Abstract
Background
Microtubules (MTs) participate in the spatial regulation of actin-based processes such as cytokinesis and cell polarization [1]. The fission yeast Schizosaccharomyces pombe is a rod-shaped cell which exhibits polarized cell growth at cell tips. MT plus ends contact and shrink from the cell tips and contribute to polarity regulation.
Results
Here, we investigate the effects of changing cell shape on MTs and cell polarization machinery. We physically bend fission yeast cells by forcing them into microfabricated femtoliter chambers. In these bent cells, MTs maintain a straight axis and contact and shrink from cortical sites at the sides of cells. At these ectopic sites, polarity factors such as bud6p, for3p (formin), and cdc42p are recruited and assemble actin cables in an MT-dependent manner. MT contact at the cortex induces the appearance of a bud6p dot within seconds. The accumulation of polarity factors leads to cell growth at these sites, when the MT-associated polarity factor tea1p is absent. This process is dependent on MTs, mal3p (EB1), moe1p (an EB1-binding protein), and for3p, but surprisingly, is independent of the tea1p–tea4p pathway.
Conclusions
These studies provide a direct demonstration for how MTs induce actin assembly at specific locations on the cell cortex and begin to identify a new pathway involved in this process. MT interactions with the cortex may be regulated by cortical attachment sites. These findings highlight the crosstalk between cell shape, polarity mechanisms, and MTs responsible for cell morphogenesis.
INTRODUCTION
Microtubules (MTs) are fundamental for the spatial regulation of cell shape in such processes as cell migration, neuronal axon guidance and cytokinesis [1]. In cytokinesis, MTs associated with the spindle dictate the site of contractile actin ring assembly, while in cell migration, MTs contribute to specifying the direction of movement. Although there has been some recent headway into identifying MT-associated factors involved in these processes [2], in general, how MTs function in molecular terms in these processes remain poorly understood.
The fission yeast Schizosaccharomyces pombe has emerged as an excellent model to study the molecular mechanisms that link MTs to actin assembly and cell polarization [3]. These are rod-shaped cells that grow by tip extension in a cell-cycle-regulated manner. During interphase, MTs are organized in 3–5 antiparallel bundles. The MT plus ends grow to the cell tips, contact the cortex for 1–2 minutes, and then exhibit catastrophe [4]. The large majority of catastrophe events occur upon contact at the cell tip. The basis of this regulation is not well understood, although different models have been proposed.
Polarized cell growth in these cells is achieved in part by the localized delivery of membranes and proteins involved in cell wall remodeling to the cell tip using myosinV-based transport on actin cables [5]. One key regulator of cell polarity is the formin for3p, which assembles the actin cables from cell tips, and is activated by the cdc42 GTPase and by the formin-activating protein bud6p [6]. Interphase MTs influence cell shape and polarity [7]. Tea1p and tea4p define one well characterized link between MT plus ends and the actin assembly machinery [8]. These proteins are carried on MT plus ends and deposited onto the cell tips at sites of MT catastrophe, where they are required for maintaining tip growth in conditions such as recovery from starvation [9]. These factors are also needed to induce a transition from monopolar growth to bipolar growth during G2-phase, a process termed New End Take Off (NETO) [3]. Tea1p and tea4p are thought to initiate cell polarization and actin assembly at the “new” cell end by recruiting proteins such as for3p and its activator bud6p.
It is increasingly clear that there is a functional interplay between cell shape and the internal organization of the cell [10, 11]. In fission yeast, the orientation of the MTs along the long axis of the cell is thought to be due to the rod cell shape and its cortical interactions [4]. Round mutant cells, for instance, display disorganized MT cytoskeletons [12]. The orientation is independent of any organizing influence of the nucleus, as MT bundles orient properly in anucleate cells [13, 14]. Thus, there are elements of a positive-feedback loop between the MT cytoskeleton and cell shape itself that reinforces the morphogenesis of the rod shape. However, because of this feedback, it has been difficult to disentangle cause-effect relationships, and it has never been carefully tested whether MTs can actively induce local actin assembly, for instance at an ectopic site.
Here, we sought to examine the effects of changing cell shape. We developed a method for physically bending fission yeast cells into microfabricated chambers. We find that this change in cell shape leads to rapid changes in the organization of the MTs and cortical polarity factors. This process provides novel insights into MT regulation and reveals a new pathway that links MTs to actin assembly.
RESULTS
Shaping single fission yeast cells in situ
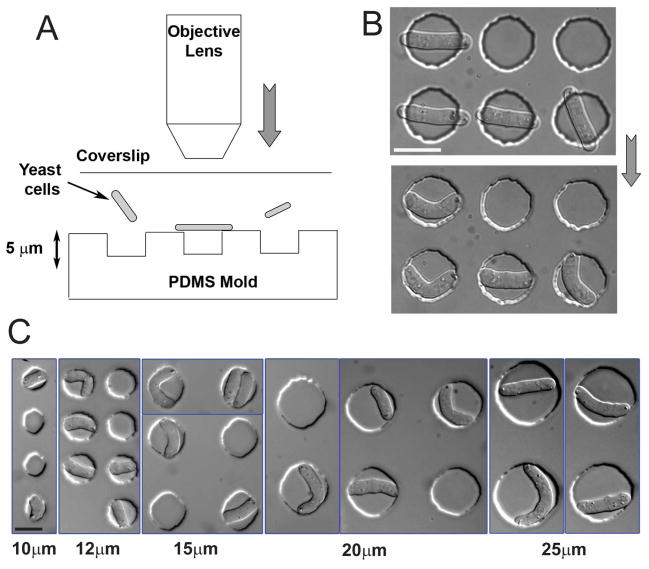
We developed a method to rapidly change the shape of live fission yeast cells in situ. Fission yeast cells were placed on a microfabricated PDMS array of cylindrical wells 5μm deep with a diameter of 10–30μm (Figure S1). Cells were then physically pushed into the wells by gently pressing the top glass coverslip by over-focusing the objective of an upright microscope. Cells longer than the diameter of the well buckled into the chamber and adopted a bent morphology (Figure 1A and 1B; Movie 1). Some cells occasionally popped out of the well and bent back into a straight rod shape, demonstrating that the cell wall is elastic (Movie 2) [15].
Figure 1. Bending fission yeast cells in situ.
(A) Schematic illustration of the technique used to bend fission yeast cells in situ: cells in liquid media are placed between a PDMS array of microwells and a glass coverslip. The coverslip is subsequently pressurized by overfocusing the objective of an upright microscope. (B) DIC images of the same fission yeast cells before and after pressurizing with the objective. Strain: FC504 (cdc25-22 at 25°C). (C) DIC images of fission yeast cells of varying sizes bent in holes having different diameters (indicated at the bottom). Strains: FC420, FC504. Scale bars: 10μm.
We could produce cells with different degrees of bending (represented by the bending ratio, γ; see supplementary material and Figure 2C) by manipulating cell length and well size (Figure 1C). A given cell length and well size reproducibly induced the same degree of bending that we could quantify and mathematically predict through simple geometrical calculations (Figure S2A–C). Cells of different lengths were identified from those in different periods of the cell cycle, or derived from delaying cell cycle progression with hydroxyurea treatment or cdc25-22 mutation.
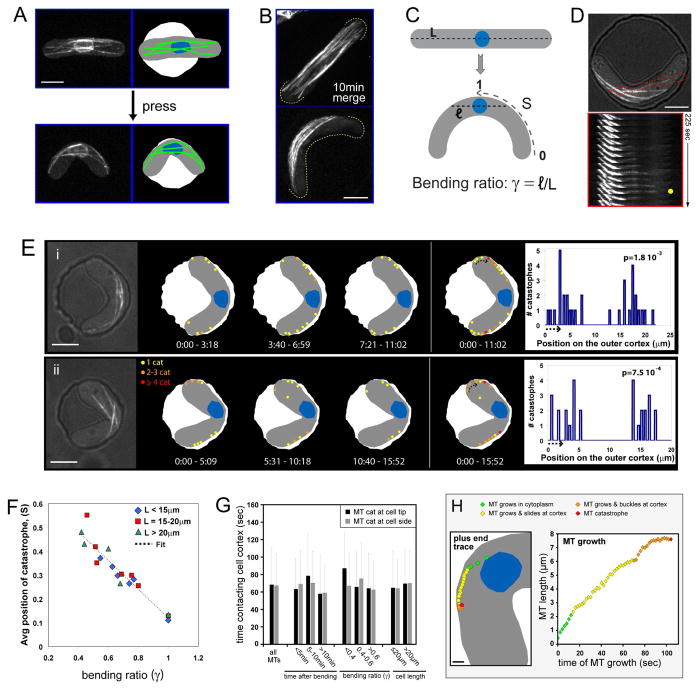
Figure 2. Effects of bent cell geometry on the dynamic interphase MTs.
(A) MT organization in a bent cell. Maximum projection confocal image of a cell expressing GFP-tubulin and cut11-GFP (nuclear pore) prior to and immediately after being pushed into a chamber. In the schematic, MTs are indicated in green and the nucleus in blue. Strain: FC1451 (cdc25-22 at 25°C). (B) MTs associate with the outer curvature of bent cells. Merge of 14 confocal stacks over 10 min from cells expressing GFP-tubulin. Strain: NM11 (cdc25-22 at 25°C). (C) Schematic representing the geometrical parameters for predicting MT sites of contact in a bent cell. L is the cell length, and ℓ is twice the distance from the center of the nucleus to the side of the cell following the former axis of the straight cell in the bent cell. The bending ratio, γ = ℓ/L describes how close the cortex is from the nucleus along the axis perpendicular to the short axis of the cell. S is the absolute coordinate along the outer curvature (S=0 at the tip and S=1 in the middle). (D) MTs undergo catastrophe at the side of bent cells. Top: DIC/GFP-tubulin merge of a bent cell with a single MT boxed in red. Bottom: time-lapse images of the boxed MT, showing MT growth to the side of the cell, catastrophe at the cortex (yellow dot), and shrinkage. Strain: NM11. (E) Schematic of MT catastrophe events in two representative bent cells—one with a centered nucleus (i), and one with an off-centered nucleus (ii). MT catastrophes were determined from time-lapse images of GFP-tubulin. Specific points on the cortex that experience multiple catastrophe events are indicated (orange and red dots) and further represented in the bar plot on the right. Dot size and bar width encompass the error made in detecting the MT + TIP. The arrow in the cell indicates the beginning of the corresponding bar plot. The number p, indicated on the top right of each bar plot represents the probability of having this particular distribution as compared to a Monte-Carlo trial (see supplementary material). p is calculated using a cortical stretch of 16μm for cell (i) and 9μm for cell (ii). Time is in min:sec. Strain: NM11. Scale bars: 5μm. (F) Average position of catastrophes (Scat) in different cells plotted as a function of the bending ratio. The linear fit returns Scat = 0.72 – 0.60γ (See supplementary material also) Strains: FC1234, FC1234 + HU and NM11. (G) Cortical contact times of MTs that undergo catastrophe at the cell tip vs. the cell side. Data represents 276 MTs (136 and 140 for the cell tip and cell side, respectively) from 19 cells. Error bars represent the standard deviations for the cortical times in each condition. Strain: NM11. (H) MTs in wildtype bent cells expressing GFP-tubulin were examined in time-lapse (single-plane, 0.5fps; n=14 MTs) as in Movie 7. Left: schematic indicating the position of a growing MT plus end in a bent cell (4s intervals). Right: entire length of this same MT as a function of time (2s intervals) as it grows to the cortex (12s), slides along the cortex (56s), and buckles (36s) before catastrophe. Strain: NM11. Scale bar: 1μm.
Upon bending, cells remained viable, continued to grow from their tips without any major effect on the growth rate and went on to divide (Figure S2D). Bending the cell did not create any notable stress response, such as the presence of large vacuoles. The cell wall on the outer curvature is stretched by 10–20% depending on cell length and degree of bending, which is well below the 80–90% degree of stretching needed to break the cell wall measured in budding yeast [15].
Changing the cell shape induces a major reorganization of the MT cytoskeleton
Changing the cell shape had immediate effects on MT behavior. In unbent control cells, GFP-labeled MTs were arranged in longitudinal bundles that reached cell tips (Figure 2A and 2B). Upon cell bending, most MTs no longer reached the cell tips, but were aligned on an axis tangential to the cell curvature and contacted the cortex primarily along the outer curvature (Figure 2A, 2B and Movies 3–6). MTs exhibited normal rates of growth and shrinkage, as well as cortical dwell times (Figure S3B and 2G), suggesting that the general mechanisms of MT regulation were not perturbed. However, sites of MT catastrophe were altered. In contrast to normal cells, in which catastrophe events occur predominately at cell tips [4], in bent cells, most catastrophe events occurred on the sides of the cells along the outer curvature (Figure 2D, 2E and S3A). MT behavior was also dependent on the position of the nucleus, which is usually attached to the MT bundles. In most cases, the nucleus was centered, and MT catastrophes occurred symmetrically on the outer curvature of the cell to both sides of the nucleus (Figure 2Ei and Movie 5). In cells that were asymmetrically bent and displayed an off-center nuclear position, catastrophes were clustered at one cell tip and one site on the side (Figure 2Eii and Movie 6). The change in catastrophe sites correlated with the degree of bending and not cell length (Figure 2F and supplementary material).
MTs undergo catastrophe at specific cortical attachment sites
Analysis of individual MTs in vivo provided additional insights into mechanisms of catastrophe. MTs grew out from the area of the nucleus, contacted the cortex at the side of the cell on the outer curvature, and slid along the cortex. When sliding, the MT growth rate was inversely correlated with the angle of contact between the MT and the cortex (Figure S3C). MTs then appeared to stop sliding and continued to grow at a slower rate with their plus end attached to the cortex. MT growth then stalled for a period prior to undergoing catastrophe (Figure 2H and Movie 7). MTs generally exhibited catastrophe at low angles of contact with the cortex, and 17% of events occurred when the MTs had been sliding practically parallel to the cell surface. Thus, prior to catastrophe MTs may attach to the cortex and accumulate compressive and also lateral strain as they progressively grow more bent being confined by the curved cell shape.
Mapping the catastrophe sites over time revealed the presence of “hot spots” on the cortex. Many MTs appeared to target to and shrink from a small number of specific sites repeatedly (within the resolution of light; see red and orange spots in Figure 2E and S4A), even those originating from different areas (Figure S4C). These observations suggest that a local property of the cortex is responsible for repeatedly inducing catastrophe at the same location. These sites appeared to persist for many minutes of observation. Comparing these behaviors with Monte Carlo simulations showed that these hot spot distributions are significantly different from random ones (Figure S4B and supplementary material).
MTs establish new ectopic sites of bud6p recruitment in bent wildtype cells
We next tested if MTs could induce a site of polarization at the sides of these cells. One of the advantages of rapidly changing cell shape in situ is that we could observe a “clear zone” on the cortex that is initially devoid of polarity factors. As a representative polarity factor, we used bud6p. Bud6p (a homolog of S. cerevisiae Bud6p) is a G-actin-binding protein that promotes actin assembly by activating formins [6, 16]. Bud6p is normally localized at cell tips (Figure 3Ai) and is not generally detectable on MTs [17, 18].
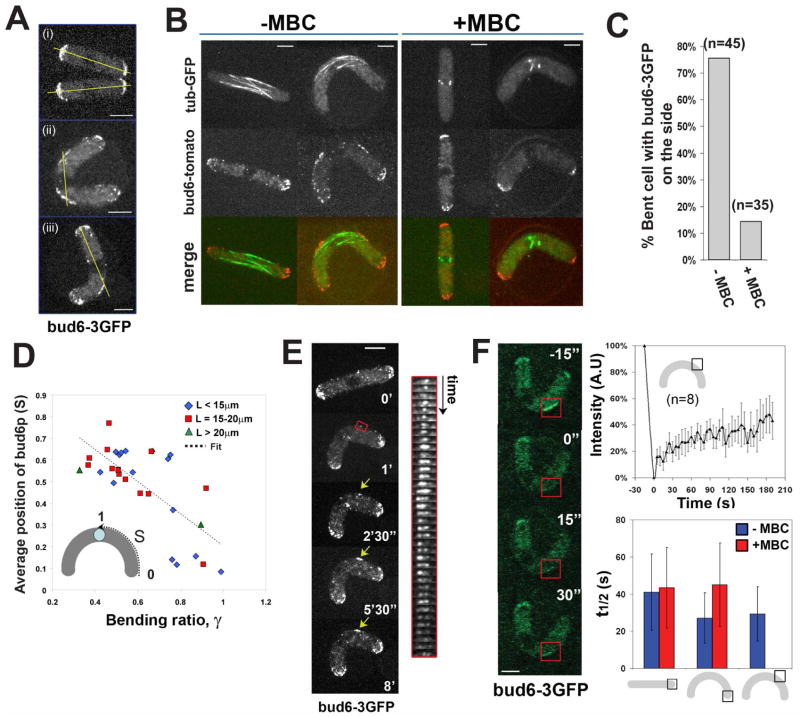
Figure 3. Initial recruitment of the polarity factor bud6p is MT-dependent.
(A) Bud6-3GFP localization in (i) straight and (ii and iii) bent fission yeast cells. The yellow line represents the predicted axis of MT growth from the nucleus. Strain: NM01 (cdc25-22 at 25°C). (B) Bud6-tomato and GFP-tubulin in straight and bent cells in the absence and in the presence of 50μM MBC. Strain: NM42 (cdc25-22 at 25°C). (C) Proportion of cells having bud6-3GFP specifically on the side, in the absence and in the presence of 50μM MBC. (D) Average position of the center of the bud6-3GFP band along the cell outer curvature as a function of the bending ratio. The linear fit returns Sbud6 = 0.95 – 0.74γ. (E) Time-lapse images of bud6-3GFP recruitment to the cell side upon bending of the cell (indicated by the yellow arrow). The kymograph follows the region of the cell in the red box, with a time interval of 30s. Strain: NM01. (F) FRAP experiments on bud6-3GFP. Time-lapse imaging of bud6-3GFP on the cell side recovering from photobleaching. Fluorescent intensity of bud6-3GFP on the cell side as a function of time during a FRAP experiments; the plot represents the average of 8 experiments on 8 different cells, and the bars represent the standard deviation for each point. Half-time recovery are shown for FRAP experiments of bud6-3GFP in the depicted configurations, in the absence and in the presence of 50μM MBC (in the presence of MBC bud6-3GFP is not targeted to the cell side). Strain: NM01. Scale bars: 5μm.
Upon cell bending, bud6-3GFP dots generally accumulated in two bands on the outer curvature of the bent cell, but not on the inner curvature (Figure 3Aii). In asymmetrically bent cells, only one band of bud6-3GFP was present (Figure 3Aiii). Two-color imaging showed that bud6p accumulated in the regions where MTs contacted the cortex (Figure 3B and Movie 9). This distribution also followed a linear trend close to the geometrical description for predicted sites of MT contact (Figure 3D). Importantly, bud6p ectopic recruitment was blocked by treatment with 50μM MBC (a MT inhibitory drug), but not with Latrunculin A (an actin inhibitory drug) (Figure 3B, 3C). Thus, the recruitment of bud6p was dependent on MTs.
We examined the dynamics of bud6p at these ectopic sites. Dots of bud6-3GFP appeared within the first minute of cell bending, and over the next few minutes, accumulated and increased in intensity, becoming a persistent cortical band (Figure 3E and Movies 8). We used these data to quantify the rate of bud6-3GFP establishment at the cell side by comparing it to the signal at cell tips. This ratio increased at a rate of 3.6 ± 1.4 %.min−1 and saturated with a t ½ of 5.8 ± 2.3 min (n=8 cells). FRAP measurements of bud6-3GFP showed that once established, bud6p continuously exchanged on the cortex with a t ½ around 30s at the side of bent cells and at the tip of straight cells (Figure 3F). MTs are thus needed to establish the ectopic site, but once established, the site can continuously recruit bud6p from the cytoplasm.
Bud6p is recruited to sites of MT contact
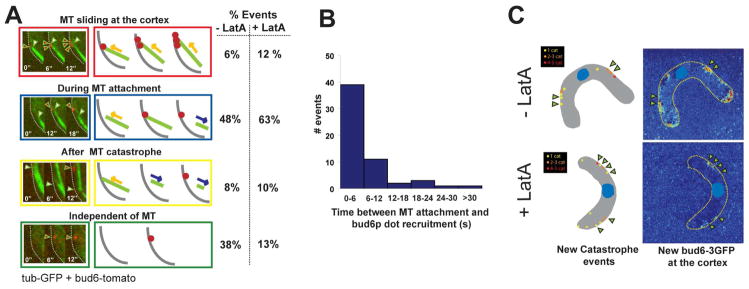
We next examined more closely if bud6p recruitment occurs precisely at the cortical sites contacted by individual MT plus ends (Movie 9 and 10). In 48 % of events (n=60), we observed that a bud6p dot appeared on the cortex within 6s after MT attachment, prior to catastrophe (Figure 4A and 4B). In 6% of cases, multiple bud6p dots appeared on the cortex during MT sliding. In 8% of cases, a bud6p dot was only seen after MT catastrophe (after 12 to 36s of MT attachment) (Figure 4B). A subset of cases (38 %) showed bud6p appearing without MT association. Some of these events may be due to actin-based movements of bud6p into the plane of focus [18], because the incidence of these MT-independent events was reduced in LatA-treated cells to 13%.
Figure 4. Recruitment of bud6p by MT contact.
(A) Dynamics of bud6p recruitment upon MT contact. Left: Single focal plane, two-color imaging (6s intervals) of GFP-tubulin and bud6-tomato on the outer curvature of a bent cell. Right: Schematics of the different observed relationships between MT contact and bud6p-dots recruitment. Far right: Proportion of these observed events in the absence (n=52) and in the presence (n=60) of LatA. (Note that in most cases, MT contact led to a bud6p dot recruitment) (B) Histogram representing the time between a MT attachment on the cortex and the appearance of a bud6p dot. Two color single focal planes were used to compute these data, the binning corresponds to the error in the measurement. (C) Colocalization studies of MT catastrophe and new bud6p deposition in bent cells in the absence and in the presence of LatA. Two-color 3D confocal time-lapse images of cells expressing GFP-tubulin and bud6-tomato were acquired for about 10 min with a 30s temporal resolution. MT catastrophe mappings in these cells are depicted on the left side. Bud6-tomato addition is computed by subtracting the first image from the average image of the entire time-lapse and represented using Matlab. Arrowheads indicate bud6p deposition colocalizing with catastrophe sites. Strain: NM42.
Mapping the cumulative sites of MT catastrophes and bud6p addition over longer time periods confirmed that these MT-based events can largely account for the spatial distribution of bud6p on the cortex (Figure 4C). There was a more precise colocalization between MT catastrophe and bud6p sites of accumulation in LatA-treated cells, showing that once bud6p is recruited to the cortex, actin-based movements may spread its distribution. These direct observations provide strong evidence that MT plus end contact induces the local recruitment of bud6p.
We also considered possible additional factors such as membrane curvature, stretching or cell wall damage. These did not appear to contribute to bud6p accumulation and MT organization in these bent cells. In particular, no bud6p was redistributed in cells that were bent only transiently (for 1–2 min), showing that the bending process itself did not lead to this effect (Figure S5B).
Formation of new actin assembly sites
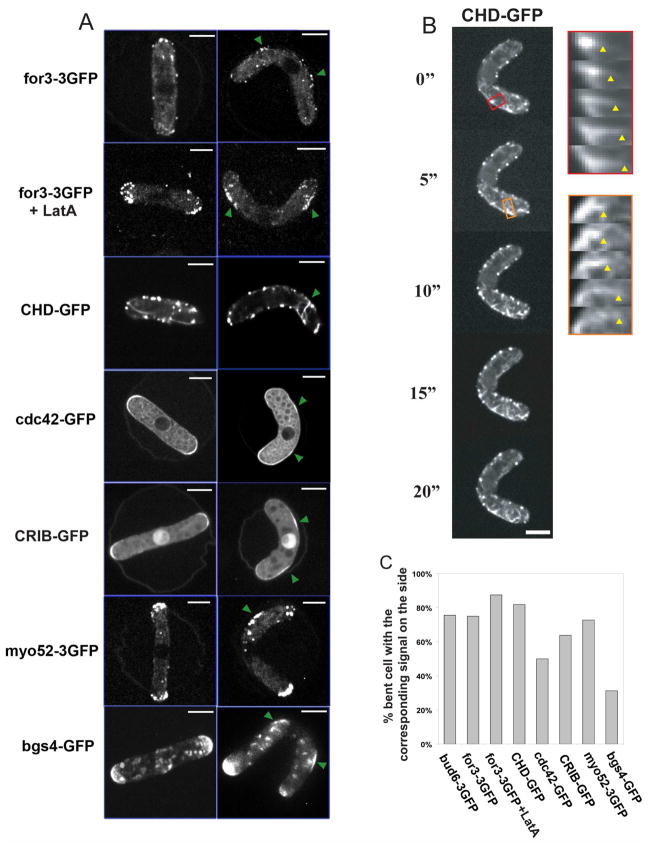
We then tested whether other polarity factors are also recruited to these ectopic sites. The formin for3p, which nucleates actin cables from cell tips [19], was redistributed to these cortical regions. As for3p dots move away from their initial cortical localization in an actin-dependent manner [20], we treated cells with LatA and found that for3p was more concentrated at these ectopic cortical sites. The small GTPase cdc42p, which regulates many aspects of cell polarization including activation of for3p [6], was also redistributed to these cortical sites (Figure 5A and 5C). A CRIB-GFP marker, a marker for active cdc42p [21], confirmed that cdc42p was active in these cortical regions. Actin cables, as visualized by the actin marker calponin-homology-domain-GFP [20], grew from ectopic sites along the outer curvature of the cells, and were often oriented abnormally, perpendicular to the long axis of the cell (Figure 5B; Movie 11). The type V myosin myo52p, which moves towards the barbed end of actin filaments [22], was also targeted to these new sites. One potential cargo for myosin V and actin cables is bgs4p, an integral membrane protein involved in cell wall assembly [23]. Bgs4p also accumulated at these ectopic sites in some bent cells.
Figure 5. Accumulation of polarity factors and actin assembly at ectopic sites in bent cells.
(A) Localization of the indicated polarity factor fused to GFPs in straight and bent cells, the green arrows point to the sites of recruitment on the sides of the bent cells. Images are maximum confocal projections except for CHD-GFP and cdc42-GFP which are single focal planes. Strains from top to bottom: NM07, NM33, NM59, NM145 (all cdc25-22 at 25°C); FC1371 + HU and NM15 (cdc25-22 at 25°C). (B) Time-lapse imaging of a bent cell expressing an F-actin marker GFP-CHD. Kymographs show that actin filaments are growing from ectopic sites of the cell. The arrowheads indicate the end of the elongating actin cable. Single-plane confocal images are shown. Strain: NM33. (C) Proportion of wildtype bent cells that show a specific location of the indicated factors to the cell’s outer curvature (n≈30 for each condition). Scale bars: 5μm.
However, under these conditions, not all events for polarized cell growth were initiated. Calcofluor and filipin staining, which mark new cell wall addition and sterol-rich membranes [24] respectively, were not increased at these sites. New sites of actual cell growth at these sites were never seen in these wildtype bent cells, even after several hours of observation.
Recruitment is independent of the tea1p-NETO pathway and MT dynamics
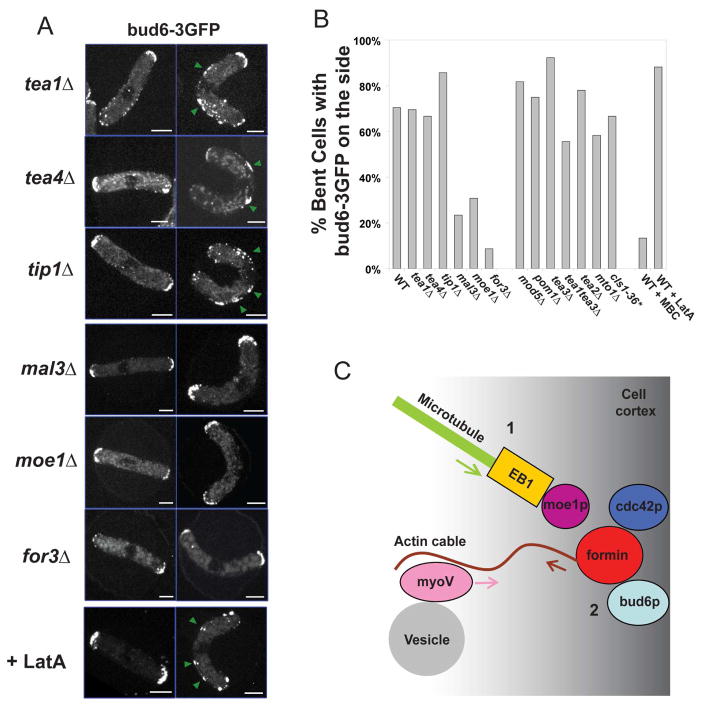
Next, we tested which factors besides MTs were needed for recruitment of these polarity factors. We examined the distribution of bud6-3GFP upon bending in a series of mutant cells. The tea1p–tea4p pathway is a prime candidate, given the molecular links between these MT plus end factors and formins [8]. In bent cells, tea1-GFP was deposited to these ectopic sites (Figure S7). Surprisingly, bud6p was still recruited in mutants of the tea1p–tea4p pathway [3]: tea1Δ, tea4Δ, tip1Δ (CLIP-170), mod5Δ (a peripheral membrane protein), pom1Δ (DYRK kinase), tea2Δ (kinesin) and tea3Δ (kelch protein) (Figure 6A, 6B and S9). The recruitment of for3p was also tea1p-independent (Figure S8). Thus, this process is independent of the tea1p–tea4p polarization pathway.
Figure 6. Recruitment of polarity factors by MTs is independent of the tea1-tea4p pathway, but requires mal3p, moe1p, for3p and the secretory pathway.
(A) Bud6-3GFP position in normal and bent cell of the indicated mutants, and in cells treated with the indicated chemicals. Strains: NM14, NM34, NM30, NM48, NM117, NM115, NM01 (all cdc25-22 at 25°C). Scale bars: 5μm. (B) Proportion of bent cells that show a specific location of bud6-3GFP to the cell side in the indicated genetic backgrounds and conditions (n≈30 for each condition). Some mutants and the corresponding strain numbers are shown in Figure S9. *For this experiment, the temperature of the objective was raised to 30°C with a Bioptechs objective heater. (C) Proposed model for how MT contact induces sites of cell polarization at the cortex: 1. A MT plus end contacts the cortex and recruits for3p (formin) bud6p and cdc42p. This recruitment is dependent on mal3p (EB1) and moe1 (EB1-binding protein). 2. The formin assembles actin cable structures from these sites. 3. Actin filaments guide the transport of myosin V and its cargo such as vesicles towards these ectopic sites.
Proper regulation of MT catastrophe also did not appear to be critical for this process. tip1 (CLIP170 homolog) mutants exhibit MTs with a shortened dwell time at the cortex [4], while mto1 (gamma tubulin regulator) mutants exhibit reduced catastrophe frequencies [25]. Recruitment of bud6p occurred normally in tip1, mto1 and cls1/peg1 (CLASP) mutants [26, 27]
Recruitment is dependent on a new pathway that includes mal3p (EB1) and moe1p
We then screened other candidates for involvement in bud6p recruitment on the cell sides (Figure 6B and S9). Notably, the targeting of bud6-3GFP to the sides was strongly reduced in mal3Δ, moe1Δ and for3Δ mutants (Figure 6A and 6B). Importantly, in all these cases, cells could still localize bud6p properly at cell tips, but could not establish bud6p at the new site on the cell sides (Figure 6A and 6B). The recruitment of for3p was also inhibited in mal3Δ and moe1Δ mutants (Figure S8).
Mal3p (EB1 homolog) is implicated in regulation of MT plus end dynamics and may function in part to load other MT-associated proteins onto MTs [28]. MTs in mal3Δ mutants are short, due to premature catastrophe events, but display normal growth and shrinkage rates and still contact the cell sides in the bent cells, albeit closer to the nucleus (Figure S10). These MT defects are very similar to those seen in tip1 mutants, which have no bud6p localization defects, suggesting that the mal3 defects are not solely due to abnormal MT behavior.
Moe1p has been shown to interact directly with mal3p [29] as well as other factors (see Discussion). moe1Δ mutants have abundant interphase MT bundles of normal length and dynamics [29] (Figure S10), showing that its defects in bud6p recruitment are downstream of MTs and probably also of mal3p. Moe1p could regulate formin for3p through its interactions with scd1p, a cdc42p GEF [6, 30]. In addition, we found that for3p co-immunoprecipitates with moe1p in yeast extracts (Figure S11A). However, the significance of this interaction between moe1p and for3p remains to be tested.
For3p may recruit bud6p directly. These proteins directly interact [6, 31]. Inhibition of actin by LatA did not inhibit bud6p recruitment, so the effects of for3p in this process are actin-independent. It is interesting to note that in normal cells, bud6p localization is independent of for3p [31], while in bent cells, it is strictly dependent. These different relationships highlight how the assembly pathways differ in these circumstances.
Taken together, these results provide the outlines of a new pathway in which MT plus ends, in concert with mal3p, moe1p and for3p, act to recruit bud6p to these cortical sites (Figure 6C). This new pathway is functionally distinct from the tea1p-based pathway. In particular, moe1 and mal3 single mutants exhibited no NETO defects. Further, characterization of mal3 tea1 and moe1 tea1 double mutants did not reveal synthetic effects on cell shape or NETO (Figure S11B). Thus, this new mal3p–moe1p pathway may play a role more specifically in regulating polarity factors on the sides of cells.
MTs specify sites of ectopic cell growth in a tea1Δ background
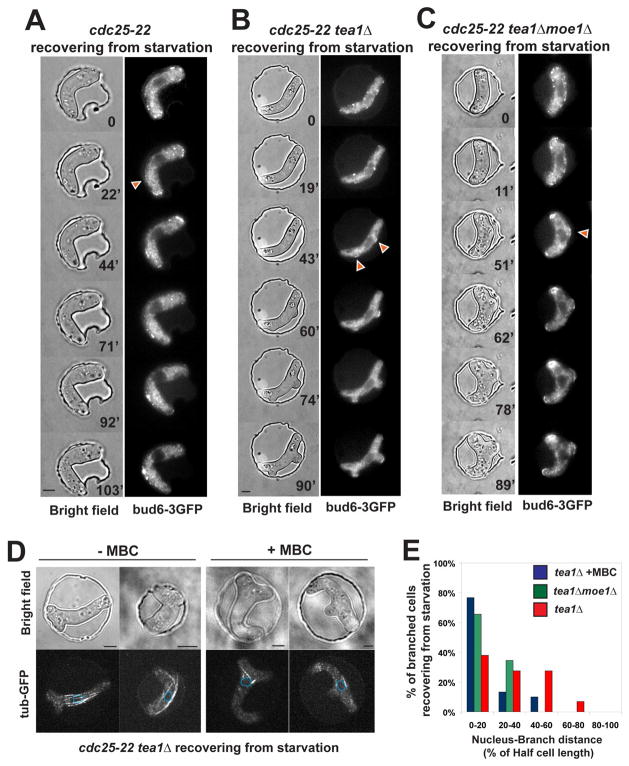
We investigated why ectopic cell growth did not occur in the bent cells. It is possible that cell growth from the sides of cells is inhibited by a stress response, local damage to the cortical regions, steric restraints from the well, or by factors in the cell that discourage growth from cell sides. Tea1p, in addition to its role in NETO, is known to have a role in inhibiting cell branching, especially upon recovery from starvation [32]. We thus examined the effect of tea1p on bent cells recovering from starvation. We observed ectopic growth in a tea1Δ cdc25 background (85.7%, n=56), but not in a tea1+ cdc25 background (0%, n>100) (Figure 7A and 7B). Cells formed branches that often pushed the cell backward into the well (Movie 12), even when the cell was initially plastered against the side of the chamber (data not shown). Thus the presence of tea1p, and not some physical constraint or some putative damage to the plasma membrane or cell wall, is responsible for repressing branching in these bent cells.
Figure 7. MTs position ectopic branching in tea1Δ cells in a moe1p dependent manner.
(A) cdc25-22 cell in a wildtype background (at 25°C) expressing bud6-3GFP recovers from starvation in a bent conformation in a chamber. The cell grows at the tips while bud6p is ectopically recruited (arrowhead). Strain: NM01. (B) cdc25-22 tea1Δ cell (at 25°C) expressing bud6-3GFP recovers from starvation in a bent conformation in a chamber. The cell grows at the ectopic sites where bud6p was recruited (arrowheads). Strain: NM14. (C) cdc25-22 tea1Δ moe1Δ cell (at 25°C) expressing bud6-3GFP recovers from starvation in a bent conformation in a chamber. Strain: NM151. (D) cdc25-22 cells in a tea1Δ background (at 25°C) expressing GFP-tubulin upon recovery from starvation in a bent conformation imposed by the chamber ±MBC. The nuclear position is indicated by the blue dotted lines. Strain: NM20. (E) Effect of MBC and moe1p on the absolute distance from the nucleus to the branching sites in cdc25-22 tea1Δ cells recovering from starvation in a bent conformation in a chamber. Strain: NM20. Scale Bars = 5μm.
These sites of cell branching in the tea1 mutants were situated at predicted sites of MT contact on the outer curvature (98%, n=49) and were anticipated by accumulation of bud6p at these sites (Figure 7B and movie 10). It is likely that this ectopic growth is driven by the collection of polarity factors at these sites, as branch formation in tea1 mutants is formin-dependent [31]. The positions of these growth sites were dependent on MTs. Similar to what was seen in straight cells [33], when the bent tea1 cells were treated with MBC, the positions of the branching were situated significantly closer to the nucleus and sometimes even on the inner curvature of the bent cell (15.8%, n=57) (Figure 7C and 7D). Similarly, moe1 tea1 double mutant cells, which have intact MTs, also formed branches (75.6%, n=41), and at sites closer to the nucleus (Figure 7B and 7D), and sometimes on the inner curvature (34.7%, n=32). These distributions indicate that the placement of these ectopic growth sites at the sites of MT contact is dependent on MTs and moe1p.
These results suggest that there are at least three competing layers of polarity regulation: 1) a MT–tea1p-dependent pathway that promotes tip growth; 2) a MT–moe1p–mal3p-dependent pathway that promotes polarization on the cell sides; 3) a “nucleus” pathway that promotes branching near the nucleus when the other two pathways are absent [33]. All three pathways ultimately act to recruit bud6p and other polarity factors to discrete sites on the cortex.
DISCUSSION
Manipulating cell shape
The physical manipulation of cells provides an important tool in cell biology. Here, we devised a new way to manipulate the shape of single living yeast cells using wells that have a volume as small as a femtoliter (10−15 l). The method is robust and simple and allows for near real-time imaging. We find that changes in cell shape produce rapid reorganization of the cytoskeleton and cortical components. The MT bundles largely maintain their straight axis, and now contact the sides of these bent cells and begin to establish new sites of actin assembly. The properties of these MT bundles—their flexural rigidity and their cortical interactions—in the context of this small cell size may begin to reveal how these cells adopt a straight axis. The influence of MTs on the cortex, and vice versa, are key processes of cell morphogenesis.
Regulation of MT catastrophe at the cortex
The distribution and orientation of MTs are dependent on spatial regulation of MT catastrophe. This work further provides new insights into this process. We find that MTs repeatedly attach to and shrink from the same cortical “hot spots” repeatedly. These cortical sites may act as transient “attachment” sites that stop a MT from sliding. They exist in bent cells on the cell sides, and preliminary data suggests that they may also exist in normal cells at cell tips. We speculate that these sites could represent simply a “ditch” or a “bump” on the cortex, or may represent clusters of protein complexes that bind to MTs. It would be interesting to test whether the tea1p protein complex itself, which marks sites of previous catastrophe, could serve in MT attachment.
The existence of these attachment sites alters views of current models for how MT catastrophe occurs selectively at cell tips in normal cells. The “force-based” model is that compressive force from MTs contacting and growing at the cortex is the primary factor in producing catastrophes [34]; this model postulates that MTs tend to exhibit catastrophe at the cell tips because that is where MTs encounter the highest angle of contact and force, due to the geometry of the rod-shape. We find in the bent cells that a near perpendicular angle at which a MT contacted the cortex is not necessary to induce catastrophe. MTs generally slide along the side of the cell at a shallow angle before stopping and shrinking. This behavior is reminiscent of in vitro experiments in which MTs were caught by some discontinuity of the surface before shrinking [34, 35]. Thus, in addition to the effects of global geometry, local nano-scale properties may also be a critical parameter for specifying catastrophe behaviors.
A “catastrophe factor” model is that there are catastrophe-promoting factors specifically at cell tips (although such factors have not been identified) [4]. Our data show that catastrophe, is not restricted to cell tips in the bent cells. It is possible that the bending process may delocalize these putative catastrophe factors, or that MTs deliver them to the ectopic sites on the cell sides. However, we note that the initial MTs upon the first minutes of cell bending already show altered sites of catastrophe.
Similar regulation of MT stability may also occur in mammalian cells [36]. Recent work shows that MTs display catastrophe upon contact with focal adhesions in a paxillin-dependent manner [37], or are transiently stabilized at cortical sites in a CLASP-dependent manner [38]. MT attachment to the cortex occurs at selective regions of the cell, for instance at the leading edge and also at the cleavage furrow [39, 40].
Polarity establishment by MTs
Here, we directly observed that the contact of a MT plus end with the cortex actively leads to the recruitment of polarity factors within seconds. Although the cortical deposition of MT-associated factors such as tea1p has been observed [31], these are the first direct observations, at least in fission yeast, to visualize and time the recruitment of a non-MT associated protein (bud6p) to a site of MT contact.
One surprising result is that rather than using the established tea1p–tea4p pathway, MTs in these bent cells can regulate formin activity through another pathway. Our initial characterization coupled with published data suggests a pathway of the MT-plus-end–mal3p–moe1p–scd1p–cdc42p–for3p–bud6p. The role of mal3p (EB1) in loading other proteins onto the MT plus end is relatively well characterized [41, 42]. Moe1p has been found to bind to mal3p in numerous assays [30]. Moe1p also binds to scd1p, the GEF for cdc42p, and thus could regulate cell polarity and formins via these proteins. moe1 mutants display slightly fatter cell shape and occasional cell bending indicative of functions in cell polarity [29]. It does not obviously affect MT dynamic behaviors (although it does affect MT bundle number), and is localized diffusely in the cytoplasm. Other studies have shown that moe1p may have additional functions, as it associates with the proteasome and with the translational initiation factor eIF3, although it does not grossly affect translation initiation [43, 44].
In animal cells, there are beginning to be outlines of similar pathways that link MTs with formins. In cytokinesis, EB1 binds to Rho-GEF at MT plus ends and may locally activate Rho1 on the cortex, possibly leading to formin activation [45]. In addition, the formin mDia has been shown to bind to EB1 and APC (an EB1-interacting protein), as well as to MTs directly [2, 46]. Therefore, although orthologs of the S. pombe tea1p–tea4p pathway have not been yet defined, the mal3p–moe1p pathway has some similarities with this animal cytokinesis pathway.
Cell bending response
These studies reveal the existence of at least two distinct pathways in S. pombe for linking MTs with cell polarity. The tea1p–tea4p pathway is needed for NETO and for maintaining cell growth at the cell tips [3]. The mal3p–moe1p pathway is not genetically redundant with the tea1p–tea4p pathway and does not appear to operate in NETO, but is required at least for guiding the site of actin assembly from the sides of the bent cells. Since MTs are not constantly inducing cell polarization at both cell tips, such as in monopolar pre-NETO cells, the mal3p–moe1p pathway may be turned on only in particular circumstances or cell-cycle phases.
One clear physiological function of the mal3p-moe1p pathway in fission yeast is to respond to cell bending. In nature, fungal cells may find itself in a hole and encounter forces that cause them to bend, for instance, from force of their own growth [47, 48]. Growth at the sides of cells may allow the cell to grow out of such a confined environment. One intriguing possibility then is that there are different pathways for responding to specific cell shapes. Further investigations into the crosstalk between cell shape, polarity mechanisms, and MTs promise to elucidate new concepts in cellular morphogenesis.
MATERIAL AND METHODS
Microchambers Fabrication and Operation
Chambers containing the arrays of microwells were fabricated by rapid prototyping and PDMS technology [49]. A master composed of a positive relief of SU-8 resin on a silicon wafer was first made by microlithography. This master is composed of several arrays containing thousands of 5μm high cylindrical posts with varying diameters (10-12-15-20-25-30-40 and 50μm). A 10:1 mixture of PDMS Sylgard 184 silicone elastomer and curing agent was then poured onto the master and subsequently baked at 50°C for 6h. The replica was finally cut and peeled off the master. A 2μL drop of cells in liquid media was then placed between the PDMS replica and a 22mm2 coverslip. Depending on the application, the coverslip was either gently pressed with the finger or by over-focusing the objective of an upright microscope, leading to an instantaneous bending of the cells.
Microscopy and Image Analysis
Microscopy was performed at room temperature (25–27°C) with either an upright wide-field fluorescence microscope or a spinning-disk confocal fluorescence microscope. Bud6-3GFP confocal stacks presented throughout the manuscript are made of 6 z-sections spaced by 1μm taken with 1s exposure. GFP-tubulin confocal stacks are made of 12 z-sections spaced by 0.5μm taken with 0.5s exposure. Images were acquired with OpenLab 4.0.4 (Improvision) and processed and analyzed with OpenLab, Image J, and Matlab. FRAP experiments were performed on a Zeiss LSM 510 confocal microscope.
Data Quantification
MT positions were tracked manually. The cell tip was considered as the zone encompassing 20% of the cell length. The percentage of bent cells having a GFP fused protein specifically targeted to the outer curvature was also counted manually. Cells were positively counted upon 15 min of bending on the chamber, when the protein was solely visible at the outer curvature of the bent cell with possible extension to the tip. Cells with a wide delocalized signal and those with a obvious kink in the inner curvature (<15% cells) were not included in the analysis.
Yeast Strains, Media, and Genetic Methods
Strains used in this study are listed in Table S1. Standard methods for S. pombe media and genetic manipulations were used.
Pharmacological Inhibitors and Drugs
Methyl-2-benzimidazole carbamate (MBC, Aldrich) was used at a final concentration of 50μg/ml from a 100X stock solution made fresh in DMSO. Latrunculin A (LatA, Sigma) was used at a final concentration of 100μM from a 200X stock in DMSO. In the data presented, these drugs were added to the media 10 min prior to introducing the cells in the chambers. Hydroxyurea (HU) was used at a final concentration of 25mM from a 60X stock solution in water and was added to the cell 3h before observation.
Supplementary Material
Acknowledgments
The authors acknowledge all members of the Chang laboratory for discussions and technical assistance. We thank K. Sawin, S. Martin, P. Perez and J.C. Ribas for strains, the Sheetz laboratory for technical support, as well as M. Merle and N. Galfard for discussions. Microfabrication was made in the Columbia CEPSR clean room. This work was supported by National Institutes of Health (NIH) GM056836 and GM069670.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes Dev. 2007;21:483–496. doi: 10.1101/gad.1511207. [DOI] [PubMed] [Google Scholar]
- 2.Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 3.Martin SG, Chang F. New end take off: regulating cell polarity during the fission yeast cell cycle. Cell Cycle. 2005;4:1046–1049. [PubMed] [Google Scholar]
- 4.Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. doi: 10.1016/s0092-8674(00)00091-x. [DOI] [PubMed] [Google Scholar]
- 5.Chang F, Peter M. Yeasts make their mark. Nat Cell Biol. 2003;5:294–299. doi: 10.1038/ncb0403-294. [DOI] [PubMed] [Google Scholar]
- 6.Martin SG, Rincon SA, Basu R, Perez P, Chang F. Regulation of the formin for3p by cdc42p and bud6p. Mol Biol Cell. 2007;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang F. Establishment of a cellular axis in fission yeast. Trends Genet. 2001;17:273–278. doi: 10.1016/s0168-9525(01)02279-x. [DOI] [PubMed] [Google Scholar]
- 8.Martin SG, McDonald WH, Yates JR, 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev Cell. 2005;8:479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 10.Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, Theriot JA. Mechanism of shape determination in motile cells. Nature. 2008;453:475–480. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thery M, Racine V, Piel M, Pepin A, Dimitrov A, Chen Y, Sibarita JB, Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc Natl Acad Sci U S A. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verde F, Mata J, Nurse P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carazo-Salas RE, Nurse P. Self-organization of interphase microtubule arrays in fission yeast. Nat Cell Biol. 2006;8:1102–1107. doi: 10.1038/ncb1479. [DOI] [PubMed] [Google Scholar]
- 14.Daga RR, Lee KG, Bratman S, Salas-Pino S, Chang F. Self-organization of microtubule bundles in anucleate fission yeast cells. Nat Cell Biol. 2006;8:1108–1113. doi: 10.1038/ncb1480. [DOI] [PubMed] [Google Scholar]
- 15.Smith AE, Zhang Z, Thomas CR, Moxham KE, Middelberg AP. The mechanical properties of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2000;97:9871–9874. doi: 10.1073/pnas.97.18.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moseley JB, Goode BL. Differential activities and regulation of Saccharomyces cerevisiae formin proteins Bni1 and Bnr1 by Bud6. J Biol Chem. 2005;280:28023–28033. doi: 10.1074/jbc.M503094200. [DOI] [PubMed] [Google Scholar]
- 17.Jin H, Amberg DC. Fission yeast Aip3p (spAip3p) is required for an alternative actin-directed polarity program. Mol Biol Cell. 2001;12:1275–1291. doi: 10.1091/mbc.12.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glynn JM, Lustig RJ, Berlin A, Chang F. Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast. Curr Biol. 2001;11:836–845. doi: 10.1016/s0960-9822(01)00235-4. [DOI] [PubMed] [Google Scholar]
- 19.Feierbach B, Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- 20.Martin SG, Chang F. Dynamics of the formin for3p in actin cable assembly. Curr Biol. 2006;16:1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- 21.Tatebe H, Nakano K, Maximo R, Shiozaki K. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr Biol. 2008;18:322–330. doi: 10.1016/j.cub.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motegi F, Arai R, Mabuchi I. Identification of two type V myosins in fission yeast, one of which functions in polarized cell growth and moves rapidly in the cell. Mol Biol Cell. 2001;12:1367–1380. doi: 10.1091/mbc.12.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cortes JC, Carnero E, Ishiguro J, Sanchez Y, Duran A, Ribas JC. The novel fission yeast (1,3)beta-D-glucan synthase catalytic subunit Bgs4p is essential during both cytokinesis and polarized growth. J Cell Sci. 2005;118:157–174. doi: 10.1242/jcs.01585. [DOI] [PubMed] [Google Scholar]
- 24.Wachtler V, Rajagopalan S, Balasubramanian MK. Sterol-rich plasma membrane domains in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 2003;116:867–874. doi: 10.1242/jcs.00299. [DOI] [PubMed] [Google Scholar]
- 25.Zimmerman S, Chang F. Effects of {gamma}-tubulin complex proteins on microtubule nucleation and catastrophe in fission yeast. Mol Biol Cell. 2005;16:2719–2733. doi: 10.1091/mbc.E04-08-0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bratman SV, Chang F. Stabilization of overlapping microtubules by fission yeast CLASP. Dev Cell. 2007;13:812–827. doi: 10.1016/j.devcel.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grallert A, Beuter C, Craven RA, Bagley S, Wilks D, Fleig U, Hagan IM. S. pombe CLASP needs dynein, not EB1 or CLIP170, to induce microtubule instability and slows polymerization rates at cell tips in a dynein-dependent manner. Genes Dev. 2006;20:2421–2436. doi: 10.1101/gad.381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beinhauer JD, Hagan IM, Hegemann JH, Fleig U. Mal3, the fission yeast homologue of the human APC-interacting protein EB-1 is required for microtubule integrity and the maintenance of cell form. J Cell Biol. 1997;139:717–728. doi: 10.1083/jcb.139.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CR, Chen J, Chang EC. A conserved interaction between Moe1 and Mal3 is important for proper spindle formation in Schizosaccharomyces pombe. Mol Biol Cell. 2000;11:4067–4077. doi: 10.1091/mbc.11.12.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen CR, Li YC, Chen J, Hou MC, Papadaki P, Chang EC. Moe1, a conserved protein in Schizosaccharomyces pombe, interacts with a Ras effector, Scd1, to affect proper spindle formation. Proc Natl Acad Sci U S A. 1999;96:517–522. doi: 10.1073/pnas.96.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feierbach B, Verde F, Chang F. Regulation of a formin complex by the microtubule plus end protein tea1p. J Cell Biol. 2004;165:697–707. doi: 10.1083/jcb.200403090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawin KE, Snaith HA. Role of microtubules and tea1p in establishment and maintenance of fission yeast cell polarity. J Cell Sci. 2004;117:689–700. doi: 10.1242/jcs.00925. [DOI] [PubMed] [Google Scholar]
- 33.Castagnetti S, Novak B, Nurse P. Microtubules offset growth site from the cell centre in fission yeast. J Cell Sci. 2007;120:2205–2213. doi: 10.1242/jcs.03464. [DOI] [PubMed] [Google Scholar]
- 34.Janson ME, de Dood ME, Dogterom M. Dynamic instability of microtubules is regulated by force. J Cell Biol. 2003;161:1029–1034. doi: 10.1083/jcb.200301147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- 36.Gundersen GG. Evolutionary conservation of microtubule-capture mechanisms. Nat Rev Mol Cell Biol. 2002;3:296–304. doi: 10.1038/nrm777. [DOI] [PubMed] [Google Scholar]
- 37.Efimov A, Schiefermeier N, Grigoriev I, Brown MC, Turner CE, Small JV, Kaverina I. Paxillin-dependent stimulation of microtubule catastrophes at focal adhesion sites. J Cell Sci. 2008;121:196–204. doi: 10.1242/jcs.012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lansbergen G, Grigoriev I, Mimori-Kiyosue Y, Ohtsuka T, Higa S, Kitajima I, Demmers J, Galjart N, Houtsmuller AB, Grosveld F, et al. CLASPs attach microtubule plus ends to the cell cortex through a complex with LL5beta. Dev Cell. 2006;11:21–32. doi: 10.1016/j.devcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Lansbergen G, Akhmanova A. Microtubule plus end: a hub of cellular activities. Traffic. 2006;7:499–507. doi: 10.1111/j.1600-0854.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- 40.Canman JC, Cameron LA, Maddox PS, Straight A, Tirnauer JS, Mitchison TJ, Fang G, Kapoor TM, Salmon ED. Determining the position of the cell division plane. Nature. 2003;424:1074–1078. doi: 10.1038/nature01860. [DOI] [PubMed] [Google Scholar]
- 41.Browning H, Hackney DD, Nurse P. Targeted movement of cell end factors in fission yeast. Nat Cell Biol. 2003;5:812–818. doi: 10.1038/ncb1034. [DOI] [PubMed] [Google Scholar]
- 42.Bieling P, Laan L, Schek H, Munteanu EL, Sandblad L, Dogterom M, Brunner D, Surrey T. Reconstitution of a microtubule plus-end tracking system in vitro. Nature. 2007;450:1100–1105. doi: 10.1038/nature06386. [DOI] [PubMed] [Google Scholar]
- 43.Bandyopadhyay A, Lakshmanan V, Matsumoto T, Chang EC, Maitra U. Moe1 and spInt6, the fission yeast homologues of mammalian translation initiation factor 3 subunits p66 (eIF3d) and p48 (eIF3e), respectively, are required for stable association of eIF3 subunits. J Biol Chem. 2002;277:2360–2367. doi: 10.1074/jbc.M107790200. [DOI] [PubMed] [Google Scholar]
- 44.Yen HC, Gordon C, Chang EC. Schizosaccharomyces pombe Int6 and Ras homologs regulate cell division and mitotic fidelity via the proteasome. Cell. 2003;112:207–217. doi: 10.1016/s0092-8674(03)00043-6. [DOI] [PubMed] [Google Scholar]
- 45.Rogers SL, Wiedemann U, Hacker U, Turck C, Vale RD. Drosophila RhoGEF2 associates with microtubule plus ends in an EB1-dependent manner. Curr Biol. 2004;14:1827–1833. doi: 10.1016/j.cub.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 46.Rogers SL, Wiedemann U, Stuurman N, Vale RD. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J Cell Biol. 2003;162:1079–1088. doi: 10.1083/jcb.200303023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bastmeyer M, Deising HB, Bechinger C. Force exertion in fungal infection. Annu Rev Biophys Biomol Struct. 2002;31:321–341. doi: 10.1146/annurev.biophys.31.091701.170951. [DOI] [PubMed] [Google Scholar]
- 48.Minc N, Boudaoud A, Chang F. Submitted. [Google Scholar]
- 49.Weibel DB, Diluzio WR, Whitesides GM. Microfabrication meets microbiology. Nat Rev Microbiol. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.