by the SCF/Slimb ubiquitin ligase complex, which recognizes a phosphorylated degron motif located within the N terminus of the kinase (Cunha-Ferreira et al., 2009; Rogers et al., 2009). Mutation of this motif, replacing serine/threonine residues to prevent phosphorylation, to produce a nondegradable form of PLK4 results in increased expression levels and a higher incidence of centriole amplification compared with the wild-type kinase. The effect of mutating this degron motif, which is conserved between species, upon the stability of human PLK4 has not been determined. We decided to mutate the degron motif in human PLK4, because it potentially offered a means of expressing a stabilized form of PLK4 without inhibiting the proteasome, to compare its stability and, more importantly, S305 phosphorylation status to wild-type PLK4. Serine 285 and threonine 289 within the degron motif of human PLK4 were mutated to alanine to generate a nondegradable form of PLK4 (PLK4 ND). The same mutations were also introduced into the kinase inactive form of PLK4 (PLK4 K41M ND). We also decided to investigate the role of S305 autophosphorylation in governing PLK4 stability and the regulation of centriole duplication. To do this, we generated PLK4 S305 mutants in which this residue was replaced with an alanine (PLK4 S305A) to prevent phosphorylation or a glutamic acid (PLK4 S305E) to mimic phosphorylation. All of the above-mentioned constructs generated where tagged with EGFP at the N terminus.
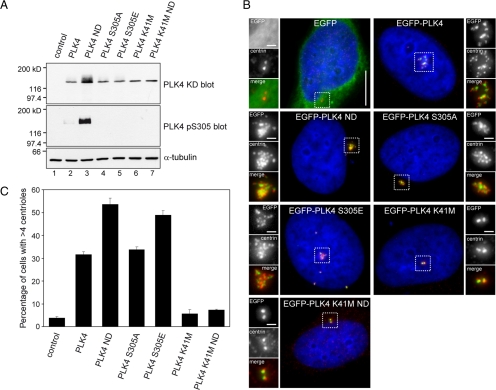
RPE1 cells were transiently transfected with EGFP-PLK4 mutants, whole cell lysates were prepared, and Western blotting was carried out using anti-α-tubulin, anti-PLK4 KD, and anti-PLK4 pS305 antibodies (Figure 7A). Comparison of PLK4 levels by Western blotting with the PLK4 KD antibody clearly showed that mutation of the degron motif had the greatest effect upon the stability of PLK4, with amount of nondegradable PLK4 being substantially higher than the other forms of PLK4. Mutation of S305 to either prevent or mimic phosphorylation had no effect upon the stability of PLK4 with levels being comparable with wild-type PLK4. These data suggest that S305 phosphorylation does not directly control the stability of the kinase. Interestingly, PLK4 K41M ND protein levels did not accumulate to a similar extent as PLK4 ND, suggesting that kinase activity influences the abundance of the kinase. To determine whether mutation of the degron motif resulted in the accumulation of active PLK4, in a manner similar to proteasome inhibition, Western blotting was carried out with the PLK4 pS305 antibody. This showed that PLK4 ND was heavily phosphorylated on S305, compared with wild-type PLK4, implying a large fraction of the nondegradable kinase was active. In vitro kinase assays with FLAG-tagged PLK4 immunoprecipitated from transfected PC3 cells showed that all the forms of PLK4 expressed were active with the exception of the kinase dead versions (Supplemental Figure S4). Interestingly, the wild-type and nondegradable forms of PLK4 seemed to migrate as a several discrete bands, suggesting the existence of multiple posttranslational modified species.
Figure 7.
Autophosphorylation enhances centriole amplification. (A) RPE1 cells were transiently transfected with pEGFP-PLK4 constructs and Western blotting was carried out using anti-PLK4 KD and anti-PLK4 pS305 antibodies to determine total and S305 phosphorylated PLK4 levels. An anti-α-tubulin blot was carried out as a loading control. The results show that mutation of S305, either to mimic or prevent phosphorylation, has no impact upon the stability of the kinase. Wild-type PLK4 ND containing two mutations in the degron motif, S285A and T289A, to prevent phosphorylation was expressed to high levels, whereas PLK4 K41M and PLK4 K41M ND were both expressed at a lower level, indicating that kinase activity may influence protein stability. Western blotting with the PLK4 pS305 antibody shows PLK4 ND is heavily S305 phosphorylated and suggests that stabilization allows active kinase to accumulate. (B) HeLa cells were transiently transfected with EGFP-PLK4 (green) expression constructs, fixed, and stained with DAPI (blue) to label DNA and anti-centrin antibody (red). Bar, 10 μm; inset, 1 μm. (C) Results of three independent experiments quantifying the number of transfected cells exhibiting centriole amplification (n = 200). The results showed that wild-type PLK4 and PLK4 S305A triggered centriole amplification to a similar extent, whereas PLK4 S305E triggered centriole amplification more frequently, indicating that mimicking S305 autophosphorylation enhances centriole amplification. In agreement with published results in Drosophila S2 cells, PLK4 ND expression resulted in a higher incidence of centriole amplification compared with wild-type PLK4.
To compare the abilities of the different PLK4 mutants to trigger centriole amplification, HeLa cells were transfected and stained with an anti-centrin antibody (Figure 7B). All of the catalytically active forms of PLK4 were able to trigger centriole amplification albeit to varying efficiencies (Figure 7C). PLK4 ND promoted centriole amplification more frequently, with 54% of transfected cells possessing supernumerary centrioles compared with 32 and 34% of cells transfected with wild-type PLK4 and PLK4 S305A, respectively. Importantly, mimicking S305 phosphorylation significantly enhanced centriole amplification in comparison with wild-type PLK4, with 49% of transfected cells possessing multiple centrioles. Similar results were obtained from transfected RPE1 cells stained with an anti-HsSAS-6 antibody (Supplemental Figure S5). This suggests S305 phosphorylation might be important in the process of centriole duplication.
Serine 305-phosphorylated PLK4 Is Sequestered at the Centrosome
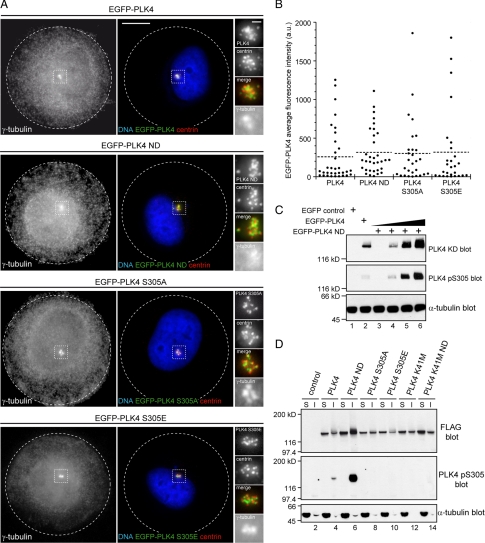
To evaluate the relevance PLK4 S305 autophosphorylation in triggering centriole amplification, we decided to carry out several different experiments. First, we decided to measure and compare the fluorescence intensities of PLK4, PLK4 ND, PLK4 S305A, and PLK4 S305E at the centrosome to determine whether there was a difference in amount of exogenously expressed protein localized at this site. To facilitate this analysis, we plated transfected HeLa cells onto circular micropatterns, because these have been shown to promote centrosome clustering and would normalize measurement (Thery et al., 2006). The cells were subsequently fixed and stained with anti-centrin and anti-γ-tubulin antibodies, to identify the cells possessing supernumerary centrioles, and the EGFP fluorescence intensity was measured within defined regions drawn around the centrosomes (Figure 8A). The results showed that PLK4 levels at the centrosome ranged greatly, but in a similar manner, in all of the PLK4 forms tested (Figure 8B). These data indicated that centriole amplification could be initiated in cells expressing a relatively low amount of PLK4 and failed to explain why nondegradable PLK4 was better than wild-type PLK4 at triggering centriole amplification. The fact that nondegradable PLK4 is heavily phosphorylated on S305 suggested that the fraction of active kinase is greater compared with wild-type PLK4. This was investigated by transfecting RPE1 cells with wild-type PLK4 and varying amounts of PLK4 ND and carrying out Western blotting with the PLK4 KD and PLK4 pS305 antibodies (Figure 8C). In wild-type PLK4, the fraction of active kinase seemed to be low, as the kinase could barely be detected with the PLK4 pS305 antibody despite there being a strong signal with the PLK4 KD antibody. Titration of the amount of PLK4 ND transfected revealed that more of nondegradable form of the kinase was phosphorylated on S305, suggesting the fraction of active kinase was greater compared with wild-type PLK4 (compare lanes 2 and 4 of Figure 8C). Measuring EGFP-PLK4 fluorescence intensity at the centrosome in cells with supernumerary centrioles showed that there was no difference in the average amount of exogenous protein present between the wild-type, nondegradable, and S305 mutants. We investigated the possibility that cytoplasmic PLK4 might be implicated in the process of centriole amplification by preparing soluble and insoluble extracts from FLAG-tagged PLK4-transfected RPE1 cells. These extracts were probed by Western blotting with anti-FLAG to detect the total amount of PLK4 expressed, anti-PLK4 pS305, and anti-α-tubulin antibodies to determine the quality of the extracts (Figure 8D). Exogenously expressed PLK4 partitioned in both the soluble and insoluble extracts, and in nearly all cases, with the exception of PLK4 ND, more was present in the soluble than insoluble extract. The same blot was reprobed with the PLK4 pS305 antibody. Strikingly, S305 phosphorylated PLK4 was found only to be present in the insoluble extracts derived from wild-type and PLK4 ND-transfected cells (lanes 4 and 6 of Figure 8D). These results suggest active PLK4 is sequestered at the centrosome and is not present within the cytoplasm.
Figure 8.
S305-autophosphorylated PLK4 is important for centriole duplication and is sequestered at the centrosome. (A) HeLa cells were transiently transfected with EGFP-PLK4 (green), plated onto 37-μm-diameter circular micropatterned coverslips, fixed, and stained DAPI (blue) to label DNA, anti-centrin (red), and anti-γ-tubulin antibodies (black and white). (B) EGFP-PLK4 fluorescence intensity was quantified in transfected cells exhibiting centriole amplification by drawing a region around the centrosomal area and measuring the fluorescence intensity (n = 28–33 cells). No significant difference was found in EGFP-PLK4 levels at the centrosome between the mutant forms and wild-type PLK4. (C) RPE1 cells were transiently transfected with EGFP, EGFP-PLK4, or varying amounts of EGFP-PLK4 ND, and Western blotting was carried out with PLK4 KD, PLK4 pS305 and α-tubulin antibodies. The results show more of the nondegradable form of PLK4 is phosphorylated compared with the wild-type kinase, indicating that the fraction of active kinase in PLK4 ND transfected cells is greater than in those transfected with wild-type PLK4. (D) Results of Western blotting using anti-PLK4 pS305, anti-FLAG, and anti-α-tubulin antibodies to probe soluble and insoluble extracts that had been prepared from RPE1 cells transiently transfected with EGFP-PLK4–3xFLAG expression constructs. Probing with the anti-FLAG antibody showed that overexpressed PLK4 was present in both the soluble and insoluble extracts, whereas S305-phosphorylated PLK4 partitioned in the insoluble fraction in extracts prepared from cells transfected with wild-type and nondegradable wild-type PLK4 (lanes 4 and 6). These data suggest that the active form of PLK4 is restricted to the centrosome because it is present in the insoluble, and not the soluble, extract.
DISCUSSION
We have presented data showing that PLK4 S305 autophosphorylation occurs as a consequence of kinase activation and that this event can used to monitor active PLK4 within the cell. PLK4 activation is first observed at the replicating mother centriole in G1/S phase and continues to increase throughout interphase until mitosis where it reaches a maximum. In contrast, PLK4 activation at the replicating daughter centriole starts considerably later in the cell cycle, in G2, but peaks to a level approximately equivalent to the replicating mother centriole in M phase. This delay in activation at the replicating daughter centriole is consistent with the proposal that procentriole assembly is initiated at the mother centriole before the daughter (White et al., 2000). It also might represent a means of restricting PLK4 activity to a discrete site, whereby PLK4 substrates are first phosphorylated at the mother centriole before being transferred to the daughter. Our data support this idea, because we found that active PLK4 is probably sequestered at centrosomes and is not present in the cytoplasm. This finding implies PLK4 is either activated locally or active PLK4 is efficiently transported to the centrosome, possibly to prevent centriole assembly from occurring at distant sites. It is also in keeping with the idea that centrioles act as scaffolds for regulatory molecules, sequestering and concentrating them at a discrete site, to facilitate procentriole assembly (Rodrigues-Martins et al., 2007).
The increase in the amount of active PLK4 from S phase onward, reaching a maximum in mitosis, also suggests that sustained kinase activity is required for procentriole assembly, probably reflecting a need to continually phosphorylate substrates to build and maintain these structures. A high mitotic PLK4 activity might be required to finalize the procentriole structure, because procentriole elongation is not completed until metaphase where its full length is attained (Rattner and Phillips, 1973; Vidwans et al., 1999). Supporting this idea, it has recently been shown that the centriolar protein hPOC5 is recruited to procentrioles at G2/M and is essential for centriole elongation (Azimzadeh et al., 2009). In addition, human POC5 is phosphorylated in G2/M and could possibly be a PLK4 substrate (data not shown; Azimzadeh et al., 2009). Another possibility is that PLK4 phosphorylates substrates in mitosis to prime for centriole duplication in the next cell cycle.
The activity of PLK4 at centrioles is probably regulated by different mechanisms depending upon cell cycle state. Published data have shown that PLK4 is subject to proteasome-dependent degradation, and, more recently, that it is a substrate of the SCF/Slimb ubiquitin ligase complex. The treatment of cells with proteasome inhibitors promotes centriole amplification in a PLK4-dependent manner and our data offer an explanation as to why this occurs. We have observed that in interphasic cells proteasome inhibition leads to an increase in the proportion of active kinase at centrioles, potentially exceeding the normal threshold of activity, which could lead to centriole amplification. In agreement with this, we found that a nondegradable form of PLK4 carrying mutations within the degron motif, which can no longer be recognized by the SCF/Slimb complex, is autophosphorylated at residue S305, suggesting that PLK4 is in its active form. Together, these data indicate that the active form of PLK4 is targeted to the proteasome during interphase. In mitosis, PLK4 abundance at centrosomes increases dramatically and its abundance might not be regulated in the same way as in interphase. This can be explained in several different ways. It has been shown in mice and Drosophila that PLK4 transcript levels are at their lowest in early G1 and at their highest in mitosis, suggesting that expression of the kinase is under transcriptional control and is augmented in mitosis (Fode et al., 1996; Rogers et al., 2009). It should also be noted that PLK4 may no longer be phosphorylated on its degron motif, thus escaping the SCF/Slimb complex and thereby contributing to the mitotic increase (Cunha-Ferreira et al., 2009; Rogers et al., 2009). Another mechanism might also exist, because in Caenorhabditis elegans the centriolar abundance of ZYG-1, the homologue of PLK4 (O'Connell et al., 2001), is regulated by a putative RNA-binding protein, SZY-20. In the absence of SZY-20 activity, ZYG-1 levels, in addition to other centrosomal proteins including SPD-2 and γ-tubulin, increase twofold at centrosomes (Song et al., 2008). It is possible that this yet to be completely characterized form of PLK4 regulation exists in other species as SZY-20 is conserved (Song et al., 2008).
PLK4 S305 autophosphorylation is not essential for centriole duplication, but it does seem to enhance this process as overexpression of a PLK4 S305E phospho-mimetic mutant promotes centriole amplification to a greater extent than the wild-type protein. One possible way in which S305 phosphorylation could influence centriole duplication is by controlling the localization of the active kinase restricting it to the centrosome. It is also possible that autophosphorylation causes a conformational change in the kinase allowing it to interact with centriolar components or increasing its affinity for them.
Our data highlight that the regulation of PLK4 at replicating centrioles in more complex than previously envisaged and raise important questions that will need to be addressed. The inequality in PLK4 abundance between the two centrioles is intriguing and will require further study to determine whether this is regulated by proteasomal degradation or whether there is another mechanism at work. One point pertinent to this is the remarkable fact that S305 is only 16–20 residues from the degron motif, opening the possibility there is a cross-talk between S305 autophosphorylation and access to the degron motif through conformational changes, which could control its localization. Identifying the factors responsible for initiating PLK4 activity in G1/S phase, and controlling it in later stages of the cell cycle, will be the next step, and the tools we have generated should be useful to do this. Another outstanding question is why some of the PLK4 present on the mother centriole is localized to the distal end. It is tempting to speculate that this is necessary for the completion of the distal end of the centriole and association of appendages, which are unique to the mother centriole. This question could perhaps be answered by using an inhibitor to block PLK4 activity late in the cell cycle after centriole duplication has been initiated. Finally, it would also be interesting to investigate whether other centriolar components are capable of influencing PLK4 activity at centrioles.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sabine Bardin, Jenny Fink, and Matthieu Piel for help making micropatterned coverslips; Jeffrey Salisbury for anti-centrin (20H5) mAb; Renata Basto for critically reading the manuscript; and Jean-Baptiste Sibarita of the imaging department (Institut Curie) for writing the MetaMorph journal to automatically calculate the average fluorescence intensity at centrosomes. We also acknowledge the Nikon Imaging Centre (Institut Curie).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-06-0505) on December 23, 2009.
REFERENCES
- Azimzadeh J., Hergert P., Delouvee A., Euteneuer U., Formstecher E., Khodjakov A., Bornens M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 2009;185:101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azioune A., Storch M., Bornens M., Thery M., Piel M. Simple and rapid process for single cell micro-patterning. Lab Chip. 2009;9:1640–1642. doi: 10.1039/b821581m. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M. K., Carmo N., Balloux F., Callaini G., Glover D. M. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Chen Z., Indjeian V. B., McManus M., Wang L., Dynlacht B. D. CP110, a cell cycle-dependent CDK substrate, regulates centrosome duplication in human cells. Dev. Cell. 2002;3:339–350. doi: 10.1016/s1534-5807(02)00258-7. [DOI] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D. M., Bettencourt-Dias M. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- Doxsey S., Zimmerman W., Mikule K. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Duensing A., Liu Y., Perdreau S. A., Kleylein-Sohn J., Nigg E. A., Duensing S. Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene. 2007;26:6280–6288. doi: 10.1038/sj.onc.1210456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink J., Thery M., Azioune A., Dupont R., Chatelain F., Bornens M., Piel M. Comparative study and improvement of current cell micro-patterning techniques. Lab Chip. 2007;7:672–680. doi: 10.1039/b618545b. [DOI] [PubMed] [Google Scholar]
- Fisk H. A., Mattison C. P., Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc. Natl. Acad. Sci. USA. 2003;100:14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C., Binkert C., Dennis J. W. Constitutive expression of murine Sak-a suppresses cell growth and induces multinucleation. Mol. Cell. Biol. 1996;16:4665–4672. doi: 10.1128/mcb.16.9.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C., Motro B., Yousefi S., Heffernan M., Dennis J. W. Sak, a murine protein-serine/threonine kinase that is related to the Drosophila polo kinase and involved in cell proliferation. Proc. Natl. Acad. Sci. USA. 1994;91:6388–6392. doi: 10.1073/pnas.91.14.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y. D., Wilkinson C. J., Nigg E. A. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hudson J. W., Kozarova A., Cheung P., Macmillan J. C., Swallow C. J., Cross J. C., Dennis J. W. Late mitotic failure in mice lacking Sak, a polo-like kinase. Curr. Biol. 2001;11:441–446. doi: 10.1016/s0960-9822(01)00117-8. [DOI] [PubMed] [Google Scholar]
- Keller L. C., Geimer S., Romijn E., Yates J., 3rd, Zamora I., Marshall W. F. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell. 2009;20:1150–1166. doi: 10.1091/mbc.E08-06-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y. D., Nigg E. A. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Ko M. A., Rosario C. O., Hudson J. W., Kulkarni S., Pollett A., Dennis J. W., Swallow C. J. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat. Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- Kuriyama R., Borisy G. G. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- Leidel S., Gonczy P. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 2003;4:431–439. doi: 10.1016/s1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- Leung G. C., Ho C. S., Blasutig I. M., Murphy J. M., Sicheri F. Determination of the Plk4/Sak consensus phosphorylation motif using peptide spots arrays. FEBS Lett. 2007;581:77–83. doi: 10.1016/j.febslet.2006.11.080. [DOI] [PubMed] [Google Scholar]
- Leung G. C., Hudson J. W., Kozarova A., Davidson A., Dennis J. W., Sicheri F. The Sak polo-box comprises a structural domain sufficient for mitotic subcellular localization. Nat. Struct. Biol. 2002;9:719–724. doi: 10.1038/nsb848. [DOI] [PubMed] [Google Scholar]
- Meraldi P., Lukas J., Fry A. M., Bartek J., Nigg E. A. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Mogensen M. M., Malik A., Piel M., Bouckson-Castaing V., Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Moudjou M., Bornens M. Isolation of centrosomes from cultured animal cells. In: Celis J. E., editor. Cell Biology: A Laboratory Handbook. London: Academic Press; 1994. pp. 595–604. [Google Scholar]
- O'Connell K. F., Caron C., Kopish K. R., Hurd D. D., Kemphues K. J., Li Y., White J. G. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/s0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- Paintrand M., Moudjou M., Delacroix H., Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J. L., Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C. L., Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J. B., Phillips S. G. Independence of centriole formation and DNA synthesis. J. Cell Biol. 1973;57:359–372. doi: 10.1083/jcb.57.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins E., Jentzsch G., Micali A. The centriole cycle in synchronized HeLa cells. J. Cell Biol. 1968;36:329–339. doi: 10.1083/jcb.36.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D. M., Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- Rogers G. C., Rusan N. M., Roberts D. M., Peifer M., Rogers S. L. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. H., Aravind L., Muller-Reichert T., O'Connell K. F. The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev. Cell. 2008;15:901–912. doi: 10.1016/j.devcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin A. M., Celati C., Paintrand M., Bornens M. Identification of an Spc110p-related protein in vertebrates. J. Cell Sci. 1997;110:2533–2545. doi: 10.1242/jcs.110.20.2533. [DOI] [PubMed] [Google Scholar]
- Thery M., Racine V., Piel M., Pepin A., Dimitrov A., Chen Y., Sibarita J. B., Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Natl. Acad. Sci. USA. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidwans S. J., Wong M. L., O'Farrell P. H. Mitotic regulators govern progress through steps in the centrosome duplication cycle. J. Cell Biol. 1999;147:1371–1378. doi: 10.1083/jcb.147.7.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I. A., Chentsov Yu S. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnke S., Kemmler S., Hames R. S., Tsai H. L., Hoffmann-Rohrer U., Fry A. M., Hoffmann I. Polo-like kinase-2 is required for centriole duplication in mammalian cells. Curr. Biol. 2004;14:1200–1207. doi: 10.1016/j.cub.2004.06.059. [DOI] [PubMed] [Google Scholar]
- White R. A., Pan Z., Salisbury J. L. GFP-centrin as a marker for centriole dynamics in living cells. Microsc. Res. Tech. 2000;49:451–457. doi: 10.1002/(SICI)1097-0029(20000601)49:5<451::AID-JEMT7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Wu J., Cho H. P., Rhee D. B., Johnson D. K., Dunlap J., Liu Y., Wang Y. Cdc14B depletion leads to centriole amplification, and its overexpression prevents unscheduled centriole duplication. J. Cell Biol. 2008;181:475–483. doi: 10.1083/jcb.200710127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y., Kajigaya S., Yoshida K., Ueno S., Ota J., Ohmine K., Ueda M., Miyazato A., Ohya K., Kitamura T., Ozawa K., Mano H. Sak serine-threonine kinase acts as an effector of Tec tyrosine kinase. J. Biol. Chem. 2001;276:39012–39020. doi: 10.1074/jbc.M106249200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.