Abstract
Our aim in this study was to provide an atomic description of ampicillin translocation through OmpF, the major outer membrane channel in Escherichia coli and main entry point for β-lactam antibiotics. By applying metadynamics simulations, we also obtained the energy barriers along the diffusion pathway. We then studied the effect of mutations that affect the charge and size at the channel constriction zone, and found that in comparison to the wild-type, much lower energy barriers are required for translocation. The expected higher translocation rates were confirmed on the macroscopic scale by liposome-swelling assays. A microscopic view on the millisecond timescale was obtained by analysis of temperature-dependent ion current fluctuations in the presence of ampicillin and provide the enthalpic part of the energy barrier. By studying antibiotic translocation over various timescales and length scales, we were able to discern its molecular mechanism and rate-limiting interactions, and draw biologically relevant conclusions that may help in the design of drugs with enhanced permeation rates.
Introduction
Bacteria develop mechanisms of resistance that render the use of antibiotics ineffective (1). Moreover, an increase in multidrug-resistant pathogens is appearing at a time when only a few novel active antibacterial compounds are in clinical trials (2). To respond to this alarming situation, we need to reinforce and reinvent antibacterial research. Microscopically based drug design, starting from molecular knowledge of resistant mechanisms, represents a potentially efficient way to bring new agents to the market (3). A key resistance mechanism in Gram-negative bacteria is the prevention of antibiotic uptake, mediated by outer-membrane porins. For example, the resistance of pathogenic bacteria to β-lactams has been attributed to alterations in the expression or the molecular structures of porins (4). The OmpF porin in Escherichia coli has an hourglass shape and the channel structure reveals a spatial constriction created by loop L3, which folds back into the channel. As shown in Fig. 1, this region is also characterized by a transversal electric field created by acidic residues on the L3 side (D113 and E117) facing a cluster of arginines (R42-R82-R132) (5). Several studies have raised questions concerning the role of these amino acids in diffusion processes through OmpF. For example, the single substitutions R132A and D113A were found to dramatically increase the uptake of β-lactams antibiotics (6,7). Such findings provide investigators with an opportunity to tune the uptake of antibiotics based on only slight chemical modifications. This attractive strategy requires the development of better-tuned quantitative methods to elucidate the rate-limiting molecular interactions between drug and channel residues. In this work, we studied antibiotic diffusion by combining atomic-level descriptions provided by molecular-dynamics (MD) simulations (8), electrophysiology techniques at the single-molecule level (9), and liposome-swelling assays (10). Our findings reveal, for the first time to our knowledge, the complete pathways of ampicillin permeation through wild-type (WT) OmpF as well as D113N and R132A mutants, from their macroscopic flux down to their molecular mechanism.
Figure 1.
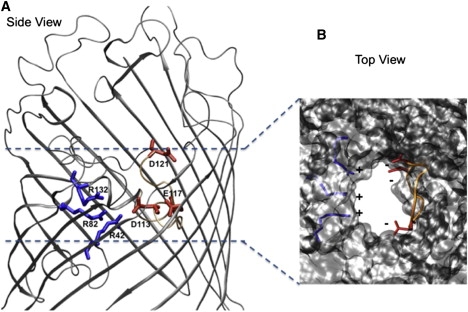
Structural details of OmpF. (A) The backbone of OmpF is displayed in gray cartoons. The charged residues at the constriction region (D113, E117, and D121 on the L3 side, and R42, R82, and R132 on the anti-L3 side) are colored by residue type (positively charged in blue, negatively charged in red). (B) The OmpF structure is displayed in gray molecular surface to highlight the space available. Loop L3 is colored in orange and the charged residues at the constriction region are colored as in A.
Materials and Methods
MD simulations
For the MD simulations, we followed a previously described protocol (8), starting from the crystal structure (Protein Data Bank code: 2OMF) and residue protonation state as described by Im and Roux (11). We used the program ORAC and the Amber force field (12) for system setup and simulation (13). The porin mutants were obtained by substituting the single amino acid residue starting from the high-resolution structure of OmpF (2OMF) using the MD package ORAC. After the molecular replacement was completed, we further equilibrated the mutant system for ∼2 ns of a standard MD simulation. All simulated systems were validated for convergence and stabilization of energy, temperature, and root mean-square deviation (RMSD) with respect to the starting structure. Based on previous findings (7,8,14), we chose the following collective variables to simulate antibiotic translocation using metadynamics (15): 1), the distance Z, defined as the difference between the center of mass of the antibiotic and the center of mass of the system (porin + detergent) along the z axis; and 2), the angle θ, defined as the orientation of the long axis of the molecule with respect to the z axis. All simulated systems were validated for convergence and stabilization of energy, temperature, and RMSD with respect to the starting structure. Our choice of OmpF as a monomer is supported by previous studies that reported a mutual independence of the three monomers (i.e., no cooperativity) for ions, small-molecule transport, and antibiotics (16–18). Using this biased simulation strategy, we obtained translocation of ampicillin through WT OMPF, D113N, and R132A after 38, 27, and 15 ns, respectively. The metadynamics algorithm enables one to reconstruct the free energy in the subspace of the collective variables by integrating the history-dependent terms (15). Because of the complexity of the process studied, we calculated the free energy after obtaining the first translocation path, which is considered to be the most probable path because it passes through the lowest saddle point, as was previously done for the unthreading of a molecule (19). We used the resulting approximated free-energy surface to select the regions of energy minima. Additional metadynamics simulations were launched starting from each minimum, which enabled us to reconstruct the one-dimensional (1D) free-energy profile for the translocation of ampicillin through WT OmpF and the two mutants. The profiles only report the energetic barriers from the time the antibiotic enters the channel to when it reaches the highest barrier (also called the main or effective barrier). In fact, once the antibiotic crosses the constriction region, we expect a diffusive regime, with no significant affinity sites. The error bars associated with the energy barrier calculations were assessed as previously described (19) and were 2 kT at most. Furthermore, to decipher the molecular details of the translocation mechanism, additional equilibrium MD simulations (1 ns length) were started from each visited minimum along the ampicillin diffusion path. In-depth analysis included the calculation of 1), atomic fluctuations; 2), hydrogen (H)-bonds (the criteria were a distance of at most 2.8 Å and a donor-hydrogen-acceptor angle of at least 130°) and hydrophobic interactions (the criterion was a distance of at least 3 Å between nonpolar atoms) of ampicillin; 3), the residence time of water molecules around ampicillin (20); and 4), the cross-sectional solvent-accessible surface area (SASA) (21) using both an in-house program and the software VMD (22).
Experiments
The chemicals used in this study were arabinose, raffinose, KCl, MES, n-pentane, hexadecane, squalene, ampicillin anhydrous, Dextran 40000 (Sigma-Aldrich, Buchs, Switzerland), Octyl-POE (Alexis, Switzerland), 1,2-diphytanoyl-sn-glycero-3-phosphatidylcholine (DPhPC), and E. coli total lipid extract (Avanti Polar Lipids, Alabaster, AL). Reconstitution experiments and single-channel analyses were performed as previously described (18). A stable planar lipid bilayer was formed on a 25-μm-thick Teflon film (aperture diameter: 40–50 μm), and spontaneous channel insertion was obtained under high applied voltage. A Peltier element was used for temperature regulation (Dagan), and ion current blockages were measured after the addition of ampicillin to the chamber. The data in Table 1 were obtained by a fluctuation, single-channel analysis as outlined in previous studies (4,9,18,23,24). The on-rate was calculated from the number of binding events (kon = υ / (3[c]), where υ is the number of events and c is the concentration of antibiotic). The off-rate (t » koff − 1) was calculated from the residence time, as described previously. The flux of antibiotic through the channel is proportional to the kon rate (J = kon Δc/2). WT OmpF and mutants (1 mg/mL) in 1% Octyl-POE were reconstituted into liposomes as described by Nikaido and Rosenberg (25). E. coli total lipid extract (Avanti Polar Lipids, Alabaster, AL) was used for liposome formation, and 17% Dextran (molecular weight: 40,000; Fluka) was used for liposome filling. After incubation, multilamellar liposomes were formed by sonication in a water bath sonicator. The size of the formed liposomes was checked with the use of a Nano-ZS ZEN3600 zetasizer (Malvern Instruments, UK). Control liposomes were prepared in the same manner but without the addition of porin. The isotonic concentration was determined by diluting the proteoliposomes into different concentrations of raffinose (Sigma) with an Osmomat 30 osmolarimeter (Gonotec). Each batch (containing WT, D113N, or R132A) was separated into smaller aliquots assuming a homogeneous distribution. One aliquot of each batch was tested for arabinose, a smaller molecule for which we expect maximum permeation, and this swelling rate was set as our reference 100% for the respective batch. By normalizing each batch separately, we were able to reduce the effects of the variable reconstitution efficiencies of different preparations. Changes in the optical density were monitored at 400 nm with a Cary 100 Scan spectrophotometer (Varian). The swelling rates were taken as averages from at least three different sets of experiments, calculated as described by Nikaido and Rosenberg (25), and then normalized to the rate obtained with arabinose.
Table 1.
Kinetic analysis of the ampicillin-binding events: kon, koff, K (binding constant), and J (flux) of ampicillin for WT OmpF and the mutant D113N
| Ampicillin at ΔC = 10 mM | Threshold gating potential [mV] | koncis [1/(s M)] | kontrans [1/(s M)] | koff [1/s] | K [1/M] | Flux J cis to trans [molecule/s] |
|---|---|---|---|---|---|---|
| OmpF50 mV−50 mV | 100–150 | 2800 ± 2803000 ± 300 | 2700 ± 2702600 ± 260 | 5300 ± 5304800 ± 480 | 1.01.2 | 1415 |
| D113N50 mV−50 mV | 200 | 17000 ± 17004100 ± 420 | 3200 ± 3202500 ±250 | 1000010000 | 20.7 | 8022 |
Results
First, we measured ion current fluctuations through single trimeric OmpF (WT and mutants R132A and D113N) reconstituted into lipid bilayers, which allowed us to extract the kinetic rates (18). As shown in Fig. 2, A–C, at 25°C ampicillin causes significant ion current fluctuations in both WT OmpF and mutant D113N, but, surprisingly, few blocking events are visible in the case of R132A. The average residence time of ampicillin is calculated to be 180 ± 20 μs for WT OmpF and as low as 100 ± 20 μs for the D113N mutant. In the case of both mutants, our findings highlight a possible underestimation of the number of events due to the resolution limit of the method. To rule out this possibility, we repeated the measurements at lower temperatures, as slowing down the diffusion should reveal potential fast events.
Figure 2.
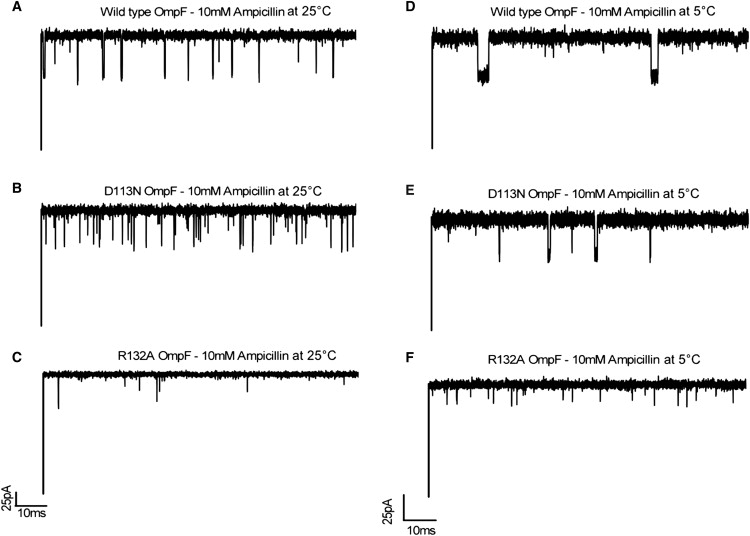
Typical tracks of ion current through a single WT OmpF channel and mutants D113N and R132A, reconstituted into DPhPC lipid membranes in the presence of 10 mM ampicillin and 1 M KCl at pH 6. Applied voltage is 50 mV.
At 5°C, as shown in Fig. 2, D–F, we measure fewer blocking events but elongated residence times for both the WT and D113N OmpF. Lowering the temperature brings the residence time of D113N well above the resolution limit (Fig. 3 B). In the case of R132A, we observe more events, but they are still partial blocking events, and thus we cannot calculate the binding kinetics. To sum up, the number of binding events is much higher in the case of D113N compared to WT OmpF at all applied temperatures (Fig. 3 A). Of interest, the fitting procedure reveals that beyond 25°C, in the range of physiological temperatures, the number of measured events is underestimated in the case of D113N, which is as expected when the residence time approaches the resolution limit of the method (inverse filter frequency: 100 μs).
Figure 3.
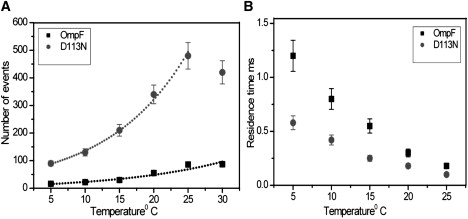
(A) Statistical analysis revealing temperature-dependent blocking events of ampicillin with WT OmpF and D113N. The continuous line represents the exponential fit. (B) Effect of temperature on the antibiotic residence time (τ) for WT OmpF and D113N mutant.
According to a simple kinetic model, at low substrate concentration the flux J is only proportional to the on-rate kon, J = (kon/2) Δc, where Δc is the concentration gradient (23,26). The kinetic analysis (Table 1) yielded kon = 3000 ± 300 s−1M−1 for WT OmpF and 17000 ± 1700 s−1M−1 for the D113N mutant, resulting in the translocation of ∼6 times more ampicillin molecules for D113N than for WT OmpF.
We then used additional methods to bridge the timescales and length scales of transport, to clarify the case of R132A in which only partial blocking events were observed at all measured temperatures.
To elucidate the energetic details of transport, we performed metadynamics simulations of ampicillin translocation through WT OmpF, D113N, and R132A mutants. From the reconstructed 1D free-energy profiles (Fig. 4), we observe that the effective barrier for ampicillin to translocate is higher in the case of WT OmpF (14 kT) and lower for R132A (9 kT) and D113N (5 kT). To compare these findings with our predictions, we quantified the in vitro macroscopic flux of ampicillin using a liposome-swelling assay, a method that has been successfully applied to such problems in previous studies (10,25). The advantage of this technique is that the penetration rates of ampicillin in proteoliposomes generally mimic those of the intact cells, and swelling rates are directly proportional to the permeability of the antibiotic. As shown in Fig. 5, we find a higher diffusion rate of ampicillin for both mutants, as it increases by 25% for R132A and 40% for D113N compared to the WT OmpF. The trend of the flux is in good agreement with that of the energy barriers obtained from the molecular simulations. Taken together, our results demonstrate that a single point mutation at the constriction region can remarkably affect the kinetics of ampicillin and thus its uptake.
Figure 4.
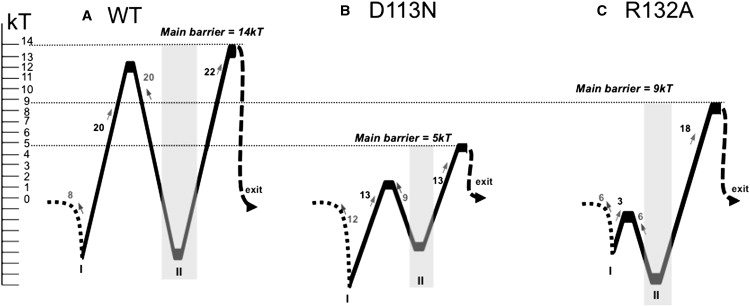
One-dimensional free-energy profiles for the translocation of ampicillin through WT OmpF (A), D113N (B), and R132A (C). The minima at the constriction region are highlighted in gray and the energy barriers are reported in kT. The “exit” label refers to the periplasmic side.
Figure 5.
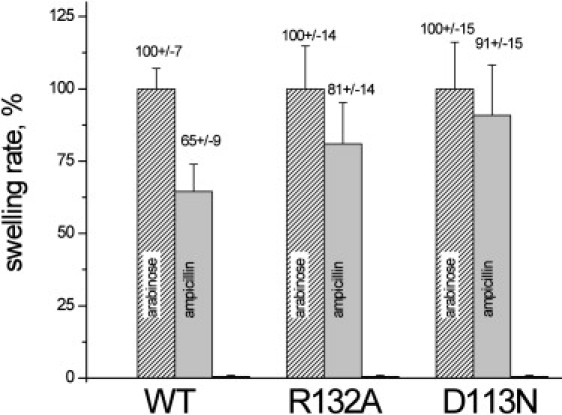
Liposome-swelling assay with proteoliposomes containing WT OmpF or mutants D113N and R132A. Arabinose, a small molecule that is able to penetrate perfectly, was used as a reference to normalize the swelling rates (=100%). A second control was performed with raffinose, which does not permeate through OmpF.
To elucidate the molecular mechanism and rationalize our findings, we then deciphered the physicochemical and structural properties of the diffusion process. Each relevant minimum identified by the metadynamics (as labeled in Fig. 4) was used as a starting point for the additional equilibrium MD simulations for which we analyzed the solvation, flexibility, SASA, and interaction patterns between ampicillin and the OmpF residues.
In the case of the WT, when ampicillin is above the constriction region (in the structures sampled along the equilibrium simulations at Minimum-I), it interacts only transiently with channel residues, as the antibiotic undergoes numerous reorientations and attempts to enter the constriction zone. This is confirmed by the averaged atomic fluctuations of ampicillin of 0.78 Å (along Minimum-I), which is close to the antibiotic fluctuation calculated in bulk water (Table 2). Such extensive rearrangements above the constriction region were also previously described in the case of diffusion of glycerol through aquaglyceroporin (27). Furthermore, when it reaches the affinity site at the constriction region (Minimum-II), the ampicillin fluctuations are as low as 0.35 Å, and such a decrease in entropy was also previously shown upon ligand binding (28). The entropy-enthalpy compensation is made possible via specific interactions between ampicillin and OmpF through both H-bonds (with D113 and R42-R82-R132) and hydrophobic contacts (Fig. 6 A). In this affinity site (Minimum-II), we also find that ampicillin has durable interactions with slow water molecules or “bound waters” (Table 2). Such a strong network of interactions of ampicillin at the constriction region explains the high-energy barrier calculated to exit the channel, which correlates well with the long residence time measured.
Table 2.
Structural details obtained from the equilibrium MD simulations
| Region of analysis | Averaged atomic fluctuations (Å) |
Interaction with “bound waters” (% of the total number of interacting water molecules) |
||||
|---|---|---|---|---|---|---|
| WT | D113N | R132A | WT | D113N | R132A | |
| Mini I (above constriction region) | 0.74 | 0.39 | 0.59 | 0 | 0 | 0 |
| Mini II (at constriction region) | 0.35 | 0.29 | 0.32 | 10 | 5 | 7 |
| Mini III (below constriction region) | 0.60 | 0.75 | 1.00 | 4 | 0 | 0 |
First column reports the averaged atomic fluctuations of ampicillin, and the second column provides the number of “bound waters”, defined as the molecules that interact with ampicillin with a residence time of >30% of the simulation time.
Figure 6.
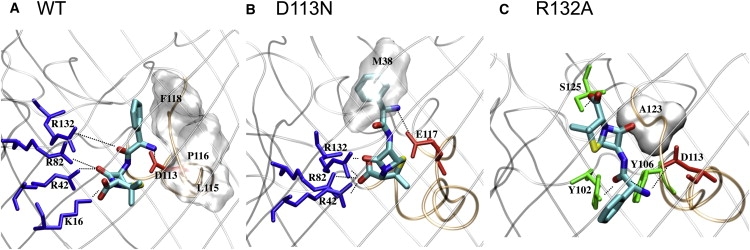
Molecular details (side views) of ampicillin at the binding site of the constriction region of (A) WT OmpF, (B) D113N, and (C) R132A. The views and orientations in this figure are the same as in Fig. 1 (the top is toward the vestibule, the bottom is toward the periplasmic space). The antibiotic is displayed in stick representation and colored by atom type (blue for nitrogen, red for oxygen, cyan for carbon) where hydrogens are not shown. The backbone of OmpF is displayed in gray cartoons to highlight its secondary structures. The constriction region is highlighted by loop L3 (colored in orange). Residues of OmpF that are seen as strongly interacting with the antibiotic are labeled using the one-letter amino acid code; those making H-bonds are colored by residue type (positively charged in blue, negatively charged in red, polar in green), and those making hydrophobic contacts are displayed with their molecular surface, highlighting their shape.
In the case of D113N, ampicillin does not reorient extensively above the constriction region, and the fluctuations calculated along Minimum-I are only of 0.29 Å (Table 2). In contrast to the WT, the antibiotic finds the optimal orientation rapidly (Fig. 6) to fit the constriction region. This explains the lower energy barrier to enter the constriction region and agrees with the higher number of measured events. At the constriction region, ampicillin maintains a favorable network of interactions (Fig. 6 B) in which the side chain of the residue E117 now repositions in the lumen of the channel to make H-bonds with the positive group of ampicillin. However, once ampicillin crosses the constriction region, its positive group does not find a salt-bridge partner, and this facilitates its diffusion further down. The lack of interactions retarding the antibiotic's exit through the channel constitutes a major difference compared to the WT and explains the lower energetic barrier and measured residence time.
Of interest, a different molecular path is found by ampicillin in the case of the mutant R132A, which explains the partial blocking events measured (Fig. 2 C). In the case of this mutant, ampicillin takes advantage of the structural and polarity changes in the channel to rapidly reach the constriction region (the barrier is reduced to 3 kT; see Fig. 4 C), and translocates with its phenyl group pointing down (Fig. 6 C). Furthermore, when it is at the constriction region, ampicillin neither accommodates in the hydrophobic pocket at the L3 side nor makes H-bonds with the basic residues at the anti-L3 side (Fig. 6 C). Compared to the WT OmpF, the only conserved interaction in this case is the salt-bridge between the amino positive group of ampicillin and D113. Still, in the case of R132A, we observe that ampicillin interacts with some novel residues (Fig. 6 C), has a low flexibility, and interacts with slow “bound waters” (Table 2). This well-defined affinity site once again explains the high-energy barrier needed to exit the channel (Fig. 4 C).
The occlusion of the channel is well illustrated by the MD simulation snapshots (Fig. 7, A–C), which show that it is only in the case of the mutant R132A that ampicillin is not positioned centrally and instead leaves a large portion of space available. We quantified the available area (Fig. 7, D–F) and found that ampicillin only significantly occludes the pore upon translocation in the cases of the WT OmpF and the D113N mutant. For the mutant R132A, there is still at least 60% of space available. This means that the antibiotic diffusion would not interfere significantly with the ionic current, which would explain the lack of well-resolved ionic current blockage at any measured temperature in this case (Fig. 2 C).
Figure 7.
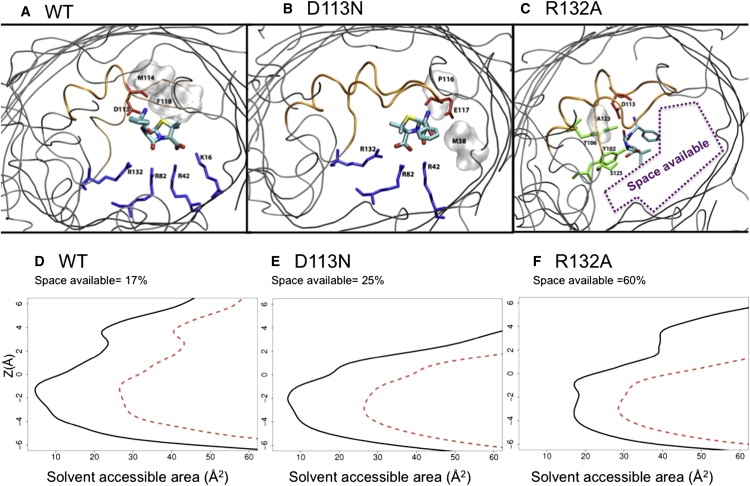
Molecular details (top view) from equilibrium simulations started at Minima-II for WT OmpF (A), D113N (B), and R132A (C) mutants. Ampicillin is displayed in stick representation and colored according to atom type. The backbone of OmpF is shown by gray cartoons (L3 is colored orange). The residues making H-bonds are colored by residue type, and those making hydrophobic contacts are displayed by gray molecular surface. Below, the average SASA is reported for WT OmpF (D), D113N (E), and R132A (F) in the presence (black) and absence (red) of ampicillin.
Discussion
In all three systems (WT OmpF, D113N, and R132A), we found an affinity site for ampicillin in OmpF, defined by specific interactions with key residues of the constriction region and with strongly ordered water molecules. For the concentration used here (far from saturation and within the limit of the physiological dose), the only parameter that makes a difference in the flux is kon. Of interest, we can conclude from our results that kon can be tuned by mutations at the constriction region that affect, very locally, the molecular interactions. The fact that ampicillin uptake can be tuned by specific interactions inside the channel is in agreement with previous theoretical and experimental studies (11,23,29–31). Cornell et al. (12) assumed that kon is only dependent on the diffusion coefficient of the molecule and the radius of attraction. This implies that, surprisingly, even very local interactions at the constriction region must be taken into account in the definition of this radius.
We conclude that the bottleneck for antibiotic translocation stems from the difficulty of overcoming the constriction region. In the case of WT OmpF, ampicillin has to deal with a particularly reduced size and a strong electrostatic field. Using computer simulations, we were able to predict when the presence of an ampicillin molecule would block the ion current, and thus rationalize the ion current fluctuations induced by antibiotics upon translocation.
By combining different approaches, we were able to follow the ampicillin translocation process over various timescales and length scales. This allowed us to reveal the complete molecular mechanism of diffusion and relate it to biologically relevant conclusions. The identification of crucial antibiotic-channel interactions will benefit the design of novel molecules with enhanced permeation rates. Finally, we believe that our multiscale approach can be conveniently employed to study porin-antibiotic interactions in other enterobacterial pathogens (1,4), such as those involved in persistent tuberculosis.
Acknowledgments
We thank Tivadar Mach, Malcom Page, and Jurg Dreier for their support and productive discussions.
This study was supported by the European Union, FP6 grant MRTN-CT-2005-019335 (Translocation), and by the computer center and consortiums Cybersar, CASPUR, and CINECA through CPU hours.
References
- 1.Arias C.A., Murray B.E. Antibiotic-resistant bugs in the 21st century—a clinical super-challenge. N. Engl. J. Med. 2009;360:439–443. doi: 10.1056/NEJMp0804651. [DOI] [PubMed] [Google Scholar]
- 2.Spellberg B., Powers J.H., Edwards J.E. Trends in antimicrobial drug development: implications for the future. Clin. Infect. Dis. 2004;38:1279–1286. doi: 10.1086/420937. [DOI] [PubMed] [Google Scholar]
- 3.Barker J.J. Antibacterial drug discovery and structure-based design. Drug Discov. Today. 2006;11:391–404. doi: 10.1016/j.drudis.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Pagès J.M., James C.E., Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat. Rev. Microbiol. 2008;6:893–903. doi: 10.1038/nrmicro1994. [DOI] [PubMed] [Google Scholar]
- 5.Cowan S.W., Schirmer T., Rosenbusch J.P. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 6.Simonet V., Malléa M., Pagès J.M. Substitutions in the eyelet region disrupt cefepime diffusion through the Escherichia coli OmpF channel. Antimicrob. Agents Chemother. 2000;44:311–315. doi: 10.1128/aac.44.2.311-315.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidal S., Bredin J., Barbe J. β-Lactam screening by specific residues of the OmpF eyelet. J. Med. Chem. 2005;48:1395–1400. doi: 10.1021/jm049652e. [DOI] [PubMed] [Google Scholar]
- 8.Ceccarelli M., Danelon C., Parrinello M. Microscopic mechanism of antibiotics translocation through a porin. Biophys. J. 2004;87:58–64. doi: 10.1529/biophysj.103.037283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozhinjampara M., Chimerel C., Winterhalter M. Antibiotic translocation through membrane channels: temperature-dependent ion current fluctuation for catching the fast events. Eur. Biophys. J. 2009 doi: 10.1007/s00249-009-0495-0. in press. [DOI] [PubMed] [Google Scholar]
- 10.Yoshimura F., Nikaido H. Diffusion of β-lactam antibiotics through the porin channels of Escherichia coli K-12. Antimicrob. Agents Chemother. 1985;27:84–92. doi: 10.1128/aac.27.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Im W., Roux B. Ions and counterions in a biological channel: a molecular dynamics simulation of OmpF porin from Escherichia coli in an explicit membrane with 1 M KCl aqueous salt solution. J. Mol. Biol. 2002;319:1177–1197. doi: 10.1016/S0022-2836(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 12.Cornell W.D., Cieplak P., Kollmann P. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 13.Procacci P., Darden T.A., Marchi M. ORAC: a molecular dynamics program to simulate complex molecular systems with realistic electrostatic interactions. J. Comput. Chem. 1997;18:1848–1862. [Google Scholar]
- 14.Danelon C., Nestorovich E.M., Bezrukov S.M. Interaction of zwitterionic penicillins with the OmpF channel facilitates their translocation. Biophys. J. 2006;90:1617–1627. doi: 10.1529/biophysj.105.075192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laio A., Parrinello M. Escaping free-energy minima. Proc. Natl. Acad. Sci. USA. 2002;99:12562–12566. doi: 10.1073/pnas.202427399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robertson K.M., Tieleman D.P. Orientation and interactions of dipolar molecules during transport through OmpF porin. FEBS Lett. 2002;528:53–57. doi: 10.1016/s0014-5793(02)03173-3. [DOI] [PubMed] [Google Scholar]
- 17.Rostovtseva T.K., Nestorovich E.M., Bezrukov S.M. Partitioning of differently sized poly(ethylene glycol)s into OmpF porin. Biophys. J. 2002;82:160–169. doi: 10.1016/S0006-3495(02)75383-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nestorovich E.M., Danelon C., Bezrukov S.M. Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc. Natl. Acad. Sci. USA. 2002;99:9789–9794. doi: 10.1073/pnas.152206799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laio A., Rodriguez-Fortea A., Parrinello M. Assessing the accuracy of metadynamics. J. Phys. Chem. B. 2005;109:6714–6721. doi: 10.1021/jp045424k. [DOI] [PubMed] [Google Scholar]
- 20.Sterpone F., Ceccarelli M., Marchi M. Dynamics of hydration in hen egg white lysozyme. J. Mol. Biol. 2001;311:409–419. doi: 10.1006/jmbi.2001.4860. [DOI] [PubMed] [Google Scholar]
- 21.Mach T., Neves P., Gameiro P. Facilitated permeation of antibiotics across membrane channels—interaction of the quinolone moxifloxacin with the OmpF channel. J. Am. Chem. Soc. 2008;130:13301–13309. doi: 10.1021/ja803188c. [DOI] [PubMed] [Google Scholar]
- 22.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14 doi: 10.1016/0263-7855(96)00018-5. 33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- 23.Berezhkovskii A.M., Bezrukov S.M. Optimizing transport of metabolites through large channels: molecular sieves with and without binding. Biophys. J. 2005;88:L17–L19. doi: 10.1529/biophysj.104.057588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weingart H., Petrescu M., Winterhalter M. Biophysical characterization of in- and efflux in Gram-negative bacteria. Curr. Drug Targets. 2008;9:789–796. doi: 10.2174/138945008785747752. [DOI] [PubMed] [Google Scholar]
- 25.Nikaido H., Rosenberg E.Y. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J. Bacteriol. 1983;153:241–252. doi: 10.1128/jb.153.1.241-252.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nekolla S., Andersen C., Benz R. Noise analysis of ion current through the open and the sugar-induced closed state of the LamB channel of Escherichia coli outer membrane: evaluation of the sugar binding kinetics to the channel interior. Biophys. J. 1994;66:1388–1397. doi: 10.1016/S0006-3495(94)80929-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hénin J., Tajkhorshid E., Chipot C. Diffusion of glycerol through Escherichia coli aquaglyceroporin GlpF. Biophys. J. 2008;94:832–839. doi: 10.1529/biophysj.107.115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jusuf S., Loll P.J., Axelsen P.H. Configurational entropy and cooperativity between ligand binding and dimerization in glycopeptide antibiotics. J. Am. Chem. Soc. 2003;125:3988–3994. doi: 10.1021/ja027780r. [DOI] [PubMed] [Google Scholar]
- 29.Berezhkovskii A.M., Pustovoit M.A., Bezrukov S.M. Channel-facilitated membrane transport: average lifetimes in the channel. J. Chem. Phys. 2003;119:3943–3951. [Google Scholar]
- 30.Berezhkovskii A.M., Hummer G., Bezrukov S.M. Identity of distributions of direct uphill and downhill translocation times for particles traversing membrane channels. Phys. Rev. Lett. 2006;97:020601. doi: 10.1103/PhysRevLett.97.020601. [DOI] [PubMed] [Google Scholar]
- 31.Tieleman D.P., Berendsen H.J. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys. J. 1998;74:2786–2801. doi: 10.1016/S0006-3495(98)77986-X. [DOI] [PMC free article] [PubMed] [Google Scholar]


