Despite their limited chemical functionality, proteins and nucleic acids dominate the solutions to many complex chemical problems because they can be evolved through iterated cycles of diversification, selection, and amplification. Researchers have demonstrated extensively that proteins and nucleic acids initially lacking desired activities can be mutated, amplified, and reselected to afford evolved molecules with greatly enhanced properties.1 We are interested in creating amplifiable and evolvable libraries of non-natural small molecules by developing methods to translate DNA into synthetic structures. Achieving this goal requires using DNA to direct chemical reactions sequence-specifically in a manner much more general than has been reported thus far. Researchers have previously demonstrated the ability of nucleic acid templates to promote the coupling of adjacently annealed oligonucleotides to form nucleic acids and nucleic acid analogues.2 We hypothesized that the proximity effect provided by DNA-templated synthesis can be used to generate libraries of synthetic small molecules unrelated in structure to the DNA backbone in one-pot, parallel reactions.
We examined the ability of two DNA architectures to support solution-phase DNA-templated synthesis (Figure 1). Both hairpin (H) and end-of-helix (E) templates bearing electrophilic maleimide groups reacted efficiently with one equivalent of thiol reagent linked to a complementary DNA oligonucleotide to yield the thioether product in minutes at 25 °C. DNA-templated reaction rates (kapp = ∼105 M−1 s−1) were similar for H and E architectures despite significant differences in the relative orientation of their reactive groups. In contrast, no product was observed when using reagents containing sequence mismatches, or when using templates pre-quenched with excess β-mercaptoethanol (Figure 1). Both H and E templates therefore support the sequence-specific DNA-templated addition of a thiol to a maleimide even though the structures of the resulting products differ markedly from the structure of the natural phosphodiester backbone. Little or no nontemplated intermolecular reaction products are produced under the reaction conditions (pH 7.5, 25 °C, 250 mM NaCl, 60 nM template and reagent).
Figure 1.
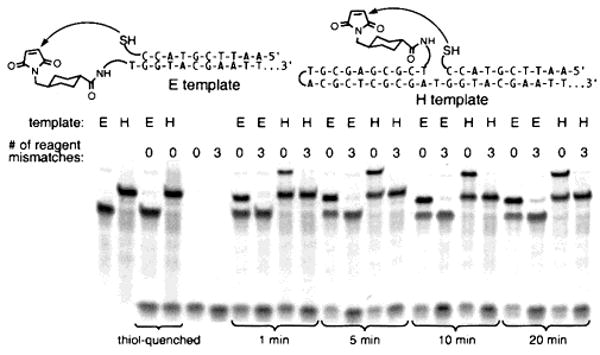
Synthesis directed by hairpin (H) and end-of-helix (E) DNA templates. Reactions were analyzed by denaturing PAGE after the indicated reaction times. Lanes 3 and 4 contained templates quenched with excess β-mercaptoethanol prior to reaction.
Surprisingly, sequence-specific DNA-templated reactions spanning a variety of reaction types (SN2 substitutions, additions to α,β-unsaturated carbonyl systems, and additions to vinyl sulfones), nucleophiles (thiols and amines), and reactant structures all proceeded in good yields with excellent sequence selectivity (Figure 2). In each case, matched but not mismatched reagents afforded product efficiently despite considerable variations in their transition-state geometry, steric hindrance, and conformational flexibility. Collectively, these findings indicate that DNA-templated synthesis is a general phenomenon capable of supporting a range of reaction types and is not limited to the creation of structures resembling nucleic acid backbones.
Figure 2.
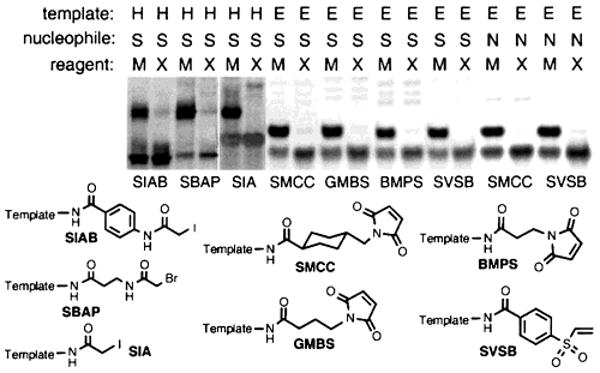
Matched (M) or mismatched (X) reagents linked to thiols (S) or primary amines (N) were mixed with 1 equiv of template functionalized with the variety of electrophiles shown. Reactions with thiol reagents were conducted at pH 7.5 under the following conditions: SIAB and SBAP: 37 °C, 16 h; SIA: 25 °C, 16 h; SMCC, GMBS, BMPS, SVSB: 25 °C, 10 min. Reactions with amine reagents were conducted at 25 °C, pH 8.5 for 75 min.
Since sequence discrimination is important for the faithful translation of DNA into synthetic structures, we measured the reaction rate of a matched reagent compared with that of a reagent bearing a single mismatched base near the center of its 10-base oligonucleotide. At 25 °C, the initial rate of reaction of matched thiol reagents with iodoacetamide-linked H templates is 200-fold faster than that of reagents bearing a single mismatch (kapp = 2.4 × 104 M−1 s−1 vs 1.1 × 102 M−1 s−1, Figure 3a). In addition, small amounts of products arising from the annealing of mismatched reagents can be eliminated by elevating the reaction temperature beyond the Tm3 of the mismatched reagents (Figure 3b). The decrease in the rate of product formation as temperature is elevated further indicates that product formation proceeds by a DNA-templated mechanism rather than a simple intermolecular mechanism.
Figure 3.
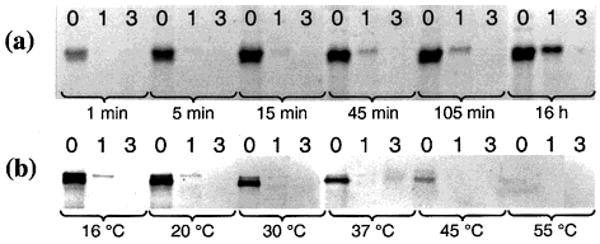
(a) H templates linked to α-iodoacetamide group were reacted with thiol reagents containing 0, 1, or 3 mismatches at 25 °C. (b) Reactions in (a) were repeated at the indicated temperature for 16 h. Calculated3 reagent Tm: 38 °C (matched), 28 °C (single mismatch).
In addition to reaction generality and sequence specificity, DNA-templated synthesis also demonstrates remarkable distance independence. Both H and E templates linked to maleimide or α-iodoacetamide groups promote the sequence-specific reaction with matched, but not mismatched, thiol reagents annealed anywhere on the templates examined thus far (up to 30 bases away from the reactive group on the template). Reactants annealed one base away react with rates similar to those annealed 2, 3, 4, 6, 8, 10, 15, 20, or 30 bases away. In all cases, templated reaction rates are at least several hundred-fold higher than the rate of untemplated reaction for both E templates (kapp = 104–105 M−1 s−1 vs 5 × 101 M−1 s−1, Figure 4a) and H templates (data not shown). At intervening distances of 30 bases, products are efficiently formed presumably through transition states resembling 200-membered rings. These findings contrast sharply with the well-known difficulty of macrocyclization in organic synthesis.4
Figure 4.
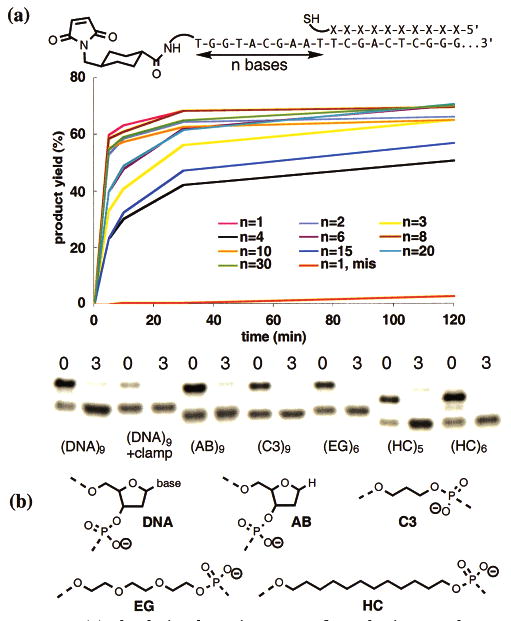
(a) The depicted reaction was performed using a 41-base E template and a 10-base reagent designed to anneal 1–30 bases from the 5′ end of the template. The kinetic profiles in the graph show the average of two trials (deviations < 10%). The “n = 1, mis” reagent contains three mismatches. (b) The n = 10 reaction in (a) was repeated using templates in which the nine bases following the 5′-NH2-dT were replaced with the backbone analogues shown. Five equivalents of a DNA oligonucleotide complementary to the intervening bases was added to the “DNA + clamp” reaction. Reagents were matched (0) or contained three mismatches (3). The gel shows reactions at 25 °C after 25 min.
To determine the basis of the distance independence of DNA-templated synthesis, we first synthesized a series of modified E templates in which the intervening bases were replaced by a series of DNA analogues designed to evaluate the possible contribution of (i) interbase interactions, (ii) conformational preferences of the DNA backbone, (iii) the charged phosphate backbone, and (iv) backbone hydrophilicity. Templates in which the intervening bases were replaced with any of the analogues in Figure 4b had little effect on the rates of product formation. These findings indicate that backbone structural elements specific to DNA are not responsible for the observed distance independence of DNA-templated synthesis. However, the addition of a 10-base DNA oligonucleotide complementary to the single-stranded intervening region significantly reduced product formation (Figure 4b), suggesting that the flexibility of this region is critical to efficient DNA-templated synthesis.
The distance-independent reaction rates may be explained if the bond-forming events in a DNA-templated format are sufficiently accelerated relative to their nontemplated counterparts such that DNA annealing, rather than bond formation, is rate-determining. If DNA annealing is at least partially rate-limiting, then the rate of product formation should decrease as the concentration of reagents is lowered because annealing, unlike templated bond formation, is a bimolecular process. Decreasing the concentration of reactants in the case of the E template with one or 10 intervening bases between reactive groups resulted in a marked decrease in the observed reaction rate (Figure 5). This observation suggests that proximity effects in DNA-templated synthesis can enhance bond-formation rates to the point that DNA annealing becomes rate-determining.
Figure 5.
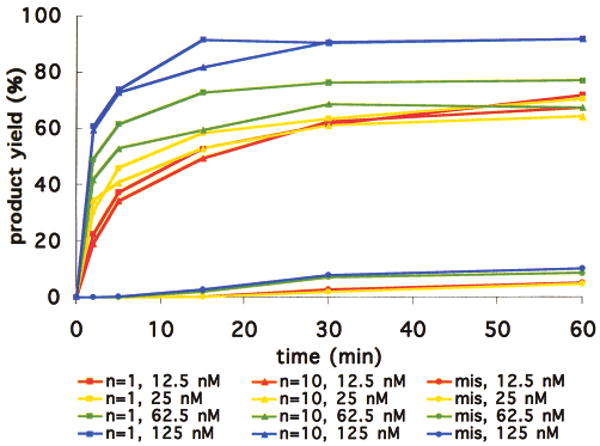
The n = 1, n = 10, and n = 1 mismatched (mis) reactions described in Figure 4a were repeated with template and reagent concentrations of 12.5, 25, 62.5, or 125 nM.
These findings raise the possibility of using DNA-templated synthesis to translate in one-pot libraries of DNA into libraries of synthetic molecules suitable for diversification, selection, and PCR amplification. The ability of DNA-templated synthesis to support a variety of transition-state geometries suggests its potential in directing a range of powerful water-compatible5 synthetic reactions. The sequence specificity described above suggests that mixtures of reagents may be able to react predictably with complementary mixtures of templates. Finally, the observed distance independence suggests that different regions of DNA (“codons”) may be used to encode different groups on the same synthetic scaffold without impairing reaction rates.
As a demonstration of this approach, we synthesized a library of 1025 maleimide-linked E templates, each with a different DNA sequence in an eight-base encoding region (Figure 6). One of these sequences, 5′-TGACGGGT-3′, was chosen to code for the attachment of a biotin group to the template. A library of thiol reagents linked to 1025 different oligonucleotides was also generated. The reagent linked to 3′-ACTGCCCA-5′ contained a biotin group, while the other 1024 reagents (linked to 3′-NGNTNGNN-5′, where N = A, C, G, or T) contained no biotin. Equimolar ratios of all 1025 templates and 1025 reagents were mixed in one pot at 60 nM total concentration for 10 min at 25 °C, and the resulting products were selected in vitro for binding to immobilized streptavidin. Molecules surviving the selection were amplified by PCR using primers binding outside of the coding region and analyzed by restriction digestion and DNA sequencing.
Figure 6.
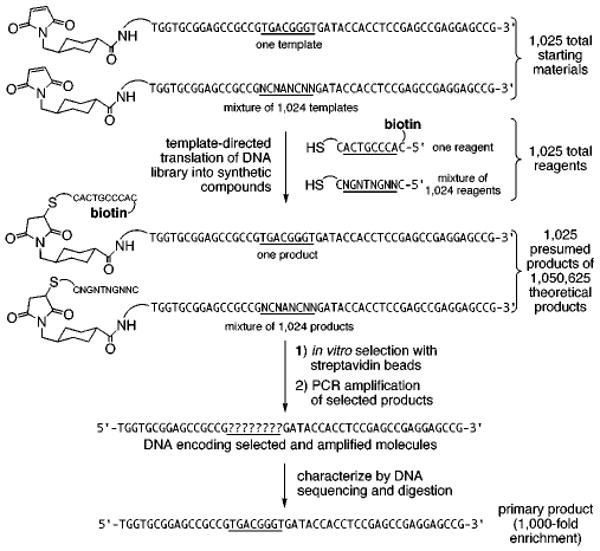
A model translation, selection, and amplification of synthetic molecules that bind streptavidin from a DNA-encoded library.
Digestion with the restriction endonuclease Tsp45I, which cleaves GTGAC and therefore cuts the biotin-encoding template but none of the other templates, revealed a 1:1 ratio of biotin-encoding to non-biotin-encoding templates following selection. This represents a 1000-fold enrichment compared with the unselected library. DNA sequencing of the PCR-amplified pool before and after selection suggested a similar degree of enrichment and indicated that the biotin-encoding template is the major product after selection and amplification (Figure 7a). The ability of DNA-templated synthesis to support the simultaneous sequence-specific reaction of 1025 reagents, each of which faces a 1024:1 ratio of nonpartner-to-partner templates, demonstrates its potential as a method to create synthetic libraries in one pot. The above proof-of-principle translation, selection, and amplification of a synthetic library member having a specific property (avidin affinity in this example) addresses several of the requirements for the evolution of non-natural small molecule libraries (Figure 7b).
Figure 7.
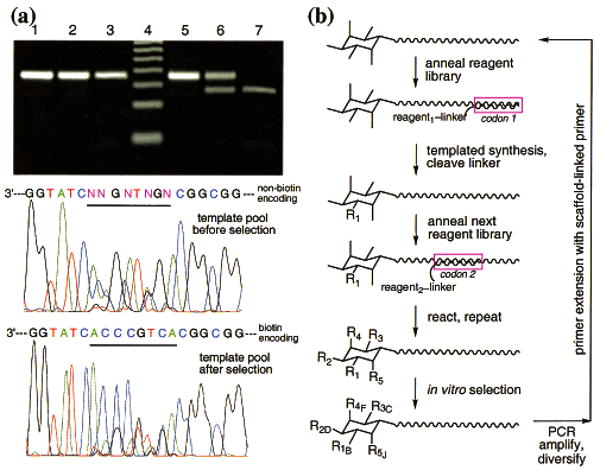
(a) Lanes 1 and 5: PCR: amplified library before stretptavidin binding selection. Lanes 2 and 6: PCR amplified library after selection. Lanes 3 and 7: PCR amplified authentic biotin-encoding template. Lane 4: 20 bp ladder. Lanes 5–7 were digested with Tsp45I. DNA sequencing traces of the amplified templates before and after seletion are also shown, together with the sequences of the non-biotin-encoding and biotin-encoding templates. (b) General scheme for the creation and evolution of libraries of non-natural molecules using DNA-templated synthesis, where –R1 represents the library of product functionality transferred from reagent library 1, and –R1B represents a selected product.
Taken together, these results suggest that DNA-templated synthesis is a surprisingly general phenomenon capable of directing, rather than simply encoding, a range of chemical reactions to form products unrelated in structure to nucleic acid backbones. For several reactions examined, the DNA-templated format accelerates the rate of bond formation beyond the rate of a 10-base DNA oligonucleotide annealing to its complement, resulting in surprising distance independence. The facile nature of long-distance DNA-templated reactions may also arise in part from the tendency of water to contract the volume of nonpolar reactants5 and from possible compactness of the intervening single-stranded DNA between reactive groups. These findings may have implications for prebiotic evolution and for understanding the mechanisms of catalytic nucleic acids, which typically localize substrates to a strand of RNA or DNA.1b,6 Previous observations of DNA-templated syntheses of nucleic acid analogues2 may in part arise from the general proximity effects described here rather than from the commonly cited reason of the template precisely aligning reactive groups.
The generality of DNA-templated synthesis provides a basis for translating the amplifiable information in a library of DNA into non-natural synthetic small molecules in one pot, as demonstrated by the translation, selection, and amplification of the model synthetic library described above. Linked to their encoding DNA, libraries of synthetic molecules generated in this manner may be subjected to powerful genetic methods previously available only to proteins and nucleic acids, including sequencing, random mutation, recombination, and in vitro selection. In contrast to existing library synthesis methods, yields of each reaction are much less important in this format because PCR amplification requires only vanishingly small quantities (<104 molecules)7 of template following selection. In addition, these libraries are selected on the basis of each individual molecule's properties, obviating the need for assaying and deconvoluting mixtures. Ongoing efforts in our group seek to further develop this new approach to generating molecular function.
Supplementary Material
Acknowledgments
We thank Pantila Vanichakarn for assistance in oligonucleotide purification and Steve Vollmer for help with automated DNA sequencing. Professor E. J. Corey and Professor Stuart Schreiber provided many helpful comments. This work was supported by an Office of Naval Research Young Investigator Award (N00014-00-1-0596), the Searle Scholars Program, a Research Corporation Research Innovation Award, and Harvard University. Zev Gartner is supported by a NSF Graduate Research Fellowship.
Footnotes
Supporting Information Available: DNA sequences and experimentals for template functionalization, DNA-templated synthesis, and in vitro selection for avidin binding (PDF). This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Arnold FH, Volkov AA. Curr Opin Chem Biol. 1999;3:54–9. doi: 10.1016/s1367-5931(99)80010-6. [DOI] [PubMed] [Google Scholar]; (b) Jaschke A, Seelig B. Curr Opin Chem Biol. 2000;4:257–62. doi: 10.1016/s1367-5931(00)00086-7. [DOI] [PubMed] [Google Scholar]; (c) Minshull J, Stemmer WP. Curr Opin Chem Biol. 1999;3:284–90. doi: 10.1016/s1367-5931(99)80044-1. [DOI] [PubMed] [Google Scholar]; (d) Wilson DS, Szostak JW. Annu Rev Biochem. 1999;68:611–47. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 2.(a) Xu Y, Karalkar NB, Kool ET. Nature Biotechnol. 2001;19:148–152. doi: 10.1038/84414. [DOI] [PubMed] [Google Scholar]; (b) Luo P, Leitzel JC, Zhan ZYJ, Lynn DG. J Am Chem Soc. 1998;120:3019–3031. [Google Scholar]; (c) Herrlein MK, Nelson JS, Letsinger RL. J Am Chem Soc. 1995;117:10151–10152. [Google Scholar]; (d) Bruick RK, Dawson PE, Kent SBH, Usman N, Joyce GF. Chem Biol. 1996;3:49–56. doi: 10.1016/s1074-5521(96)90084-8. [DOI] [PubMed] [Google Scholar]; (e) Gat Y, Lynn DG. Biopolymers. 1998;48:19–28. doi: 10.1002/(SICI)1097-0282(1998)48:1<19::AID-BIP3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]; (f) Orgel LE. Acc Chem Res. 1995;28:109–118. doi: 10.1021/ar00051a004. [DOI] [PubMed] [Google Scholar]; (g) Luther A, Brandsch R, von Kiedrowski G. Nature. 1998;396:245–8. doi: 10.1038/24343. [DOI] [PubMed] [Google Scholar]
- 3.Meinkoth J, Wahl GM. Anal Biochem. 1984;138:267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- 4.(a) Illuminati G, Mandolini L. Acc Chem Res. 1981;14:95–102. [Google Scholar]; (b) Woodward RB, Logusch E, Nambiar KP, Sakan K, Ward DE, Auyeung BW, Balaram P, Browne LJ, Card PJ, Chen CH, Chenevert RB, Fliri A, Frobel K, Gais HJ, Garratt DG, Hayakawa K, Heggie W, Hesson DP, Hoppe D, Hoppe I, Hyatt JA, Ikeda D, Jacobi PA, Kim KS, Kobuke Y, Kojima K, Krowicki K, Lee VJ, Leutert T, Malchenko S, Martens J, Matthews RS, Ong BS, Press JB, Babu TVR, Rousseau G, Sauter HM, Suzuki M, Tatsuta K, Tolbert LM, Truesdale EA, Uchida I, Ueda Y, Uyehara T, Vasella AT, Vladuchick WC, Wade PA, Williams RM, Wong HNC. J Am Chem Soc. 1981;103:3210–3213. [Google Scholar]
- 5.Li CJ, Chan TH. Organic Reactions in Aqueous Media. Wiley and Sons; New York: 1997. [Google Scholar]
- 6.Joyce GF. Curr Biol. 1996;6:965–7. doi: 10.1016/s0960-9822(02)00640-1. [DOI] [PubMed] [Google Scholar]
- 7.Kramer MF, Coen DM. In: Current Protocols in Molecular Biology. Ausubel FM, editor. Vol. 3. Wiley; 1999. pp. 15.1–15.3. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


