Abstract
Telomerase plays a critical role in cancer, prompting the pursuit of various telomerase-based therapeutic strategies. One such strategy, telomerase-interference, exploits the high telomerase activity in cancer cells and reprograms telomerase to encode “toxic” telomeres. To date, telomerase interference has been tested in human cancer cells xenografted into mice, an approach that does not recapitulate spontaneous malignancy and offers few insights about host toxicities, because human telomerase is targeted in a mouse host. To address these limitations, we designed and validated two new gene constructs specifically targeting mouse telomerase: Mutant template mouse telomerase RNA (MT-mTer) and siRNA against wild type mouse telomerase RNA (α-mTer-siRNA). Using lentiviral delivery in mouse prostate cancer cells, we achieved α-mTer-siRNA-mediated knockdown of wild type mTer (80% depletion) and concurrent overexpression of MT-mTer (50-fold). We showed that the two constructs effectively synergize to reprogram murine telomerase to add mutant instead of wild-type telomeric repeats, resulting in rapid telomeric uncapping (5-fold increase in DNA damage foci). This, in turn, led to rapid and significant apoptosis (>90% of cells) and growth inhibition in vitro (90% reduction in viable cell mass) and in vivo (75% reduction in tumor allograft wet weight). In summary, we have demonstrated that mouse cancer cells are vulnerable to direct telomerase interference using novel murine telomerase targeting constructs; this approach can now be used to study the true therapeutic potential of telomerase interference in mouse spontaneous cancer models.
Keywords: telomerase targeting, prostate cancer, mouse cancer models
Introduction
The enzyme telomerase preserves cells’ proliferative capacity by lengthening and protectively “capping” telomeres, the tandem repetitive DNA sequences at the ends of human chromosomes (1). It is a ribonucleoprotein consisting of two core components: a reverse transcriptase protein (TERT) and telomerase RNA (Ter). Ter contains a short template sequence used by TERT to synthesize telomeric DNA (2). Whereas benign, terminally differentiated tissues have extremely low telomerase levels (3), malignant cells from a variety of cancers have significant telomerase expression and activity levels that correlate directly with malignant/metastatic potential (4–6). Attenuation of human telomerase function in cancer cells has produced apoptosis and growth inhibition (7–9), underscoring the great clinical promise of this therapeutic approach.
Previously, we and others have explored various telomerase-based strategies against human cancer cells (7, 10–13) and tested them in vivo by targeting human telomerase in cancer cells xenografted into mice (7, 10, 11, 13). Telomerase depletion also has been studied in knockout models that lack telomerase entirely (14, 15). Such xenograft and knockout models necessitate a leap of faith: that the observed sequelae of telomerase manipulation will accurately predict efficacy and toxicity in an actual host with spontaneous malignancy and normal telomerase function. Such an assumption is particularly tenuous in the field of telomerase targeting, because telomerase is known to play important roles both in tumors and in normal progenitor tissue compartments (5, 16–18).
In this study, we set out to engineer and validate two new gene constructs that can effectively reprogram mouse telomerase: 1. Short hairpin RNA against wild-type mouse telomerase RNA (α-MTer-siRNA), and 2. Mutant-template mouse telomerase RNA (MT-mTer) which encodes incorrect mouse telomeric repeats. When co-expressed in a mouse prostate cancer cell line derived from the cPten−/− mouse (19–21), α-MTer-siRNA and MT-mTer co-opt the activity of mouse telomerase and reprogram it to encode an altered telomeric sequence, eliciting a rapid cascade of telomeric uncapping, cellular apoptosis, and growth inhibition in vitro and in vivo. These experiments validate the mechanism of action and biologic efficacy of α-mTer-siRNA and MT-mTer in mouse cancer cells, and these new constructs can now be used to study the systemic efficacy and toxicities of telomerase targeting in mouse models of spontaneous malignancy.
Materials and Methods
Plasmid Construction
The siRNA lentiviral vector was generated by PCR with U6 promoter as the template using the 5′ primer 5′-GGACTAGTAAGG TCGGGCAGGA AGAGGGC-3′ and the 3′ primer 5′-AAAACTGCAGAAAAATTA CCTAACCCTGATTTTCATTCTCTTGAAATGAAAATCAGGGTTAGGTGGTGTT TCGTCCTTTCCACAAG-3′ and inserted into SpeI/PstI sites of pHR’CMVPuroWsin18 (11, 22). The mutant mouse Ter expression construct (pIU1-MT-mTer) was PCR cloned using the 5′ primer 5′-GGATCC ACCAAACCC AGATTTTCATTAGCT-3′ (mutated sites in bold) and the 3′ primer 5′-CTC GAGG GTTGTGAGAACCGAGTTCCG-3′, subcloned into BglII/SalI sites in pIU1-T7 vector and then into pHR’CMVPuroWsin18 vector to generate Lenti-MT-mTer. To generate MT-mTer/siRNA vector, f1 origin from pBluescript (Promega, Madison, WI) was PCR amplified using the 5′ primer 5′-AGATCTTTGTTCCAGTTTGGAA CAAGAGT-3′ and the 3′ primer 5′-GAATTCGCATTAAGCGCGGCG-3′, and inserted into BamHI/EcoRI sites of pIU1-MT-mTer. The DNA fragment containing f1 origin and MT-mTer was subcloned into α-mTer-siRNA.
Cell culture
Mouse prostate cancer cell line, E4 was generously provided by Dr. Pradip Roy-Burman (University of Southern California, Los Angeles, CA). Cells were cultured at 37°C, 5% CO2 in DMEM (Cellgro, Manassas, VA) with 10% fetal bovine serum (Omega Scientific, Tarzana, CA), 5 μg/ml Insulin (Sigma, St. Louis, MO), 25 μg/ml bovine pituitary extract, 6 ng/ml rEGF, 100 units/ml penicillin and 100 μg/ml streptomycin (Invitrogen, Carlsbad CA).
Virus Production and infection
Lentivirus was generated as previously described (23). Virus-containing supernatant from several different preps was combined and concentrated by centrifugation at 28,000 RPM (Beckman, SW32T rotor, Fullerton, CA) for 2 hr and resuspended in culture medium and used to infect E4 cells as described previously (10).
RNA extraction, reverse transcription, and PCR
Total RNA from infected cells was extracted using RNA-Bee reagent (TEL-TEST, Friendswood, TX). First strand cDNA was synthesized using the RETROscript reverse transcription kit (Applied Biosytems Inc, Foster City, CA). To differentiate MT-mTer expression from wild type mTer, PCR was designed using one primer against the mTer coding region and one against a sequence that is downstream of the mTer coding region in the construct. This extra sequence resulted from pIU1-T7 vector and was located before the transcription termination signal; thus it was transcribed in MT-mTer while not included in endogenous WT-mTer (see Supplementary Table 1 Online for PCR primer sequences).
Real-time PCR
Real-time PCR for mTer quantitation was performed using B-R SYBR Green Supermix for iQ (Quanta BioSciences, Gaithersburg, MD; see Supplementary Table 1 Online for real-time primer sequences) on a MyiQ single color real-time PCR detection system (Bio-Rad, Hercules, CA) for 40 cycles at 95°C for 10s, 55 °C for 45s. The iQ5 optical system software version 2.0 was used to analyze results as normalized to β-actin internal controls.
Telomeric Repeat Amplification Protocol (TRAP) assay
Telomerase activity from cell extracts was analyzed using real-time PCR based telomeric repeat amplification protocol (TRAP) on a Bio-Rad MyiQ system as described (24). For the second (amplification) step, two reverse primers were used: ACX reverse primer specific for wild-type TTAGGG encoded by endogenous WT-mTer, and 4A10A reverse primer specific for mutant TTTGGG encoded by MT-mTer (see Supplementary Table 2 Online for oligo sequences). The reactions were run for 40 cycles at 95°C, 0″; 50°C, 5″; 72°C, 10″ to amplify WT-mTer product (TTAGGG) or MT-mTer product (TTTGGG), respectively. The iQ5 optical system software version 2.0 was used to analyze the results.
Telomere length assay
Genomic DNA was extracted from infected E4 cells using Qiagen DNeasy Blood & Tissue Kit (Qiagen), and relative telomere lengths were analyzed in triplicate by real time PCR (Biorad MyIQ) as described previously using T and S primers (25), (see Supplementary Table 1 Online for T and S primer sequences).
Cell viability and apoptosis assays
Assays were performed on E4 cells 3 and 4 days post infection using MTS ((Promega, Madison, WI), In Situ Cell Death Detection Kit (TUNEL) and Fluorescein and Homogeneous Caspases Assay Kit (Roche Applied Science, Indianapolis, IN) per the manufacturer protocols. For TUNEL assays, fluorescence activated cell sorting (FACS) was performed on BD LSR-II (BD Biosciences). For MTS and caspase assays, the optical density (OD) was measured at 490 nm by Plate Chameleon Multi-technology plate-reader (Hidex, Turku, Finland).
Immunofluorescence microscopy
104 E4 cells were plated on coverslips in 12 well plates and infected with lentivirus overnight in the presence of 8 μg/ml polybrene. On day 4, cells were stained as described previously (10) using primary antibodies against p53BP1 (Bethyl Laboratories) and TRF2 (BD Biosciences), secondary antibodies conjugated to Alexa Fluor 647 (Invitrogen) and Alexa Fluor 568 (Invitrogen), and DAPI for DNA visualization. p53BP1 foci were enumerated using a Zeiss Imager.Z1 microscope with Axiovision software at 63X magnification. Confocal colocalization was analyzed using Zeiss LSM 510 with Zeiss LSM 510 software at 63X magnification.
Subcutaneous tumor allografts
6–8 week old, male NOD-SCID mice were purchased from NIH. E4 cells were infected overnight with control or MT-mTer/siRNA lentiviruses and cultured at 37°C, 5% CO2 for 1 day after changing media. Before inoculation into mice, total RNA and genomic DNA from E4 cells was isolated as described above to check the expression of mTer. For each mouse, 106 cells were resuspended in media, mixed with 50 μl ice-cold matrigel (BD biosciences, San Jose, CA) and placed on ice until inoculation. 1 ml insulin syringe was used for subcutaneous inoculation onto the flank of each mouse (5 mice per treatment group). All experiments were approved and performed following the rules of the Institutional Animal Care and Use Committees at University of Southern California. Thirty days after inoculation, mice were sacrificed, and tumors were resected and weighed. A portion of tumor tissues was fixed and used to make paraffin-embedded slides for Hematoxylin and Eosin (H&E) staining. Another portion of tumors from three mice were also digested with 1mg/ml collagenase (Sigma), 1 μg/ml DNAse I (Invitrogen) and 1 mg/ml Hyaluronidase (Sigma) at 37°C, 1 hr to obtain a single cell suspension. These cells were then stained with biotinylated antibodies against CD13, CD45, and Ter119 (BD biosciences), Streptavidin-PE/Cy5 secondary antibody against biotinylated antibodies, Sca-1-PE and CD49f-Alexa Fluor 647 (Biolegen). Lin-Sca-1+CD49f+ E4 cells were sorted by BD FACSAria (BD Biosciences). Genomic DNAs from FACS purified E4 cells were isolated and levels of genomically-integrated MT-mTer were measured by PCR, using the single copy gene 36B4 to control for DNA loading (see Supplementary Table 1 Online for PCR primer sequences).
Results
α-mTer-siRNA knocks down WT-mTer and inhibits telomerase activity in E4 mouse prostate cancer cells
We synthesized a short hairpin α-mTer-siRNA specifically targeting the template region of wild type mouse telomerase RNA (WT-mTer) component and cloned it into a three-plasmid-based lentiviral system under the U6 promoter (Figure 1A). Lentivirus bearing the α-mTer-siRNA was introduced into E4 cells, a mouse prostate cancer cell line derived from prostate tumors arising in the transgenic cPten−/− mouse prostate cancer model (generously provided by the laboratory of Dr. Pradip Roy-Burman). The E4 cells bear a Lin-Sca-1+CD49f+ surface marker signature, stain positive for androgen receptor (AR), strongly express telomerase, and are tumorigenic, based on data presented in this report, which are consistent with the detailed phenotypic and biologic characteristics of this novel cell line determined by the Roy-Burman group (manuscript in preparation).
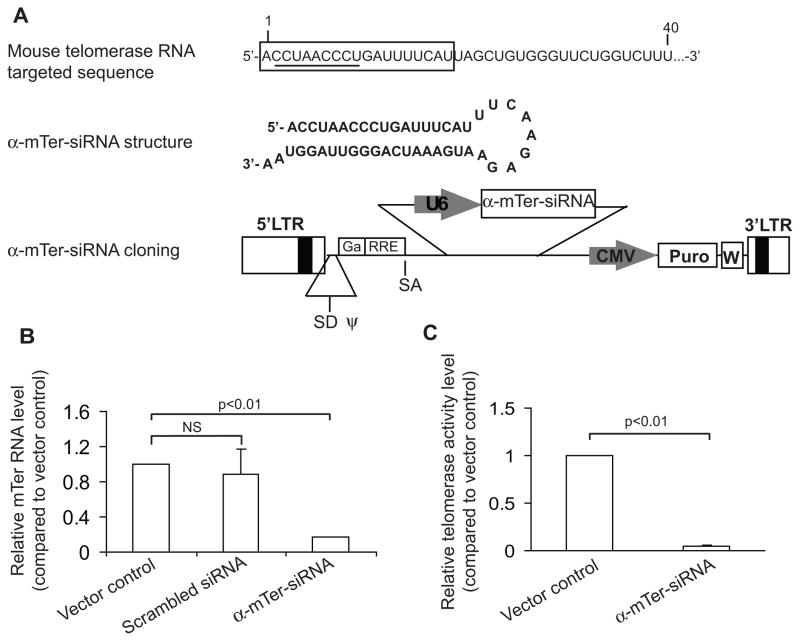
Figure 1. α-mTer-siRNA knocks down WT-mTer and inhibits telomerase activity in E4 mouse prostate cancer cells.
(A) α-mTer-siRNA structure and cloning into lentiviral vector system (target sequence is boxed, template region is underlined). (B) α-mTer-siRNA knocks down WT-mTer by 80% three days after expression in E4 cells, as quantified by real-time PCR (RT-PCR). (C) α-mTer-siRNA inhibits telomerase activity by 95% three days after expression in E4 cells, as quantified by real-time PCR telomeric repeat amplification protocol.
To confirm lentiviral infection and expression efficiency, we conducted an initial control experiment in which lentivirus containing only a puromycin selection marker (“vector control”) was used to infect E4 cells. After 4 days of puromycin selection, >95% of these cells remained viable, versus <5% viable cells in mock-infected E4 cells. Having confirmed >95% lentiviral infection and expression efficiency in this manner, we dispensed with the 4 day puromycin selection period in subsequent experiments and thus were able to measure the immediate short-term effects of telomerase interfering constructs relative to vector control.
Using real-time PCR, we found that cells infected with α-mTer-siRNA had an 80% depletion of endogenous WT-mTer 3 days after infection compared to cells infected with vector control or with a nonspecific scrambled siRNA (Figure 1B). Real-time PCR-based telomeric repeat amplification protocol (RT-PCR-TRAP) assays performed on cell protein extracts 3 days after infection showed a 95% reduction of telomerase activity in α-mTer-siRNA infected cells compared to cells infected with vector control (Figure 1C).
Mutant template telomerase RNA (MT-mTer) incorporates into mouse telomerase and reprograms it to add TTTGGG mutant telomeric repeats
Using PCR-based mutagenesis, we introduced two mutations (T→A) at the 4 and 10 positions of the mouse telomerase RNA gene. This new MT-mTer construct was cloned into lentiviral vector under the IU1 small nuclear RNA promoter and introduced into E4 target cells, where it was transcribed to generate mouse telomerase RNA with two mutations (U→A) in the template region (Figure 2A). The MT-mTer construct also was cloned into a lentiviral vector already containing α-mTer-siRNA for co-expression, so-called “MT-mTer/siRNA” (Figure 2A).
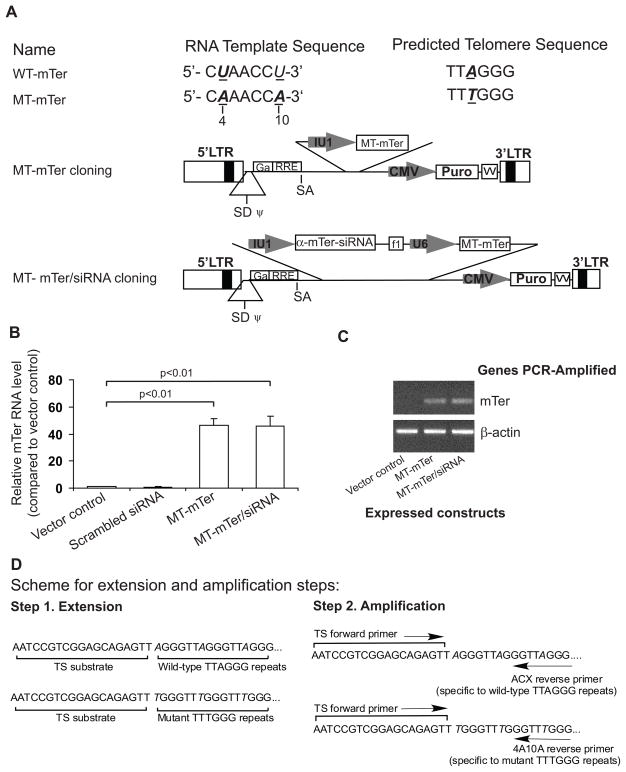
Figure 2. Mutant template telomerase RNA (MT-mTer) incorporates into active telomerase enzyme and reprograms it to add mutated telomeric repeats.
(A) MT-mTer and MT-mTer/siRNA structure and cloning into lentiviral vector system. (B) Ectopic over-expression (50 fold) of MT-mTer in E4 mouse prostate cancer cells three days after lentiviral infection, as quantified by RT-PCR. (C) MT-mTer levels by PCR are unaffected by co-expresseed α-mTer-siRNA, which is designed to knock down only the endogenous wild type mTer. (D) Experiment scheme of modified RT-PCR TRAP assay designed to specifically detect addition of wild-type TTAGGG vs. mutated TTTGGG nucleotide repeats.
Lentiviral introduction of the MT-mTer and MT-mTer/siRNA constructs into E4 cells achieved 50-fold over-expression of MT-mTer by real-time PCR relative to vector control or a scrambled constructs after 3 days (Figure 2B). The real-time PCR primers detected both MT-mTer and WT-mTer; therefore, we further confirmed the specific expression of MT-mTer by regular PCR using primers designed specifically for MT-mTer. This approach demonstrated that the α-mTer-siRNA component of MT-mTer/siRNA had no effect on the expression of MT-mTer and specifically knocked down only endogeneous WT-mTer (Figure 2C).
We investigated whether over-expressed MT-mTer RNA could successfully incorporate into an active telomerase enzyme and serve as a template for the addition of TTTGGG repeats in place of TTAGGG. For this purpose, we employed a modified RT-PCR-TRAP assay using the ACX and 4A10A reverse primers, which were specific for the telomeric repeats generated by endogenous WT-mTer or ectopically-expressed MT-mTer, respectively (experiment scheme depicted in Figure 2D and results summarized in Table 1). In TRAP assays for the detection of wild-type TTAGGG repeats (Table 1, left half), expression of MT-mTer alone had no effect on telomerase activity detected with the ACX wild-type-specific primer, while expression of MT-mTer/siRNA reduced wild-type telomerase activity to 5% of vector control as expected from α-Ter-siRNA knock-down of WT-mTer. In TRAP assays for the detection of mutated TTTGGG repeats (Table 1, right half), expression of either MT-mTer or MT-mTer/siRNA increased telomerase activity detected with the 4A10A mutant-specific primer by 2.7- and 3.6- fold, respectively, indicating that MT-mTer RNAs incorporated with mouse telomerase reverse transcriptase to form active telomerase enzyme capable of adding mutant TTTGGG tandem repeats. This effect was potentiated by concurrent siRNA-depletion of competing WT-mTer (hence 3.5-fold increase in mutant telomerase activity with MT-mTer/α-Ter-siRNA and only 2.7-fold with MT-mTer alone).
Table 1. Mutant template telomerase RNA (MT-mTer) reprograms telomerase enzyme to add mutated telomeric repeats.
When MT-mTer and MT-mTer/siRNA are over-expressed, significantly more telomeric products (270% and 350%, respectively) are detected using a TTTGGG-specific (“4A10A”) reverse primer in the second (amplification) step of RT-PCR TRAP, an effect which is preserved even when just dGTP and dTTP are added in the first (extension) step of the reaction. Activity values are % of vector control and are means of triplicates; all values differ from vector control with statistical significance (p<0.01).
| Telomerase activity (as % of vector control)* | ||||
|---|---|---|---|---|
| ** Amplification: ACX reverse primer (specific to wild-type TTAGGG) | ** Amplification: 4A10A reverse primer (specific to mutant TTTGGG) | |||
| ** Extension: 4 dNTPs | ** Extension: dGTP & dTTP | ** Extension: 4 dNTPs | ** Extension: dGTP & dTTP | |
| Vector Control | 100 | 0 | 0 | 5 |
| MT-mTer | 100 | 0 | 270 | 270 |
| MT-mTer/siRNA | 5 | 0 | 350 | 350 |
All activity values are means of triplicates
Extension and amplification steps illustrated in Figure 2D
We confirmed these findings by performing additional TRAP reactions using only dTTP and dGTP instead of all 4 dNTPs, thus taking advantage of the sequence difference between the wild-type (TTAGGG) and mutant-template (TTTGGG). This was accomplished by providing only dTTP and dGTP in the extension (first) step of the TRAP reaction, then supplementing dATP and dCTP for the amplification (second) step (illustrated in Figure 2D). As predicted, addition of dTTP and dGTP alone produced no detectable telomerase activity when the ACX (wild-type-specific) reverse primer was used. In contrast, telomerase activity was readily detectable from cells expressing MT-mTer when the 4A10A (mutant-specific) reverse primer was used in the presence of dTTP and dGTP alone (Table 1).
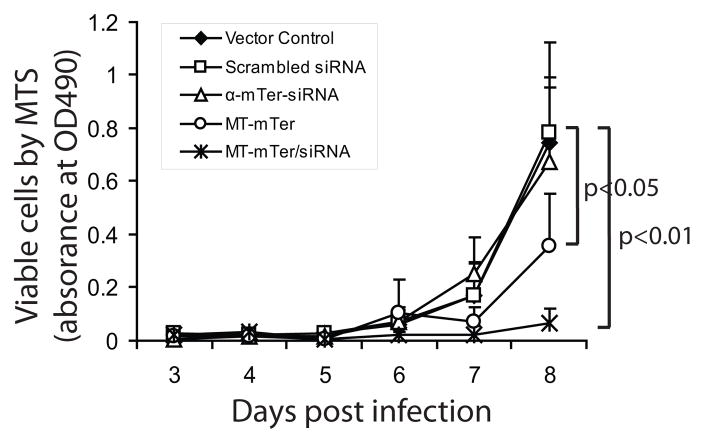
MT-mTer and α-mTer-siRNA inhibit E4 mouse prostate cancer proliferation in vitro and in vivo
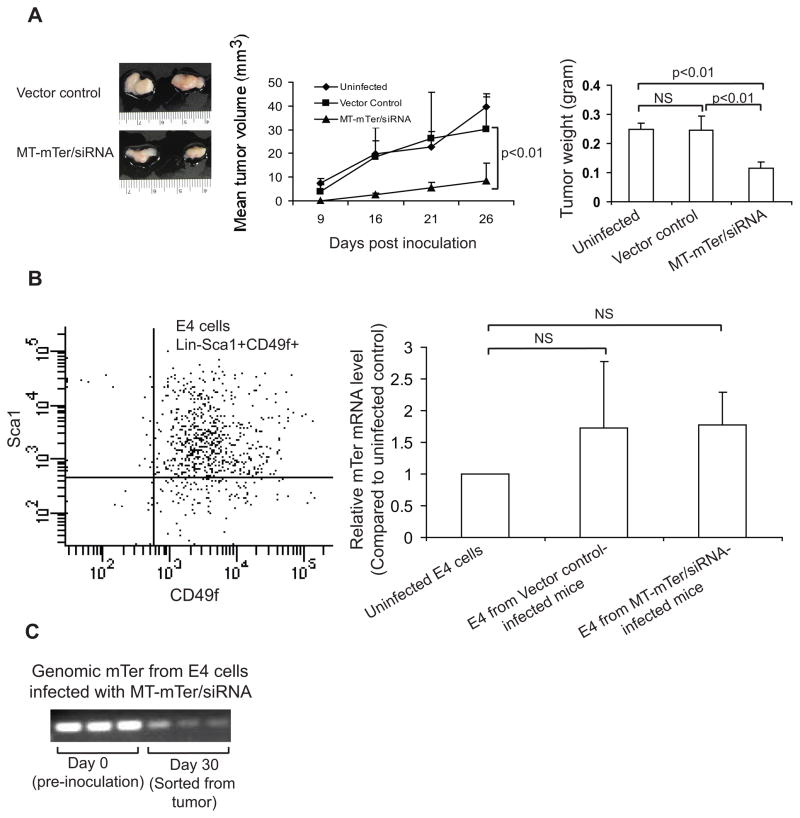
We introduced MT-mTer and α-mTer-siRNA by lentiviral infection into E4 cells and measured cell numbers daily by MTS cell viability assay. By day 7, α-mTer-siRNA alone had a modest effect (non-significant) on proliferation, while MT-mTer and MT-mTer/siRNA inhibited proliferation by 50% and 90%, respectively (Figure 3). As MT-mTer/siRNA had yielded the strongest mutant template TRAP activity (Table 1) and most significant in vitro inhibition of proliferation (Figure 3), we next tested if this construct could inhibit the growth of tumors in vivo as well. We infected E4 cells in vitro with lentivirus expressing MT-mTer/siRNA and subcutaneously allografted these cells into NOD-SCID mice; this treatment group was compared to 2 control groups inoculated either with vector control-infected cells or with uninfected cells (total of three groups, 5 mice per group). The growth of tumors was observed and recorded as tumor volume by caliper measurement (Figure 4A). Thirty days after inoculation, we sacrificed the mice and excised and recorded the wet weight of tumors, as this was a more accurate readout than the caliper measurements. Mice inoculated with E4 cells expressing MT-mTer/siRNA formed smaller tumors at all time points, and their excised final tumor weights were 50% smaller than those of the control groups (mean wet weight 0.12 g versus 0.25 g, p < 0.01, Figure 4A).
Figure 3. MT-mTer and MT-mTer/siRNA inhibit E4 mouse prostate cancer proliferation in vitro.
Cell growth by MTS colorimetric cell viability assay is significantly inhibited by day 7 after expression of MT-mTer or MT-mTer/siRNA constructs (50% and 90% reduction, respectively).
Figure 4. MT-mTer/siRNA inhibits E4 tumor growth.
(A) Left: Representative samples of MT-mTer/siRNA or vector control tumors. E4 cells were infected in vitro with lentivirus expressing active or control constructs and then allografted subcutaneously into NOD/SCID mice. Middle: Growth of tumors was inhibited by E4 cells infected by MT-mTer/siRNA lentivirus. Right: Wet weights of tumors resected 4 weeks after inoculation. (B) E4 cells (Lin-Sca-1+CD49f+) were sorted by FACSAria from freshly-resected, disaggregated and digested tumors (left). MT-mTer RNA levels from sorted E4 cells were quantified by RT-PCR (right). (C) Genomic mTer DNA levels from sorted E4 cells were quantified by PCR using the single copy gene 36B4 to control for DNA loading. (bottom). Representative bands from three different samples are shown.
Although E4 cells expressing MT-mTer/siRNA formed significantly smaller tumors, we were curious as to how they were able to form tumors at all. To gain insight into this question, we first isolated the E4 cells from mouse tumors using FACS sorting for Lin- Sca-1+ CD49f+ cells, the known, unique surface marker signature of the cPten−/− mouse prostate tumor cells (20) (Figure 4B Left panel). Using RT-PCR, we found no significant difference in MT-mTer RNA levels between MT-mTer/siRNA tumor cells and vector control tumor cells at day 30 (Figure 4B). At the same time, MT-mTer genomic DNA levels were significantly lower in MT-mTer/siRNA tumor cells at day 30 than in E4 cells at day 0, that is immediately prior to inoculation into the mice (Figure 4C). Taken together, these experiments suggested that the development of MT-mTer/siRNA tumors, albeit significantly smaller tumors, could be attributed to outgrowth of cells lacking genomic MT-mTer/siRNA (failed integration or subsequent loss), as well as to down-regulation of MT-mTer/siRNA expression in cells that did possess the construct.
MT-mTer/siRNA induces rapid DNA damage and apoptosis without significantly altering telomere lengths
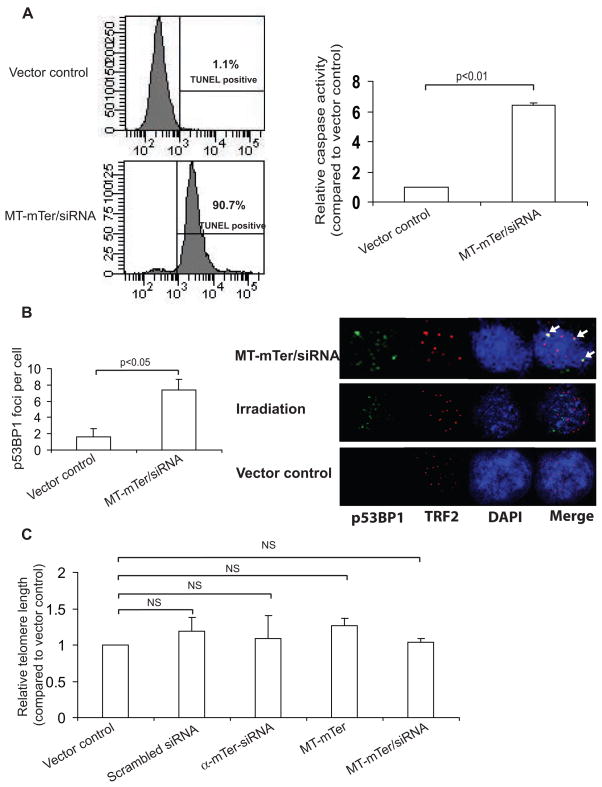
TUNEL assay performed 4 days after expression of MT-mTer/siRNA demonstrated that 90% of cells were TUNEL positive, versus <5% in cells expressing vector control (Figure 5A). Similarly, caspase activity after 4 days increased more than 4-fold in cells expressing MT-mTer/siRNA relative to cells expressing vector control (Figure 5A). Together, these assays strongly suggested that ectopic over-expression of MT-mTer in combination with knockdown of WT-mTer inhibited proliferation by inducing rapid and marked apoptosis. To verify whether cellular apoptosis was generated by DNA damage and “uncapped telomeres” as shown in human systems (10, 11, 26, 27), we analyzed p53BP1 foci by immunofluorescent staining and found significantly more p53BP1 foci in MT-mTer/siRNA-infected cells than in vector control infected cells (Figure 5B). Moreover, the p53BP1 foci appeared to localize at telomeres by TRF2 co-localization, in contrast to p53BP1 foci generated by gamma irradiation, which presumably induced DNA breaks indiscriminately throughout the genome (Figure 5B), suggesting that the DNA damage induced by MT-mTer/siRNA indeed occurred preferentially at telomeres. Having observed rapid onset of DNA damage, apoptosis, and growth inhibition within days, we suspected that minimal bulk telomere shortening had occurred during this short time period. Indeed, RT-PCR-based telomere length assay on E4 cells 3 days after MT-mTer/siRNA expression showed no significant changes in telomere lengths relative to control cells (Figure 5C).
Figure 5. MT-mTer/siRNA induces rapid apoptosis and DNA damage in E4 cells without altering bulk telomere length.
(A) Left: TUNEL assay performed 4 days after lentiviral expression of MT-mTer/siRNA demonstrates brisk apoptotic cell death, 90.7% versus 1.1% with vector control by FACS analysis; shown are representative plots of three different assays with statistical significance (p < 0.01). Right: Caspase assay performed at the same day 4 time point reveals significantly increased caspase activity consistent with apoptosis in cells expressing MT-mTer/siRNA versus control. (B) Left: Quantitation of p53BP1 DNA damage foci in MT-mTer/siRNA or vector control infected cells. Cells were grown on glass cover slips, fixed and stained on day 4 post lentiviral infection, and foci were counted under 63X magnification using a Zeiss Imager.Z1. Right: Representative fluorescence micrographs depicting p53BP1 foci, TRF2, DAPI, and merge. Cells were grown, infected, fixed, and stained as before, then photographed at 63x magnification using a Zeiss LSM 510 confocal scope. MT-mTer/siRNA-infected cells appear to have more p53BP1 DNA damage foci than vector control-infected cells and more co-localization of p53BP1 DNA damage foci with telomeres (TRF2) than irradiated cells (co-localization indicated by arrows, right). (C) Cells infected with lentivirus expressing the various constructs were harvested 3 days after infection and assayed for bulk telomere lengths using RT-PCR (mean of triplicate experiments).
Discussion
The role of telomerase in cancer and progenitor cells has been studied intensely; however, current xenograft and knockout mouse models cannot recapitulate the effects of telomerase targeting in an actual host with spontaneous malignancy and normal telomerase function. To address this limitation, we designed and validated two novel gene constructs – α-mTer-siRNA and MT-mTer – that specifically target murine telomerase and can be used to modulate telomerase activity in mouse models. The constructs were validated in vitro and in vivo and were shown to effectively reprogram mouse telomerase and induce telomeric uncapping, cellular apoptosis, and growth inhibition.
We performed our studies in E4 mouse prostate cancer cells, which are derived from prostate tumors arising in the transgenic cPten−/− mouse, regarded as perhaps the best mouse model of spontaneous prostate cancer currently available (19–21, 28). The cPten−/− mouse accurately recapitulates human disease, progressing from prostatic hyperplasia, to prostate intraepithelial neoplasia (PIN), to locally advanced prostate adenocarcinoma, to eventual micrometastatic and hormone-refractory disease. We were confident that the Pten−/− phenotype of the E4 cells would not unduly influence their susceptibility to treatment, as telomerase interference had previously been found equally efficacious in human cancer cell lines possessing wild-type or null Pten status (11, 29). Hence, the E4 cells constituted not only a reliable in vitro model for construct validation, but also a direct link to our future in vivo studies of systemic telomerase interference in the cPten−/− spontaneous prostate cancer mouse model.
When ectopically expressed in the E4 cells, the first construct, α-mTer-siRNA, achieved significant knock-down of mouse telomerase RNA and telomerase activity. We engineered the α-mTer-siRNA construct using a hairpin loop structure targeting the 5′ mTer region that encompasses the template sequence, as this strategy had previously proven effective for knock-down of human telomerase RNA (10, 11, 30). Notably, we observed that co-expression of α-mTer-siRNA with MT-mTer (so-called MT-mTer/siRNA) did not affect MT-mTer levels (Figure 2B, 2C); that is, the α-mTer-siRNA specifically depleted only WT-mTer and thus potentiated the “substitution” of MT-mTer for WT-mTer.
The second gene construct, MT-mTer, contained a mutated template sequence intended to alter the telomeric repeat sequence in the target cells. This approach was first introduced in ciliates and yeast (31, 32) and subsequently accomplished in human cell lines (10, 11, 29, 33–35). In the human studies, U → A template mutations were used successfully, prompting us to engineer analogous mutations in the mTer template. Successful incorporation of MT-Ter into active telomerase enzyme had been assessed previously either by blotting genomic DNA with a probe specific for the predicted telomere sequence (31, 34, 35) or by traditional PCR TRAP assay with amplification primers specific to the predicted altered sequences added to a telomeric substrate (29, 33, 35, 36). In our current study, we adapted the latter approach to next generation RT-PCR-TRAP, which showed that MT-mTer successfully partnered with mTERT to form enzymatically active telomerase that added incorrect telomeric repeats (TTTGGG instead of TTAGGG), and that this effect was potentiated by co-expression of siRNA against WT-mTer.
Expression of MT-mTer alone induced a significant (50%) growth inhibitory effect, consistent with previous reports of mutant template telomerase RNA expression in other model systems like ciliates, yeast, and human immortal or cancer cell lines (11, 29, 32, 34, 35, 37); these earlier studies utilized a spectrum of mutant template strategies and growth readouts, but all observed some degree of growth disruption. In contrast to the effects of MT-mTer, introduction of α-mTer-siRNA alone produced no significant short-term growth inhibition despite inhibition of telomerase activity. This was consistent with past reports in human cancer cell lines, where hTer depletion achieved either no significant growth inhibition or inhibition only with long-term culture over a period of weeks, attributed to a necessary “lag” period of telomere shortening in the absence of telomerase activity (10, 11, 30, 38). In the current study, co-expression of α-mTer-siRNA and MT-mTer induced marked (90%) growth inhibition, suggesting once again that α-mTer-siRNA potentiated the effects of MT-mTer by removing competing WT-mTer.
We tested the effects of MT-mTer/siRNA on tumor growth in SCID mice. This experiment potentially could have been conducted in immune competent syngeneic mice, but such an approach would have risked lower engraftment rates while still not addressing the ultimate question of efficacy in spontaneously arising tumors. Therefore, we elected to use immune deficient SCID mice in order to maximize engraftment, thereby enabling a more robust validation of tumor inhibition by telomerase interference relative to vector control. When allografted into mice, E4 cells expressing MT-mTer/siRNA formed tumors that were 50% smaller compared with tumors from mock- or vector-infected cells. We found a significant reduction in MT-mTer RNA and DNA levels in MT-mTer/siRNA tumor cells, suggesting that these small tumors likely formed from E4 cells that had down-regulated MT-mTer expression or genomically lacked the construct altogether. From a therapeutic standpoint, such escape mechanisms are to be expected. Even highly efficacious therapies rarely if ever achieve total eradication of tumor growth with a single treatment; rather, systemic treatment (in mouse spontaneous malignancy models or in patients) will necessitate repeat dosing to ensure maximal delivery and cell kill with successive cycles.
We investigated whether the growth inhibition mediated by MT-mTer/siRNA was the result of “uncapped” telomeres – an inability to assume a protected configuration, presumably because the altered telomere sequence cannot appropriately interact with members of the protective shelterin complex (11, 26, 27, 29, 35). Indeed, we found that cells expressing MT-mTer/siRNA accumulated significantly higher numbers of p53BP1 DNA damage foci that appeared to localize preferentially at telomeres, which in turn led to marked increases in apoptosis (over 90% of cells). Though quite promising from an anti-tumor perspective, such a high rate of apoptosis in telomerase+ cells raises the specter of toxicity in the in vivo setting. Two factors that mitigate this concern are: 1. We have shown previously that telomerase interference is dependent on the presence of active telomerase (TERT), which generally is expressed at much higher levels in tumor cells than in host tissues, thus conferring mechanistic specificity to this therapeutic approach. 2. As with all antineoplastic strategies, efficacy and toxicity will be titrated to an optimal ratio (therapeutic index) by modulating the dose intensity and frequency, first in animal models and ultimately in human clinical trials. Such optimization requires the ability to systemically deliver telomerase interference in a host with cancer – precisely the rationale for designing these constructs.
We measured the effects of telomerase interference on the length of mouse telomeres, which are significantly longer than their human counterparts (39, 40). In the first report (in ciliates) of mutant template telomerase RNA expression (31), Blackburn and colleagues found an increase in bulk telomere length, possibly due to “inability to bind some length-regulating factor”. A subsequent study performed by that group in yeast found variable telomere length effects depending on the specific template mutations induced (32). In human immortalized cells or cancer cell lines, a preponderance of studies found no significant telomere length changes with short-term (days to weeks) expression of mutant template hTer (10, 11, 29, 34, 35). Similarly, studies depleting human telomerase RNA with siRNA found no short term telomere length changes (10, 30), and a study of long term telomerase inhibition with an oligonucleotide that binds hTer noted telomere shortening only after a period of several weeks (38). A recent study was reported wherein two mouse immortal cell lines (telomerase + or mTer−) were directly transfected with a mutant template mTer (41); however, that study employed direct transfection and long term clone selection to introduce the mutant template mTer, resulting in low levels of expression. Consequently, no mutant telomerase activity or biologic effects were demonstrable in the telomerase+ line, and the effects noted in the mTer- line were not consistent among clones (41).
In our study, MT-mTer/siRNA induced telomere dysfunction without bulk telomere shortening in mouse cancer cells. The length-independent, rapid effects of these new constructs may prove particularly useful in mice, which typically possess very long telomeres (39, 40). Alternative telomerase strategies that directly inhibit the enzyme can induce cell growth inhibition only after a prolonged “lag phase” of cell division and progressive telomere shortening (38). Such approaches may be of limited utility in the setting of very long mouse telomeres, which must progressively shorten over numerous cell divisions and several generations in order to manifest a phenotype (14, 15, 42, 43). In contrast, reprogramming mouse telomerase with MT-mTer/siRNA can effectively elicit telomere dysfunction, apoptosis and growth inhibition in a matter of days without the need for significant telomere shortening.
Given the recent enthusiasm and substantial efforts allocated to telomerase-based therapies, it is critical to develop more informative models which accurately reflect the potential efficacy and toxicity of these strategies. Current models may not accurately predict the efficacy of telomerase-based strategies. While telomerase targeting may appear efficacious against homogeneous, rapidly-dividing cancer cells xenografted into mice, it may be less effective against spontaneously-arising tumors consisting of sub-populations of tumor cells with varying degrees of differentiation and telomerase activation. Indeed, such phenotypic heterogeneity within tumors has been observed in a large spectrum of common malignancies, ranging from leukemia, to breast cancer, to prostate cancer, to glioblastoma (16–18). At the same time, existing models provide minimal insight regarding the potential toxicity of telomerase manipulation. Most telomerase-targeting agents are engineered specifically against human telomerase; therefore, when tested in a mouse xenograft model, many of these human telomerase-targeting agents may be “blind” to the active mouse telomerase present in renewable host tissue compartments such as bone marrow, skin and gut. Several studies have demonstrated that telomerase activation within these compartments plays a critical role in cycling, proliferation, and differentiation of progenitor cells, and that telomerase knockout in mice eventually leads to defects in these very same compartments (15, 43–48). While telomerase knock-out mice do illuminate some of these progenitor cell toxicities, they are not cancer models and do not recapitulate the scenario of acute telomerase targeting in a telomerase-wild-type host with cancer.
MT-mTer/siRNA will help to surmount the limitations of current xenograft and knockout models, providing a ready set of new tools for directly studying the effects of telomerase disruption on tumors and progenitor compartments in mouse models of spontaneous malignancy. As with all mouse models, the implications of MT-mTer/siRNA for human therapy should be interpreted with a recognition of important differences between human and mouse telomere and telomerase biology, notably the greater length of mouse telomeres and the higher expression of telomerase in normal mouse tissues (42, 49). One speculates that both of these factors would endow telomerase interference with an even better efficacy/toxicity profile in humans than in mice, though such conclusions would have to await direct testing. Even with their acknowledged differences from human biology, mouse models inarguably have yielded many of our seminal insights about mammalian telomere and telomerase function, and they continue to provide a valuable and necessary preclinical setting for studying and manipulating telomeres and telomerase. Systemic delivery of MT-mTer/siRNA in mice will enable – for the first time – controlled depletion of telomerase activity (α-mTer-siRNA) and uncapping of telomeres (MT-mTer) in tumor cells and in progenitor compartments of a an immune competent mammalian host.
Having validated rapid telomerase interference in mouse cancer cells, we are currently packaging MT-mTer/siRNA for targeted systemic delivery by repeat dosing in the cPten−/− spontaneous prostate cancer mouse. Notably, although our current studies employ a prostate cancer model, we expect telomerase interference to achieve significant efficacy in a wide spectrum of malignancies, as >90% of human cancers upregulate telomerase activity by way of TERT expression, thus rendering them susceptible to targeting with MT-Ter/siRNA. It is our profound hope that introduction of these new constructs will open the door to systemic studies in mice, in larger mammals, and ultimately in human clinical trials to fully explore and develop this powerful and near-universal anti-cancer strategy.
Supplementary Material
Acknowledgments
We thank Dr. Pradip Roy-Burman for generously providing the E4 cells used in this work.
Financial Support: Studies were funded in part by grant support from NIH/National Cancer Institute K08 CA126983-01 (AG), R01 CA113392 (PR-B), Tower Foundation (AG), and Wright Foundation (AG).
Abbreviations list
- mTERT
mouse telomerase reverse transcriptase
- mTer
mouse telomerase RNA
- α-mTer-siRNA
anti-mTer short interfering RNA
- MT-mTer
mutant-template mTer
- TRAP
Telomeric Repeat Amplification Protocol
References
- 1.T De Lange VL. EH Blackburn Telomere. 2. New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 2.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 3.Wright WE, Piatyszek MA, Rainey WE, Byrd W, Shay JW. Telomerase activity in human germline and embryonic tissues and cells. Dev Genet. 1996;18:173–179. doi: 10.1002/(SICI)1520-6408(1996)18:2<173::AID-DVG10>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Meeker AK. Telomeres and telomerase in prostatic intraepithelial neoplasia and prostate cancer biology. Urol Oncol. 2006;24:122–130. doi: 10.1016/j.urolonc.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Shay JW, Bacchetti S. A survey of telomerase activity in human cancer. Eur J Cancer. 1997;33:87–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 6.Tomoda R, Seto M, Tsumuki H, et al. Telomerase activity and human telomerase reverse transcriptase mRNA expression are correlated with clinical aggressiveness in soft tissue tumors. Cancer. 2002;95:1127–1133. doi: 10.1002/cncr.10793. [DOI] [PubMed] [Google Scholar]
- 7.Dikmen ZG, Gellert GC, Jackson S, et al. In vivo inhibition of lung cancer by GRN163L: a novel human telomerase inhibitor. Cancer Res. 2005;65:7866–7873. doi: 10.1158/0008-5472.CAN-05-1215. [DOI] [PubMed] [Google Scholar]
- 8.Hahn WC, Stewart SA, Brooks MW, et al. Inhibition of telomerase limits the growth of human cancer cells. Nat Med. 1999;5:164–1170. doi: 10.1038/13495. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Mar V, Zhou W, Harrington L, Robinson MO. Telomere shortening and apoptosis in telomerase-inhibited human tumor cells. Genes Dev. 1999;13:2388–2399. doi: 10.1101/gad.13.18.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldkorn A, Blackburn EH. Assembly of mutant-template telomerase RNA into catalytically active telomerase ribonucleoprotein that can act on telomeres is required for apoptosis and cell cycle arrest in human cancer cells. Cancer Res. 2006;66:5763–5771. doi: 10.1158/0008-5472.CAN-05-3782. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Rosenberg JE, Donjacour AA, et al. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- 12.Shay JW, Wright WE. Telomerase therapeutics for cancer: challenges and new directions. Nat Rev Drug Discov. 2006;5:577–584. doi: 10.1038/nrd2081. [DOI] [PubMed] [Google Scholar]
- 13.Gowan SM, Harrison JR, Patterson L, et al. A G-quadruplex-interactive potent small-molecule inhibitor of telomerase exhibiting in vitro and in vivo antitumor activity. Mol Pharmacol. 2002;61:1154–1162. doi: 10.1124/mol.61.5.1154. [DOI] [PubMed] [Google Scholar]
- 14.Blasco MA, Lee HW, Hande MP, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 15.Lee HW, Blasco MA, Gottlieb GJ, Horner JW, 2nd, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 16.Collins AT, Maitland NJ. Prostate cancer stem cells. Eur J Cancer. 2008;42:1213–1218. doi: 10.1016/j.ejca.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 17.Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 18.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 19.Wang S, Gao J, Lei Q, Rozengurt N, et al. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Garcia AJ, Wu M, Lawson DA, Witte ON, Wu H. Pten deletion leads to the expansion of a prostatic stem/progenitor cell subpopulation and tumor initiation. Proc Natl Acad Sci U S A. 2006;103:1480–1485. doi: 10.1073/pnas.0510652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao CP, Zhong C, Saribekyan G, et al. Mouse models of prostate adenocarcinoma with the capacity to monitor spontaneous carcinogenesis by bioluminescence or fluorescence. Cancer Res. 2007;67:7525–7533. doi: 10.1158/0008-5472.CAN-07-0668. [DOI] [PubMed] [Google Scholar]
- 22.Naldini L, Blomer U, Gallay P, et al. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 23.Ramezani A, Hawley RG. Generation of HIV-1-based lentiviral vector particles. Curr Protoc Mol Biol. 2002;Chapter 16(Unit 16) doi: 10.1002/0471142727.mb1622s60. [DOI] [PubMed] [Google Scholar]
- 24.Herbert BS, Hochreiter AE, Wright WE, Shay JW. Nonradioactive detection of telomerase activity using the telomeric repeat amplification protocol. Nat Protoc. 2006;1:1583–1590. doi: 10.1038/nprot.2006.239. [DOI] [PubMed] [Google Scholar]
- 25.Cawthon RM, Smith KR, O’Brien E, Sivatchenko A, Kerber RA. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet. 2003;361:393–395. doi: 10.1016/S0140-6736(03)12384-7. [DOI] [PubMed] [Google Scholar]
- 26.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 27.Xu L, Blackburn EH. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad I, Sansom OJ, Leung HY. Advances in mouse models of prostate cancer. Expert Rev Mol Med. 2008;10:e16. doi: 10.1017/S1462399408000689. [DOI] [PubMed] [Google Scholar]
- 29.Kim MM, Rivera MA, Botchkina IL, Shalaby R, Thor AD, Blackburn EH. A low threshold level of expression of mutant-template telomerase RNA inhibits human tumor cell proliferation. Proc Natl Acad Sci U S A. 2001;98:7982–7987. doi: 10.1073/pnas.131211098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li S, Crothers J, Haqq CM, Blackburn EH. Cellular and gene expression responses involved in the rapid growth inhibition of human cancer cells by RNA interference-mediated depletion of telomerase RNA. J Biol Chem. 2005;280:23709–23717. doi: 10.1074/jbc.M502782200. [DOI] [PubMed] [Google Scholar]
- 31.Yu GL, Bradley JD, Attardi LD, Blackburn EH. In vivo alteration of telomere sequences and senescence caused by mutated Tetrahymena telomerase RNAs. Nature. 1990;344:126–132. doi: 10.1038/344126a0. [DOI] [PubMed] [Google Scholar]
- 32.Lin J, Smith DL, Blackburn EH. Mutant telomere sequences lead to impaired chromosome separation and a unique checkpoint response. Mol Biol Cell. 2004;15:1623–1634. doi: 10.1091/mbc.E03-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng J, Funk WD, Wang SS, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- 34.Guiducci C, Cerone MA, Bacchetti S. Expression of mutant telomerase in immortal telomerase-negative human cells results in cell cycle deregulation, nuclear and chromosomal abnormalities and rapid loss of viability. Oncogene. 2001;20:714–725. doi: 10.1038/sj.onc.1204145. [DOI] [PubMed] [Google Scholar]
- 35.Marusic L, Anton M, Tidy A, Wang P, Villeponteau B, Bacchetti S. Reprogramming of telomerase by expression of mutant telomerase RNA template in human cells leads to altered telomeres that correlate with reduced cell viability. Mol Cell Biol. 1997;17:6394–6401. doi: 10.1128/mcb.17.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerone MA, Londono-Vallejo JA, Bacchetti S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum Mol Genet. 2001;10:1945–1952. doi: 10.1093/hmg/10.18.1945. [DOI] [PubMed] [Google Scholar]
- 37.Blackburn EH. Cell biology: telomeres sans frontieres. Nature. 1990;343:122. doi: 10.1038/343122a0. [DOI] [PubMed] [Google Scholar]
- 38.Herbert B, Pitts AE, Baker SI, et al. Inhibition of human telomerase in immortal human cells leads to progressive telomere shortening and cell death. Proc Natl Acad Sci U S A. 1999;96:14276–14281. doi: 10.1073/pnas.96.25.14276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zijlmans JM, Martens UM, Poon SS, et al. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc Natl Acad Sci U S A. 1997;94:7423–7428. doi: 10.1073/pnas.94.14.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hemann MT, Greider CW. Wild-derived inbred mouse strains have short telomeres. Nucleic Acids Res. 2000;28:4474–4478. doi: 10.1093/nar/28.22.4474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marie-Egyptienne DT, Brault ME, Nimmo GA, Londono-Vallejo JA, Autexier C. Growth defects in mouse telomerase RNA-deficient cells expressing a template-mutated mouse telomerase RNA. Cancer lett. 2009;275:266–276. doi: 10.1016/j.canlet.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 42.Prowse KR, Greider CW. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc Natl Acad Sci U S A. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Samper E, Fernandez P, Eguia R, et al. Long-term repopulating ability of telomerase-deficient murine hematopoietic stem cells. Blood. 2002;99:2767–2775. doi: 10.1182/blood.v99.8.2767. [DOI] [PubMed] [Google Scholar]
- 44.Forsyth NR, Wright WE, Shay JW. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- 45.Allsopp RC, Morin GB, DePinho R, Harley CB, Weissman IL. Telomerase is required to slow telomere shortening and extend replicative lifespan of HSCs during serial transplantation. Blood. 2003;102:517–520. doi: 10.1182/blood-2002-07-2334. [DOI] [PubMed] [Google Scholar]
- 46.Ferron S, Mira H, Franco S, et al. Telomere shortening and chromosomal instability abrogates proliferation of adult but not embryonic neural stem cells. Development. 2004;131:4059–4070. doi: 10.1242/dev.01215. [DOI] [PubMed] [Google Scholar]
- 47.Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 48.Gonzalez-Suarez E, Samper E, Ramirez A, et al. Increased epidermal tumors and increased skin wound healing in transgenic mice overexpressing the catalytic subunit of telomerase, mTERT, in basal keratinocytes. Embo J. 2001;20:2619–2630. doi: 10.1093/emboj/20.11.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis T, Kipling D. Telomeres and telomerase biology in vertebrates: progress towards a non-human model for replicative senescence and ageing. Biogerontology. 2005;6:371–385. doi: 10.1007/s10522-005-4901-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.